Abstract
The biological methyl donor S-adenosyl-l-methionine (AdoMet) is spontaneously degraded by inversion of its sulfonium center to form the R,S diastereomer. Unlike its precursor, (S,S)-AdoMet, (R,S)-AdoMet has no known cellular function and may have some toxicity. Although the rate of (R,S)-AdoMet formation under physiological conditions is significant, it has not been detected at substantial levels in vivo in a wide range of organisms. These observations imply that there are mechanisms that either dispose of (R,S)-AdoMet or convert it back to (S,S)-AdoMet. Previously, we identified two homocysteine methyltransferases (Mht1 and Sam4) in yeast capable of recognizing and metabolizing (R,S)-AdoMet. We found similar activities in worms, plants, and flies. However, it was not established whether these activities could prevent R,S accumulation. In this work, we show that both the Mht1 and Sam4 enzymes are capable of preventing R,S accumulation in Saccharomyces cerevisiae grown to stationary phase; deletion of both genes results in significant (R,S)-AdoMet accumulation. To our knowledge, this is the first time that such an accumulation of (R,S)-AdoMet has been reported in any organism. We show that yeast cells can take up (R,S)-AdoMet from the medium using the same transporter (Sam3) used to import (S,S)-AdoMet. Our results suggest that yeast cells have evolved efficient mechanisms not only for dealing with the spontaneous intracellular generation of the (R,S)-AdoMet degradation product but for utilizing environmental sources as a nutrient.
Keywords: Aging, Enzymes, Homocysteine, Metabolic Regulation, Methionine, S-adenosylmethionine (SAM), Transport Amino Acids, Yeast Metabolism
Introduction
Aging can be seen as the accumulation of damaged biomolecules over time (1–3). As such, understanding the mechanisms by which organisms can slow such accumulation, as well as how these mechanisms may themselves eventually break down and fail, is crucial to an understanding of the aging process. Repair pathways for damaged DNA have been well established (4); damaged proteins can be removed by a combination of proteolytic and repair pathways (3, 5–8). However, we only are beginning to understand how cells can prevent the accumulation of damaged small molecules.
To date, there are only a few pathways known for metabolizing damaged or unwanted small molecules. trans-Aconitate, the spontaneous degradation product of the citric acid cycle intermediate cis-aconitate, can be detoxified by the action of a specific methyltransferase (9). l-2-Hydroxyglutarate is formed as an abnormal byproduct when l-malate dehydrogenase uses α-ketoglutarate rather than oxalacetate as a substrate. The accumulation of the toxic l-2-hydroxyglutarate product is prevented, however, by the action of an enzyme that converts it back to α-ketoglutarate. Defects in this repair enzyme in humans leads to the brain disorder l-2-hydroxyglutaric aciduria (10). A third example, the diastereomer of S-adenosylmethionine ((R,S)-AdoMet), forms spontaneously from (S,S)-AdoMet (11–14) and is recognized by specific homocysteine methyltransferases in yeast and other organisms for conversion to the normal metabolites methionine and S-adenosylhomocysteine (15).
AdoMet is the most common methyl donor in the cell and is second only to ATP as the most widely used cofactor (16–19). Over time, however, AdoMet can degrade in several ways. Briefly, there is an intramolecular reaction that produces homoserine lactone and 5′-methylthioadenosine, a hydrolysis reaction that produces adenine and S-pentosylmethionine, and the racemization reaction described above that produces (R,S)-AdoMet (13, 14, 20). (R,S)-AdoMet forms from the inversion of the configuration of the sulfonium center of the molecule from S to R. The full extent of (R,S)-AdoMet toxicity is still unknown. In addition to simply taking up valuable cell space, it has been shown to lead to the production of cellular inhibitors (21, 22) as well as to directly inhibit some methyltransferase reactions (23). At least one of the latter inhibitory effects, however, is still controversial (24).
Interestingly, (R,S)-AdoMet levels are low to undetectable in the several tissues that have been assayed (13, 14, 24). (R,S)-AdoMet levels were undetectable in immature soybeans, soybean callus culture, radish leaves, yeast, and rat liver (13); an (R,S)/(S,S)-AdoMet ratio of 0.03 was found in mouse liver (14); whereas (R,S)-AdoMet was found to be only 3% of total AdoMet in rat brain (24). These low (R,S)-AdoMet levels are in contrast to those predicted based on the measured racemization rate indicating that ∼20% of AdoMet should be in the R,S form in cells that maintain a steady state level of AdoMet (14). These findings suggest that there are mechanisms responsible for keeping (R,S)-AdoMet levels at low levels in cells.
There are several possible ways that (R,S)-AdoMet can be depleted from cells. At least one radical AdoMet enzyme, the HemN coproporphyrin III oxidase in Escherichia coli, has been found to bind both (R,S)- and (S,S)-AdoMet (25). However, it is unclear whether the R,S form is converted to methionine and the deoxyadenosyl radical. Another depletion pathway involves homocysteine methyltransferase activity that transfers methyl groups from a methyl donor to homocysteine to create methionine. We previously found that two homocysteine methyltransferases in the yeast Saccharomyces cerevisiae, Mht1 and Sam4, are capable of using (R,S)-AdoMet as the methyl donor, thus metabolizing this molecule (15). Sam4 was found to metabolize both the S,S and the R,S form of AdoMet, with a higher affinity for the R,S form, while Mht1 was found to metabolize only the R,S form. Homologs for Mht1 and Sam4, as well as (R,S)-AdoMet dependent homocysteine methyltransferase activities, were also found in worms, plants, and flies (15).
In this work, we now demonstrate that these enzymes are capable of limiting the accumulation of (R,S)-AdoMet in intact yeast cells. Furthermore, we show that yeast cells are capable of transporting (R,S)-AdoMet from the external environment, suggesting that yeast can metabolize both endogenously formed and environmental (R,S)-AdoMet.
EXPERIMENTAL PROCEDURES
Yeast Strains and Growth for Accumulation Assays
Strains used are described in Table 1. Strains CVY1, -2, and -3 were prepared by mating the mht1− (BY4741) and sam4− (BY4742) strains followed by tetrad analysis as described previously (15). The presence or absence of the MHT1 and SAM4 genes was determined by PCR analysis and sensitivity to kanamycin as described previously (15). Cells were grown after an initial inoculation in a 6-ml YPD (1% bacto-yeast extract, 2% bacto-peptone, 2% dextrose) starter culture and shaken overnight at 30 °C. These cultures were then diluted in 250 ml of YPD to an optical density of 0.01 at 600 nm and then incubated with shaking at 225 rpm at 30 °C. For the accumulation experiments, 25 ml of the culture was collected at various time points. A 100-μl aliquot was diluted with 900 μl of water to measure the optical density at 600 nm with a Beckman DU640B spectrophotometer. The rest of the 25 ml was centrifuged at 3,000 rpm for 5 min at 4 °C in a Beckman Coulter Allegra X-15R swinging bucket centrifuge. The resulting pellets were washed once after resuspension in 5 ml of deionized water and centrifugation as before. The final pellet was then stored at −80 °C until needed for extract preparation.
TABLE 1.
Yeast strains
| Strain | Relevant genotype | Source |
|---|---|---|
| BY4741 | MATahis3Δ leu2Δ0 met15Δ0 ura3Δ0 | SGDPa |
| mht1− (BY4741) | BY4741, Δyll062c::Kanr | SGDPa |
| BY4742 | MATα his3Δ leu2Δ0 lys2Δ0 ura3Δ0 | SGDPa |
| sam4− (BY4742) | BY4742, Δypl273w::Kanr | SGDP |
| mht1− (BY4742) | BY4742, Δyll062c::Kanr | SGDPa |
| CVY1 mht1−/sam4− | MATα Δypl273w::Kanr Δyll062c::Kanr Lys− Met− | This study |
| CVY2 mht1−/sam4− | MATα Δypl273w::Kanr Δyll062c::Kanr Lys+ Met+ | This study |
| mmp1− (BY4742) | BY4742, Δyll061w::Kanr | SGDPa |
| sam3− (BY4742) | BY4742, Δypl274w::Kanr | SGDPa |
| CVY3 | BY4742, Lys+ | This study |
a Strains were prepared by the Saccharomyces Genome Deletion Project and purchased from Invitrogen.
Preparation of Yeast Extracts for AdoMet Analysis
Cell pellets for each time point prepared as described above were melted on ice. 100 μl of each resulting wet cell pellet was then combined with 200 μl of deionized water and 100 μl of glass beads (0.55 mm soda lime; BioSpec Products, Bartlesville, OK) in a 1.6-ml S3 low retention polymer microcentrifuge tube. The resuspended cells were then alternately vortexed and iced for 1 min for seven cycles as described previously (15). Broken cells were then transferred to new tubes and centrifuged for 10 min at 20,800 × g at 4 °C. Supernatants were then transferred to new tubes and combined with 200 μl of 20% (w/v) trichloroacetic acid. After vortexing, the tubes were incubated on ice for 10 min and then centrifuged for 10 min as above. The supernatants were then stored at −80 °C until needed for HPLC analysis.
Measurement of (R,S)- and (S,S)-AdoMet in Yeast Extracts
50 μl of each extract was injected on a Partisil SCX column eluted at 1 ml/min with a 60/40 ratio of buffers A and B as described previously (15). Elution times for (R,S)- and (S,S)-AdoMet were determined using a 50/50 (R,S)/(S,S)-AdoMet standard prepared as described previously (15) and injected prior to each set of experiments.
Measurement of AdoMet Uptake in Intact Yeast
Single colonies of the different yeast strains were inoculated in 6 ml of YPD medium. They were grown overnight with shaking at 30 °C. Samples were then diluted 10-fold in 5 ml of fresh medium and grown to an optical density at 600 nm of 1.0. Depending on the experiment, a specified amount of radiolabeled (R,S)- or (S,S)-[3H]AdoMet,2 prepared as described previously (15), was combined with an aliquot from each culture for a total volume of 1 ml each and was incubated with shaking at 30 °C for the specified amount of time. At each time point, the tubes were removed from shaking and spun at 3,000 × g for 5 min at 4 °C. The resulting pellets were separated from the supernatants, and extracts were prepared by adding an equal volume of glass beads and two volumes of water and lysed as described above. Radioactivity was measured for the supernatants and extracts by combining each with 5 ml of fluor (Safety-Solve, Research Products International, Mount Prospect, IL) and counting them on a Beckman LS6500 counter. The configuration of the internalized (R,S)-AdoMet 9 h after uptake by BY4742 yeast was determined by injection of the extract onto the Partisil SCX column as described above along with the 50/50 R,S/S,S standard.
NMR Analysis of AdoMet Racemization
AdoMet (chloride salt; purity ∼70% with 1 mol/mol H2O and 4.6% methanol; Sigma) was dissolved in 0.1 m HCl at a concentration of 30 mg/ml and incubated at 30 °C and 37 °C. At specified time points, 100 μl aliquots were collected, dried, and dissolved in D2O to final concentrations of 6 mg/ml. The 1H NMR spectrum for 500 μl of each aliquot was determined using a Bruker ARX400 spectrometer operating at 400.13 MHz as described previously (15, 26). Relative levels of (R,S)- and (S,S)-AdoMet were determined using the integrals of NMR peaks at 2.89 and 2.93 ppm, respectively.
RESULTS
(R,S)-AdoMet Accumulates in mht1−/sam4− Yeast Grown in Stationary Phase
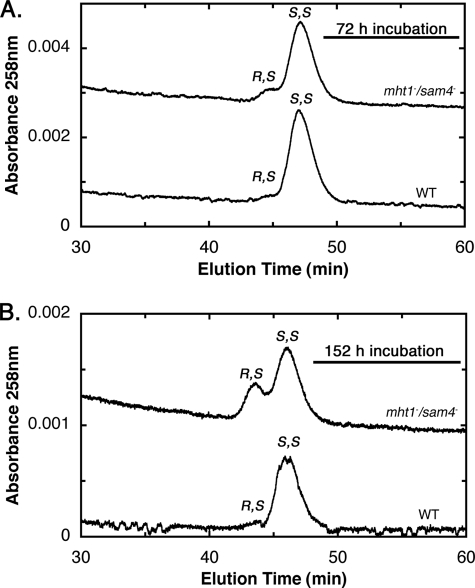
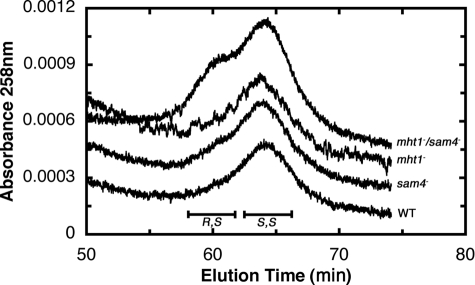
We first measured the relative levels of (R,S)-AdoMet and (S,S)-AdoMet after HPLC separation from extracts of yeast in stationary phase. For the BY4742 wild type parent strain, we found that almost all of AdoMet is in the S,S configuration at both the 72- and 152-h incubation points (Fig. 1). However, for the CVY1 mutant strain deleted in both the MHT1 and SAM4 genes, we found an accumulation of (R,S)-AdoMet at 72 h in stationary phase and an even greater accumulation at 152 h (Fig. 1). Analysis of a similar experiment with an independently prepared double mutant (CVY2) and wild type parent strain (CVY3) gave similar results (data not shown). When we measured (S,S)- and (R,S)-AdoMet levels in single mutant strains of either the MHT1 or SAM4 genes in stationary phase, we found that the presence of either gene was sufficient to reduce R,S levels to nearly those of wild type cells (Fig. 2).
FIGURE 1.
(R,S)-AdoMet accumulates more rapidly in the S. cerevisiae mht1−/sam4− CVY1 strain than in the wild type parent BY4742 strain. Wild type or mht1−/sam4− yeast were grown for either 72 h (optical density at 10.5 and 8.4, respectively) (upper panel) or 152 h (optical density at 13.2 and 9.5, respectively) (lower panel) as described under the “Experimental Procedures.” (R,S)- and (S,S)-AdoMet were separated from extracts by cation exchange HPLC and quantitated by their UV absorbance as described under “Experimental Procedures.” The position of R,S and S,S elution was determined using R,S/S,S racemic AdoMet prepared and analyzed by HPLC as described previously (Ref. 15, Fig. 4). Similar results were obtained in three replicate samples. Baseline absorbance values of the mht1−/sam4− traces were increased by 0.0025 and 0.001 absorbance units for A and B, respectively, to separate the traces.
FIGURE 2.
MHT1 or SAM4 can prevent accumulation of (R,S)-AdoMet. Wild type (WT), mht1−, sam4−, and mht1−/sam4− strains were grown for 190 h (16.8, 16.6, 16.7, and 14.8 optical density at 600 nm, respectively) as in Fig. 1. (R,S)- and (S,S)-AdoMet in cell extracts were separated and analyzed as described in Fig. 1. Baseline absorbance values were adjusted to separate the individual traces.
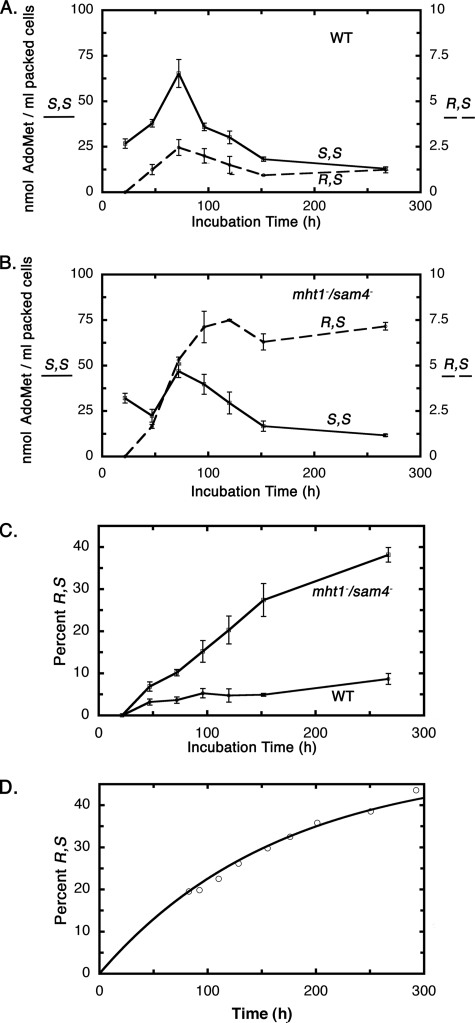
To determine the kinetics of R,S accumulation, we measured the concentration of (R,S)-AdoMet and (S,S)-AdoMet as a function of incubation time for the BY4742 wild type strain and the CVY1 mht1−/sam4− double mutant strain (Fig. 3). We found that the total levels of AdoMet measured at times up to 72 h for both strains were similar to those found previously in other strains (27). For the wild type cells shown in Fig. 3A, the level of (S,S)-AdoMet peaked at 72 h and then declined as cells continue in stationary phase. A similar rise and fall was found for the level of (R,S)-AdoMet that was present at 0 to 7% of the total level of AdoMet (Fig. 3C). For the mutant strain, the levels of (S,S)-AdoMet were similar to those found for the wild type strain (Fig. 3B). However, the level of (R,S)-AdoMet in the mutant cells continued to rise with incubation time in stationary phase, resulting in its net accumulation (Fig. 3B). These data are summarized in Fig. 3C, which shows how the percent of AdoMet in the R,S form increases with time in both the wild type and mutant strains but much more extensively in the mutant strain.
FIGURE 3.
Quantification of (R,S)- and (S,S)-AdoMet in wild type and in mht1−/sam4− double knock-out yeast incubated for various times. Levels of (S,S)- and (R,S)-AdoMet as a function of incubation time for wild type (WT) yeast (A) and for mht1−/sam4− yeast (B). In each case, the results of triplicate experiments are shown with error bars representing ± S.D. Note that the scale for (R,S)-AdoMet has been expanded 10-fold for A and B. C, the percentage of AdoMet in the R,S form in wild type and mht1−/sam4− yeast is given as an average of three experiments ± S.D. D, chemical racemization of AdoMet was determined as described under “Experimental Procedures.” The percent of total AdoMet in the R,S form is shown as a function of time for AdoMet incubated at pH 1.0 and 30 °C (open circles). A rate constant of 8.3 × 10−7 s−1 was calculated from the data points using a sum of least squares method; the line was determined from this value. This value is ∼3-fold less than that determined using similar methods at 37 °C (2.6 × 10−6 s−1; not shown) and comparable to previously determined literature rates at 37 °C of 1.8 × 10−6 s−1 (14), 2.4 × 10−6 s−1 (13), and 8 × 10−6 s−1 (20).
To compare the rate of accumulation of (R,S)-AdoMet in the mutant strain with the rate of spontaneous chemical formation, we measured the racemization rate of AdoMet in solution at the growth temperature of 30 °C (Fig. 3D). We found that the accumulation of (R,S)-AdoMet in yeast cells and in chemical solution was similar (compare Fig. 3, C and D). The in vivo formation of (R,S)-AdoMet in the mht1−/sam4− double knock-out strain was somewhat less that that of chemical formation, possibly due to the continuous production of (S,S)-AdoMet that occurs up until the 72-h incubation point (Fig. 3B). In both cases, the R,S:S,S ratio approached a 50:50 equilibrium.
In these experiments, we noted no difference in the growth of the parent, the mht1− or sam4− knock-out strains or the double knock-out mht1−/sam4− strain as determined by viable cell counts (data not shown).
(R,S)-AdoMet Is Transported from the Medium into the Cell via Sam3
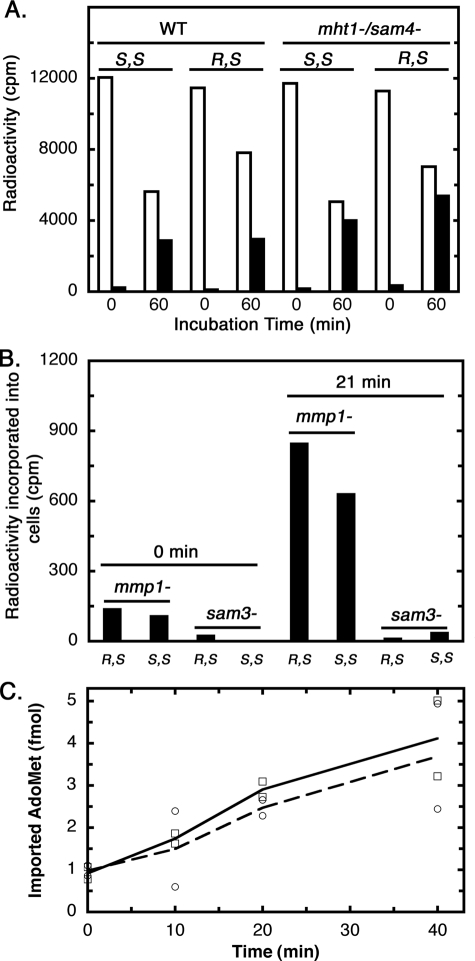
It was shown previously that a racemic mixture of AdoMet could be taken up by yeast as readily as (S,S)-AdoMet (28). We now have measured directly the uptake of (R,S)-AdoMet in both the BY4742 parent and the mht1−/sam4− double knock-out strains (Fig. 4). We showed that the uptake of (R,S)-AdoMet was similar to that of (S,S)-AdoMet (Fig. 4A). We then asked whether this transport was facilitated by one or both of the known transporters Mmp1 (specific for S-methylmethionine (29)) and Sam3 (specific for AdoMet (30)). We found that the transport of both (S,S)- and (R,S)-AdoMet was blocked when the SAM3 gene was knocked out, whereas transport continued when the MMP1 gene was knocked out (Fig. 4B). These results indicate that Sam3 is a transporter for both (R,S)- and (S,S)-AdoMet. To compare the efficiency of transport of each stereoisomer, the initial rate of uptake was measured at low concentrations, where differences in either the Km or Vmax of transport would be apparent (Fig. 4C). We found that the rate was similar for each isomer, supporting the previous study showing similar rates of uptake of racemic and (S,S)-AdoMet (28).
FIGURE 4.
Both (R,S)- and (S,S)-AdoMet are imported into yeast via the Sam3 transporter. A, wild type (BY4742) and mht1−/sam4− (CVY1) yeast were incubated with 17,000 cpm/ml (200 pm) of either (S,S)- or (R,S)-[3H]AdoMet for 0 or 60 min at 30 °C as described under “Experimental Procedures.” Cells were centrifuged, and radioactivity was measured in either the supernatant (open bars) or the cell extract prepared from the pellet (filled bars). B, a similar experiment was performed as in A but with yeast cells lacking the Mmp1 S-methylmethionine transporter or the Sam3 AdoMet transporter. AdoMet concentrations for this experiment were 9,000 cpm/ml (100 pm). C, comparison of the initial rate of uptake of (S,S)- or (R,S)-[3H]AdoMet in wild type BY4742 cells. Cells (7 ml at an optical density of 1.0 at 600 nm) were incubated in duplicate with either isotopically labeled isomer of AdoMet at a final concentration of 28 pm. At the indicated time points, 1.0 ml was taken for analysis as above. The uptake into cells of (S,S)-[3H]AdoMet is shown by the solid line (open squares); the uptake of (R,S)-[3H]AdoMet is shown by the dotted line (open circles).
Utilization of Exogenous (R,S)-AdoMet in Yeast
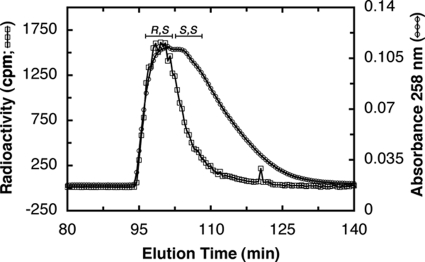
We then attempted to trace the metabolism of radiolabeled (R,S)-AdoMet when added to intact yeast cells in log phase for 9 h (Fig. 5). We were surprised to find it remained in the R,S configuration. This suggests that either the in vivo homocysteine methylation reaction is slow or that (R,S)-AdoMet is stored, possibly into the vacuole. Previous studies have provided evidence for the transport of both (R,S)- and (S,S)-AdoMet into this organelle (28). This result suggests that yeast cells may store exogenous (R,S)-AdoMet as a reserve and only utilize it in during stationary phase when nutrients are more scarce, presumably metabolizing it with the Mht1 and Sam4 enzymes once released into the cytosol.
FIGURE 5.
(R,S)-AdoMet is not metabolized immediately in yeast cells. Yeast cells (strain BY4742) were incubated in 51,600 cpm/ml (573 pm) (R,S)-[3H]AdoMet for 9 h at 30 °C in YPD medium. The configuration of the incorporated AdoMet was determined by injecting the extract along with unlabeled R,S/S,S standard on a cation exchange column as described in Fig. 1. The unlabeled AdoMet standard (circles) was determined via absorbance at 258 nm. The incorporated AdoMet (squares) was determined via radioactive counts of 500-μl fractions.
DISCUSSION
This study demonstrates that (R,S)-AdoMet can form in yeast from (S,S)-AdoMet and that both the Mht1 and Sam4 enzymes are capable of metabolizing it. The finding that (R,S)-AdoMet accumulates when both the SAM4 and MHT1 genes are deleted suggests that these are the major enzymes involved in limiting its buildup. It thus appears that YMR321C, encoding a protein that is 99% identical to Sam4 over the last 103 amino acids, may not be involved in (R,S)-AdoMet metabolism (31–33). These results also suggest that (R,S)-AdoMet may not be simply excreted, or converted back to the S,S form via radical SAM enzymes, or spontaneously degraded. Thus, Mht1 and Sam4 are the major factors that maintain low levels of cellular (R,S)-AdoMet in yeast.
It was shown previously that yeast are capable of taking up exogenous (S,S)-AdoMet (34) and that a single protein Sam3 is the transporter responsible for this uptake (29, 30, 35). It has also been shown that the racemic mixture of (R,S)/(S,S)-AdoMet is taken up as readily as the S,S form, suggesting that the R,S form can be transported as well (28). Our study confirms that the Sam3 transporter can take up (R,S)- as well as (S,S)-AdoMet. The fact that that R,S is taken up by yeast suggests that these cells may be capable of not only utilizing (R,S)-AdoMet that breaks down from (S,S)-AdoMet intracellularly, but also that R,S-AdoMet that may be available extracellularly. It is likely that AdoMet present in the environment from decaying organisms may be largely racemized, and the ability of yeast cells to use R,S-AdoMet may given them a significant advantage in obtaining carbon, sulfur, and nitrogen. The capability to use the R,S as well as the S,S form of AdoMet as a nutrient source may, in fact, provide an evolutionary advantage to the yeast, as well as to other lower organisms containing (R,S)-AdoMet specific homocysteine methyltransferases (36). Interestingly, we have not detected any such activity in mammalian cells; how these cells limit (R,S)-AdoMet accumulation is still unknown (36).
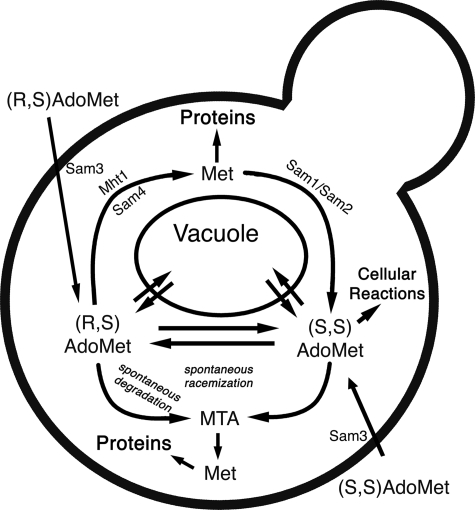
In yeast cells, AdoMet can be transported and stored in the vacuole (34). The identity of these transporters remains elusive (37). Both the R,S and S,S forms of AdoMet are apparently transported into the vacuoles (28). These results provide a rationale for our observation here that exogenously supplied (R,S)-AdoMet remains in the R,S configuration for at least 9 h after uptake from the medium; this material may be rapidly taken up into the vacuole and sequestered from the Mht1 and Sam4 enzymes localized in the cytosol (38, 39). For cells growing in log phase, the ready availability of nutrients may not require the utilization of vacuolar stores of AdoMet. However, when cells are in stationary phase, they may draw upon the stored (R,S)-AdoMet and use the Mht1 and Sam4 proteins present in the cytoplasm to metabolize it to methionine. We summarize the metabolism of (R,S)-AdoMet in Fig. 6.
FIGURE 6.
(R,S)-AdoMet metabolism in S. cerevisiae.
Several recent studies have suggested that Mht1 and Sam4 may play a role in the ability of yeast to survive under stressful conditions. For example, a 2-fold amplification at the left end of chromosome 16 that contains the SAM3 and SAM4 genes was found in the JAY270 strain of yeast that demonstrates unusual tolerance to high temperature and oxidative stress (40). It has been speculated that these genes may play a role in this tolerance (40). Additionally, Sam4 protein levels were found to be increased under menadione-induced oxidative stress (41). Both MHT1 and SAM4 have been linked to the arsenic response network (42), and MHT1 was found to be up-regulated 2-fold in response to methanol (43). Finally, SAM4 mRNA has been shown to decrease in aging yeast (44). Although the specific mechanisms by which Mht1 and Sam4 protect cells have not been determined, their role in preventing (R,S)-AdoMet accumulation may be important, especially under conditions such as heat stress where the spontaneous formation of this material would be enhanced.
Acknowledgments
We thank Giancarlo Costaguta (UCLA) for assistance with yeast tetrad analysis and Gregory Payne (UCLA) for helpful advice. We are also grateful to Albert Chan for contributions to this study and to Timothy Garrow for continuing helpful advice.
This work was supported, in whole or in part, by National Institutes of Health Grants GM026020 and AG032303. This work was also supported by the Ellison Medical Foundation.
- [3H]AdoMet
- S-adenosyl-L-[methyl-3H]-methionine
- HPLC
- high performance liquid chromatography
- YPD
- yeast peptone dextrose.
REFERENCES
- 1.Baynes J. W. (2000) Biogerontology 1, 235–246 [DOI] [PubMed] [Google Scholar]
- 2.Hipkiss A. R. (2001) Biogerontology 2, 173–178 [DOI] [PubMed] [Google Scholar]
- 3.Clarke S. (2003) Ageing Res. Rev. 2, 263–285 [DOI] [PubMed] [Google Scholar]
- 4.Lombard D. B., Chua K. F., Mostoslavsky R., Franco S., Gostissa M., Alt F. W. (2005) Cell 120, 497–512 [DOI] [PubMed] [Google Scholar]
- 5.Breusing N., Grune T. (2008) Biol. Chem. 389, 203–209 [DOI] [PubMed] [Google Scholar]
- 6.Cuervo A. M. (2008) Trends Genet. 24, 604–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ugarte N., Petropoulos I., Friguet B. (2010) Antioxid. Redox Signal., in press [DOI] [PubMed] [Google Scholar]
- 8.Veiga da-Cunha M., Jacquemin P., Delpierre G., Godfraind C., Théate I., Vertommen D., Clotman F., Lemaigre F., Devuyst O., Van Schaftingen E. (2006) Biochem. J. 399, 257–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai H., Strouse J., Dumlao D., Jung M. E., Clarke S. (2001) Biochemistry 40, 2210–2219 [DOI] [PubMed] [Google Scholar]
- 10.Rzem R., Vincent M. F., Van Schaftingen E., Veiga-da-Cunha M. (2007) J. Inherit. Metab. Dis. 30, 681–689 [DOI] [PubMed] [Google Scholar]
- 11.De La Haba G., Jamieson G. A., Mudd S. H., Richards H. H. (1959) J. Am. Chem. Soc. 81, 3975–3980 [Google Scholar]
- 12.Cornforth J. W., Reichard S. A., Talalay P., Carrell H. L., Glusker J. P. (1977) J. Am. Chem. Soc. 99, 7292–7300 [DOI] [PubMed] [Google Scholar]
- 13.Creason G. L., Madison J. T., Thompson J. F. (1985) Phytochemistry 24, 1151–1155 [Google Scholar]
- 14.Hoffman J. L. (1986) Biochemistry 25, 4444–4449 [DOI] [PubMed] [Google Scholar]
- 15.Vinci C. R., Clarke S. G. (2007) J. Biol. Chem. 282, 8604–8612 [DOI] [PubMed] [Google Scholar]
- 16.Cantoni G. L. (1975) Annu. Rev. Biochem. 44, 435–451 [DOI] [PubMed] [Google Scholar]
- 17.Lu S. C. (2000) Int. J. Biochem. Cell Biol. 32, 391–395 [DOI] [PubMed] [Google Scholar]
- 18.Fontecave M., Atta M., Mulliez E. (2004) Trends Biochem. Sci. 29, 243–249 [DOI] [PubMed] [Google Scholar]
- 19.Loenen W. A. (2006) Biochem. Soc. Trans. 34, 330–333 [DOI] [PubMed] [Google Scholar]
- 20.Wu S. E., Huskey W. P., Borchardt R. T., Schowen R. L. (1983) Biochemistry 22, 2828–2832 [DOI] [PubMed] [Google Scholar]
- 21.McCarthy D. L., Capitani G., Feng L., Gruetter M. G., Kirsch J. F. (2001) Biochemistry 40, 12276–12284 [DOI] [PubMed] [Google Scholar]
- 22.Satoh S., Yang S. F. (1989) Arch. Biochem. Biophys. 271, 107–112 [DOI] [PubMed] [Google Scholar]
- 23.Borchardt R. T., Wu Y. S. (1976) J. Med. Chem. 19, 1099–1103 [DOI] [PubMed] [Google Scholar]
- 24.Beaudouin C., Haurat G., Laffitte J. A., Renaud B. (1993) J. Neurochem. 61, 928–935 [DOI] [PubMed] [Google Scholar]
- 25.Layer G., Moser J., Heinz D. W., Jahn D., Schubert W. D. (2003) EMBO J. 22, 6214–6224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanna G. M. (2004) Pharmazie 59, 251–256 [PubMed] [Google Scholar]
- 27.Chan S. Y., Appling D. R. (2003) J. Biol. Chem. 278, 43051–43059 [DOI] [PubMed] [Google Scholar]
- 28.Nakamura K. D., Schlenk F. (1974) J. Bacteriol. 120, 482–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spence K. D. (1971) J. Bacteriol. 106, 325–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rouillon A., Surdin-Kerjan Y., Thomas D. (1999) J. Biol. Chem. 274, 28096–28105 [DOI] [PubMed] [Google Scholar]
- 31.López-Villar E., Monteoliva L., Larsen M. R., Sachon E., Shabaz M., Pardo M., Pla J., Gil C., Roepstorff P., Nombela C. (2006) Proteomics 6, S107–S118 [DOI] [PubMed] [Google Scholar]
- 32.Katju V., Farslow J. C., Bergthorsson U. (2009) Genome Biol. 10, R75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petrossian T., Clarke S. (2009) Epigenomics 1, 163–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barnett J. A. (2008) Yeast 25, 689–731 [DOI] [PubMed] [Google Scholar]
- 35.Petrotta-Simpson T. F., Talmadge J. E., Spence K. D. (1975) J. Bacteriol. 123, 516–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vinci C. R., Clarke S. G. (2010) Rejuvenation Res., 13, 362–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wiederhold E., Gandhi T., Permentier H. P., Breitling R., Poolman B., Slotboom D. J. (2009) Mol. Cell Proteomics 8, 380–392 [DOI] [PubMed] [Google Scholar]
- 38.Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. (2003) Nature 425, 686–691 [DOI] [PubMed] [Google Scholar]
- 39.Woo D. K., Phang T. L., Trawick J. D., Poyton R. O. (2009) Biochim. Biophys. Acta 1789, 135–145 [DOI] [PubMed] [Google Scholar]
- 40.Argueso J. L., Carazzolle M. F., Mieczkowski P. A., Duarte F. M., Netto O. V., Missawa S. K., Galzerani F., Costa G. G., Vidal R. O., Noronha M. F., Dominska M., Andrietta M. G., Andrietta S. R., Cunha A. F., Gomes L. H., Tavares F. C., Alcarde A. R., Dietrich F. S., McCusker J. H., Petes T. D., Pereira G. A. (2009) Genome Res. 19, 2258–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim I., Yun H., Jin I. (2007) J. Microbiol. Biotechnol. 17, 207–217 [PubMed] [Google Scholar]
- 42.Haugen A. C., Kelley R., Collins J. B., Tucker C. J., Deng C., Afshari C. A., Brown J. M., Ideker T., Van Houten B. (2004) Genome Biol. 5, R95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yasokawa D., Murata S., Iwahashi Y., Kitagawa E., Nakagawa R., Hashido T., Iwahashi H. (2010) Appl. Biochem. Biotechnol. 160, 1685–1698 [DOI] [PubMed] [Google Scholar]
- 44.Yiu G., McCord A., Wise A., Jindal R., Hardee J., Kuo A., Shimogawa M. Y., Cahoon L., Wu M., Kloke J., Hardin J., Mays Hoopes L. L. (2008) J. Gerontol. A. Biol. Sci. Med. Sci. 63, 21–34 [DOI] [PMC free article] [PubMed] [Google Scholar]