Abstract
Carbohydrate active enzymes (CAZymes) are a large class of enzymes, which build and breakdown the complex carbohydrates of the cell. On the basis of their amino acid sequences they are classified in families and clans that show conserved catalytic mechanism, structure, and active site residues, but may vary in substrate specificity. We report here the identification and the detailed molecular characterization of a novel glycoside hydrolase encoded from the gene sso1353 of the hyperthermophilic archaeon Sulfolobus solfataricus. This enzyme hydrolyzes aryl β-gluco- and β-xylosides and the observation of transxylosylation reactions products demonstrates that SSO1353 operates via a retaining reaction mechanism. The catalytic nucleophile (Glu-335) was identified through trapping of the 2-deoxy-2-fluoroglucosyl enzyme intermediate and subsequent peptide mapping, while the general acid/base was identified as Asp-462 through detailed mechanistic analysis of a mutant at that position, including azide rescue experiments. SSO1353 has detectable homologs of unknown specificity among Archaea, Bacteria, and Eukarya and shows distant similarity to the non-lysosomal bile acid β-glucosidase GBA2 also known as glucocerebrosidase. On the basis of our findings we propose that SSO1353 and its homologs are classified in a new CAZy family, named GH116, which so far includes β-glucosidases (EC 3.2.1.21), β-xylosidases (EC 3.2.1.37), and glucocerebrosidases (EC 3.2.1.45) as known enzyme activities.
Keywords: Archaebacteria, Carbohydrate Metabolism, Carbohydrate Processing, Enzyme Catalysis, Enzyme Mechanisms, Polysaccharide
Introduction
Carbohydrates, whose structural diversity exceeds by far the number of protein folds, are ubiquitous molecules that alone, or in form of glycoconjugates, mediate many biological processes (1). This extreme variety results from the diverse stereochemistry of the monosaccharide building blocks, from the enormous number of intersugar linkages they can form and to the fact that these molecules can decorate cell surface, large macromolecules (sugars themselves, proteins, nucleic acids), or small metabolites (lipids, antibiotics, etc). The breadth of the biological functions of carbohydrates, from the classical energetic and structural roles, is now well acknowledged although the mechanisms of the sugar code are not known in detail. They include the control of the correct protein folding and activity (2), the mediation of molecular recognition events regulating cell-cell interactions (such as host-pathogen, cancer metastasis, etc.), cell signal transduction (nucleo-cytoplasm communication, differentiation, immune response, etc.) (for reviews see Refs. 1, 3, 4).
The ability of carbohydrates in functioning in intermolecular interactions as encoders of biological information is made possible by a large class of enzymes, collectively known as carbohydrate-active enzymes (CAZymes),2 including glycoside hydrolases (GH), glycosyltransferases (GT), polysaccharide lyases, carbohydrate esterases, and carbohydrate binding modules, which are in charge of catalyzing the metabolism and the correct shape of the sugars of the cell. CAZymes have been classified on the basis of their amino acid sequences in families sharing the same catalytic mechanism, structure, and active site residues; in addition, families with similar three-dimensional structure are further grouped in clans (5).
The number of CAZyme families is continuously increasing, thanks to the advent of new DNA sequencing techniques and subsequent sophisticated computer-aided sequence annotation procedures combined with new biochemical characterization. Interestingly, although the number of CAZy sequences increased 14-fold in the last 8 years, the number of enzymatic and structural characterization only doubled in the same time span, with at present <10% of total proteins in the CAZy database that have been characterized enzymatically (5). This contrast clearly shows that, in comparison with highly automated sequencing techniques, enzymatic characterization of novel CAZymes is a longer and laborious process representing the limiting step for the full exploitation of genome sequencing efforts.
Here, we report the cloning, the heterologous expression and the detailed enzymatic characterization of a novel GH from the hyperthermophilic Archaeon S. solfataricus. This enzyme, encoded by the ORF SSO1353 displays sequence similarity to several unknown proteins from the three domains of life (Archaea, Bacteria, and Eukarya) and, more distantly, to human non-lysosomal glucosylceramidase, also known as β-glucosidase 2 (GBA2) (6), an enzyme involved in an alternative catabolic pathway of glucosylceramide (7).
The enzymatic characterization of the product of gene sso1353 allowed us to demonstrate the retaining reaction mechanism followed by the enzyme, to evaluate its substrate specificity toward β-linked aromatic glucosides and xylosides, and to identify the catalytic amino acids in the active site. These results allowed us propose the role possibly played in vivo by this enzyme, which is expressed from a gene situated downstream of that coding for an endoglucanase in different Sulfolobus species. Finally, by virtue of the established commonalities within glycoside hydrolase families, the mechanistic data obtained with SSO1353 can be extended to all members of the newly created GH116 family including human GBA2.
EXPERIMENTAL PROCEDURES
Reagents
All commercially available substrates were purchased from Sigma and Carbosynth. The GeneTailor Site-Directed Mutagenesis system was from Invitrogen, and the synthetic oligonucleotides were from PRIMM (Milan, Italy).
Plasmid Preparation
The SSO1353 ORF was cloned by amplification of S. solfataricus, strain P2, chromosomal DNA via PCR by using the following synthetic oligonucleotides: 1353Fw, 5′-ggaattccatatggttacatatactgataagg-3′, 1353Rv, 5′-tacatgccatggctagaataggaagctcc-3′, which introduce an NdeI and NcoI sites at the 5′, just before the first ATG, and at the 3′-ends of the ORF, respectively. The program was as follows: 5 min at 95 °C, 1,5 min at 50 °C, and 4 min at 72 °C; 30 cycles at 95 °C for 45 s, 50 °C for 1,5 min, and 72 °C for 4 min; final extension at 72 °C for 10 min. The resulting DNA fragment was cloned in the pET29a plasmid (Novagen), obtaining the vector pET1353, in which the SSO1353 ORF is under the control of the isopropyl-1-thio-β-d-galactopyranoside inducible T7 RNA polymerase promoter that drives high expression levels in bacterial hosts. The ORF obtained after amplification was controlled by DNA sequencing.
Site-directed Mutagenesis
The mutants E335G, D406G, D458G, D462G were prepared by site-directed mutagenesis from the pET1353 plasmid, by following the instructions of the manufacturer. The mutagenic oligonucleotides were the following (mismatches are underlined): E335Gmut, 5′-ATGGACAGTTCTGTGGTGCTCCGTAAATCGCA-3′; E335Grev, 5′-AGCACCACAGAACTGTCCATATTTAGGTAC-3′; D406Gmut, 5′-TTCTCCTCCCAGATGGAAGGGTATGAATCCGA-3′; D406Grev, 5′-CCTTCCATCTGGGAGGAGAAGTAGTACCAT-3′; D458Gmut, 5′-ATTCATGGAGGGAGAGATGGGCAATGCTTTTG-3′; D458Grev, 5′-CCATCTCTCCCTCCATGAATGGTAAATTCC-3′; D462Gmut, 5′-AGAGATGGACAATGCTTTTGGCGCTACCATCA-3′; D462Grev, 5′-CAAAAGCATTGTCCATCTCTCCCTCCATGA-3′. The genes containing the desired mutation were identified by direct sequencing and completely resequenced.
Expression and Purification of SSO1353 Wild Type and Mutants
Escherichia coli BL21(DE3)Ril/pET1353 wild type and mutants were grown in 2 liters of LB at 37 °C supplemented with kanamycin (50 μg/ml) and chloramphenicol (30 μg/ml). Gene expression was induced by the addition of 0.5 mm isopropyl-1-thio-β-d-galactopyranoside when the culture reached an A600 of 1.0. Growth was allowed to proceed for 16 h, and cells were harvested by centrifugation at 5,000 × g. The resulting cell pellet was thawed, resuspended in 3 ml g−1 cells of 20 mm sodium phosphate buffer, pH 7.4, 150 mm NaCl, 1% (v/v) Triton X-100 and homogenized by French cell pressure treatment. After centrifugation for 30 min at 10,000 × g, the crude extract was incubated with Benzonase (Novagen) for 1 h at room temperature and then heat-fractionated for 30 min at 55 and 75 °C and for 20 min at 85 °C. The supernatant obtained after heat fractionations, equilibrated in 1 m ammonium sulfate, was applied to a HiLoad 26/10 phenyl Sepharose high performance (Amersham Biosciences), which had been equilibrated with 20 mm sodium phosphate buffer, pH 7.3, 1 m ammonium sulfate. After washing with 2 column volumes with the loading buffer, the protein was eluted with a linear gradient of water at a flow rate of 3 ml min−1; the protein eluted in 100% water. Active fractions were pooled, equilibrated in 20 mm sodium phosphate buffer, pH 7.4, 150 mm NaCl and concentrated by ultrafiltration on an Amicon YM30 membrane (cut off 30,000 Da). For the wild-type enzyme, after concentration, the sample was loaded onto a HiLoad 26/60 Superdex 200 prep grade column (Amersham Biosciences). Active fractions were pooled and concentrated; protein concentration was determined with the method of Bradford (8). The SSO1353 wild type and mutants were 95% pure by SDS-PAGE and were stored at 4 °C.
Characterization of SSO1353 Wild Type and Mutants
The molecular mass of native SSO1353 wild type was determined by gel filtration on a Superdex 200 HR 10/30 FPLC column (Amersham Biosciences); molecular weight markers were albumin (66,000), alcohol dehydrogenase (150,000), β-amylase (200,000), and apoferritin (443,000).
The standard assay for SSO1353 activity was performed in 50 mm sodium citrate buffer at pH 5.5 at 65 °C on the indicated substrates. Typically, in each assay we used 1–10 μg of SSO1353 in the final volume of 1.0 ml. Kinetic constants of SSO1353 wild type and D462G mutant on aryl glycosides 4Np-Glc, 4Np-Xyl, and 2Np-Glc were measured at standard conditions at 65 °C by using concentrations of substrate ranging between 1 and 150 mm. The ϵmm extinction coefficients at 405 nm for 2- and 4-nitrophenol under standard conditions and 65 °C were 1.1 and 3.3 mm−1 cm−1, respectively. One unit of enzyme activity was defined as the amount of enzyme catalyzing the hydrolysis of 1 μmol of substrate in 1 min at the conditions described. The metal dependence of the wild-type enzyme was evaluated using 5 mm 4Np-Xyl as substrate in the presence of 1 mm EDTA and 5 mm MgCl2 or MnCl2, in standard conditions.
The activity of the wild-type enzyme on different aryl glycosides was tested in 50 mm sodium citrate buffer at pH 5.5 at 65 °C. Typically, in each assay we used 70 μg of SSO1353 in a final volume of 0.2 ml. The reaction was started by adding the enzyme, and it was stopped by adding 0.8 ml of 1 m iced sodium carbonate. The optical density of the solution was measured at 420 nm at room temperature. The molar extinction coefficients of 4-nitrophenol and 2-nitrophenol, measured at 420 nm, at room temperature and in 1 m sodium carbonate buffer were 17.2 and 4.7 mm−1 cm−1, respectively. In all of the assays, spontaneous hydrolysis of the substrate was subtracted by using appropriate blank mixtures without the enzyme.
The chemically rescued activity of the D462G mutant was measured in 50 mm sodium citrate buffer at pH 5.5 on 40 mm 2Np-Glc at 65 °C as described above; where indicated, the assay mixture was supplemented with the indicated concentrations of sodium azide as external nucleophile. In all of the assays, spontaneous hydrolysis of the substrate was subtracted by using appropriate blank mixtures without the enzyme. Aliquots of the reaction mixtures were analyzed on a silica gel 60 F254 TLC by using ethyl acetate/methanol/water (70:20:10 v/v) as eluant and were detected by exposure to 4% α-naphthol in 10% sulfuric acid in ethanol followed by charring. The β-d-glucosyl azide isolated from the enzymatic reaction mixture of the D462G mutant was identified by 1H and 13C NMR spectroscopy. NMR: δ 4.75 H-1 (d, 3JH1,H2 = 8.8 Hz), δ 91.4 C-1; δ 3.25 H-2 (t), δ 74.0 C-2; δ 3.51 H-3 (t), δ 76.9 C-3; δ 3.41 H-4 (t), δ 70.5 C-4; δ 3.53 H-5 (m), δ 79.0 C-5; δ 3.73 H-6a (dd), δ 3.90 H-6b (dd), 61.6 C-6.
The activity of the wild-type enzyme in the presence of 0.5–2.0 mg ml−1 Triton X-100 or CHAPS was determined on 20 mm 4Np-Glc as described above. The activity of the wild type on β-d-oligosaccharides of glucose (G3 to G5) (5 mm) and xylose (X2 to X5) (2.5 mm) was tested in 50 mm sodium citrate buffer at pH 5.5, by using 12–63 μg of enzyme, at 65 °C in a final volume of 0.2 ml. Aliquots of the reaction mixtures were analyzed by TLC by using ethyl acetate/acetic acid/isopropanol/formic acid/water (50:20:10:2:30 v/v) (for G3-G5) or acetone/isopropylic alcohol/water (60:30:15 v/v) (for X2–X5) as the eluant and were detected as described above. The activity of the wild type on glucocerebroside (Matreya), octyl-β-d-glucopyranoside (Sigma), and gangliosides, measured at the same conditions was analyzed by TLC by using chloroform/methanol/CaCl2 15 mm (60:40:9 v/v).
The steady-state kinetic constants of the wild type on MU-Glc and MU-Xyl were calculated by following a slight modified method already described (9). Briefly, the enzyme activity was determined fluorimetrically with MU-Glc (0.1–6 mm) and MU-Xyl (0.025–5 mm) as substrates in 50 mm sodium citrate buffer at pH 5.5, by using 1 μg of the enzyme. Assays (0.25 ml final volume) were conducted for 1–5 min at 65 °C, and the reaction was stopped with 0.5 ml of 0.1 m glycine-NaOH buffer, pH 10.3. The formation of the methylumbelliferone was measured by emission at 450 nm with excitation at 384 nm. In all of the assays, spontaneous hydrolysis of the substrate was subtracted by using appropriate blank mixtures without the enzyme. All kinetic data were calculated as the average of at least two experiments and were plotted and refined with the program GraFit (10).
Inhibition of SSO1353 Wild Type
The effect of the inhibitor 2,4-dinitrophenyl-β-d-2-deoxy-2-fluoro-glucopyranoside (2,4DNp-2F-Glc) (Sigma) was analyzed as previously performed by Shaikh et al. (11). Briefly, wild-type SSO1353 (0.1 μg/μl) was incubated at 45 °C in mixtures containing 0.3, 3.0, 7.0, and 18.0 mm concentrations of inhibitor and 50 mm sodium citrate buffer, pH 5.5. An identical mixture containing all the reagents with the exception of the enzyme was prepared as control. At time intervals, aliquots from the two mixtures were withdrawn and used to measure the enzymatic activity and as blank, respectively. Assays were performed on 60 mm 2Np-Glc in standard conditions. Initial rates at each time point were elaborated as described in Ref. 11 to measure the inactivation parameters Ki and ki with GraFit (10).
To determine the effect of the inhibitors N-butyldeoxynojirimycin (NB-DNJ) and conduritol β-epoxide (CBE), SSO1353 (11 μg) was incubated in the presence of increasing concentrations of the inhibitors (0.1–5 mm for NB-DNJ and 0.1–2 mm for CBE) in 50 mm sodium citrate buffer, pH 5.5, for 30 min at 45 °C, in a final volume of 80 μl. Identical mixtures containing all the reagents with the exception of the inhibitor were used to determine the 100% of activity. After incubation, the samples were diluted 3-fold in 5 mm MU-Glc, 50 mm sodium citrate buffer, pH 5.5 in a final volume of 0.25 ml, and assayed as described above.
Transglycosylation Reactions
Wild-type SSO1353 (7–148 μg), was incubated in 50 mm sodium citrate buffer pH 5.5 with 5 mm 4Np-Xyl or 4Np-Glc, or both, at 5 mm each, at 65 °C for 16 h in a final volume of 0.2–1 ml. Identical mixtures containing all the reagents but the enzyme were prepared as control. Where specified, after incubation, 50 μg of the β-glycosidase from S. solfataricus (Ssβ-gly) were added to aliquots (90 μl) of the reaction mixtures and further incubated 2 h at 65 °C. All the reactions were examined by TLC by using ethyl acetate/methanol/water (70:20:10 v/v) as eluant as described above.
Identification of the Reaction Products
The 1H and 13C NMR spectra were recorded in D2O at 600 MHz with a spectrometer equipped with a cryo probe, in the FT mode at 303 K. 1H chemical shifts are expressed in δ relative to HOD signal (4.72 ppm).
The linkage analysis of the oligosaccharide was obtained by methylation according to Ciucanu procedure, as already reported (12). The sample obtained was injected into GC-MS and partially methylated alditol acetates were recognized from their EI-MS spectra and by comparison with pure synthetic standards. Partially methylated alditol acetates were analyzed on an Agilent Technologies gas chromatograph 6850A equipped with a mass selective detector 5973N and a Zebron ZB-5 capillary column (Phenomenex, 30 m × 0.25 mm i.d., flow rate 1 ml/min, He as carrier gas). The temperature program was: 90 °C for 1 min, 90 °C → 140 °C at 25 °C min−1, 140 °C → 200 °C at 5 °C min−1, 200 °C → 280 °C at 10 °C min−1, 280 °C for 10 min.
The reaction mixture was purified by reverse phase chromatography (Polar-RP 80A, Phenomenex, 4 μ, 250 × 10 mm) on an Agilent HPLC instrument 1100 series, using H2O/CH3OH 6/4 with 20 mm trifluoroacetic acid (final concentration) as eluant. The eluted products were first analyzed by positive ions reflectron MALDI-TOF mass spectrometry.
The trisaccharide β-Xyl-(1→4)-β-Xyl-O-4-Np showed a pseudomolecular ion (M+Na)+ at m/z 425.94 (calculated m/z 426.10) and the methylation analysis indicated the presence of terminal xylopyranose and 4-substituted xylopyranose. NMR: δ 5.24 H-1A (d, JH-1,H-2 = 7.3 Hz), δ 101.0 C-1A; 3.66 H-2A (t), δ 73.7 C-2A; δ 3.72 H-3A (t), δ 74.5 C-3A; δ 3.89 H-4A (m), δ 77.4 C-4A; δ 4.19 Ha-5A (dd), δ 3.62 Hb-5A (t), δ 64.3 C-5A; δ 4.49 H-1B (d, JH-1,H-2 = 7.3 Hz), δ 103.1 C-1B; 3.29 H-2B (t), δ 74.0 C-2A; δ 3.44 H-3B (t), δ 76.9 C-3B; δ 3.62 H-4B (m), δ 70.4 C-4B; δ 3.99 Ha-5B (dd), δ 3.32 Hb-5B (t), δ 66.5 C-5B.
The trisaccharide β-Xyl-(1→4)-β-Xyl-(1→4)-β-Xyl-O-4-Np showed a pseudomolecular ion in a MALDI-TOF-MS (M+Na)+ at m/z 557.82 (calculated m/z 558.15); methylation analysis: terminal xylopyranose and 4-substituted xylopyranose.
NMR: δ 5.25 H-1A (d, JH-1,H-2 = 7.3 Hz), δ 101.2 C-1A; 3.66 H-2A (t), δ 73.8 C-2A; δ 3.72 H-3A (t), δ 74.6 C-3A; δ 3.90 H-4A (m), δ 77.3 C-4A; δ 4.19 Ha-5A (dd), δ 3.62 Hb-5A (t), δ 64.3 C-5A; δ 4.51 H-1B (d, JH-1,H-2 = 7.3 Hz), δ 103.0 C-1B; 3.31 H-2B (t), δ 74.0 C-2A; δ 3.56 H-3B (t), δ 75.0 C-3B; δ 3.80 H-4B (m), δ 77.6 C-4B; δ 4.12 Ha-5B (dd), δ 3.39 Hb-5B (t); δ 64.3 C-5B; δ 4.47 H-1C (d, JH-1,H-2 = 7.3 Hz), δ 103.1 C-1C; δ 3.26 H-2C (t), δ 74.1 C-2C; δ 3.43 H-3C (t), δ 76.9 C-3C; δ 3.63 H-4C (m), δ 70.4 C-4C; δ 3.97 Ha-5C (dd), δ 3.31 Hb-5C (t); δ 66.5 C-5C.
Nano-ESI-MS of Intact Protein Samples
Samples were analyzed using a triple quadrupole time of flight instrument (QSTAR Elite, Applied Biosystems, Foster City, CA/Toronto, Canada) equipped with a nanoflow electrospray ion source. Pulled silica capillary (170 μm outer diameter/100 μm inner diameter, tip 30 μm inner diameter) was used as nanoflow tip. For the analysis of intact proteins, 4 μg of samples were purified using ZipTip C4 (Millipore, Billerica, MA). Proteins were eluted by 50% acetonitrile and 0.1% formic acid. Purified proteins (10 μm) were loaded into the ion source at 300 nl/min flow rate using a syringe pump. Single-stage ESI mass spectra were acquired in the range of m/z 300–2000. For protein molecular mass determination three independent measurements were performed. The expected mass error on the average molecular mass of intact proteins was about ±0.01%. For data acquisition and Bayesian protein reconstruction the Analyst QS 2.0 software (Applied Biosystems, Foster City, CA/Toronto, Canada) was used.
Nano-HPLC-ESI-MS/MS Experiments
Wild-type SSO1353 (22 μg, 0.3 nmol) was incubated with 2.9 mm 2,4-dinitrophenyl-β-d-2-deoxy-2-fluoro-glucopyranoside (2,4DNp-2F-Glc) (Sigma) at 1:1000 enzyme/inhibitor ratio in 50 mm sodium citrate buffer, pH 5.5 at 45 °C. An identical mixture containing all the reagents with the exception of the inhibitor was prepared as control. At time intervals, aliquots from the two mixtures were withdrawn and assayed on 60 mm 2Np-Glc in standard conditions.
Samples (0.086 μg/μl) were enzymatically digested in the acidified inhibition buffer (formic acid to 5% (v/v) final concentration, pH 2) using pepsin from porcine stomach mucosa (3,260 units/mg, Sigma-Aldrich) at 1:20 enzyme to substrate ratio at 37 °C for 30 min. Resulting peptide mixtures (5 μl) were loaded, purified, and concentrated on a monolithic trap column (200 μm inner diameter × 5 mm, LCPackings, Sunnyvale, CA) at 25 μl/min flow rate and separated by nanoflow reverse-phase chromatography on a PS-DVB monolithic column (200 μm inner diameter × 5 cm, LCPackings) at 300 nl/min using an UltiMateTM 3000 HPLC (Dionex, Sunnyvale, CA). The following solvents and gradient conditions were used: solvent A: 2% acetonitrile in 0.1% formic acid and 0.025% trifluoroacetic acid, solvent B: 98% acetonitrile in 0.1% formic acid and 0.025% trifluoroacetic acid, gradient: 5–50% B in 40 min, 50–98% B in 6 s. Eluting peptides were directly analyzed by nano-ESI-MS in positive ion mode using information-dependent acquisition (IDA). The two most abundant multiply charged ions were automatically selected and subjected for collision induced dissociation experiments. Nitrogen was used as collision gas. Tandem mass spectra were analyzed by manual inspection and by the use of Mascot Server (version 2.2). Peak lists for Mascot containing all acquired MS/MS spectra were generated by Analyst QS 2.0 software using the default parameters. Mascot was set up to search database containing a single protein (SSO1353) sequence extracted from NCBInr and was run with a fragment ion mass tolerance of 0.1 Da and a parent ion tolerance of 50 ppm. MS/MS ion score cut-off was set to 10. No enzyme was specified. 2F-Glc was defined as variable modification in Mascot searches. Three independent inhibition experiments were performed and on each resulting samples two analytical measurements were run.
Definition and Analysis of a New Glycoside Hydrolase Family
Gapped BLAST searches (13) were performed against the non-redundant protein set at the NCBI and against classified and unclassified sequences present in carbohydrate-active enzymes database (CAZy) (5). A total of 90 sequences were used to define new family. This family includes proteins from archaeal, bacterial, and eukaryotic origin, several already collected in CAZy by similarity to human non-lysosomal bile acid β-glucosidase 2 (Gba2), but not yet assigned to a family. This family was designated as glycoside hydrolase family 116 and will be released in CAZy. The sequences were aligned with Muscle 3.7 (14), and the resulting alignments were subsequently manipulated and analyzed with an in-house modified version of Jalview (15).3 Estimated sequence distances were determined by maximum likelihood using LG distances (16) and constructed a distance tree using the Ward hierarchical clustering method (17).
RESULTS
Isolation of ORF SSO1353
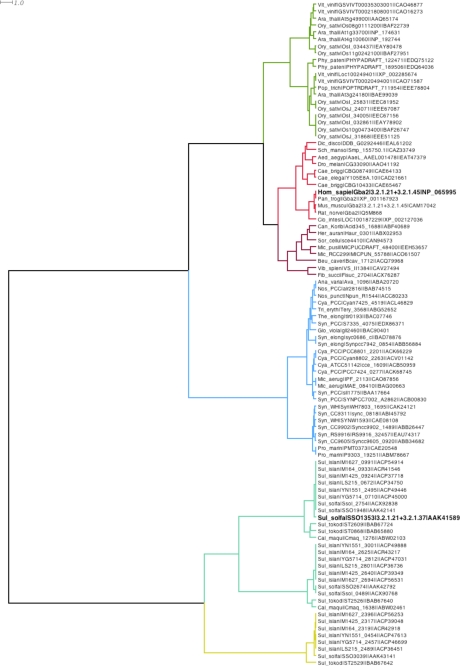
The inspection of the genomic sequence of the archaeon S. solfataricus, strain P2, revealed an ORF downstream of the gene sso1354 encoding for an endoglucanase (Fig. S1) (18). Sso1353 is presently annotated as an hypothetical protein while the other ORFs in this cluster, sso1351, sso1352, and sso1355, are a putative permease, a transcriptional regulator, and a carboxypeptidase, respectively. ORFs sso1354 and sso1353 are transcribed in the same direction and are separated by 57 bp in which the latter ORF is preceded by a putative promoter formed by an AT-rich box A (centered at −30 nt from ATG) and a TFB-responsive element (centered at −38 nt) (not shown). Northern blot analysis showed that sso1353 is expressed as an isolated gene (not shown) and the absence of a clear Shine-Dalgarno-like motif in the intergenic region suggests that sso1353 gene is translated as a leaderless gene (19). Initial gapped BLAST searches (13) revealed that SSO1353 is similar to proteins of unknown function from Archaea, Bacteria, and Eukarya and, to a lesser extent, to eukaryotic non-lysosomal bile acid β-glucosidases. The higher sequence identity scores (>31%) were with archaeal proteins with the highest (86%) with loci sso1948 from S. solfataricus, strain P2, and M1425_0924 and M1627_099 from S. islandicus, strains M.14.25 and L.S.2.15, respectively. Interestingly, these highly similar genes also lie downstream to a locus encoding for an endoglucanase (SSO1949 in S. solfataricus (20)), suggesting that gene duplication occurred in these organisms. Among non-lysosomal bile acid β-glucosidases, the scores were much lower, the best ones being with human and Ciona intestinalis enzymes (19% identity). Sequences from the NCBI were searched to complement the set of unclassified glycoside hydrolase sequences already present in CAZy previously collected based on the bile acid-glycosidases, and integrated to create a new family, that we have designated as Glycoside Hydrolase family 116 (GH116). Once the conserved catalytic regions were aligned, a distances tree was obtained (Fig. 1).
FIGURE 1.
Phylogenetic tree of family GH116 using Ward hierarchical clustering distances. The leaves of the tree indicate the organism genus and species information (e.g. Hom_sapie corresponds to Homo sapiens), the gene or Locus name, the EC activities if characterized experimentally, and a database accession number. Tree branches were colored according to identified significant subgroups using Dendroscope 2.3 (21).
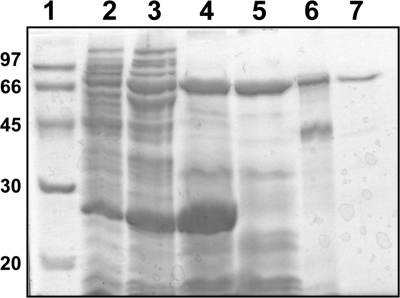
To ascertain if the sso1353 encodes for a novel glycoside hydrolase, the corresponding gene was cloned by PCR from the genomic DNA of S. solfataricus, strain P2. The primer at the 5′ of the gene was designed starting from the first Met. Attempts to express SSO1353 fused to glutathione S-transferase (at the N terminus) or to a His tag (at the C terminus) were unsuccessful; therefore, the gene was cloned in pET29a without any purification tag, obtaining the plasmid vector pET1353. The resulting recombinant SSO1353 protein was successfully expressed in the soluble fraction and purified to homogeneity by performing three subsequent heating steps followed by a hydrophobic chromatography and a gel filtration (Fig. 2). After the last purification step we obtained about 1.5 mg of pure protein per liter of E. coli culture.
FIGURE 2.
SDS-PAGE analysis of SSO1353. Lane 1, molecular weight markers; lane 2, E. coli BL21(DE3)Ril/pET1353 soluble protein extract (45 μg); lanes 3–5, protein extract after heat treatment at 55, 75, and 85 °C, respectively (70, 120 and 84 μg, respectively); lane 6, typical sample after hydrophobic chromatography (21 μg); lane 7, sample after gel filtration (3 μg).
Properties of SSO1353
A gel filtration run with the suitable molecular weight standards revealed that SSO1353 was a monomer of about 76 kDa in native conditions (not shown). The purified enzyme was optimally active at pH 5.5 (50 mm sodium citrate) when assayed on 5 mm 4-nitrophenyl-β-d-gluco- and -xylopyranoside (4Np-Glc, 4NP-Xyl) substrates at 65 °C, with specific activities of 0.3 and 0.8 units mg−1, respectively. The activity on 4Np-Xyl was not affected by Triton X-100 (0.5–2.0 mg ml−1), CHAPS (0.5–2.0 mg ml−1), Mn2+, Mg2+ (5 mm), and 1 mm EDTA.
To determine the substrate specificity of this enzyme, the hydrolytic activity of SSO1353 toward various substrates was investigated at 65 °C in 50 mm sodium citrate, pH 5.5. In addition to 4Np-Glc and -Xyl, also 4Np-Gal, 2Np-Glc, -Gal, -Xyl, methylumbellyferyl-β-d-glucopyranoside (MU-Glc), and MU-Xyl were substrates of the enzyme. Instead, no activity was observed on 4Np-Man and -GlcNAc; X-Glc and X-Gal; 2Np-cellobioside; 4Np-α-d-Gal,-Glc, and, -Man; 4Np-α-l-Fuc, -arabinoside; 4Np-β-l-Fuc, β-d-oligosaccharides of glucose and xylose (di-, tri-, tetra- and pentaose), and glucocerebroside, octyl-β-d-glucopyranoside and gangliosides even after prolonged incubations. Steady-state kinetic parameters of SSO1353 were measured for the glycosides that were substrates of the enzyme (Table 1). SSO1353 showed similar kinetic constants for the substrates tested: the highest specificity constant for MU-glycosides results from a reduced Km. Therefore, this kinetic characterization showed that SSO1353 is a β-d-glycosidase specific for gluco- and xylosides (EC 3.2.1.21/37) showing increased affinity for substrates having hydrophobic leaving groups. Finally, interestingly, the activity of SSO1353 on MU-Glc was inhibited by both N-butyl-deoxynojirimycin (NB-DNJ) and conduritol β-epoxide (CBE). The former showed an IC50 of 1.5 mm while 2 mm CBE gave 93% inhibition after 30 min of incubation. The sensitivity to this irreversible inhibitor differentiates SSO1353 from human non-lysosomal glucosylceramidase, which is insensitive to CBE (see below) (6).
TABLE 1.
Steady-state kinetic constants of SSO1353
| Substrate | kcat | Km | kcat/Km |
|---|---|---|---|
| s−1 | mm | s−1mm−1 | |
| 2Np-Glc | 4.7 ± 0.3 | 13 ± 2 | 0.37 |
| 4Np-Glc | 4.9 ± 0.5 | 54 ± 12 | 0.09 |
| 4Np-Xyl | 4.3 ± 0.4 | 25 ± 8 | 0.17 |
| MU-Glc | 1.2 ± 0.1 | 2.6 ± 0.5 | 0.47 |
| MU-Xyl | 0.8 ± 0.1 | 1.2 ± 0.3 | 0.71 |
SSO1353 Has Transglycosylation Activity and Follows a Retaining Reaction Mechanism
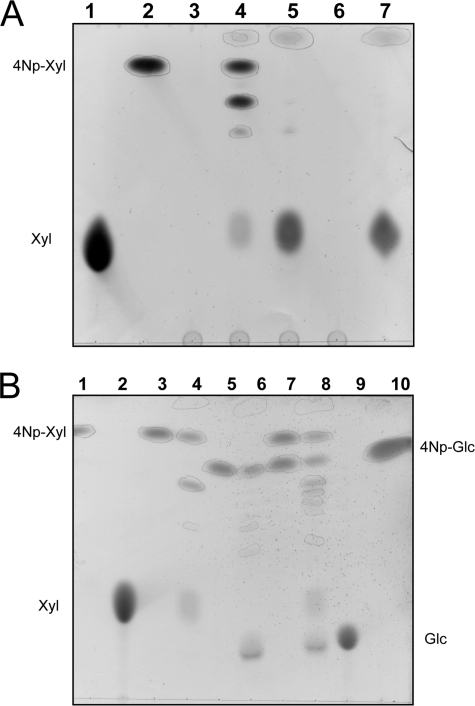
The products of the reaction mixtures containing SSO1353 and 4Np-Xyl were examined by thin layer chromatography (TLC) (Fig. 3). Interestingly, the products include not only xylose, but also oligosaccharides with a higher degree of polymerization than the 4Np-Xyl substrate, indicating that the enzyme performed transglycosylation reactions (Fig. 3A, lane 4). The transglycosylation products could be completely hydrolyzed (Fig. 3A, lane 5) by the addition of limiting amounts of β-glycosidase from S. solfataricus (Ssβ-gly), which shows broad substrate specificity for β-d-glycosides (22), indicating that SSO1353 catalyzed the formation of β-d-xylo-oligosaccharides. Remarkably, the enzyme catalyzed transglycosylation reactions also by using 4Np-Glc (5 mm) as a substrate and an increased number of transglycosylation products were found when both substrates were included in the reaction mixture: at least five compounds are easily observable by TLC (Fig. 3B, lane 8).
FIGURE 3.
Reaction products of SSO1353. Thin layer chromatography of the transxylosylation reactions with 4Np-Xyl. (A) In each lane were loaded 20 μl of the following reaction mixtures: lane 1, xylose standard 25 mm; lane 2, blank mixture containing 50 mm sodium citrate buffer pH 5.5, 4Np-Xyl 5 mm; lane 3, SSO1353 after TP85 °C (30 μg); lane 4, reaction mixture same as lane 2 with added SSO1353 after TP85 °C (150 μg); lane 5, sample loaded in lane 4 with added Ssβ-gly (50 μg); lane 6, Ssβ-gly (25 μg); lane 7, sample loaded in lane 2 with added Ssβ-gly (25 μg). Transxylosylation reactions with 4Np-Xyl and 4Np-Glc. (B) In each lane were loaded 20 μl of the following reaction mixtures: lane 1, 4Np-Xyl 5 mm; lane 2, xylose standard 25 mm; lane 3, blank mixture containing 50 mm sodium citrate buffer, pH 5.5, 4Np-Xyl 5 mm; lane 4, reaction mixture same as lane 3 with added pure SSO1353 (7 μg); lane 5, blank mixture containing 50 mm sodium citrate buffer, pH 5.5, 4Np-Glc 5 mm; lane 6, same as lane 5 with added pure SSO1353 (7 μg); lane 7, blank mixture containing 50 mm sodium citrate buffer, pH 5.5, 4Np-Xyl 5 mm and 4Np-Glc 5 mm; lane 8, same as lane 7 with added pure SSO1353 (7 μg); lane 9, glucose standard 25 mm; lane 10, 4Np-Glc 5 mm.
To determine the stereo- and regioselectivity of the transglycosylation activity of SSO1353 in the presence of 4Np-Xyl we scaled up the reaction to purify the products. The reverse phase HPLC purification revealed the presence of two transglycosylation products corresponding to 4NP-disaccharide (4Np-Xyl2) and 4NP-trisaccharide (4NP-Xyl3). Each product was identified by MALDI-TOF mass spectrometry, methylation analysis, 1H and 13C-NMR spectroscopy. The positive ions MALDI-TOF mass spectrum of 4Np-Xyl2 showed a pseudomolecular ion (M+Na)+ at m/z 425.84, which accounted for the presence of a 4-phenyl glycoside of xylose disaccharide.
The 1H-NMR spectrum showed two anomeric doublets in the ratio of 1:1 of xylose at 5.24 ppm (H-1 of β anomer A, 1,2JH,H 7.3 Hz) and 4.49 ppm (H-1 of β anomer B 1,2JH,H 7.3 Hz), respectively, together with signals of protons geminal to hydroxyl groups in the region between 3.0 and 4.0 ppm. Moreover signals having the same intensity as anomeric signals and attributable to the 4-nitrophenyl moiety, occurred at 7.25, and 8.28 ppm. The chemical shifts of these signals were in agreement with those of the disaccharide β-Xyl-(1→4)-β-Xyl-O-4-Np, as obtained by analysis of two-dimensional 1H and 13C NMR spectroscopy. In particular the (1→4) linkage was deduced from the C-4 glycosylation shift at 77.4 ppm of xylose unit A respect to the value of 70.4 ppm for an unsubstituted xylopyranoside (23). Further support to this structure derived from the methylation analysis, which showed the presence of 1,4,5-tri-O-acetyl-2,3-di-O-methyl xylitol, corresponding to 4-substituted xylopyranose unit, and of 1,5-di-O-acetyl-2,3,4-tri-O-methyl xylitol, corresponding to a terminal non-reducing end xylopyranose unit.
The positive ions MALDI-TOF mass spectrum of 4NP-Xyl3 showed a pseudomolecular ion (M+Na)+ at m/z 557.52, which accounted for the presence of a nitro-phenyl glycoside of xylose trisaccharide. The 1H NMR spectrum showed the presence of three anomeric proton signals at 5.25, 4.51, and 4.47 ppm, respectively. The beta anomeric configuration for all the xylose units was inferred from the 7.3 Hz value of 1,2JH,H coupling constants. The methylation analysis revealed the same residues as for 4Np-Xyl2, suggesting a linear trisaccharide structure β-Xyl-(1→4)-β-Xyl-(1→4)-β-Xyl-O-4-Np for 4NP-Xyl3. The complete assignment of the 1H and 13C values (see “Experimental Procedures”) confirmed the above structure. These results unequivocally demonstrate that SSO1353 promoted the transglycosylation reaction by following a retaining reaction mechanism.
Identification of the Catalytic Residues of SSO1353 by Site-directed Mutagenesis
Retaining glycosidases generally utilize a double displacement mechanism catalyzed by two enzymatic carboxylates and in which a glycosyl intermediate is formed and hydrolyzed. In the first step of the reaction, one of the carboxylic acids functions as a general acid catalyst protonating the glycosidic oxygen while the nucleophile residue attacks the sugar anomeric center to form the glycosyl enzyme intermediate (glycosylation step) (Fig. S2). In the second step (de-glycosylation step), the group previously acting as an acid now works as a base catalyst deprotonating the water and resolving the glycosyl enzyme intermediate. Both steps proceed via transition states with substantial oxocarbenium ion character (24).
The identification of key active site residues in a glycoside hydrolase is crucial to determine the catalytic machinery for the classification of this class of enzymes (5, 25, 26). These residues can be identified by using several different techniques, site-directed mutagenesis followed by kinetic analysis of the mutants being one of the approaches most used. Briefly, conserved aspartic/glutamic acid residues identified by sequence analysis are mutated with non-nucleophilic amino acids; the reduction or even abolition of the enzymatic activity is a strong indication that the mutation removed catalytic residues. The activity of the mutants can be chemically rescued in the presence of external nucleophiles such as sodium azide. The characterization of the anomeric configuration of the glycosyl-azide products allows the assignment of the mutated residue as the nucleophile or the acid/base of the reaction (Fig. S2B) (27).
We aligned the amino acid sequence of SSO1353 to eight other hypothetical proteins identified by BLAST analysis with an identity ≥22%. The multi-alignment led to the identification of 15 Asp/Glu residues highly conserved (Fig. S3); among these, Glu-335, Asp-406, and Asp-426 were invariant and, together with Asp-458, were mutated by site-directed mutagenesis obtaining the mutants E335G, D406G, D458G, and D462G. These SSO1353 mutants were expressed and purified as described above. During this procedure the proteins showed identical behavior, suggesting that the mutations did not affected the stability of the enzymes. These purification steps yielded proteins with similar concentrations and purification degrees (Fig. S4). The mutants assayed at 65 °C on 2Np-Glc 40 mm in 50 mm sodium citrate buffer pH 5.5 were completely inactive indicating that the mutations affected the catalytic machinery of SSO1353. When 1 m sodium azide was included in the assay on 40 mm 2Np-Glc we observed the reactivation of the D462G mutant, which showed a specific activity of 0.5 units mg−1, which is about 7-fold lower than that of the wild type (3.6 units mg−1) assayed in the same conditions. Instead, the external ion did not modify the specific activity of the wild type and did not reactivate the E335G, D406G, and D458G mutants.
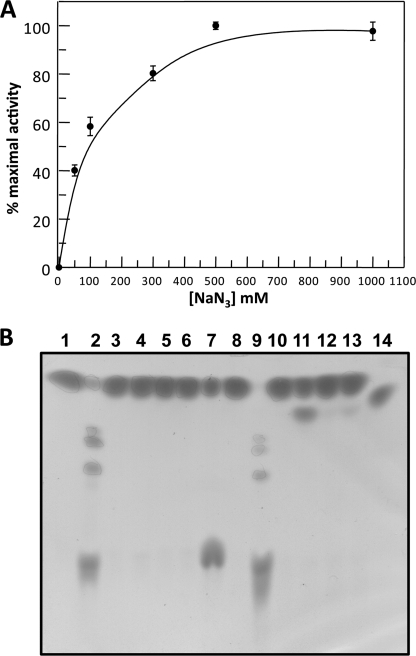
The mutant D462G was assayed at standard conditions on 40 mm 2Np-Glc in the presence of increasing concentrations of sodium azide: the maximal activity was observed at 0.5 m sodium azide (Fig. 4A). At these conditions the kinetic constants were kcat of 0.64 ± 0.1 s−1, Km of 16.2 ± 6 mm, and kcat/Km of 0.04 s−1 mm−1, showing that D462G maintained a similar affinity for the substrate, but a specificity constant 10-fold lower than the wild type. Reaction mixtures prepared at these conditions and containing the wild type and the mutants were analyzed by TLC after prolonged incubation. The D462G produced a novel compound, which was observed only in trace amounts with the other mutants. Instead, the wild type completely converted the substrate producing transglycosylation products (Fig. 4B). D462G reaction mixtures in preparative scale allowed the isolation and structural characterization of this product that was unequivocally identified as β-glucosyl azide. The reactivation in the presence of the external ion and the anomeric configuration of this product strongly indicate that Asp-462 is the acid/base of the reaction.
FIGURE 4.
Chemical rescue of the activity of SSO1353 mutants. (A) Dependence of activity of D462G mutant on different concentrations of sodium azide. (B) TLC analysis of the reaction mixtures of SSO1353 wild type and mutants in the absence (lanes 1–6) and in the presence of 0.1 m sodium azide (lanes 8–13). Standard assays were performed overnight at 65 °C on 40 mm 2NP-Glc by using 11 μg of enzyme. Lane 1, blank with no enzyme; lane 2, wild type; lane 3, E335G mutant; lane 4, D462G; lane 5, D458G; lane 6, D406G; lane 7, standards (2NP-Glc and Glc); lane 8, blank with no enzyme; lane 9, wild type; lane 10, E335G; lane 11, D462G; lane 12, D458G; lane 13, D406G; lane 14, β-glucosyl-azide standard.
Identification of the Catalytic Nucleophile of SSO1353
To identify the nucleophile of the reaction we used the mechanism-based inhibition approach in combination with nano-electrospray ionization tandem mass spectrometry (nano-ESI-MS/MS) analysis. Mechanism-based inhibitors are ligands that bind to the active site by competing with the substrate of retaining glycosidases and require mechanism-based activation to react covalently with the enzyme (for a review see Ref. 28). One group of these inhibitors includes activated 2-deoxy-2-fluoro-glycosides; the presence of fluorine substituent at C2 slows both the glycosylation and the deglycosylation steps of the reaction by destabilizing the transitions states. The incorporation of good leaving groups (as 2,4-dinitrophenol or fluoride) accelerates the glycosylation step relative to the deglycosylation step of the reaction with the effect that the incubation of the enzyme with its corresponding 2-deoxy-2-fluoro-glycosides results in a time dependent inactivation with the accumulation of the 2-deoxy-2-fluoro-glycoside enzyme intermediate. Consequently, the nucleophile of the reaction labeled with the inhibitor can be identified by mass spectrometry (29).
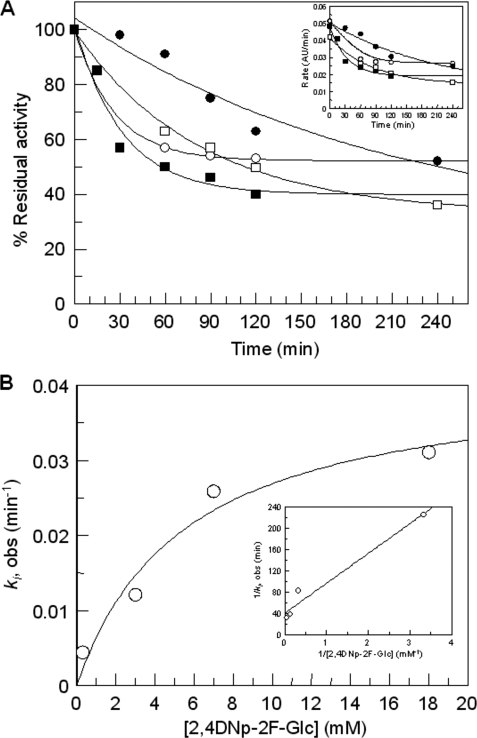
Time-dependent inactivation of SSO1353 was observed upon incubation of the enzyme with 2,4DNp-2F-Glc (Fig. 5). Inhibition was incomplete after 4 h of incubation (about 40%) even at the highest concentration of inhibitor used (18 mm). This is not surprising as the catalytic competence of GH inactivated by mechanism-based inhibitors, occurring via turnover of the intermediate via hydrolysis or transglycosylation, has been well documented (28). At these conditions we obtained the following inactivation parameters: ki = (6.9 ± 1.3) × 10−4 s−1; Ki = 5.5 ± 2.7 mm; ki/Ki = 1.2 × 10−4 s−1 mm−1.
FIGURE 5.
Time-dependent inactivation of SSO1353 using 2,4DNp-2F-Glc inhibitor. A plot of % residual activity versus time at four 2,4DNp-2F-Glc concentrations (● 0.3 mm, □ 3 mm, ○ 7 mm, and ■ 18 mm) is reported in A with the plot of rate versus time shown as inset. ki, obs were obtained from fitting the curves in A to a single exponential decay with offset because time-dependent inactivation did not decay to zero, and plotted versus inhibitor concentration (B), to determine Ki and ki. The reciprocal plot is shown as inset in B.
SSO1353 samples incubated in the absence and the presence of 2.9 mm 2,4DNp-2F-Glc for 2 h were analyzed by single-stage nano-ESI-MS to monitor alteration in the molecular mass of the protein. Nano-ESI mass spectra of intact proteins yield series of multiply charged molecular ion peaks with 40–100 positive charges under the experimental condition applied. Molecular mass of SSO1353 in the absence of inhibitor was measured to be 75914 ± 6 Da, which is comparable to the theoretical average molecular mass (75907.7 Da) within experimental error (0.009%) (Fig. S5A). After inhibition, molecular ion peaks shift toward higher m/z values leading a molecular mass of 76077 ± 5 Da and accounting for 163 ± 5.5 Da difference between the two species (Fig. S5B). To gain further evidence and a more detailed structural insight into the site-directed inhibition, samples were proteolytically digested by pepsin and the resulting peptide mixtures were analyzed by nano-HPLC-ESI-MS/MS in IDA mode. Based on MS/MS sequence data, 91 and 87% protein sequence coverage were respectively obtained in the absence and in the presence of inhibitor (Table S1A and Table S1B respectively). Interestingly, in the inhibited sample five peptides comprising residues 332–345, 332–343, 332–347, 332–348, and 332–349 showed considerable decrease in intensity, and in the same time, six new peptide molecular ions appeared in the corresponding scans (Table 2). The peptide molecular ion pairs corresponding to the normal and the modified sequences eluted at the same retention time and thus they were detected in the same survey scan in the sample containing 2,4DNp-2F-Glc. Therefore, they are likely due to in-source ion fragmentation process indicating a relatively labile bond between amino acid and inhibitor. These peptides showed an increase of 164.05 Da in molecular mass which corresponds well to the difference between unmodified and 2F-Glc modified peptides, and indicated that the ligand was likely bound to one of the amino acids present in peptide 332–349. To confirm the site of modification, nano-ESI-MS/MS spectra of the unmodified/modified peptide pairs were analyzed (Table 2, Fig. 6). MS/MS spectra show a very similar fragmentation pattern yielding characteristic b-type N-terminal fragment ions at the low m/z range. Based on these ions, and in particular, on the appearance of bn* (n≥3) modified fragment ions at m/z 641.28 (b3*), 712.32 (b4*) and 809.37 (b5*) in the inhibited sample, modification was unequivocally localized on amino acid Glu-335. Therefore, it was concluded that Glu-335 is the nucleophile of the reaction of SSO1353. Though we had no direct evidence from the chemical rescue experiment, we deduce that the invariant residue Asp-462 is the acid/base of the reaction.
TABLE 2.
Peptide mapping of SSO1353
Characteristic peptide molecular ions containing residue E at position 335 observed during nano-HPLC-ESI-MS/MS IDA analyses of SSO1353 incubated in the absence and in the presence of 2,4DNp-2F-Glc inhibitor and digested by pepsin. Peptide sequences (both unmodified and modified) were elucidated by the interpretation of nano-ESI-MS/MS spectra acquired on the doubly charged (z = 2) precursor ions (Fig. 4). Modification corresponds to the covalent attachment of 2F-Glc ligand at E335 (indicated in the sequence as E*).
| From-To | Unmodified and modified peptide sequences | Mw (calc.) | SSO1353 |
SSO1353 inhibited |
||||
|---|---|---|---|---|---|---|---|---|
| Rt | m/z | Intensity | Rt | m/z | Intensity | |||
| min | cps | min | cps | |||||
| 332–345 | AIYEAPQNCPYLGT | 1538.708 | 20.7 | 770.36 | 26 | 21.2 | 770.36 | 10 |
| AIYE*APQNCPYLGT | 1703.188 | – | n.d.a | – | 852.39 | 34 | ||
| 332–343 | AIYEAPQNCPYL | 1380.638 | 21.8 | 691.33 | 30 | 22.0 | 691.33 | 17 |
| AIYE*APQNCPYL | 1544.689 | – | n.d. | – | 773.35 | 44 | ||
| 332–347 | AIYEAPQNCPYLGTIG | 1708.813 | 23.2 | 855.42 | 42 | 23.2 | 855.42 | 16 |
| AIYE*APQNCPYLGTIG | 1873.293 | – | n.d. | – | 937.44 | 37 | ||
| 332–348 | AIYEAPQNCPYLGTIGA | 1779.85 | 23.5 | 890.94 | 280 | 23.7 | 890.93 | 63 |
| AIYE*APQNCPYLGTIGA | 1943.898 | – | n.d. | – | 972.96 | 240 | ||
| 332–349 | AIYEAPQNCPYLGTIGAC | 1882.859 | 24.8 | 942.46 | 320 | 24.6 | 942.44 | 106 |
| AIYE*APQNCPYLGTIGAC | 2046.07 | – | n.d. | – | 1024.46 | 384 | ||
| 332–340 | AIYEAPQNC | 1007.447 | – | n.d. | – | 25.2 | n.d. | – |
| AIYE*APQNC | 1171.495 | – | n.d. | – | 586.76 | 94 | ||
a n.d., not determined.
FIGURE 6.
Identification of the catalytic nucleophile site of SSO1353 by nano-HPLC-ESI mass spectrometry. (A) Nano-ESI mass spectrum of peptides eluted at 24.3–24.9 min of SSO1353 incubated with 2,4DNp-2F-Glc for 2 h. Doubly charged peptide ions at m/z 942.44 and 1024.46 correspond to the unmodified and the modified peptide 339–349, respectively. Tandem mass spectra on the unmodified (B) and modified peptides 339–349 (C) reveal modification on E4 residue.
DISCUSSION
We report here the molecular cloning, the expression in E. coli and the functional characterization of the product of the gene sso1353 from the hyperthermophilic archaeon S. solfataricus. The molecular characterization revealed the specificity of the enzyme for gluco- and -xylosides β-bound to hydrophobic groups that are hydrolyzed by following a retaining reaction mechanism. In addition, site-directed mutagenesis of conserved glutamic/aspartic amino acids and the chemical rescue of the β-glycosidase activity of the mutants, combined with the use of mechanism based inhibitors and mass spectrometric analysis, allowed us to identify Asp-462 and Glu-335 as the acid/base and the nucleophile of the reaction, respectively. Mutagenic studies also suggested that Asp-406 and Asp-458 residues play a role in catalysis, but elucidation of their function requires further investigations. Amino acid sequence analysis showed that SSO1353 shared identity with other hypothetical proteins and, remarkably, with eukaryotic non-lysosomal bile acid β-glucosidases.
So far, SSO1353 was not assigned to a defined glycoside hydrolase family in the carbohydrate active enzyme database. On the basis of our findings we propose that SSO1353 and its homologs define a new sequence-based family, namely GH116, which presently includes enzymes with β-glucosidases (EC 3.2.1.21), β-xylosidases (EC 3.2.1.37), or glucocerebrosidases (EC 3.2.1.45) activity. As for the other GH families, the retaining reaction mechanism and the catalytic role for the acid/base and the nucleophile, experimentally determined here, can be easily extended to all the enzymes belonging to this new family.
Interestingly, all the archaeal putative enzymes belonging to this new family are from Crenarchaea, and the vast majority originates from the genus Sulfolobus. A PSI-BLAST search conducted using SSO1353 as the query sequence retrieved (with low scores) uncharacterized bacterial glycosidases belonging to families GH15, GH63, and GH78. The latter families include mainly glucoamylases, α-glucosidases, and α-l-rhamnosidases, respectively, and are characterized by an (α/α)6 fold. Although SSO1353 is inactive on α-glycosides, this perhaps hints at structural similarities with enzymes from family GH116. Similar structural similarity between (α/α)6 fold glycoside hydrolase families degrading both α and β glycosidic bonds have already been described (30).
The phylogenetic analysis (Fig. 1) shows that sequences from the new family GH116 can be subdivided into two major groups, one containing sequences from Archaea and another one composed mostly of sequences from Cyanobacteria and Eukaryotes. The archaeal subgroup can be further subdivided into at least two subgroups, in which, interestingly, all the archaeal homologs of SSO1353 are present as multiple copies in the genomes of Caldivirga maquilingensis, S. tokodaii, S. solfataricus, and in the six strains of S. islandicus. The sso1353 homologs with identity >80%, lie downstream of genes encoding endoglucanases, and, interestingly, in S. solfataricus, this gene arrangement occurs twice. Presumably, the β-glycosidase activity of SSO1353 is involved, in combination with the secreted endoglucanase, in the degradation of exogenous glucans used as carbon energy source or, possibly, of the exo-polysaccharides (EPS) that are produced by S. solfataricus itself (31–33). Other sso1353 homologs, with identities in the range 21–33% exemplified by sso2674 and sso3039 in S. solfataricus, flank a putative peptidase or a putative gluconolactonase, respectively. These two other subgroups of enzymes similar to SSO1353 are present also in C. maquilingensis, S. tokodaii, and S. islandicus showing a remarkable identity (>80%) within each subgroup. The observation that the archaeal β-glycosidases from this novel GH family can be subgrouped according to their identity suggests they are present in multiple copies for functional purposes, possibly, for the degradation/modification of different substrates. A more detailed characterization of these enzymes is needed to understand their function in vivo.
The other major subdivision of the family is prone to be subdivided into several subgroups, one containing sequences from Cyanobacteria, the other having plant, animal, and mixed bacterial subdivisions. One of the members of the animal subgroup in this newly proposed family is human non-lysosomal glucosylceramidase or β-glucosidase 2 (GBA2). This enzyme, previously described as bile acid β-glucosidase (34), is involved in the catabolism of glucosylceramide, which is then converted to sphingomyelin (6). Glucocerebrosidases are important enzymes involved in the metabolism of gangliosides and globosides. Deficiency of this enzymatic activity is the cause of the most common lysosomal storage disorder named Gaucher disease (35) resulting from a defect in the lysosomal acid β-glucosidase (GBA1) belonging to GH30. This deficiency leads to the accumulation of glycosylceramides in certain organs, typically spleen, kidney, lungs, brain, and bone marrow (36). The finding that other cell types of Gaucher patients did not show accumulation of glycosylceramides suggested the existence of an alternative catabolic pathway that later was demonstrated to be catalyzed by GBA2 (6). This enzyme is ubiquitously expressed and it is associated to the cell surface. GBA2 is inactive on MU-Xyl, is inhibited by hydrophobic deoxynojirimycin (DNJ), and it is relatively insensitive to CBE (6, 34, 37). In humans, no known pathologies related to defects of GBA2 have been reported so far while only in certain mice strains treatments with NB-DNJ or gba2 gene knock-outs led to impaired spermatogenesis (38). However, such deleterious effects were not observed in other organisms including humans (6, 39, 40). These studies demonstrate the importance of understanding at the molecular level the reaction mechanism and the catalytic machinery of carbohydrate active enzymes for the development of specific inhibitors for bio-medical applications. The experimental identification of the catalytic amino acids of SSO1353 reported here, allows to easily identifying the catalytic machinery of human GBA2 despite the low sequence identity (18%) between the two enzymes. In GBA2 the nucleophile and the acid/base of the reaction are Glu-528 and Asp-678, respectively, which, as observed in a multi-alignment of putative glucocerebrosidases from mammals, plants, and tunicates belonging to this new GH family, are located in two conserved motifs (Fig. 7). In particular, amino acids with hydrophobic side chains are almost invariant in the position preceding the catalytic glutamic and aspartic acids in the enzymes belonging to the new family GH116 (Fig. S3 and Fig. 7). Our findings can now allow the planning of more detailed site-directed mutagenesis studies to better understand the molecular bases of the substrate recognition of GBA2.
FIGURE 7.
Multi-alignment of SSO1353 with glucosylceramidases. Invariant residues are indicated with “*”; increased level of conservation is indicated with “:” and “.” The residues corresponding to the nucleophile Glu-335 and acid/base Asp-462 of SSO1353 are boxed. Pan is XP_001167952.1 from Pan troglodytes; Homo is NP_065995.1 from Homo sapiens; Ciona is XP_002127036.1 from Ciona intestinalis; Sulfolobus is SSO1353 from S. solfataricus P2.
SSO1353 has substrate specificity and inhibitor sensitivity slightly different from those of GBA2. In fact, the archaeal enzyme can hydrolyze both aryl β-gluco and β-xylosides and it is inhibited with mm affinity by both NB-DNJ and CBE. Instead, GBA2 is inactive on MU-Xyl and it is relatively insensitive to CBE (6). These differences presumably reflect the different function of the two enzymes in vivo: the wider substrate specificity of the archaeal enzyme might allow to degrade a variety of substrates ensuring an efficient availability of sugars as energy source while GBA2 is involved in a well defined catabolic pathway. The purification of GBA2 is made difficult by its instability to detergents precluding its production in abundant and homogeneous form (6). Instead, robust GBA2 homologs from hyperthermophilic Archaea can be more easily expressed and purified from conventional hosts allowing more simple structural studies that might be easily extended to the human counterpart.
Supplementary Material
Acknowledgments
We thank Gerard W. Dougherty for the editing of the manuscript and Giovanni D'Angelo and Antonella De Matteis for the generous gift of the gangliosides. The IBP-CNR belongs to the Centro Regionale di Competenza in Applicazioni Tecnologico-Industriali di Biomolecole e Biosistemi.
This work was supported by Project MoMa 1/014/06/0 of the Agenzia Spaziale Italiana.
P. M. Coutinho and B. Henrissat, unpublished data.
- CAZyme
- carbohydrate active enzyme
- GBA1
- lysosomal acid β-glucosidase
- GH
- glycoside hydrolases
- GT
- glycosyltransferases
- ORF
- open reading frame
- GBA2
- non-lysosomal glucosylceramidase
- 4Np-Glc
- 4-nitrophenyl-β-d-glucopyranoside
- 4Np-Xyl
- 4-nitrophenyl-β-d-xylopyranoside
- MU-Glc
- methylumbellyferyl-β-d-glucopyranoside
- MU-Xyl
- methylumbellyferyl-β-d-xylopyranoside
- NB-DNJ
- N-butyl-deoxynojirimycin
- CBE
- conduritol β-epoxide
- TLC
- thin layer chromatography
- Ssβ-gly
- β-glycosidase from S. solfataricus
- 4Np-Xyl2
- 4NP-disaccharide
- 4NP-Xyl3
- 4NP-trisaccharide
- nano-ESI-MS/MS
- nano-electrospray ionization tandem mass spectrometry
- EPS
- exo-polysaccharides
- 2,4DNp-2F-Glc
- 2,4-dinitrophenyl-β-d-2-deoxy-2-fluoro-glucopyranoside
- IDA
- information-dependent acquisition
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid
- MALDI-TOF
- matrix-assisted laser desorption/ionization-time of flight.
REFERENCES
- 1.Varki A. (1993) Glycobiology 3, 97–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caramelo J. J., Parodi A. J. (2007) Semin Cell Dev. Biol. 18, 732–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saxon E., Bertozzi C. R. (2001) Annu. Rev. Cell Dev. Biol. 17, 1–23 [DOI] [PubMed] [Google Scholar]
- 4.DeMarco M. L., Woods R. J. (2008) Glycobiology 18, 426–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009) Nucleic Acids Res. 37, D233–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boot R. G., Verhoek M., Donker-Koopman W., Strijland A., van Marle J., Overkleeft H. S., Wennekes T., Aerts J. M. (2007) J. Biol. Chem. 282, 1305–1312 [DOI] [PubMed] [Google Scholar]
- 7.van Weely S., Brandsma M., Strijland A., Tager J. M., Aerts J. M. (1993) Biochim. Biophys. Acta 1181, 55–62 [DOI] [PubMed] [Google Scholar]
- 8.Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 9.Mackenzie L. F., Brooke G. S., Cutfield J. F., Sullivan P. A., Withers S. G. (1997) J. Biol. Chem. 272, 3161–3167 [DOI] [PubMed] [Google Scholar]
- 10.Leatherbarrow R. J. (1992) Erithacus Software Ltd., Staines, UK [Google Scholar]
- 11.Shaikh F. A., Müllegger J., He S., Withers S. G. (2007) FEBS Lett. 581, 2441–2446 [DOI] [PubMed] [Google Scholar]
- 12.Perugino G., Falcicchio P., Corsaro M. M., Matsui I., Parrilli M., Rossi M., Moracci M. (2006) Biocat. Biotrans. 24, 23–29 [Google Scholar]
- 13.Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edgar R. C. (2004) Nucleic Acids Res. 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., Barton G. J. (2009) Bioinformatics 25, 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le S. Q., Gascuel O. (2008) Mol. Biol. Evol. 25, 1307–1320 [DOI] [PubMed] [Google Scholar]
- 17.Ward J. H. (1963) J. Am. Stat. Assoc. 58, 236–244 [Google Scholar]
- 18.Maurelli L., Giovane A., Esposito A., Moracci M., Fiume I., Rossi M., Morana A. (2008) Extremophiles. 12, 689–700 [DOI] [PubMed] [Google Scholar]
- 19.Torarinsson E., Klenk H. P., Garrett R. A. (2005) Environ Microbiol. 7, 47–54 [DOI] [PubMed] [Google Scholar]
- 20.Huang Y., Krauss G., Cottaz S., Driguez H., Lipps G. (2005) Biochem. J. 385, 581–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huson D. H., Richter D. C., Rausch C., Dezulian T., Franz M., Rupp R. (2007) BMC Bioinformatics 8, 460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moracci M., Ciaramella M., Rossi M. (2001) Methods Enzymol. 330, 201–215 [DOI] [PubMed] [Google Scholar]
- 23.Bock K., Pedersen C. (1983) Adv. Carbohydr. Chem. Biochem. 41, 27–66 [Google Scholar]
- 24.Koshland D. E. (1953) Biol. Rev. Camb. Philos. Soc. 28, 416–436 [Google Scholar]
- 25.McCarter J. D., Withers S. G. (1994) Curr. Opin. Struct. Biol. 4, 885–8892 [DOI] [PubMed] [Google Scholar]
- 26.Zechel D. L., Withers S. G. (2000) Acc. Chem. Res. 33, 11–18 [DOI] [PubMed] [Google Scholar]
- 27.Ly H. D., Withers S. G. (1999) Annu. Rev. Biochem. 68, 487–522 [DOI] [PubMed] [Google Scholar]
- 28.Withers S. G., Aebersold R. (1995) Protein Sci. 4, 361–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Williams S. J., Withers S. G. (2000) Carbohydr. Res. 327, 27–46 [DOI] [PubMed] [Google Scholar]
- 30.Stam M. R., Blanc E., Coutinho P. M., Henrissat B. (2005) Carbohydr. Res. 340, 2728–2734 [DOI] [PubMed] [Google Scholar]
- 31.Elferink M. G., Albers S. V., Konings W. N., Driessen A. J. (2001) Mol. Microbiol. 39, 1494–1503 [DOI] [PubMed] [Google Scholar]
- 32.Nicolaus B., Manca M., Romano I., Lama L. (2003) FEMS Microbiol. Lett. 109, 203–206 [Google Scholar]
- 33.Zolghadr B., Klingl A., Koerdt A., Driessen A. J., Rachel R., Albers S. V. (2010) J. Bacteriol. 192, 104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matern H., Boermans H., Lottspeich F., Matern S. (2001) J. Biol. Chem. 276, 37929–37933 [DOI] [PubMed] [Google Scholar]
- 35.Grabowski G. A. (2008) Lancet 372, 1263–1271 [DOI] [PubMed] [Google Scholar]
- 36.Grabowski G. A., Gatt S., Horowitz M. (1990) Crit. Rev. Biochem. Mol. Biol. 25, 385–414 [DOI] [PubMed] [Google Scholar]
- 37.Overkleeft H. S., Renkema G. H., Neele J., Vianello P., Hung I. O., Strijland A., van der Burg A. M., Koomen G. J., Pandit U. K., Aerts J. M. (1998) J. Biol. Chem. 273, 26522–26527 [DOI] [PubMed] [Google Scholar]
- 38.van der Spoel A. C., Jeyakumar M., Butters T. D., Charlton H. M., Moore H. D., Dwek R. A., Platt F. M. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 17173–17178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amory J. K., Muller C. H., Page S. T., Leifke E., Pagel E. R., Bhandari A., Subramanyam B., Bone W., Radlmaier A., Bremner W. J. (2007) Hum. Reprod. 22, 702–707 [DOI] [PubMed] [Google Scholar]
- 40.Bone W., Walden C. M., Fritsch M., Voigtmann U., Leifke E., Gottwald U., Boomkamp S., Platt F. M., van der Spoel A. C. (2007) Reprod. Biol. Endocrinol. 5, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.