Abstract
GABAB receptors function as heterodimeric G-protein-coupled receptors for the neurotransmitter γ-aminobutyric acid (GABA). Receptor subtypes, based on isoforms of the ligand-binding subunit GABAB1, are thought to involve a differential set of associated proteins. Here, we describe two mouse lines that allow a straightforward biochemical isolation of GABAB receptors. The transgenic mice express GABAB1 isoforms that contain sequences for a two-step affinity purification, in addition to their endogenous subunit repertoire. Comparative analyses of purified samples from the transgenic mice and wild-type control animals revealed two novel components of the GABAB1 complex. One of the identified proteins, potassium channel tetramerization domain-containing protein 12, associates with heterodimeric GABAB receptors via the GABAB2 subunit. In transfected hippocampal neurons, potassium channel tetramerization domain-containing protein 12 augmented axonal surface targeting of GABAB2. The mice equipped with tags on GABAB1 facilitate validation and identification of native binding partners of GABAB receptors, providing insight into the molecular mechanisms of synaptic modulation.
Keywords: G-protein-coupled Receptors (GPCR), Mass Spectrometry (MS), Protein Purification, Protein-Protein Interactions, Trafficking, Transgenic, GABAB Receptor, KCTD12, T1 Domain, Tandem Affinity Tag
Introduction
GABAB receptors convey the metabotropic action of GABA,2 the main inhibitory neurotransmitter in the brain (1). They are concentrated at axonal boutons of GABAergic and glutamatergic neurons as well as in dendritic spines and shafts at extrasynaptic sites (2). Presynaptic GABAB receptors primarily inhibit calcium channels regulating evoked neurotransmitter release, whereas postsynaptic GABAB receptors mainly open G-protein-regulated inwardly rectifying potassium channels and thereby elicit slow inhibitory postsynaptic potentials. Additionally, GABAB receptor activation decreases the local cAMP level. GABAB receptors function as heteromeric G-protein-coupled receptors consisting of two subunits, GABAB1 and GABAB2. Heterodimerization is a prerequisite for both surface trafficking of the receptor and ligand-stimulated activation of a Gi/o-protein (3).
Two N-terminal GABAB1 variants, termed GABAB1a and GABAB1b differing only by a pair of sushi repeats that is exclusively present in GABAB1a, have been identified (4). They originate from the use of alternative transcriptional start sites (3, 5, 6) and underlie functional subtypes of the GABAB receptor with differential localization to pre- and postsynaptic sites in hippocampus and cortex (7–9). Because the sushi repeats are the only difference between GABAB1a and GABAB1b, it is to be expected that proteins interacting with these domains target or retain the GABAB1a isoform to specific microdomains. Consistently, a soluble recombinant protein of the two sushi repeats binds to neuronal membranes with low nanomolar affinity (10). An interaction of one of the two sushi repeats with fibulin-2 has been described (11), but the functional implication remains elusive.
Further subtypes of the GABAB receptor may be formed by a compartment-specific interaction of additional proteins, e.g. effector channels, with the core GABAB receptor complex (12). Furthermore, GABAB1 can associate with seven-transmembrane proteins differently from GABAB2 and may thereby modulate the signal transduction of other receptor systems (13, 14). Using the intracellular C-terminal domains of the GABAB receptor subunits as bait in genetic screens for interactors like the yeast two-hybrid system, a number of proteins ranging from transcription factors and RNA-binding proteins over trafficking factors to cytoskeletal elements and scaffolding proteins were identified (3, 15, 16). However, the native interactions of the GABAB receptor as an integral membrane protein complex may only be readily uncovered by biochemical isolation of the protein complex from brain membrane preparations followed by mass spectrometry analysis. The direct approach, immunoprecipitation of solubilized GABAB receptor complexes, relies on antisera that recognize the native conformation of GABAB1 or GABAB2 with high efficiency and specificity. As an alternative, tandem affinity purification (TAP) may be applied with the advantage of two consecutive purification steps under native conditions (17). In Saccharomyces cerevisiae the TAP strategy allowed the generation of a large protein interaction map (18). TAP proved useful in mammalian approaches as well (19–21). The first TAP-based transgenic mouse proteomics approach targeted interactions of 14-3-3 proteins, very abundant cytoplasmic regulators of cell signaling, and revealed almost 40 novel 14-3-3ζ-binding proteins (22). A TAP search for proteins binding to melatonin receptors stably expressed in HEK293 cells showed that this approach is also suitable to isolate G-protein-coupled receptor complexes from a cellular environment (23).
Here, we describe TAP of GABAB receptor complexes from mouse brain including genetic engineering of mice and analyses of purified fractions by immunoblotting and high accuracy mass spectrometry. The identification of two novel components of native GABAB receptors validated the approach.
EXPERIMENTAL PROCEDURES
DNA Constructs
Constructs for heterologous expression of rat GABAB1a (GenBankTM accession number Y10369), HA-GABAB1a, HA-SBP-GABAB1a, rat GABAB2 (GenBankTM accession number AF109405), HA-GABAB2, HA- GABAB2Δ921–940, and HA-GABAB2Δ817–940, were generated using a cytomegalovirus expression vector (24). Constructs for the expression of GABAB1a-CBP-TEV-ProtA, rat GABAB1b(Y10370)-CBP-TEV-ProtA, mouse KCTD12 (GenBankTM accession number NM_177715), and HA-GABAB2(817–920) were generated in pcDNA3. The N-terminal tags HA (amino acid sequence YPYDVPDYA) and HA-SBP (25) directly follow the signal sequence or the start methionine, the C-terminal tags, CBP-TEV-ProtA (17), and HA, the last amino acids of the wild-type proteins. pEGFP-C2 was used for the generation of fusion proteins of EGFP with KCTD12, KCTD12-T1 (amino acids 1–134), and KCTD12-CT (amino acids 135–327).
Antibodies
The purchased primary antibodies used for Western blotting were: CASK (610782, BD Biosciences, 1:1000), GABAB1 (sc-14006, Santa Cruz Biotechnology; 1:1000; AB1531, Millipore, 1:2000), GFP (ab6556, Abcam, 1:5000), GRP78 (ab21685, Abcam, 1:200), NSF (612272, BD Biosciences, 1:5000), PSD-95 (MA1-046, ABR, 1:2000), and HA (HA.11, Covance, 1:1000). The antibody to GABAB2 has been described previously (26). Polyclonal antibody to KCTD12 was raised in rabbits against a synthetic peptide corresponding to amino acid residues 254–267 in mouse KCTD12 coupled to Limulus polyphemus hemocyanin (Biogenes). The antiserum was affinity-purified using the immunogenic peptide.
Cell Culture
Growth media and additives were purchased from Invitrogen. The cell lines were grown following standard protocols. Primary hippocampal neurons were prepared from newborn rats (postnatal day (P) 0–P1) and plated on 12-mm glass coverslips coated with laminin and poly-l-lysine (Sigma-Aldrich) at a density of 105 cells/well in 24-well plates.
Bacterial Artificial Chromosome (BAC) Modification by ET Cloning
BAC citb544e14 (AF532114), kindly provided by Kirsten Fischer Lindahl, was modified by homologous recombination in Escherichia coli HS996 following previously described protocols (27). A cassette consisting of the CBP-TEV-ProtA tag sequence and a kanamycin resistance gene was inserted into exon 18 of the GABAB1 gene (6), directly 5′ to the stop codon (between nucleotides 146573 and 146574 of BAC citb544e14), using pBAD-ETγ (27). Independently, a cassette encompassing the counterselection genes SacB (28) and RpsL (29) and a kanamycin resistance gene was inserted into GABAB1a exon 1a2 (6), directly 3′ to the sequence encoding the signal peptide (between nucleotides 120,273 and 120,274 of BAC citb544e14) and replaced with an HA-SBP tag coding sequence using pGETrec (30), which was kindly provided by Panayiotis A. Ioannou. The integrity of recombinant clones was confirmed by Southern blotting and DNA sequencing.
Generation of Transgenic Mouse Lines
Transgenic mice were generated essentially as described (31). BACs were purified (Nucleobond AX500, Machery-Nagel), diluted at a concentration of approximately 0.5 μg/μl (10 mm Tris, pH 7.5, 0.5 mm EDTA, 30 nm spermine, 20 nm spermidine, 0.1 m NaCl), and microinjected into pronuclei resulting from F1-C57BL/6/CBA matings. The embryos were transferred to the oviduct of pseudopregnant F1-C57BL/6/CBA foster mothers. Progeny were genotyped by PCR analysis, and founders, in which the modified BAC randomly inserted into the genome, were mated onto a C57BL/6 background.
Aequorin Assay
Chinese hamster ovary/Gα16/mtAEQ cells (32), kindly provided by Atanas Ignatov, permanently expressing both mitochondria-targeted apoaequorin and the G-protein α-subunit Gα16, were plated into 96-well white/opaque microtiter plates (Nunc) and cotransfected with expression vectors for GABAB2 and wild-type or modified GABAB1a using Lipofectamine (Invitrogen). Two days later, the cells were loaded with coelenterazine (Perkin-Elmer Life Sciences) for 4 h and stimulated with (R)-baclofen (Tocris) at concentrations ranging from 0.01 to 100 μm. The luminescent response was measured 15 s after stimulation using a microluminometer (MicroLumat Plus, Perkin-Elmer Life Sciences). The half-maximal effective concentration (EC50) values for baclofen-stimulated G-protein coupling were calculated by fitting the relative light units into a sigmoidal dose-response curve using Origin 7.0 (Microcal software).
In Situ Hybridization
In situ hybridization on frozen mouse brain sections was performed as described previously (33). Briefly, the brains were frozen on solid CO2, and 16-μm sections were prepared on a cryostat (CM 3050-S, Leica Microsystems) and collected onto microscope slides (SuperFrost Plus, Menzel). Sense and antisense [35S]dUTP-labeled cRNA probes were generated by in vitro transcription (MAXIscript in vitro transcription kit, Ambion) of subcloned cDNA fragments. The probe sequences used were: B1a, nucleotides 1–488 of rat GABAB1a (Y10369); B1a/b pan, nucleotides 560–1062 of rat GABAB1a; HA-SBP, sequence encoding HA-SBP; and CBP-TEV-ProtA, nucleotides 36–555 of the sequence for the TAP tag (17). The hybridized sections were exposed to Kodak Biomax MR film for 3–5 days, dipped in Kodak NTB-3 nuclear emulsion (both from GE Healthcare), exposed for 8 days to 3 weeks, and counterstained with Mayer's hemalaun.
Electrophysiology
Acute transverse hippocampal slices (250 μm) were prepared from young (P21–P31) transgenic and wild-type mice and recovered for 1 h (first at 31 °C for 30 min and then at room temperature for at least 30 min) before starting whole cell recordings at 31 °C. The slices were submerged and perfused at 2 ml/min with artificial cerebrospinal fluid (125 mm NaCl, 25 mm NaHCO3, 2.5 mm KCl, 1.25 mm NaH2PO4, 1 mm MgCl2, 2 mm CaCl2, 25 mm glucose, pH 7.4, with 95% O2, 5% CO2). Excitatory or inhibitory postsynaptic currents (EPSCs or IPSCs) were electrically stimulated every 10 s by stimulating Schaffer collaterals in stratum radiatum ∼150 μm from the pyramidal layer with a glass pipette filled with artificial cerebrospinal fluid. Patch pipettes had resistances of 4–6 megohm when filled with internal solution (130 mm cesium gluconate, 5.5 mm KCl, 10 mm HEPES, 1 mm EGTA, 4 mm MgATP, 0.5 mm Na3GTP, 10 mm sodium phosphocreatine, 5 mm QX-314, pH 7.25, 280–290 mosmol). The liquid junction potential (16.3 mV) was corrected. EPSCs or IPSCs were recorded with an EPC9 (HEKA) at the reversal potential for GABA (−80 mV) or AMPA (0 mV), respectively. Baclofen (50 μm) and CGP54626 (5 μm) were kept as aliquots (stocks of 10 and 5 mm), and the solutions were freshly prepared on the day of the experiment. Holding currents were recorded in the presence of tetrodotoxin (1 μm) with patch electrodes containing 125 mm potassium gluconate, 10 mm sodium gluconate, 4 mm NaCl, 10 mm HEPES, 0.2 mm EGTA, 4 mm MgATP, 0.3 mm Na3GTP, 10 mm sodium phosphocreatine (pH 7.25, 280–290 mosmol). The recordings were excluded if input resistance and/or series resistance changed by more than 20%. The values are the means ± S.E., and statistical significance between the three genotypes was tested using one-way analysis of variance.
Extraction of Membrane Proteins from Mouse Brains
All of the protein work was performed at 4 °C if not stated otherwise. Whole brains were homogenized (S. Potter, B. Braun Biotech) in sucrose buffer (320 mm sucrose, 1 mm NaHCO3, 1 mm MgCl2, 50 μm CaCl2, protease inhibitors; Complete from Roche Applied Science), and the nuclei were removed (1,400 × g for 10 min). The membranes were pelleted (20,000 × g for 20 min), resuspended in solubilization buffer (400 mm NaCl, 10 mm Tris, pH 8.0, 1% Triton X-100, protease inhibitors), and incubated overnight on a rotator. The membrane debris was removed by ultracentrifugation (100,000 × g for 60 min). The resulting supernatant represents the solubilized membrane protein fraction.
Tandem Affinity Purification from NTAP-GABAB1a Mice
40–70 mg of solubilized membrane proteins were prepared from juvenile (P10–P16) mouse brains, and the 2-mercaptoethanol and EDTA concentrations were adjusted to 5 and 2 mm, respectively. The extract was rotated overnight with 720 μl of streptavidin-coated magnetic beads (Dynabeads MyOne Streptavidin T1, Invitrogen). Afterward, the beads were collected using a magnetic device (Invitrogen) and washed three times with 1 ml of solubilization buffer, and the bound material was eluted with 2 mm biotin in 1600 μl of solubilization buffer for 20 min on a rotating wheel. The biotin eluate was incubated with 160 μl of anti-HA affinity matrix (Roche Applied Science) rotating for 4 h. After the beads were washed three times with 1 ml of solubilization buffer, the bound material was eluted in SDS loading buffer (50 mm Tris-Cl, pH 8.0, 200 mm dithiothreitol, 6% (w/v) SDS, 10% glycerol, 7 m urea, 0.01% (w/v) bromphenol blue) for 10 min at 42 °C, separated on NuPage Novex gels (Invitrogen), and stained with silver nitrate as previously described (34).
Tandem Affinity Purification from GABAB1a/b-CTAP Mice
The purification was basically performed as previously described (17). 40–70 mg of solubilized membrane proteins prepared from juvenile (P10–P16) mouse brains were incubated with 100 μl of IgG-Sepharose beads (GE Healthcare) rotating overnight. The beads were collected by centrifugation (1 min at 1,000 × g), washed three times with 1 ml of solubilization buffer, and resuspended in 300 μl of the same buffer. Bound material was eluted from the IgG matrix using 80 units of TEV protease (Invitrogen) shaking for 2 h at 16 °C. The supernatant was collected and mixed with 900 μl of calmodulin binding buffer (1% Triton X-100, 150 mm NaCl, 10 mm Tris-Cl, pH 7.5, 7.5 mm β-mercaptoethanol, 0.75 mm MgAc, 0.75 mm imidazole, 2 mm CaCl2), and the CaCl2 concentration of the suspension was adjusted to 2 mm. The suspension was incubated with 50 μl of calmodulin-agarose (Stratagene) rotating for 2 h. After washing the beads three times with 1 ml of calmodulin binding buffer, the bound material was eluted in SDS loading buffer and analyzed as described above.
Mass Spectrometry Analysis
The applied work flow followed an established protocol (35). Briefly, the protein bands were cut out of the gel and subjected to in-gel reduction of protein disulfide bonds using dithiothreitol and subsequent alkylation by the addition of iodoacetamide. Next, the protein content was digested with trypsin overnight at 37 °C, and the peptides were extracted from the gel and then concentrated and desalted by stop and go extraction (36, 37). The samples were analyzed by liquid chromatography-tandem mass spectrometry using an Agilent 1200 high performance liquid chromatography system connected online to an LTQ-Orbitrap XL (Thermo Fisher Scientific). Fragment spectra were recorded in the LTQ and high resolution and mass accuracy spectra on the intact peptides in the Orbitrap (38). Centroided peak lists were searched with Mascot v2.2 (Matrix Science) against the mouse international protein index (IPI) database (39).
Coimmunoprecipitation
Solubilized membrane proteins (6–18 mg) of juvenile wild-type C57BL/6 mouse brains were supplemented with 10 μg of monoclonal mouse anti-GABAB1 antibody (ab55051, Abcam) or mouse IgG as a control (Sigma) and rotated overnight. The immune complexes were collected for 4 h on 50 μl of protein A beads (GE Healthcare), washed three times with 0.5 ml of solubilization buffer, and eluted in SDS loading buffer.
Purification of HA-tagged Proteins from Transfected HEK293 Cells
Two days after transfection with calcium phosphate, the cells were washed, harvested, resuspended in 600 μl of solubilization buffer, and rotated overnight. The extract was centrifuged for 1 h at 100,000 × g, and the supernatant was incubated with 20 μl of anti-HA affinity matrix (Roche Applied Science) rotating for 4 h. The matrix was washed three times with 0.5 ml of solubilization buffer, and the bound protein was eluted in SDS loading buffer.
Immunofluorescence of Cultured Neurons
Dissociated rat hippocampal neurons were transfected using calcium phosphate at day 7 in vitro. Two days later, the neurons were incubated with mouse anti-HA antibody (HA.11, Covance, 1:500) in the growth medium for 30 min at 10 °C. The neurons were washed with Dulbecco's modified Eagle's medium (Invitrogen), fixed with 4% (w/v) paraformaldehyde, 4% (w/v) sucrose/PBS for 10 min, permeabilized with 0.25% Triton X-100/PBS for 10 min, and blocked with 1% (w/v) BSA/PBS for 30 min, and rabbit anti-mictrotubule-associated protein 2 (MAP2) antibody (Synaptic Systems, 1:1000) was applied in 1% (w/v) BSA/PBS for 1 h. The fixation and all subsequent steps were performed at room temperature. After three brief washes with 1% (w/v) BSA/PBS, species-specific secondary antibodies (donkey anti-mouse Alexa 546, Molecular Probes, 1:1000; donkey anti-rabbit Cy5, Jackson ImmunoResearch, 1:500) were applied in 1% (w/v) BSA/PBS for 1 h. The neurons were washed three times in PBS, mounted on microscope slides (Aqua Poly/Mount, Polysciences), and observed under a confocal laser scanning microscope (Fluoview 1000, Olympus). The average HA-GABAB2 and EGFP fluorescence signal intensities within each of 10 rectangles covering a MAP2-negative neurite were determined using Olympus Fluoview 1.7b software. The mean HA-GABAB2/EGFP ratio was calculated as an estimate of the relative axonal HA-GABAB2 surface expression. The results from at least 38 MAP2-negative neurites are presented as the means ± S.E. A Student's t test was applied to calculate the statistical differences.
RESULTS
N- and C-terminal Tandem Affinity Tags on GABAB1
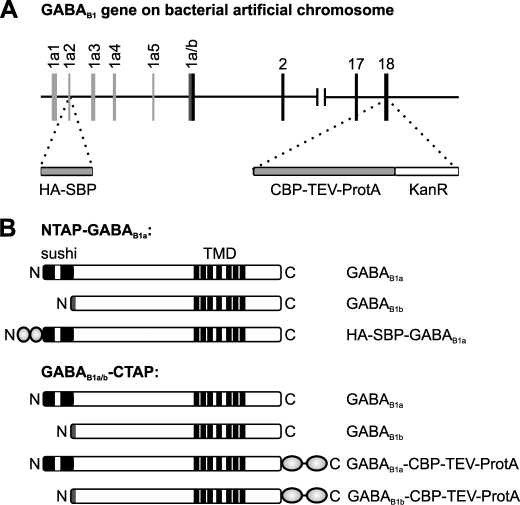
We set out to generate transgenic mice equipped with affinity tags for the purification of GABAB receptors. To this end, DNA sequences encoding two different TAP tags were independently inserted into BAC citb544e14 (40), which contains a fragment of mouse chromosome 17 including the gene for GABAB1, by homologous recombination in E. coli (27, 41) (Fig. 1A). The classical TAP tag consisting of CBP, the TEV protease recognition site, and protein A was appended to the C terminus of GABAB1 (GABAB1-CBP-TEV-ProtA). Alternatively, we added a tandem tag consisting of an HA epitope and a SBP to the N terminus of the mature GABAB1a polypeptide (HA-SBP-GABAB1a). The purpose of the former construct was to allow purification of GABAB1 complexes irrespective of the isoform, whereas the latter should help isolating specifically GABAB1a-containing receptors (Fig. 1B).
FIGURE 1.
Targeting TAP sequences into the GABAB1 gene on BAC citb544e14. A, sequences coding for the HA-SBP tag were introduced into the GABAB1a-specific exon 1a2 (6), 3′ to the signal sequence, whereas sequences coding for the CBP-TEV-ProtA tag and a kanamycin resistance gene were inserted into exon 18 (6), 5′ to the stop codon. GABAB1a-specific exons are shown in light gray, and the GABAB1b-specific region of exon 1a/b is in dark gray. B, scheme of tagged and untagged GABAB1 isoforms in the BAC transgenic mice. TMD, transmembrane domain; sushi, sushi repeat.
To check for the functional integrity of receptors containing these tags, we employed an aequorin assay. GABAB1a carrying the two different tandem affinity tags at either the N or the C terminus was coexpressed with GABAB2 in Chinese hamster ovary/Gα16/mtAEQ cells (32). The manipulated receptors were indistinguishable from wild-type versions with respect to baclofen-stimulated calcium mobilization (EC50(GABAB1a/GABAB2), 3.3 ± 2.8 μm; EC50(HA-SBP-GABAB1a/GABAB2), 2.7 ± 2.2 μm; EC50(GABAB1a-CBP-TEV-ProtA/GABAB2), 3.3 ± 2.5 μm), indicating that the tags on GABAB1a did not compromise surface expression, ligand binding, or G-protein coupling of the heterodimeric receptor.
BAC Transgenic Mice Expressing TAP-tagged GABAB1
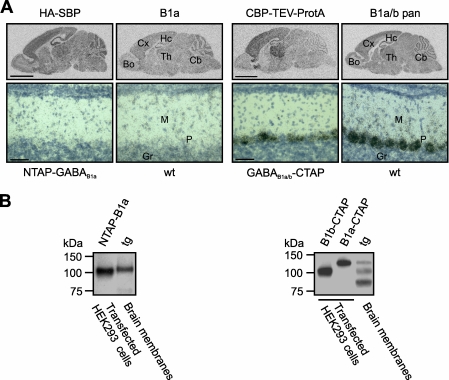
The modified BACs were injected into oocytes, and the transgenic mouse lines were established (NTAP-GABAB1a, GABAB1a/b-CTAP). The distribution of the transgenic brain mRNAs detected by in situ hybridization at P12 was largely identical to that of the endogenous transcripts in wild-type mice (Fig. 2A). However, in GABAB1a/b-CTAP mice the transgenic expression appeared slightly more pronounced in cortex and thalamus. Importantly, the BAC-derived messages recapitulated the cell type-specific expression of GABAB1 isoforms in the cerebellum known from wild-type mice (42, 43). Furthermore, expression of the TAP-tagged GABAB1 proteins was readily detectable on the Western blots of brain membranes from the transgenic mice (Fig. 2B; see also Fig. 4A).
FIGURE 2.
Expression of the transgenes in mouse brain. A, autoradiographic images (upper panels) and photomicrographs of the cerebellum (lower panels) depicting in situ hybridization of radioactively labeled cRNA probes detecting either the transgenic (HA-SBP, ProtA-TEV-CBP) or the wild-type (B1a, B1a/b pan) mRNAs at P12. Hc, hippocampus; Cx, cortex; Th, thalamus; Cb, cerebellum; Bo, bulbus olfactorius; Gr, granule cell layer; M, molecular layer; P, Purkinje cell layer. Scale bars, 3 mm in the upper panels; 50 μm in the lower panels. B, immunoblots detecting the tagged proteins in NTAP-GABAB1a (left panel, anti-HA) and GABAB1a/b-CTAP (right panel, anti-GABAB1 pan sc-14006) transgenic mice. The anti-GABAB1 blot also reveals GABAB1b (bottom band in the right lane), whereas GABAB1a comigrates with GABAB1b-CBP-TEV-ProtA. wt, wild-type; tg, transgenic; NTAP-B1a, HA-SBP-GABAB1a; B1b-CTAP, GABAB1b-CBP-TEV-ProtA; B1a-CTAP, GABAB1a-CBP-TEV-ProtA.
FIGURE 4.
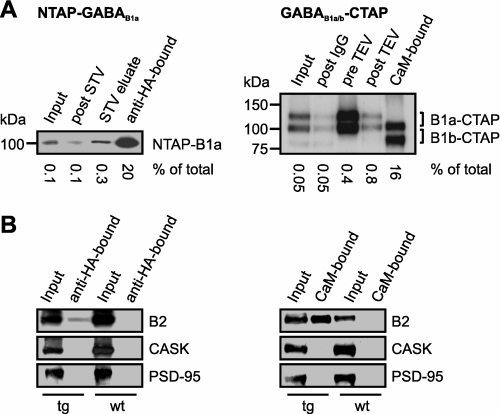
Tandem affinity purification of GABAB receptors from transgenic mouse brains. A, left panel, immunoblot of NTAP-GABAB1a purification steps stained with an anti-HA antibody; post STV, extract after incubation with the streptavidin (STV) matrix; STV eluate, protein eluted with biotin from the STV matrix. A, right panel, immunoblot of GABAB1a/b-CTAP purification steps stained with an anti-GABAB1 pan antibody (AB1531). post IgG, extract after incubation with the IgG matrix; pre TEV, IgG-bound protein prior to TEV protease cleavage; post TEV, eluate of the IgG matrix after incubation with TEV protease. Anti-GABAB1 AB1531 strongly reacted with protein A revealing primarily the TAP-tagged variants (compare with Fig. 2B). B, immunoblots of input and final TAP fractions prepared from the transgenic lines (tg) and wild-type (wt) control animals. B2, GABAB2; PSD-95, postsynaptic density protein 95.
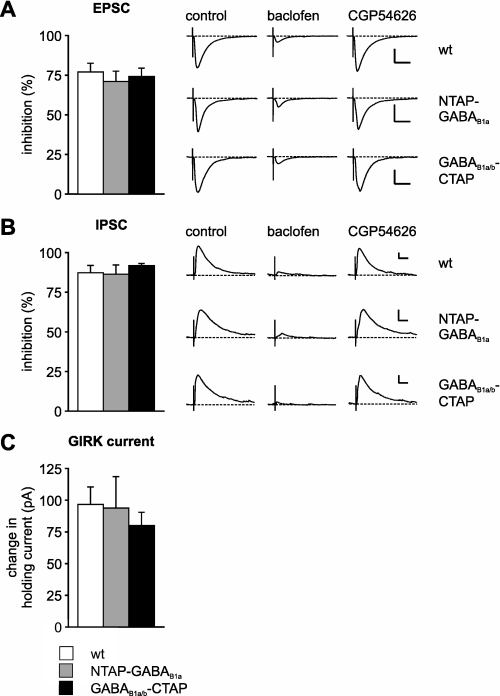
To confirm that the genetic manipulation did not interfere with physiological GABAB receptor functions, we assessed the baclofen effects in acute hippocampal slices of NTAP-GABAB1a, GABAB1a/b-CTAP, and wild-type mice (Fig. 3). Baclofen via activation of presynaptic GABAB receptors strongly reduced excitatory and inhibitory postsynaptic currents (EPSCs and IPSCs) recorded in CA1 pyramidal cells in all genotypes to similar extents (Fig. 3, A and B). Comparable increases in holding currents by baclofen in all three genotypes indicated normal postsynaptic GABAB receptor function (Fig. 3C).
FIGURE 3.
GABAB receptor activation in the hippocampus of wild-type and BAC transgenic mice. A, peak amplitudes of EPSCs recorded in CA1 neurons at −80 mV were similarly (p > 0.05) reduced in the presence of baclofen (50 μm) in wild-type (n = 8), NTAP-GABAB1a (n = 5), and GABAB1a/b-CTAP (n = 5) transgenic mice, as summarized in the bars and illustrated by representative current traces. Scale bars, 100 pA, 20 ms. B, peak amplitudes of IPSCs recorded at 0 mV were similarly (p > 0.05) reduced by baclofen (50 μm) in wild-type (n = 6), NTAP-GABAB1a (n = 6), and GABAB1a/b-CTAP (n = 4) transgenic mice. Scale bars for representative current traces, 50 pA, 20 ms. CGP54626 (5 μm) reversed the baclofen-mediated effects on EPSCs and IPSCs. All of the current traces are the averages of six consecutive traces. C, summary of changes in holding current recorded in the presence of tetrodotoxin (1 μm) at −50 mV following baclofen perfusion, reflecting activation of G-protein-regulated inwardly rectifying potassium channel currents in wild-type (n = 7), NTAP-GABAB1a (n = 3), and GABAB1a/b-CTAP (n = 4) transgenic mice. wt, wild-type.
Tandem Affinity Purification of GABAB1
Membrane proteins were solubilized from the brains of transgenic and wild-type mice and subjected to the respective TAP protocol. Western blot analysis of the final fractions (Fig. 4) revealed transgene-specific isolation not only of the tagged proteins, but also of GABAB2, indicating that the integrity of the receptors was conserved throughout the purification. Two prominent synaptic proteins not implicated with GABAB receptors, CASK and PSD-95, were not detectable in the final fractions.
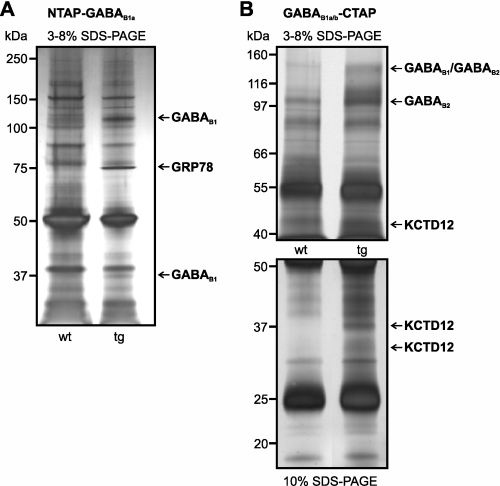
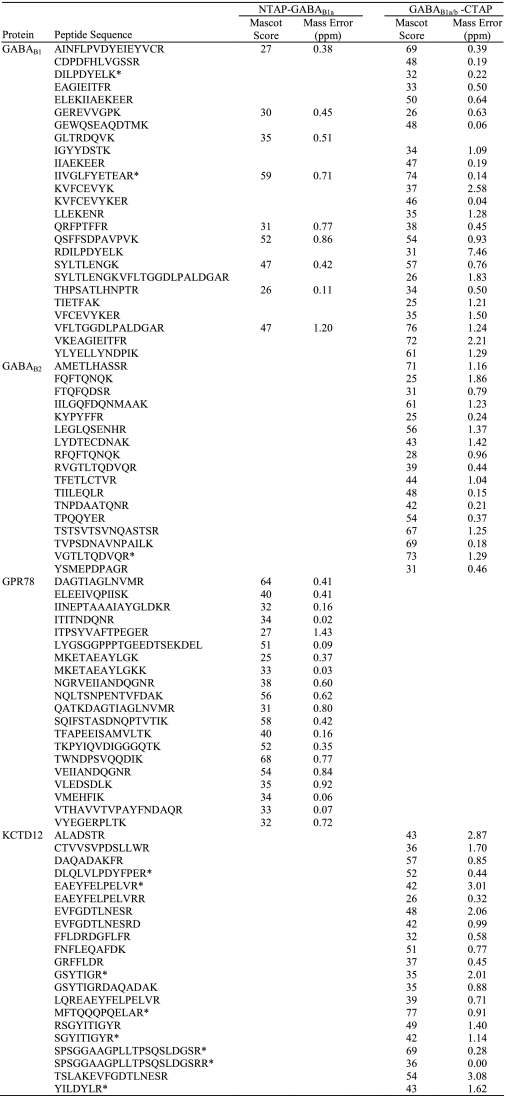
Silver staining of final TAP fractions separated by SDS-PAGE revealed complex patterns but allowed identification of bands obviously enriched in the transgenic samples by visual inspection (Fig. 5). These bands and their wild-type counterparts were isolated and subjected to trypsin digestion followed by liquid chromatography-tandem mass spectrometry. The peptide analysis revealed, besides GABAB receptor subunits, glucose-regulated protein 78 (GRP78/BiP) (44) in samples from NTAP-GABAB1a and potassium channel tetramerization domain-containing protein 12 (KCTD12/pfetin) (45) in samples from GABAB1a/b-CTAP mice (Table 1).
FIGURE 5.
Mass spectrometric identification of GRP78/BiP and KCTD12 as potential GABAB1-binding proteins. Shown are silver-stained SDS-PAGE of the final TAP fractions prepared from NTAP-GABAB1a (A) or GABAB1a/b-CTAP (B) mice and wild-type controls. A 50% excess of wild-type material was loaded to facilitate the identification of protein bands specifically enriched in the transgenic fractions. These bands, which are marked by arrows, and the corresponding regions from the wild-type control lanes were analyzed using mass spectrometry (Table 1).
TABLE 1.
Proteins identified from liquid chromatography-tandem mass spectrometry analysis of TAP fractions
Protein bands enriched in NTAP-GABAB1a or GABAB1a/b-CTAP fractions (Fig. 4) were analyzed by liquid chromatography-tandem mass spectrometry. All of the unambiguously identified peptide sequences from the corresponding proteins are listed along with the score of the identification reported from the Mascot search engine and the deviation of the experimentally determined mass from the theoretical in parts per million. The asterisk indicates peptides also identified in wild-type control samples.
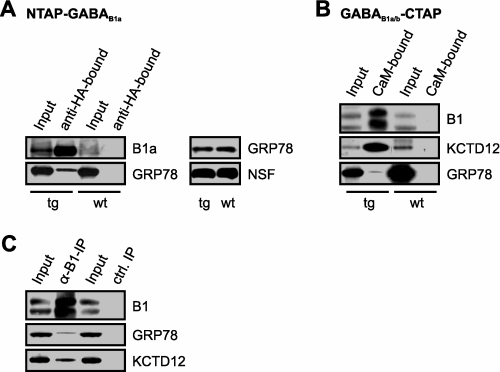
Western blots of the final TAP fractions confirmed the mass spectrometric identification of the two candidate GABAB receptor interactors (Fig. 6, A and B). Moreover, anti-GABAB1 immunoprecipitates from wild-type mice contained both GRP78 and KCTD12, corroborating them as constituents of native GABAB1 receptor complexes (Fig. 6C).
FIGURE 6.
GRP78 and KCTD12 are components of mouse brain GABAB1 complexes. A and B, immunoblots of input and final TAP fractions prepared from transgenic (tg) and wild-type (wt) animals. B1a, GABAB1a; B1, GABAB1. A, right panel, immunoblot with GRP78 antibody showing that extracts of NTAP-GABAB1a and wild-type mouse brains contain comparable amounts of GRP78; as a loading control the blot was incubated with an antibody to N-ethylmaleimide-sensitive factor (NSF). C, immunoblots of GRP78 and KCTD12 coimmunoprecipitated (IP) with GABAB1 from wild-type mouse brains. Input: 0.1% in A,B; 0.5% in C. Control (ctrl).
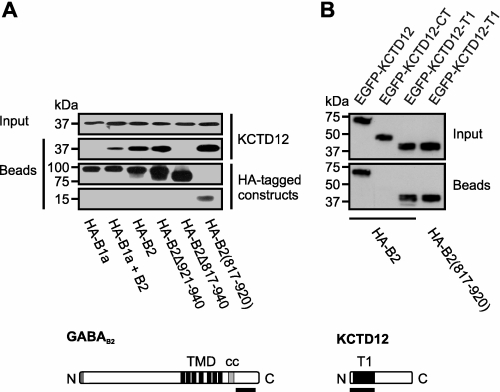
The T1 Domain of KCTD12 Interacts with GABAB Receptors via the C Terminus of GABAB2
The interaction between KCTD12 and the GABAB receptor was further assessed in transfected HEK293 cells (Fig. 7). KCTD12 bound to GABAB2 and not to GABAB1 and allowed formation of a ternary complex consisting of GABAB1, GABAB2, and KCTD12. We mapped the KCTD12 interaction site in GABAB2 to a region of the C terminus distal to the membrane (amino acids 817–920). GABAB2 binding was mediated by an N-terminal fragment of KCTD12 encompassing a voltage-gated potassium channel tetramerization (T1) domain (45). Thus, KCTD12 appears to interact with functional GABAB receptors by T1 domain binding to the C-terminal region of GABAB2.
FIGURE 7.
The C terminus of GABAB2 engages the T1 domain of KCTD12. Shown are analyses of anti-HA affinity purifications from HEK293 cells coexpressing HA-tagged receptor constructs with either KCTD12 (A) or EGFP-tagged KCTD12 mutants (B). Immunoblots with anti-KCTD12 (A) or anti-GFP (B) antibody reveal the KCTD12 interaction; anti-HA detection (A) confirms the integrity of the HA fusion proteins. Input, 0.125%. Bars beneath the protein schemes (lower panels) illustrate the interacting regions in GABAB2 (A) and KCTD12 (B). cc, coiled-coil domain.
Influence of KCTD12 on the Neuronal Distribution of GABAB Receptors
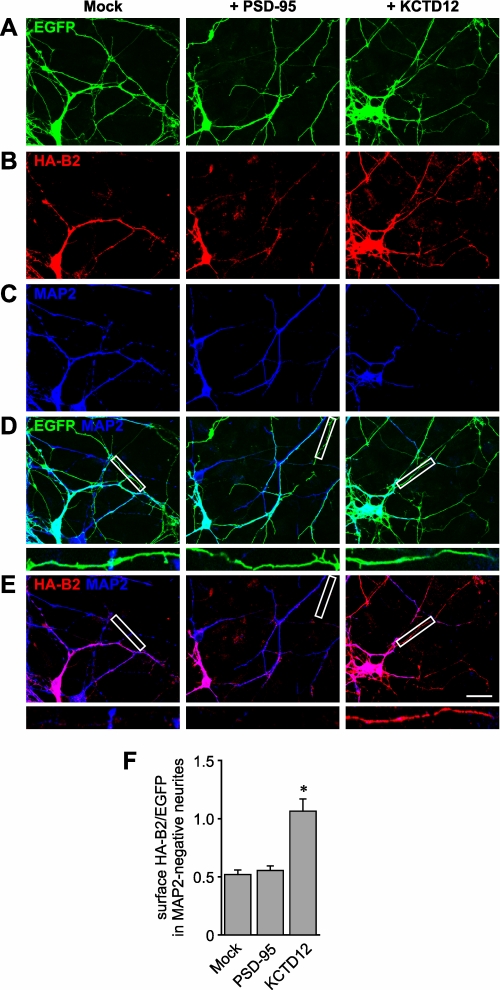
Transfection of KCTD12 constructs into primary neurons from rat hippocampus altered the distribution of coexpressed HA-GABAB2 (Fig. 8). KCTD12 increased the amount of HA-GABAB2 at the surface of neurites. This effect was largely confined to regions that were negative for the dendritic marker MAP2. These data suggest that KCTD12 supports GABAB2 surface expression primarily in axonal domains. Thus, KCTD12 may have an impact on the axonal transport or surface stability of GABAB receptors.
FIGURE 8.
Increased axonal GABAB2 expression upon coexpression of KCTD12 in transfected hippocampal neurons. Primary rat hippocampal neurons were transfected with expression vectors for EGFP, HA-GABAB2, and either PSD-95 (+ PSD-95), KCTD12 (+ KCTD12), or no additional protein (empty vector, Mock). The neurons were stained under nonpermeabilizing conditions with an anti-HA antibody and after permeabilization with an antibody directed against the dendritic marker MAP2. Fluorescent signals for EGFP, HA-B2, and MAP2 are shown individually (A–C) and as overlays (D and E). Examples for MAP2-negative neurites of transfected cells are indicated by white boxes (enlarged in the insets) in D and E. Scale bars, 25 μm for large panels; 7 μm for enlarged frames. HA-B2, HA-GABAB2. F, quantitative analysis of the ratio of the axonal red and green fluorescence intensities. PSD-95, n = 38 axons; Mock, KCTD12, n = 40 axons each. *, p < 0.0001.
DISCUSSION
Here we used tandem affinity purification from mouse brain to identify proteins associated with the GABAB receptor subunit GABAB1. As expected from a BAC approach, mRNAs for the tagged receptor subunits were distributed similarly to their endogenous counterparts, and the proteins were not strongly overexpressed by the genetic manipulation. In addition, the physiological pre- and postsynaptic effects of GABAB receptors were preserved. Thus, GABAB1 complexes isolated from these mice have a good chance to mimic the in vivo situation. The mice should therefore offer a convenient way to unravel native GABAB1 complexes. However, visual inspection of silver-stained gels loaded with purified fractions from transgenic and wild-type mice yielded a small number of complex-specific protein bands. This limitation is attributed to unspecific binding to the affinity matrices under the described conditions, which probably reflects the tagged target protein GABAB1 being only a minute constituent within the solubilized brain membranes used as source. The highest expression of GABAB1a occurs during the first postnatal days (3). We therefore tried purification from newborn NTAP-GABAB1a mice but observed a pattern of silver-stained protein bands virtually identical to the one from P10-P16 mice.
The HA-SBP tag combination introduced here may offer advantages as compared with the established CBP-TEV-ProtA tag. The HA-SBP tag is rather small (56 amino acids; ∼6 kDa) and might therefore show less interference with the trafficking and function of tagged target molecules. Moreover, substitution of the CBP tag may be preferred for purification from tissues with significant expression of calmodulin. However, we noted an unexpected loss of GABAB2 specifically during purifications from NTAP-GABAB1a mice (Fig. 4B). It is possible that the HA tag is better accessible in unassembled HA-SBP-GABAB1a than in the heterodimer. The fact that we did not find KCTD12 in the NTAP-GABAB1a samples may result from the loss of GABAB2.
Our data show that the chaperone protein GRP78 bound to GABAB1 in vivo. Purified samples from NTAP-GABAB1a mice routinely showed a conspicuously strong band migrating slightly more slowly than a 75-kDa marker protein, which was identified as GRP78 by mass spectrometry. GRP78 is a luminal endoplasmic reticulum protein mainly involved with chaperone functions and the unfolded protein response (44, 46, 47). However, a control Western blot suggested that the transgenic receptor subunits did not lead to an obvious induction of GRP78, which would have been expected in the case of an endoplasmic reticulum stress response (Fig. 6A). In addition, we detected GRP78 not only in purified fractions from both transgenic mouse lines but also in GABAB1 complexes immunoprecipitated from wild-type mice. GRP78 binding may mediate the maturation of the GABAB receptor, possibly by supporting the proper folding of the GABAB1 subunit and avoiding its aggregation until the heterodimer is formed. The particularly large amount of GRP78 found in the final fractions of the NTAP-GABAB1a mice may reflect that our purification scheme favored isolation of the unassembled subunit (see above). The detection of a luminal endoplasmic reticulum protein as a binding partner shows that the strategy used here is not limited to the isolation of cytoplasmic components and may therefore ultimately also allow identification of extracellular interactions of GABAB receptors.
Our analysis of GABAB1a/b-CTAP mice revealed KCTD12 as a novel complex constituent. However, a few peptides for GABAB1, GABAB2, and KCTD12 were also detected in the samples from wild-type mice, although in small numbers and at lower intensities. Because final fractions of GABAB1a/b-CTAP purifications from wild-type mice did not show bands in Western blots with antibodies to GABAB1, GABAB2, or KCTD12, we ascribe these peptides to contaminations that possibly occurred during isolation of the corresponding bands or the subsequent peptide separation. KCTD12 has been identified as a gene with predominant fetal expression, most prominently in cochlea and brain (45). KCTD12/pfetin protein was reported to be a prognostic marker for gastrointestinal stromal tumors showing an inverse relation to tumor metastasis (48). The KCTD12 protein contains a T1 domain (45) that belongs to the family of BTB/POZ protein-protein interaction motifs with various cellular functions (49). The T1 domain tetramer of voltage-gated potassium channels subserves β-subunit association controlling both channel properties and axonal/dendritic targeting (50–55). Interestingly, KCTD12 peptides have also been identified in synaptosomal preparations from mouse brain (56), supporting a role for KCTD12 in synaptic transmission.
With the C-terminal region of GABAB2 we report the first binding site for the T1 domain in KCTD12. Importantly, this domain interaction allows association of KCTD12 with the heteromeric, functional GABAB receptor. Our experiments in transfected dissociated neurons suggest that KCTD12 binding may play a role in transport of the GABAB receptor to axonal plasma membrane sites reminiscent of the T1 domain in Kv channels (52, 54), implying that T1 domain interactions mediate subcellular targeting of selected neuronal membrane proteins. Given that GABAB receptors subserve various different functions in dendritic shafts, dendritic spines, and axonal boutons in neurons of the brain, their precise localization is of pivotal importance. In summary, our data introduce the T1 domain protein KCTD12 as an integral part of GABAB receptors. The TAP-tagged mice may help to unravel the native protein composition of GABAB1 complexes in brain and other tissues.
Acknowledgments
We are grateful to Julia Kuhlmann, Stefanie Wilhelm, Janina Sülflow, and Andrea Zaisser for excellent technical assistance. We thank Irm Hermans-Borgmeyer for oocyte injections, Susanne Fehr for in situ hybridization, Chudamani Rasa Raithore for cultured neurons, and Michaela Schweizer for help with microscopy.
This work was supported in part by the Deutsche Forschungsgemeinschaft (SFB444).
- GABA
- γ-aminobutyric acid
- AMPA
- α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid
- BAC
- bacterial artificial chromosome
- CBP
- calmodulin-binding peptide
- EPSC
- excitatory postsynaptic current
- GRP78
- glucose-regulated protein 78
- HA
- hemagglutinin
- IPSC
- inhibitory postsynaptic current
- KCTD12
- potassium channel tetramerization domain-containing protein 12
- MAP2
- microtubule-associated protein 2
- Pn
- postnatal day n
- ProtA
- protein A
- SBP
- streptavidin-binding peptide
- T1 domain
- voltage-gated potassium channel tetramerization domain
- TAP
- tandem affinity purification
- TEV
- tobacco etch virus
- EGFP
- enhanced green fluorescent protein
- PBS
- phosphate-buffered saline
- BSA
- bovine serum albumin.
REFERENCES
- 1.Bowery N. G., Bettler B., Froestl W., Gallagher J. P., Marshall F., Raiteri M., Bonner T. I., Enna S. J. (2002) Pharmacol. Rev. 54, 247–264 [DOI] [PubMed] [Google Scholar]
- 2.Kulik A., Vida I., Luján R., Haas C. A., López-Bendito G., Shigemoto R., Frotscher M. (2003) J. Neurosci. 23, 11026–11035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bettler B., Kaupmann K., Mosbacher J., Gassmann M. (2004) Physiol. Rev. 84, 835–867 [DOI] [PubMed] [Google Scholar]
- 4.Kaupmann K., Huggel K., Heid J., Flor P. J., Bischoff S., Mickel S. J., McMaster G., Angst C., Bittiger H., Froestl W., Bettler B. (1997) Nature 386, 239–246 [DOI] [PubMed] [Google Scholar]
- 5.Peters H. C., Kämmer G., Volz A., Kaupmann K., Ziegler A., Bettler B., Epplen J. T., Sander T., Riess O. (1998) Neurogenetics 2, 47–54 [DOI] [PubMed] [Google Scholar]
- 6.Lamp K., Humeny A., Nikolic Z., Imai K., Adamski J., Schiebel K., Becker C. M. (2001) Cytogenet. Cell Genet. 92, 116–121 [DOI] [PubMed] [Google Scholar]
- 7.Vigot R., Barbieri S., Bräuner-Osborne H., Turecek R., Shigemoto R., Zhang Y. P., Luján R., Jacobson L. H., Biermann B., Fritschy J. M., Vacher C. M., Müller M., Sansig G., Guetg N., Cryan J. F., Kaupmann K., Gassmann M., Oertner T. G., Bettler B. (2006) Neuron 50, 589–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pérez-Garci E., Gassmann M., Bettler B., Larkum M. E. (2006) Neuron 50, 603–616 [DOI] [PubMed] [Google Scholar]
- 9.Guetg N., Seddik R., Vigot R., Turecek R., Gassmann M., Vogt K. E., Bräuner-Osborne H., Shigemoto R., Kretz O., Frotscher M., Kulik A., Bettler B. (2009) J. Neurosci. 29, 1414–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tiao J. Y., Bradaia A., Biermann B., Kaupmann K., Metz M., Haller C., Rolink A. G., Pless E., Barlow P. N., Gassmann M., Bettler B. (2008) J. Biol. Chem. 283, 31005–31011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blein S., Ginham R., Uhrin D., Smith B. O., Soares D. C., Veltel S., McIlhinney R. A., White J. H., Barlow P. N. (2004) J. Biol. Chem. 279, 48292–48306 [DOI] [PubMed] [Google Scholar]
- 12.Kulik A., Vida I., Fukazawa Y., Guetg N., Kasugai Y., Marker C. L., Rigato F., Bettler B., Wickman K., Frotscher M., Shigemoto R. (2006) J. Neurosci. 26, 4289–4297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tabata T., Araishi K., Hashimoto K., Hashimotodani Y., van der Putten H., Bettler B., Kano M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 16952–16957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang W., Tu C., Cheng Z., Rodriguez L., Chen T. H., Gassmann M., Bettler B., Margeta M., Jan L. Y., Shoback D. (2007) J. Biol. Chem. 282, 25030–25040 [DOI] [PubMed] [Google Scholar]
- 15.Bettler B., Tiao J. Y. (2006) Pharmacol. Ther. 110, 533–543 [DOI] [PubMed] [Google Scholar]
- 16.Kornau H. C. (2006) Cell Tissue Res. 326, 517–533 [DOI] [PubMed] [Google Scholar]
- 17.Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Séraphin B. (1999) Nat. Biotechnol. 17, 1030–1032 [DOI] [PubMed] [Google Scholar]
- 18.Gavin A. C., Bösche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J. M., Michon A. M., Cruciat C. M., Remor M., Höfert C., Schelder M., Brajenovic M., Ruffner H., Merino A., Klein K., Hudak M., Dickson D., Rudi T., Gnau V., Bauch A., Bastuck S., Huhse B., Leutwein C., Heurtier M. A., Copley R. R., Edelmann A., Querfurth E., Rybin V., Drewes G., Raida M., Bouwmeester T., Bork P., Seraphin B., Kuster B., Neubauer G., Superti-Furga G. (2002) Nature 415, 141–147 [DOI] [PubMed] [Google Scholar]
- 19.Häcker H., Redecke V., Blagoev B., Kratchmarova I., Hsu L. C., Wang G. G., Kamps M. P., Raz E., Wagner H., Häcker G., Mann M., Karin M. (2006) Nature 439, 204–207 [DOI] [PubMed] [Google Scholar]
- 20.Bürckstümmer T., Bennett K. L., Preradovic A., Schütze G., Hantschel O., Superti-Furga G., Bauch A. (2006) Nat. Methods 3, 1013–1019 [DOI] [PubMed] [Google Scholar]
- 21.Gregan J., Riedel C. G., Petronczki M., Cipak L., Rumpf C., Poser I., Buchholz F., Mechtler K., Nasmyth K. (2007) Nat. Protoc. 2, 1145–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Angrand P. O., Segura I., Völkel P., Ghidelli S., Terry R., Brajenovic M., Vintersten K., Klein R., Superti-Furga G., Drewes G., Kuster B., Bouwmeester T., Acker-Palmer A. (2006) Mol. Cell. Proteomics 5, 2211–2227 [DOI] [PubMed] [Google Scholar]
- 23.Daulat A. M., Maurice P., Froment C., Guillaume J. L., Broussard C., Monsarrat B., Delagrange P., Jockers R. (2007) Mol. Cell. Proteomics 6, 835–844 [DOI] [PubMed] [Google Scholar]
- 24.Schall T. J., Lewis M., Koller K. J., Lee A., Rice G. C., Wong G. H., Gatanaga T., Granger G. A., Lentz R., Raab H., Kohr W. J., Goeddel D. V. (1990) Cell 61, 361–370 [DOI] [PubMed] [Google Scholar]
- 25.Keefe A. D., Wilson D. S., Seelig B., Szostak J. W. (2001) Protein Expression Purif. 23, 440–446 [DOI] [PubMed] [Google Scholar]
- 26.Grünewald S., Schupp B. J., Ikeda S. R., Kuner R., Steigerwald F., Kornau H. C., Köhr G. (2002) Mol. Pharmacol. 61, 1070–1080 [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y., Buchholz F., Muyrers J. P., Stewart A. F. (1998) Nat. Genet. 20, 123–128 [DOI] [PubMed] [Google Scholar]
- 28.Gay P., Le Coq D., Steinmetz M., Ferrari E., Hoch J. A. (1983) J. Bacteriol. 153, 1424–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dean D. (1981) Gene 15, 99–102 [DOI] [PubMed] [Google Scholar]
- 30.Narayanan K., Williamson R., Zhang Y., Stewart A. F., Ioannou P. A. (1999) Gene Ther. 6, 442–447 [DOI] [PubMed] [Google Scholar]
- 31.Nagy A. (2003) Manipulating the Mouse Embryo: A Laboratory Manual, 3rd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 32.Stables J., Green A., Marshall F., Fraser N., Knight E., Sautel M., Milligan G., Lee M., Rees S. (1997) Anal. Biochem. 252, 115–126 [DOI] [PubMed] [Google Scholar]
- 33.Hartmann D., Fehr S., Meyerhof W., Richter D. (1995) Dev. Neurosci. 17, 246–255 [DOI] [PubMed] [Google Scholar]
- 34.Shevchenko A., Wilm M., Vorm O., Mann M. (1996) Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 35.Prokhorova T. A., Rigbolt K. T., Johansen P. T., Henningsen J., Kratchmarova I., Kassem M., Blagoev B. (2009) Mol. Cell. Proteomics 8, 959–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rappsilber J., Ishihama Y., Mann M. (2003) Anal. Chem. 75, 663–670 [DOI] [PubMed] [Google Scholar]
- 37.Blagoev B., Ong S. E., Kratchmarova I., Mann M. (2004) Nat. Biotechnol. 22, 1139–1145 [DOI] [PubMed] [Google Scholar]
- 38.Olsen J. V., de Godoy L. M., Li G., Macek B., Mortensen P., Pesch R., Makarov A., Lange O., Horning S., Mann M. (2005) Mol. Cell. Proteomics 4, 2010–2021 [DOI] [PubMed] [Google Scholar]
- 39.Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. (1999) Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 40.Takada T., Kumánovics A., Amadou C., Yoshino M., Jones E. P., Athanasiou M., Evans G. A., Fischer Lindahl K. (2003) Genome Res. 13, 589–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nefedov M., Williamson R., Ioannou P. A. (2000) Nucleic Acids Res. 28, E79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Billinton A., Upton N., Bowery N. G. (1999) Br. J. Pharmacol. 126, 1387–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bischoff S., Leonhard S., Reymann N., Schuler V., Shigemoto R., Kaupmann K., Bettler B. (1999) J. Comp. Neurol. 412, 1–16 [PubMed] [Google Scholar]
- 44.Lee A. S. (2001) Trends Biochem. Sci. 26, 504–510 [DOI] [PubMed] [Google Scholar]
- 45.Resendes B. L., Kuo S. F., Robertson N. G., Giersch A. B., Honrubia D., Ohara O., Adams J. C., Morton C. C. (2004) J. Assoc. Res. Otolaryngol. 5, 185–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li J., Lee A. S. (2006) Curr. Mol. Med. 6, 45–54 [DOI] [PubMed] [Google Scholar]
- 47.Goloubinoff P., De Los Rios P. (2007) Trends Biochem. Sci. 32, 372–380 [DOI] [PubMed] [Google Scholar]
- 48.Suehara Y., Kondo T., Seki K., Shibata T., Fujii K., Gotoh M., Hasegawa T., Shimada Y., Sasako M., Shimoda T., Kurosawa H., Beppu Y., Kawai A., Hirohashi S. (2008) Clin. Cancer Res. 14, 1707–1717 [DOI] [PubMed] [Google Scholar]
- 49.Stogios P. J., Downs G. S., Jauhal J. J., Nandra S. K., Privé G. G. (2005) Genome Biol. 6, R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gulbis J. M., Zhou M., Mann S., MacKinnon R. (2000) Science 289, 123–127 [DOI] [PubMed] [Google Scholar]
- 51.Minor D. L., Lin Y. F., Mobley B. C., Avelar A., Jan Y. N., Jan L. Y., Berger J. M. (2000) Cell 102, 657–670 [DOI] [PubMed] [Google Scholar]
- 52.Gu C., Jan Y. N., Jan L. Y. (2003) Science 301, 646–649 [DOI] [PubMed] [Google Scholar]
- 53.Long S. B., Campbell E. B., Mackinnon R. (2005) Science 309, 897–903 [DOI] [PubMed] [Google Scholar]
- 54.Rivera J. F., Chu P. J., Arnold D. B. (2005) Eur. J. Neurosci. 22, 1853–1862 [DOI] [PubMed] [Google Scholar]
- 55.Xu M., Cao R., Xiao R., Zhu M. X., Gu C. (2007) J. Neurosci. 27, 14158–14170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Munton R. P., Tweedie-Cullen R., Livingstone-Zatchej M., Weinandy F., Waidelich M., Longo D., Gehrig P., Potthast F., Rutishauser D., Gerrits B., Panse C., Schlapbach R., Mansuy I. M. (2007) Mol. Cell. Proteomics 6, 283–293 [DOI] [PubMed] [Google Scholar]