Abstract
Viruses initiate infection by attaching to molecules or receptors at the cell surface. Hepatitis C virus (HCV) enters cells via a multistep process involving tetraspanin CD81, scavenger receptor class B member I, and the tight junction proteins Claudin-1 and Occludin. CD81 and scavenger receptor class B member I interact with HCV-encoded glycoproteins, suggesting an initial role in mediating virus attachment. In contrast, there are minimal data supporting Claudin-1 association with HCV particles, raising questions as to its role in the virus internalization process. In the present study we demonstrate a relationship between receptor active Claudins and their association and organization with CD81 at the plasma membrane by fluorescence resonance energy transfer and stoichiometric imaging methodologies. Mutation of residues 32 and 48 in the Claudin-1 first extracellular loop ablates CD81 association and HCV receptor activity. Furthermore, mutation of the same residues in the receptor-inactive Claudin-7 molecule enabled CD81 complex formation and virus entry, demonstrating an essential role for Claudin-CD81 complexes in HCV infection. Importantly, Claudin-1 associated with CD81 at the basolateral membrane of polarized HepG2 cells, whereas tight junction-associated pools of Claudin-1 demonstrated a minimal association with CD81. In summary, we demonstrate an essential role for Claudin-CD81 complexes in HCV infection and their localization at the basolateral surface of polarized hepatoma cells, consistent with virus entry into the liver via the sinusoidal blood and association with basal expressed forms of the receptors.
Keywords: Fluorescence Resonance Energy Transfer (FRET), Receptor Structure-Function, Receptors, Tight Junction, Virus Entry
Introduction
Hepatitis C Virus (HCV)3 poses a global health problem, with over 170 million infected individuals worldwide, and establishes a persistent infection in the majority of individuals that leads to progressive liver injury, frequently culminating in fibrosis and hepatocellular carcinoma. The major cell types supporting HCV replication in vivo are hepatocytes in the liver. The observation, that retroviral pseudoparticles bearing HCV E1E2 glycoproteins (HCVpp) infect hepatocytes and hepatoma derived cell lines, suggests that virus glycoprotein-receptor interactions may in part define HCV tropism for the liver (1–3). Inhibiting HCV entry into host cells is one obvious target for therapeutic intervention, and the recent development of HCV strains that replicate and assemble infectious particles in cultured cells (4–6) has allowed studies on HCV particle internalization and replication.
Current evidence suggests that CD81 (1, 2, 7–9), scavenger receptor class B member I (10–13), and tight junction proteins Claudin-1 (CLDN1) (14–17) and Occludin (18–20) are critical factors defining HCV entry (reviewed in Ref. 21). Two additional Claudin family members, CLDN6 and CLDN9, have been reported to confer HCVpp entry (15, 17). CD81 and scavenger receptor class B member I interact with HCV-encoded E1E2 glycoproteins (gps), suggesting an initial role in mediating high affinity virus attachment to the cell (reviewed in Ref. 22). In contrast, there is minimal data supporting an interaction of CLDN1 with the HCV particle that may reflect the multistep and sequential nature of the entry process or that tight junction proteins facilitate particle internalization by indirect routes.
CD81 is a member of the tetraspanin superfamily of type III transmembrane proteins that contain four transmembrane domains and several conserved signature amino acid residues, including a conserved CCG motif and 4–6 cysteine residues that form critical disulfide bonds in the second extracellular loop (EC2). Tetraspanins associate with tetraspanin and non-tetraspanin proteins at cholesterol enriched microdomains and exert an array of biological functions, including cell-cell adhesion, cell migration, and proliferation (reviewed in Ref. 23). CD81 interacts with HCV-encoded E2 glycoprotein via a series of discontinuous amino acid residues in EC2 (1, 2, 7–9, 24). Kitadokoro and colleagues reported on the dimeric structure of CD81 EC2, suggesting that motifs within this domain drive CD81 dimerization (24, 25). CD81 is known to associate with the related tetraspanin CD9 and the immunoglobulin superfamily proteins EWI-2 and EWI-F (26), however, the one or more mechanisms underlying these protein-protein interaction(s) are unknown.
CLDNs are critical components of tight junctions that regulate paracellular permeability of endothelia and epithelia (27). Although CLDN polymerization is critical for the establishment of membranous tight junction strands (28), additional transmembrane (Occludin, tricellulin, and junctional adhesion molecule) and scaffold proteins are required for the spatial and functional organization of tight junctions in polarized epithelia (reviewed in Ref. 29). CLDNs exhibit a similar topology to the tetraspanins and have the capacity to associate with themselves and other CLDNs both within a cell and between apposing cells via interactions between their extracellular loops (27, 28). Piontek and colleagues recently reported FRET between tagged forms of CLDN5, suggesting that dimers are the primary building block(s) of tight junction strands (28).
We (30) and others (16, 31, 32) have reported that CLDN1 associates with CD81 in a variety of cell types. To ascertain the role of CLDN1-CD81 complexes in HCV entry, we investigated the relationship between various members of the CLDN family and CD81 in 293T human embryonal kidney (HEK) cells. Only CLDNs 1, 6, 9, and 12 co-localized with CD81 in a defined organization at the plasma membrane, consistent with a 1:1 molar relationship or stoichiometry. FRET studies confirmed interaction(s) between CLDN1, -6, and -9 and CD81, suggesting a relationship between CLDN association with CD81 and HCV receptor activity. In contrast, several members of the CLDN family associated with Occludin independent of viral receptor activity. Previous studies have highlighted a critical role for the first extracellular loop (EC1) of CLDN1 in HCV entry (14–17). Mutation of residues 32 and 48 in CLDN1 EC1 ablated its association with both CD81 and Occludin and viral receptor activity. Importantly, mutation of the same residues in the receptor inactive CLDN7 molecule allowed association with CD81 and facilitated viral entry into 293T cells in the absence of any detectable Occludin interaction. These data demonstrate an essential role for CLDN-CD81 complexes in HCV infection.
Because CLDNs oligomerize and associate with other cytoplasmic and signaling proteins to form tight junctions in polarized cells we investigated the presence and location of CLDN1-CD81 complexes in polarized HepG2 hepatoma cells. CLDN1 associated with basolateral pools of CD81 in polarized hepatoma cells in a comparable manner to that observed in non-polarized 293T cells. In contrast, CLDN1-CD81 complexes were absent from apically located tight junctions. The observation that HCV enters the liver via the sinusoidal blood and will encounter receptors expressed on the basolateral hepatocyte surface is consistent with a model where virus engagement of basolateral pools of CD81 and CLDN1 may initiate the particle internalization process.
EXPERIMENTAL PROCEDURES
Cell Lines and Reagents
293T and HepG2 cells were propagated in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 1% non-essential amino acids. HepG2 polarity was determined by MRP2 staining as previously described (33). The primary antibodies used were: anti-CLDN1, JAY.8 (Invitrogen), and 1C5-D9 (Novus); anti-CD81, 2s20, and 2s66 (University of Birmingham); anti-CD9 and TS9 (Abcam, UK); anti-VAP1 (gift from David Adams, University of Birmingham); and anti-ZO-1 (Invitrogen). Secondary labeled antibodies were obtained from Invitrogen: Alexa Fluor 488 goat anti-mouse IgG, 488 goat anti-mouse IgG2a, 633 goat anti-mouse IgG, 633 goat anti-mouse IgG1, 488 goat anti-rabbit IgG, and 633 goat anti-rabbit IgG.
HCVpp Genesis and Infection
Pseudoviruses expressing luciferase were generated as previously reported (30). Briefly, 293T cells were transfected with a 1:1 ratio of plasmids encoding HIV provirus expressing luciferase and HCV strain H77 E1E2 envelope gps, MLV gp, or empty vector (Env-pp), as previously described (2). Supernatants were harvested 48 h post transfection, pooled, and filtered. Virus-containing medium was added to target cells plated at 1.5 × 104 cells/cm2 for 4 h, unbound virus was removed by washing, and the cells were refed with growth media and incubated at 37 °C. At 72h post-infection the cells were lysed, the substrate was added, and luciferase activity was measured for 10 s in a luminometer (Lumat LB 9507). Specific infectivity was calculated by subtracting the mean Env-pp relative light unit signal from the HCVpp or MLVpp signals.
Generation of Fluorescence-tagged CLDN Proteins
The CLDN proteins used in this study were kindly supplied as a panel of pBABE-CLDN constructs by HongKui Deng (17) and were fused to Aequorea coerulescens green fluorescent protein (AcGFP) or Discosoma sp. red-monomer fluorescent (DsRED) at their N terminus using previously described methods (30). CLDN1 and CLDN7 mutants were kindly provided by Charles Rice (14) and amplified using the following primers: 5′-GGATCCGCCACCATGGCCAACGCGGGGCTG (CLDN1 FWD), 5′-CTCGAGTCACACGTAGTCTTTCCCGCTGG (CLDN1 REV), 5′-GGATCCGCCACCATGGCCAATTCGGGCCTGCA (CLDN7 FWD), and 5′-AAGCTTTCACACATACTCCTTGGAAGAGTTGG (CLDN7 REV) with the Phusion High Fidelity DNA polymerase (New England Biolabs) according to the manufacturer's instructions. PCR products were inserted into the pBABE vector using BamHI/XhoI (CLDN1) or BamHI/HindIII (CLDN7) restriction sites and sequenced.
Laser Scanning Confocal Microscopy
293T and HepG2 cells transfected to express AcGFP- and DsRED-tagged CLDNs or CD81 were grown on 13-mm glass coverslips and fixed in ice-cold methanol. The cells were imaged on a Zeiss Meta head laser scanning confocal microscope with settings optimized for each fluorescent protein to obtain the highest signal-to-noise ratio. Both 63 × 1.3 numerical aperture (NA) water immersion and 100 × 1.4 NA oil immersion objectives were employed to provide optimal resolution. CLDN and CD81 expression at the plasma membrane and intracellular locations was quantified using the overlay, profiling, and intensity frequency tools of Zeiss laser scanning confocal microscope software. Arbitrary fluorescence units (AFUs) were calculated by combining the number of pixels multiplied by the average intensity for the whole cell and was defined as 100%. The relative fluorescent intensities at the plasma membrane and intracellular locations were calculated relative to the whole cell signal.
Antibody Staining
293T cells grown on poly-l-lysine-coated glass coverslips were fixed in ice-cold methanol for 5 min. Primary antibodies were diluted in PBS/0.5% BSA (PBS-BSA buffer) and incubated with cells for 1 h at room temperature. Cells were washed 3× in PBS-BSA before addition of labeled secondary Abs for a further hour at room temperature. Finally, cells were washed 3× in PBS-BSA before counterstaining with 4′,6-diamidino-2-phenylindole in PBS for 5 min. Coverslips were mounted on glass slides for imaging (ProLong Gold Antifade, Invitrogen).
Fluorescence Intensity Ratio Analysis
The AcGFP and DsRED fluorophores were quantified using the profile function of the Zeiss laser scanning confocal microscope software, which measures the pixel intensity of the two fluorophores along the cell periphery (30). The brightness or intensity values of each pixel in the red and green channels were recorded to generate a series of paired values around the cell periphery. In the case of HepG2 cells, this series could be stratified according to whether or not pixels were located at the basolateral membrane or tight junction, as defined by ZO-1 staining. These paired fluorescent intensity values were used to generate a scatter plot. The degree of association between the signals can be described by a simple linear regression, where the slope y/x (red signal/green signal) describes the relative fluorescent signal and has been termed the fluorescence intensity ratio (FIR) (34, 35). The correlation coefficient (r2) determines how well the data fit the linear regression slope. Theoretically a 1:1 ratio of proteins would result in a FIR of 1, but as the absolute intensity of the DsRED fluorophore is 60% of AcGFP a FIR of 0.6 is consistent with a 1:1 ratio. To confirm this we exchanged the fluorophores fused to CD81 and CLDN1 and performed the analysis for both g.CD81/r.CLDN1- and g.CLDN1/r.CD81-transfected cells. In both cases similar gradients of ∼0.6 were obtained for the red and green fluorophores (data not shown).
FRET
The gradual acceptor photobleaching method of FRET was used, which entailed photobleaching the DsRED fluorophore gradually over time while monitoring AcGFP fluorescent intensity, as previously described (30). The profile of AcGFP- and DsRED-tagged proteins at the plasma membrane provided up to 1000 pixels for this analysis. After background and cross-talk correction, any increase in AcGFP intensity following DsRED photobleaching could be ascribed to FRET between the proteins, implying a distance of <10 nm. The percent FRET was defined as the proportion of pixels showing a measurable increase in AcGFP intensity (30).
Antibody Treatments and Neutralization Assays
293T cells grown on poly-l-lysine-coated glass coverslips were incubated with anti-CD81 or control monoclonal antibody (mAb) at 37 °C for 1 h. Cells were fixed immediately in ice-cold methanol with duplicate samples undergoing secondary antibody staining to verify primary mAb binding. Samples were counterstained with 4′,6-diamidino-2-phenylindole (Invitrogen) in PBS for 5 min. Coverslips were mounted (ProLong Gold Antifade, Invitrogen) on glass slides, and the cells were imaged with a meta head confocal microscope (Zeiss, LSM510) and a 100 × 1.3 NA oil immersion objective. In parallel, mAb-treated cells were infected with HCVpp, MLVpp, or Env-pp for 4 h at 37 °C, unbound virus and agents were removed by washing, and the cells were cultured for 72h. Viral infection was assessed by measuring luciferase activity, as previously described (2).
Cholesterol Modifications
293T cells grown on poly-l-lysine-coated glass coverslips were incubated in Dulbecco's modified Eagle's medium containing 10 mm methyl-β-cyclodextrin (MβCD, Sigma) for 1 h at 37 °C. Dose-response assays showed that 10 mm MβCD treatment for 1 h removed ∼50% of cholesterol without affecting cell viability (data not shown). Cholesterol was partially replenished in the MβCD-treated cells by incubating with 1 mm MβCD-cholesterol complex for 1 h (cholesterol, water-soluble, Sigma). Following these treatments the cells were fixed immediately in ice-cold methanol, counterstained with 4′,6-diamidino-2-phenylindole, and mounted. The cholesterol content before and after the various treatments was determined using an Amplex Red Assay kit (Invitrogen) in accordance with the manufacturer's instructions. In parallel, MβCD-treated cells were infected with HCVpp, MLVpp, or Env-pp for 4 h at 37 °C, unbound virus and agents were removed by washing, and the cells were cultured for 72 h. Viral infection was assessed by measuring luciferase activity, as previously described (2).
Analysis of CLDN1-CD81 Interactions by Surface Plasmon Resonance
The first extracellular domain(s) (EC1) of CLDN1 and CLDN7 between amino acid residues 29–81 were constructed using overlapping primers, introducing terminal NotI and SalI restriction sites. The EC1 sequences were cloned into the pMAL-c2 expression plasmid (New England Biolabs) downstream of the maltose-binding protein (MBP) sequence and a triple-alanine linker. Sequences were confirmed by big-dye terminator sequencing. The proteins were expressed in BL21(DE3)-competent cells (Stratagene), isolated from the soluble fraction, and batch purified using amylose resin (New England Biolabs) as previously described (36). Monomeric MBP-EC1 fusion proteins were further purified by size-exclusion chromatography (Superdex 75 26/60, Amersham Biosciences), and their purity was confirmed by analytical size-exclusion chromatography (Superdex 75 3.2/30) and SDS-PAGE analysis. The molecular masses of each protein were confirmed by mass spectrometry.
Recombinant protein interactions were assessed using a Biacore 3000 system (Biacore, Amersham Biosciences). MBP-CLDN1 EC1 or MBP-CD81 EC2 were immobilized to a CM5 chip using a standard amine-coupling method (Biacore, Amersham Biosciences) and MBP-CLDN1, MBP-CLDN7, MBP-CD81 and control MBP flowed over the chip surface at 1 mg/ml. To control for nonspecific protein interactions all recombinant proteins were flowed over an “empty” (activated 1 m ethanolamine-blocked) channel, and any response units observed were subtracted from test channels. All interactions were performed in HBS-EP buffer (Biacore, Amersham Biosciences) at 25 °C at a flow rate of 5 μl/min for 10 min, and dissociation was recorded for 2 min.
Statistics
The association between fluorescent proteins was assessed by linear regression; we chose to use the median and interquartile range of the values obtained to take into account the non-Gaussian distribution of the data. The %FRET values fit a Gaussian distribution and are presented as mean ± S.D. Differences in %FRET observed between samples were compared by Fisher's exact test. Corrections for multiple sampling (Bonferroni method and Dunnett's multiple comparison test) were used when appropriate. Statistical analyses were carried out in the Prism 4 package (GraphPad software, San Diego, CA), and probabilities are represented as p < 0.05 (*), p < 0.01 (**), and p < 0.001 (***).
RESULTS
Stoichiometry of CLDN1-CD81 Complexes
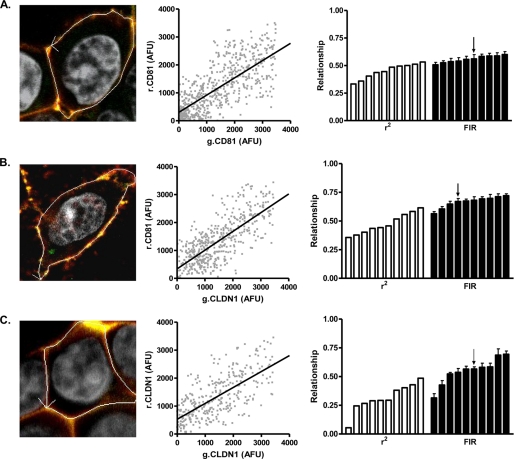
HEK 293T cells do not express CLDNs and are the preferred cell type to study homotypic and heterotypic CLDN interactions (28). Furthermore, expression of exogenous CLDN1 in 293T cells facilitates HCV entry and replication (14–17, 30). We therefore selected 293T cells to study the relationship between plasma membrane expressed forms of AcGFP- and DsRED-tagged CLDN1 and CD81. Several reports have quantified the ratio of fluorescent proteins at defined locations to assess protein subunit stoichiometry of cyclic nucleotide-gated channels and epithelial sodium channels in mammalian cells (34, 35). CD81 has been reported to dimerize (3, 31, 37); we therefore measured AcGFP.CD81 (g.CD81) and DsRED.CD81 (r.CD81) relative expression at the plasma membrane of ten individual cells. We observed a significant association between g.CD81 and r.CD81 AFUs with a median FIR of 0.6 with an interquartile range (IQR) of 0.58–0.62 (r2 = 0.5, 0.4–0.5) (Fig. 1A). If one corrects for the 60% lower fluorescence of the DsRED fluorophore compared with AcGFP, the ratio increases to 1.0, implying an equimolar association between g.CD81 and r.CD81, consistent with protein dimerization.
FIGURE 1.
Fluorescent intensity ratio of CLDN1 and CD81 in 293T cells. 293T cells were transfected to express AcGFP (g) and DsRED (r) fluorescence-tagged g.CD81-r.CD81 (A); g.CLDN1-r.CD81 (B), or g.CLDN1-r.CLDN1 (C). AcGFP and DsRed arbitrary fluorescence units (AFUs) at the cell periphery were determined by laser scanning confocal microscopy and used to generate a scatter plot, allowing one to calculate a correlation coefficient (r2) and fluorescent intensity ratio (FIR) for each cell analyzed. A representative scatter plot is depicted in the middle column, and the r2 correlation coefficient (white bars) and FIR (black bars) values for ten cells are summarized as a bar chart in the final column, where each bar depicts a single cell, and the arrow denotes the respective values for the presented image and scatter plots. In summary, the median FIR values from analyzing ten cells expressing g.CD81-r.CD81, g.CLDN1-r.CD81, and g.CLDN1-r.CLDN1 were 0.56 (IQR 0.53–0.59 and r2 0.47 (IQR 0.38–0.51)), 0.68 (IQR 0.63–0.74 and r2 0.45 (IQR 0.39–0.57)), and 0.56 (IQR 0.47–0.64 and r2 0.29 (IQR 0.25–0.42)), respectively.
Analysis of 293T cells expressing g.CLDN1 and r.CD81 demonstrated a significant association with a median FIR of 0.7 (r2 = 0.5), suggesting that CLDN1 and CD81 associate in a similar 1:1 relationship to that seen for g.CD81-r.CD81 (Fig. 1B). A similar relationship was noted for 293T cells co-expressing g.CLDN1 and r.CLDN1 (median FIR of 0.6), however a greater level of heterogeneity was apparent (r2 = 0.3, IQR 0.3–0.4), with some cells showing little or no evidence for protein association (Fig. 1C), possibly reflecting the more dynamic nature of CLDN1 homotypic associations required during strand formation (28, 38–41).
We previously reported that FRET occurs between g.CLDN1 and r.CD81 (30), leading to a transfer of energy from donor to recipient proteins and a concomitant loss in AcGFP fluorescence which could bias the regression analysis and alter the FIR. To assess whether FRET altered FIR values, we performed separate regression analyses for FRET and non-FRET data points. FRET occurred between CLDN1 and CD81 and had a minimal effect on FIR (supplemental Fig. S1), consistent with the linear relationship observed between the fluorescent proteins over a range of protein expression (AFUs). In summary, quantification of fluorescence-tagged proteins has allowed us to define the stoichiometry of the plasma membrane-expressed CLDN1-CD81 receptor complex.
Association of CLDN Family Members with CD81
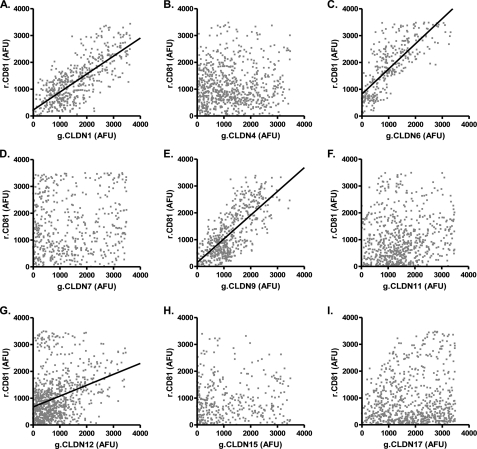
To investigate whether other CLDN proteins could associate with CD81 we generated a panel of AcGFP N-terminal-tagged fusion proteins representing classic (CLDN4, -6, -7, -9, -15, and -17) and non-classic (CLDN11 and -12) members of the CLDN family. All of the g.CLDN proteins localized to a comparable level at the plasma membrane and demonstrated enhanced expression at areas of cell-cell contact, consistent with trans interaction(s) between CLDNs expressed on adjacent cells (data not shown) (28). HCVpp only infected 293T cells expressing CLDN1, -6, and -9, whereas MLVpp infected parental and CLDN-expressing cells with comparable efficiency (supplemental Fig. S2), confirming earlier reports that a restricted number of CLDNs support HCV entry (15, 17). To investigate whether CLDN association with CD81 defines HCV co-receptor activity we measured the FIR of g.CLDN-r.CD81 complexes in co-transfected 293T cells. A significant association was observed between AcGFP and DsRED signals in cells expressing g.CLDN1, g.CLDN6, or g.CLDN9 and r.CD81 (Fig. 2). There was limited evidence for an association between g.CLDN12 and r.CD81 in a minority of cells, however the variability of the data set and scatter of points around the line of best fit suggest that, if present, this relationship is less well defined (Table 1).
FIGURE 2.
Analysis of CLDN-CD81 interactions. 293T cells were transfected to express DsRED-CD81 (r.CD81) and a panel of AcGFP-tagged CLDN (g.CLDN) constructs (CLDN1–17, panels A–I). Ten cells were imaged as described in Fig. 1, and estimates of r.CD81 association with g.CLDNs were evaluated by regression analysis and summarized in Table 1. The images represent scatter plots closest to the median FIR for each g.CLDN-r.CD81 studied.
TABLE 1.
Fluorescence-tagged CLDN protein association with CD81, Occludin, and CLDN1
| r2 (IQR)a | FIR (IQR) | %FRET ± S.D.b | |
|---|---|---|---|
| CD81 association with | |||
| CLDN1 | 0.5 (0.42–0.61) | 0.7 (0.67–0.74) | 60 ± 4 |
| CLDN4 | 0.2 (0.17–0.24) | 0.3 (0.21–0.41) | 5 ± 8 |
| CLDN6 | 0.6 (0.54–0.62) | 1.0 (0.80–1.10) | 50 ± 6 |
| CLDN7 | 0.0 (0.00–0.09) | NDc | ND |
| CLDN9 | 0.4 (0.36–0.50) | 0.5 (0.40–0.67) | 63 ± 7 |
| CLDN11 | 0.2 (0.16–0.23) | 0.3 (0.19–0.41) | 7 ± 4 |
| CLDN12 | 0.4 (0.28–0.48) | 0.5 (0.26–0.83) | 15 ± 12 |
| CLDN15 | 0.0 (0.00–0.04) | ND | ND |
| CLDN17 | 0.2 (0.17–0.30) | 0.3 (0.22–0.52) | 6 ± 8 |
| Occludin association with | |||
| CLDN1 | 0.4 (0.29–0.54) | 0.7 (0.60–0.82) | 48 ± 1 |
| CLDN4 | 0.2 (0.14–0.34) | 0.3 (0.29–0.53) | 30 ± 7 |
| CLDN6 | 0.2 (0.04–0.36) | 0.8 (0.57–0.53) | 27 ± 9 |
| CLDN7 | 0.1 (0.04–0.18) | ND | ND |
| CLDN9 | 0.3 (0.13–0.48) | 0.7 (0.39–0.79) | 18 ± 8 |
| CLDN11 | 0.4 (0.12–0.45) | 0.7 (0.62–0.83) | 23 ± 7 |
| CLDN12 | 0.3 (0.19–0.37) | 0.6 (0.51–0.71) | 38 ± 6 |
| CLDN15 | 0.1 (0.08–0.21) | ND | ND |
| CLDN17 | 0.5 (0.28–0.51) | 0.9 (0.74–1.07) | 40 ± 5 |
| CLDN1 association with | |||
| CLDN1 | 0.7 (0.67–0.74) | 0.5 (0.43–0.58) | 41 ± 4 |
| CLDN4 | 0.3 (0.22–0.41) | 0.2 (0.18–0.24) | 18 ± 12 |
| CLDN6 | 1.0 (0.82–1.11) | 0.6 (0.51–0.59) | 31 ± 6 |
| CLDN7 | 0.1 (0.00–0.10) | ND | ND |
| CLDN9 | 0.4 (0.36–0.42) | 0.5 (0.44–0.63) | 18 ± 3 |
a Interquartile range.
b Standard deviation.
c ND, not determined.
The FIR method gives an accurate measure of protein-protein association but is limited by the resolution of the microscope. To demonstrate protein-protein interactions (within 10 nm) we measured FRET between CLDNs and CD81 using a previously reported acceptor-photobleaching method (30). FRET occurred between g.CLDN1, -6, and -9 and r.CD81 (60%, 50, and 63% of plasma membrane pixels, respectively) at a significantly higher frequency than with other g.CLDNs (Table 1). The level of g.CLDN or r.CD81 expression had no effect on FRET (34) (supplemental Fig. S1). Because Occludin is also important for HCV entry, we investigated the stoichiometry and association of different CLDN molecules with Occludin. The majority of CLDNs tested (CLDN1, -6, -9, -11, -12, and -17) demonstrated a median FIR with an IQR of between 0.6 and 0.9 (r2 = 0.2–0.5) with variable FRET, demonstrating no association between CLDN receptor activity and ability to interact with Occludin (Table 1). To ascertain whether CLDN1 could associate with other CLDNs we measured FIR and FRET values in 293T cells transfected to express g.CLDN1, -4, -6, -7, or -9 and r.CLDN1. Associations were detected between g.CLDNs 1, 4, 6, and 9 and r.CLDN1 by both FIR and FRET analysis, demonstrating CLDN1 homotypic and heterotypic cis-interactions; in contrast there was no demonstrable association between g.CLDN7 and r.CLDN1 (Table 1). In summary, we observed a significant relationship between the association of CLDN with CD81 and HCV receptor activity, suggesting that a close physical association between CLDN and CD81 is essential for HCV entry.
Analysis of CLDN1-CD81 Interactions by Surface Plasmon Resonance
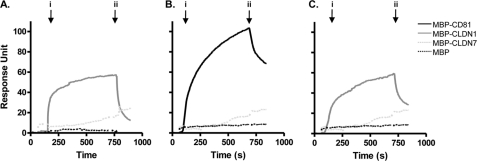
To independently assess CLDN1-CD81 protein interactions, recombinant MBP-CLDN1 EC1 or MBP-CD81 EC2 proteins were immobilized onto a BIAcore sensor chip, as detailed under “Experimental Procedures.” Homotypic CLDN1 and CD81 protein interactions were observed, with minimal detection of either recombinant loop protein interacting with MBP (Fig. 3, A and B). Importantly, MBP-CD81 EC2 demonstrated a specific interaction with MBP-CLDN1 EC1 and minimal interaction with MBP or MBP-CLDN7 (Fig. 3C). These data are consistent with our FRET imaging-based data (Table 1) and demonstrate a role for the EC loops in driving CLDN1-CD81 protein interactions.
FIGURE 3.
Analysis of CLDN1-CD81 extracellular loop interactions by surface plasmon resonance. MBP-CLDN1 EC1 (A) and MBP-CD81 EC2 (B and C) were immobilized onto the bio-sensor chip surface. Homotypic protein interactions were demonstrated by flowing MBP-CLDN1 EC1 (solid gray line) and MBP-CD81 EC2 (solid black line) over the respective chip surfaces (A and B) with both MBP-CLDN7 EC1 (dotted light gray line) and MBP (dotted black line) as negative controls at a concentration of 1 mg/ml. Heterotypic interaction between MBP-CLDN1-EC1 and MBP-CD81 EC2 is depicted in C. To control for nonspecific interactions, all MBP fusion proteins were flowed over an activated and blocked “empty” channel, and the response unit(s) were subtracted from the test channels. The arrow indicates the “association time” i when proteins are flowed over the respective chip surfaces and the “dissociation phase” begins at time ii when protein injection is stopped. Data are representative of two independent experiments.
Perturbation of CLDN1-CD81 Association
Tetraspanin protein interactions are known to be modulated by cholesterol (42–44). Kapadia and colleagues (45) reported that HCV entry is dependent on the cholesterol content of the host cell membrane, however, the underlying mechanism was not defined. To investigate the role of cholesterol in CLDN1-CD81 and CD81-CD81 association, 293T cells co-expressing g.CLDN1-r.CD81 or g.CD81-r.CD81 were treated with MβCD, a cyclic oligosaccharide that depletes cholesterol from the plasma membrane (46). Treatment with 10 mm MβCD removed ∼50% of total cholesterol and reduced g.CLDN1 localization at the plasma membrane, leading to an altered g.CLDN1-r.CD81 FIR and reduced FRET (from 50 ± 1.7% to 35 ± 9.5%) (supplemental Fig. S3). In contrast, MβCD had no detectable effect on CD81 expression at the plasma membrane or g.CD81-r.CD81 FIR. However, the frequency of pixels where FRET occurred between g.CD81 and r.CD81 was reduced, from 63% ± 5.0 to 55% ± 1.8 (supplemental Fig. S3). To further investigate the effects of cholesterol, MβCD-treated cells were incubated with MβCD-cholesterol complexes that restored cellular cholesterol to 75% of the initial value. This treatment restored both g.CLDN1-r.CD81 FIR or FRET and g.CD81-r.CD81 FRET values to pretreatment levels (supplemental Fig. S3). As previously reported (44) we found that MβCD treatment reduced HCVcc-cultured cells and HCVpp infection (data not shown). These data demonstrate that CLDN1 localization at the plasma membrane and its association with CD81 is cholesterol-dependent, which may in part contribute to the reduced susceptibility of MβCD-treated hepatoma cells to HCV infection.
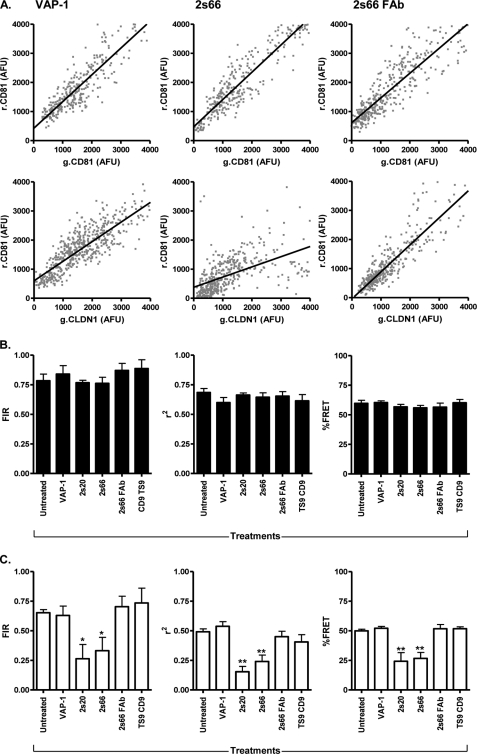
CD81 is a critical molecule defining HCV infection and treatment of hepatoma cells with anti-CD81 mAbs inhibits both recombinant HCV E2 binding (1, 2, 7–9) and virus infectivity (reviewed in Ref. 47). However, the one or more mechanisms by which the antibodies neutralize virus infectivity are not known. We were interested in studying the effect(s) of two HCV-neutralizing anti-CD81 mAbs on CD81 homotypic and heterotypic protein-protein interactions. 293T cells co-expressing g.CD81-r.CD81 or g.CLDN1-r.CD81 were treated with mAbs 2s20 and 2s66 specific for CD81 large extracellular loop or a control mAb specific for vascular adhesion protein 1. Both CD81-specific mAbs significantly reduced g.CLDN1-r.CD81 FIR and FRET values and yet had minimal effect(s) on g.CD81/r.CD81 FIR and FRET (Fig. 4). In contrast, anti-vascular adhesion protein 1 had no effect on g.CD81-r.CD81 or g.CLDN1-r.CD81 FIR/FRET values (Fig. 4). To investigate whether bivalent engagement of CD81 is critical to modulate lateral protein-protein interactions, we studied the effect of monovalent 2s66 FAb fragment on g.CD81-r.CD81 and g.CLDN1-r.CD81 FIR. Although 2s66 FAb bound to CD81 LEL with a similar affinity to the complete IgG molecule, it had no detectable effect on g.CD81-r.CD81 or g.CLDN1-r.CD81 FIR or FRET values (Fig. 4), suggesting that bivalent engagement and possible cross-linking of CD81 are essential to modulate lateral protein interactions. Of note, 5-fold increased concentrations of 2s66 FAb were required to neutralize HCV infectivity compared with the bivalent 2s66 IgG (IC50 values for 2s66 IgG are 0.8 μm and 2s66 FAb are 4.3 μm). Because CD81 is known to associate with tetraspanin CD9, we assessed the ability of anti-CD9 mAb TS9 to influence CD81 protein associations. Anti-CD9 TS9 bound to 293T cells but had no detectable effect on g.CD81-r.CD81 or g.CLDN1-r.CD81 FIR or FRET values, consistent with its inability to inhibit HCV entry (Fig. 4). In summary, bivalent antibody engagement of CD81 perturbs protein interactions with CLDN1 and may contribute to the neutralizing activity of these mAbs.
FIGURE 4.
Anti-CD81 modulation of CD81-CD81 and CLDN1-CD81 association(s). 293T cells were transfected to express AcGFP (g) and DsRED (r) fluorescence-tagged g.CD81-r.CD81 or g.CLDN1-r.CD81 and were treated with control anti-VAP1, anti-CD81 mAbs 2s20 and 2s66, 2s66 FAb fragment, and anti-CD9 TS9 at equimolar concentrations (13 μm) for 1 h at 37 °C. Representative g.CD81-r.CD81 and g.CLDN1-r.CD81 scatter plots of transfected cells treated with anti-VAP-1, anti-CD81 2s66 IgG, and FAb are shown (A). The effect of mAb treatments on g.CD81-r.CD81 (B) and g.CLDN1-r.CD81 (C) mean correlation coefficient (r2) and fluorescent intensity ratio (FIR) of ten cells is presented. One way analysis of variance and Dunnett's multiple comparison test were used to determine the degree of significance (*, p < 0.05; **, p < 0.01).
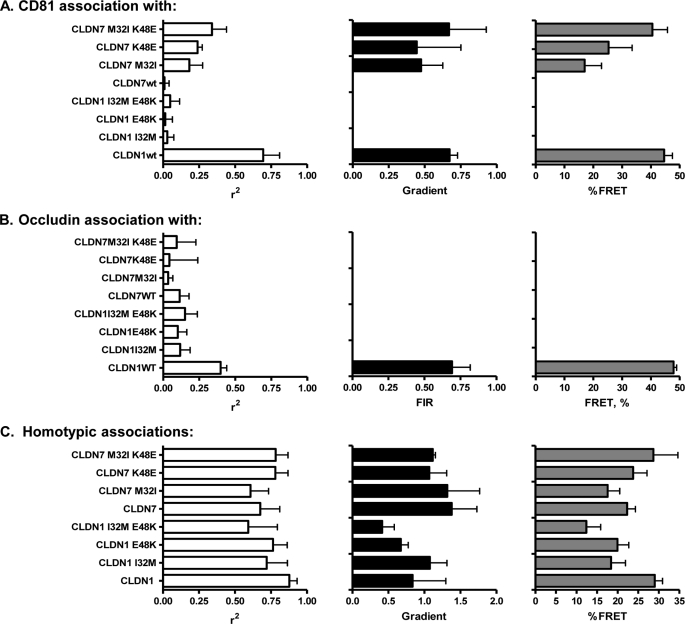
Mutations in CLDN EC1 Modulate CD81 Interaction(s) and HCV Receptor Activity
CLDN1 and CLDN7 differ in five residues within their first extracellular loop, and Evans and colleagues reported that the interchange of amino acid residues at positions 32 (Ile/Met) and 48 (Glu/Lys) rendered CLDN1 inactive and CLDN7 active for infection by unknown mechanism(s) (14). We hypothesized that these residues could play a role in the association of CLDN1 with CD81. To evaluate this 293T cells were co-transfected with plasmids expressing r.CD81 or r.Occludin with either g.CLDN1, g.CLDN1-I32M, g.CLDN1-E48K, g.CLDN1-I32M/E48K, g.CLDN7, g.CLDN7-M32I, g.CLDN7-K48E, or g.CLDN7-M32I/K48E (Fig. 5). Confocal imaging demonstrated that all proteins were expressed at comparable levels at the plasma membrane (data not shown). Analysis of the panel of g.CLDNs for their association(s) with r.CD81 and r.Occludin was performed by both FIR and FRET methodologies. g.CLDN1 and g.CLDN7 wild-type proteins showed the expected high and low FIR values with CD81 and Occludin, as previously observed (Table 1). Interestingly, the two mutations in CLDN1 (32M and 48K), either alone or in combination, abrogated any association with CD81 or Occludin. In contrast, both of the complementary changes in CLDN7 (32I and 48K) resulted in a mutated CLDN7 molecule that showed clear association with CD81 (16 and 25% FRET for 32I and 48E, respectively), and this was even more marked in the double mutant CLDN7 whose levels of association with CD81 were indistinguishable from that of CLDN1 (Fig. 5). Neither of the mutations promoted CLDN7 association with Occludin, suggesting that CLDN-Occludin interactions are not required for CLDN receptor activity. When tested for their susceptibility to infection these cells recapitulated known phenotypes (14), where the single and double mutation in CLDN1 abrogated HCVpp entry and CLDN7 mutants showed 36% (M32I), 41% (E48K), and 47% (double mutant) of the infectivity signal obtained for g.CLDN1 (supplemental Fig. S4).
FIGURE 5.
Effect of mutations in CLDN1 and CLDN7 EC1 on CD81 and Occludin association. 293T cells were transfected to express AcGFP (g) and DsRED (r) fluorescence-tagged wild-type and mutant forms of g.CLDN and r.CD81 (A) or r.Occludin (B), and the degree of association between fluorophore-tagged proteins was assessed by FIR and FRET analysis. C, 293T cells were transfected with AcGFP- and DsRED-tagged versions of wild-type and mutant CLDN constructs to assess the effect of EC1 mutations on CLDN-CLDN cis-interactions. Median FIR and FRET values from ten individual cells are presented (*, p < 0.05; **, p < 0.01).
To assess whether the mutations in CLDN1 and CLDN7, which modulate CD81 association, alter CLDN-CLDN homotypic interactions, 293T cells were co-transfected with AcGFP- and DsRED-tagged versions of parental and mutant CLDNs and assessed by FIR and FRET analysis (Fig. 5C). As noted earlier (Fig. 1C), parental CLDN1 molecules associated with each other (Fig. 5C), however, when either of the residues at codons 32 or 48 were changed, there was a modest but non-significant reduction in FIR, suggesting that these residues have a minor role in the genesis or stabilization of CLDN1 dimers. In contrast neither of the alterations in CLDN7 altered the degree of homotypic association (Fig. 5C). Thus, we infer that it is the heterotypic interaction between CLDNs and CD81, rather than the homotypic CLDN-CLDN interactions, that is important for HCV entry.
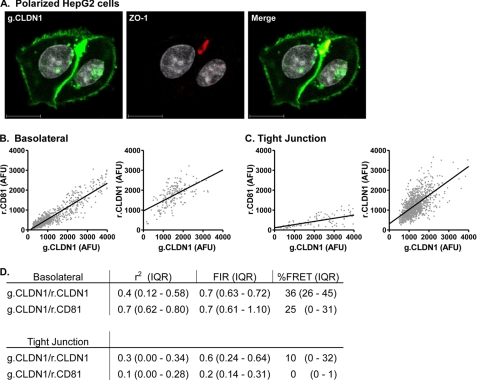
The Effect(s) of Cell Polarization on CD81-CLDN1 Association
293T cells do not polarize or form functional tight junctions; we therefore sought to confirm our observations on CLDN1-CD81 association in a polarized cell type. This is particularly relevant to our understanding of HCV infection, because hepatocytes, the major target cell supporting HCV replication in the liver, are highly polarized (48). The human HepG2 hepatoblastoma line has been shown to polarize in culture, forming spheroid structures at sites of cell-cell contact that contain tight junctions and multiple membrane protein markers that resemble bile canaliculi in the liver (reviewed in Ref. 48) (Fig. 6A). HepG2 cells were transduced to express g.CLDN1-r.CLDN1 or g.CLDN1-r.CD81 and allowed to proliferate for 72 h, whereupon polarized cells were identified by staining for the apical tight junction marker ZO-1 (Fig. 6A) (33). g.CLDN1-r.CLDN1 at the basolateral membrane associated with a median FIR of 0.67 (r2 = 0.36) (Fig. 6), comparable to the values observed in 293T cells. Similar observations were made for g.CLDN1-r.CLDN1 at tight junctions demonstrating a median FIR of 0.56 (r2 = 0.28) (Fig. 6). FRET between g.CLDN1-r.CLDN1 at basolateral and tight junctions, 36 and 10%, respectively, occurred at a lower frequency than that observed in 293T cells, most likely reflecting the presence of endogenous CLDN1 in HepG2 cells that may compete for CD81 and CLDN1 association(s) (30). Basolateral membrane pools of g.CLDN1-r.CD81 demonstrated a median FIR of 0.7 (r2 = 0.7) and 25% FRET (Fig. 6). In contrast, r.CD81 was largely excluded from tight junctions with no detectable CLDN1-CD81 complexes (r2 = 0.1 and <1% FRET) (Fig. 6). In summary, CLDN1 demonstrated comparable association(s) with CLDN1 at basolateral and tight junction membranes in polarized HepG2 cells, similar to our observations in non-polarized 293T cells. In contrast, significant differences were noted for g.CLDN1-r.CD81 association at the basolateral and tight junction domains of HepG2 cells, with minimal evidence for complex formation at the tight junction. These data are consistent with our previous report demonstrating a role for basolateral pools of CLDN1 in HCV infection (33).
FIGURE 6.
Effect of cell polarization on CLDN1-CD81 and CLDN1-CLDN1 association. HepG2 cells transfected to express AcGFP (g) and DsRED (r) fluorescence-tagged g.CLDN-r.CD81 and g.CLDN1-r.CLDN1 were allowed to polarize over a period of 3 days. Apical bile canalicular structures were identified by staining with anti-ZO-1 and visualized with alexa-633-conjugated secondary anti-rabbit Ig (A). Representative scatter plots of g.CLDN-r.CD81 and g.CLDN1-r.CLDN1 at basolateral (B) and tight junction (C) membrane domains are shown, and the cumulative data from ten cells are summarized below (D).
DISCUSSION
HCV entry into host cells is most likely to occur through a multistep process. Current evidence suggests that scavenger receptor class B member I may define the attachment of HCV virolipoparticles to the cell surface, and this may prime particle interaction(s) with CD81 and Claudin co-receptors that are essential for subsequent particle internalization events (49, 50). In the present study we demonstrate a relationship between receptor-active CLDNs and their association and organization with CD81 at the plasma membrane. Given the reported role of Occludin in HCV entry, we investigated the stoichiometry and FRET between CLDN proteins and Occludin. The majority of CLDN proteins interacted with Occludin, and there was no discernable relationship between CLDN receptor activity and ability to associate with Occludin (Table 1). Mutation of residues 32 and 48 in CLDN1 EC1 ablated CD81 and Occludin association and HCV receptor activity (Fig. 5), which may reflect an altered EC1 conformation. Importantly, mutation of the same residues in CLDN7 enabled CD81 complex formation and virus entry without any detectable Occludin association, demonstrating an essential role of CLDN-CD81 receptor complexes in HCV infection.
Tagging CLDN proteins with AcGFP or DsRED enabled us to assess protein localization in the absence of CLDN-specific mAbs. The majority of AcGFP-tagged CLDNs localized to the plasma membrane, with only CLDN15 showing a dominant intracellular staining pattern. As viral receptor activity is dependent on cell surface expression (51), the inability of CLDN4, -7, -11, -12, and -17 to mediate HCVpp entry is unlikely to be ascribed to low expression levels. The observation that only receptor-active CLDNs associate with CD81 (Fig. 2) provides supporting evidence for a role for these protein complexes in the HCV entry process.
CLDNs oligomerize to form tight junction strands, and several reports demonstrate homotypic and heterotypic interactions between different members of the CLDN family (52–54). FIR and FRET analysis demonstrated interactions between CLDN1 and CLDNs 4, 6, and 9 in 293T cells (Table 1). In contrast there was no significant association between CLDN1 and CLDN7 (r2 = 0.11) (Table 1). These data are supported by surface plasmon resonance data showing CLDN1 EC1-EC1 interaction in the absence of any significant association between CLDN1 and CLDN7 EC1 regions (Fig. 3). It was interesting to note that overexpression of non-receptor CLDNs with g.CLDN1 in 293T cells had minimal effects on CLDN1-CLDN1 association or HCV entry (data not shown). Mutation of residues 32 and 48 in EC1 of CLDN1 and CLDN7 had no significant effects on homotypic cis-interactions (Fig. 5), consistent with a recent report that residues in the EC2 of CLDN5 define homotypic cis-interactions (28). We hypothesize that CLDN1 may associate with both CD81 and CLDN1 via interactions through its EC1 and EC2 loops, respectively, suggesting the presence of receptor heterodimers.
Anti-CD81 mAbs 2s20 and 2s66 reduced CLDN1-CD81 FIR and FRET values and yet had minimal impact on CD81-CD81 association (Fig. 4), reflecting potential differences in protein conformation or temporal association of CD81-CD81 and CLDN1-CD81 complexes. 2s66 FAb had a minimal effect on CLDN1-CD81 association, despite showing a comparable Kd for recombinant CD81 LEL to the complete IgG molecule (2s66 IgG Kd 1.7E−07; 2s66 FAb Kd 8.6E−06), suggesting that bivalent cross-linking of CD81 may be necessary to perturb CD81 association with CLDN1. It is interesting to note that 2s66 FAb demonstrated a 5-fold reduced capacity to neutralize HCV infection compared with 2s66 IgG, suggesting that antibody-induced disruption of CLDN1-CD81 complexes may contribute to the neutralizing activity of anti-CD81 antibodies.
To investigate whether CLDN association with CD81 is critical for viral receptor activity we investigated the effect of known receptor inactivating mutations in CLDN1 EC1 on protein association with CD81. We selected two previously identified amino acids (residues 32 and 48) in EC1, where the reciprocal interchange between CLDN1 and CLDN7 sequences abrogated receptor activity of CLDN1 and conferred receptor activity to CLDN7 by unknown mechanism(s) (14). Neither mutation had an effect on CLDN1/CLDN7 expression or transport to the cell surface, consistent with a recent report by Cuikerman and colleagues demonstrating that alanine substitution at residue 32 ablated receptor activity with minimal perturbation of protein localization (32). In contrast Liu and colleagues reported that CLDN1 I32M was not expressed at the cell surface (19). These differences may reflect the use of different vectors to express CLDN1 in the various studies, because we and others have noted that high level expression of CLDNs can lead to their intracellular accumulation (30, 32). Importantly, we found that introduction of both mutations in CLDN1 destroyed lateral associations with CD81, whereas substitution of both residues in CLDN7 promoted heterotypic association with CD81 (Fig. 5A). In contrast, mutation of both residues in CLDN1 or CLDN7 had a modest to negligible effect on homotypic cis-associations (Fig. 5C), respectively, demonstrating a critical role for CLDN-CD81 complexes in HCV entry.
The major reservoir supporting HCV replication in vivo is thought to be hepatocytes in the liver. Hepatocytes polarize with at least two basal surfaces facing the circulation and a branched network of grooves between adjacent cells that constitute the apical or bile canalicular surface (55). Tight junction strands encircle the apical region and comprise multiple transmembrane, scaffolding, and signaling proteins (reviewed in Ref. 56). We recently reported CLDN1 expression at basolateral and apical hepatocyte membranes in normal liver tissue (57) and in polarized HepG2 cells, with an enrichment at tight junction-associated apical sites (33). To extend our studies on CLDN1-CD81 association in 293T cells and to investigate the presence and location of receptor complexes in polarized HepG2 cells, we transduced HepG2 cells to stably express AcGFP- and DsRED-tagged versions of CLDN1 and CD81. Overexpression of either molecule had no effect on the polarization of HepG2 cells (data not shown). CLDN1 was found to associate with basolateral pools of CLDN1 and CD81 with reduced FRET values (36 and 25%, respectively) compared with 293T cells, most likely representing competition from endogenous CLDNs, as previously reported (30). In contrast, CD81 was largely excluded from the tight junction and exhibited a minimal association with CLDN1 (Fig. 6).
HCV enters the liver via the sinusoidal blood, and therefore the virus will encounter receptors expressed on the sinusoidal or basal surface of the hepatocyte. Our current data, showing that CLDN1 receptor activity correlates with the formation of CLDN1-CD81 complexes that localize at the basolateral surface of polarized hepatoma cells, support a model where virus engagement of CD81-CLDN1 at the basal membrane may initiate the particle internalization process (33). The role of Occludin in HCV entry into polarized cells is poorly defined. Our current data fail to demonstrate a role for CLDN-Occludin complexes in HCV infection. HCV internalizes via a clathrin-dependent process and fusion is believed to occur within the early endosomes (49, 50, 58). At present it is unknown whether any of the viral receptors, including CD81 and CLDN1, are endocytosed with HCV, and further research on the trafficking and endocytic routing of receptor complexes and virus particles in polarized hepatocytes is required to fully appreciate the complex entry process of HCV in the liver.
Supplementary Material

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- HCV
- hepatitis C virus
- MLV
- murine leukemia virus
- CLDN1
- Claudin-1
- HCVpp
- pseudoparticles bearing HCV E1E2 or MLV glycoprotein, respectively
- gp
- glycoprotein
- AcGFP
- Aequorea coerulescens green fluorescent protein
- DsRED
- Discosoma sp. red-monomer fluorescent
- AFU
- arbitrary fluorescence unit(s)
- PBS
- phosphate-buffered saline
- BSA
- bovine serum albumin
- Ab
- antibody
- mAb
- monoclonal antibody
- FIR
- fluorescence intensity ratio
- MβCD
- methyl-β-cyclodextrin
- MBP
- maltose-binding protein
- IQR
- interquartile range
- HCVcc
- HCV cell culture adapted.
REFERENCES
- 1.Bartosch B., Vitelli A., Granier C., Goujon C., Dubuisson J., Pascale S., Scarselli E., Cortese R., Nicosia A., Cosset F. L. (2003) J. Biol. Chem. 278, 41624–41630 [DOI] [PubMed] [Google Scholar]
- 2.Hsu M., Zhang J., Flint M., Logvinoff C., Cheng-Mayer C., Rice C. M., McKeating J. A. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 7271–7276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drummer H. E., Maerz A., Poumbourios P. (2003) FEBS Lett. 546, 385–390 [DOI] [PubMed] [Google Scholar]
- 4.Wakita T., Pietschmann T., Kato T., Date T., Miyamoto M., Zhao Z., Murthy K., Habermann A., Kräusslich H. G., Mizokami M., Bartenschlager R., Liang T. J. (2005) Nat. Med. 11, 791–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhong J., Gastaminza P., Cheng G., Kapadia S., Kato T., Burton D. R., Wieland S. F., Uprichard S. L., Wakita T., Chisari F. V. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 9294–9299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lindenbach B. D., Evans M. J., Syder A. J., Wölk B., Tellinghuisen T. L., Liu C. C., Maruyama T., Hynes R. O., Burton D. R., McKeating J. A., Rice C. M. (2005) Science 309, 623–626 [DOI] [PubMed] [Google Scholar]
- 7.Flint M., von Hahn T., Zhang J., Farquhar M., Jones C. T., Balfe P., Rice C. M., McKeating J. A. (2006) J. Virol. 80, 11331–11342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cormier E. G., Tsamis F., Kajumo F., Durso R. J., Gardner J. P., Dragic T. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 7270–7274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pileri P., Uematsu Y., Campagnoli S., Galli G., Falugi F., Petracca R., Weiner A. J., Houghton M., Rosa D., Grandi G., Abrignani S. (1998) Science 282, 938–941 [DOI] [PubMed] [Google Scholar]
- 10.Scarselli E., Ansuini H., Cerino R., Roccasecca R. M., Acali S., Filocamo G., Traboni C., Nicosia A., Cortese R., Vitelli A. (2002) EMBO J. 21, 5017–5025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grove J., Huby T., Stamataki Z., Vanwolleghem T., Meuleman P., Farquhar M., Schwarz A., Moreau M., Owen J. S., Leroux-Roels G., Balfe P., McKeating J. A. (2007) J. Virol. 81, 3162–3169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barth H., Schnober E. K., Neumann-Haefelin C., Thumann C., Zeisel M. B., Diepolder H. M., Hu Z., Liang T. J., Blum H. E., Thimme R., Lambotin M., Baumert T. F. (2008) J. Virol. 82, 3466–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dreux M., Dao Thi V. L., Fresquet J., Guérin M., Julia Z., Verney G., Durantel D., Zoulim F., Lavillette D., Cosset F. L., Bartosch B. (2009) PLoS Pathog. 5, e1000310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans M., von Hahn T., Tscherne D. M., Syder A. J., Panis M., Wölk B., Hatziioannou T., McKeating J. A., Bieniasz P. D., Rice C. M. (2007) Nature 446, 801–805 [DOI] [PubMed] [Google Scholar]
- 15.Meertens L., Bertaux C., Cukierman L., Cormier E., Lavillette D., Cosset F. L., Dragic T. (2008) J. Virol. 82, 3555–3560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang W., Qiu C., Biswas N., Jin J., Watkins S. C., Montelaro R. C., Coyne C. B., Wang T. (2008) J. Biol. Chem. 283, 8643–8653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng A., Yuan F., Li Y., Zhu F., Hou P., Li J., Song X., Ding M., Deng H. (2007) J. Virol. 81, 12465–12471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ploss A., Evans M. J., Gaysinskaya V. A., Panis M., You H., de Jong Y. P., Rice C. M. (2009) Nature 457, 882–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu S., Yang W., Shen L., Turner J. R., Coyne C. B., Wang T. (2009) J. Virol. 83, 2011–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benedicto I., Molina-Jiménez F., Bartosch B., Cosset F. L., Lavillette D., Prieto J., Moreno-Otero R., Valenzuela-Fernández A., Aldabe R., Lüpez-Cabrera M., Majano P. L. (2009) J. Virol. 83, 8012–8020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stamataki Z., Grove J., Balfe P., McKeating J. A. (2008) Clin. Liver Dis. 12, 693–712 [DOI] [PubMed] [Google Scholar]
- 22.Cocquerel L., Voisset C., Dubuisson J. (2006) J. Gen. Virol. 87, 1075–1084 [DOI] [PubMed] [Google Scholar]
- 23.Hemler M. E. (2008) Nat. Rev. Drug Discov. 7, 747–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drummer H. E., Wilson K. A., Poumbourios P. (2005) Biochem. Biophys. Res. Commun. 328, 251–257 [DOI] [PubMed] [Google Scholar]
- 25.Kitadokoro K., Bordo D., Galli G., Petracca R., Falugi F., Abrignani S., Grandi G., Bolognesi M. (2001) EMBO J. 20, 12–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rocha-Perugini V., Montpellier C., Delgrange D., Wychowski C., Helle F., Pillez A., Drobecq H., Le Naour F., Charrin S., Levy S., Rubinstein E., Dubuisson J., Cocquerel L. (2008) PLoS One 3, e1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krause G., Winkler L., Mueller S. L., Haseloff R. F., Piontek J., Blasig I. E. (2008) Biochim. Biophys. Acta 1778, 631–645 [DOI] [PubMed] [Google Scholar]
- 28.Piontek J., Winkler L., Wolburg H., Müller S. L., Zuleger N., Piehl C., Wiesner B., Krause G., Blasig I. E. (2008) FASEB J. 22, 146–158 [DOI] [PubMed] [Google Scholar]
- 29.Hartsock A., Nelson W. J. (2008) Biochim. Biophys. Acta 1778, 660–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harris H. J., Farquhar M. J., Mee C. J., Davis C., Reynolds G. M., Jennings A., Hu K., Yuan F., Deng H., Hubscher S. G., Han J. H., Balfe P., McKeating J. A. (2008) J. Virol. 82, 5007–5020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kovalenko O. V., Yang X. H., Hemler M. E. (2007) Mol. Cell Proteomics 6, 1855–1867 [DOI] [PubMed] [Google Scholar]
- 32.Cukierman L., Meertens L., Bertaux C., Kajumo F., Dragic T. (2009) J. Virol. 83, 5477–5484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mee C. J., Harris H. J., Farquhar M. J., Wilson G., Reynolds G., Davis C., van IJzendoorn S. C., Balfe P., McKeating J. A. (2009) J. Virol. 83, 6211–6221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng J., Zagotta W. N. (2004) Neuron 42, 411–421 [DOI] [PubMed] [Google Scholar]
- 35.Staruschenko A., Adams E., Booth R. E., Stockand J. D. (2005) Biophys. J. 88, 3966–3975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drummer H. E., Wilson K. A., Poumbourios P. (2002) J. Virol. 76, 11143–11147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seigneuret M. (2006) Biophys. J. 90, 212–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morita K., Furuse M., Fujimoto K., Tsukita S. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 511–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ikenouchi J., Furuse M., Furuse K., Sasaki H., Tsukita S., Tsukita S. (2005) J. Cell Biol. 171, 939–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Furuse M., Hirase T., Itoh M., Nagafuchi A., Yonemura S., Tsukita S., Tsukita S. (1993) J. Cell Biol. 123, 1777–1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blasig I. E., Winkler L., Lassowski B., Mueller S. L., Zuleger N., Krause E., Krause G., Gast K., Kolbe M., Piontek J. (2006) Cell Mol. Life Sci. 63, 505–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Charrin S., Le Naour F., Labas V., Billard M., Le Caer J. P., Emile J. F., Petit M. A., Boucheix C., Rubinstein E. (2003) Biochem. J. 373, 409–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Silvie O., Charrin S., Billard M., Franetich J. F., Clark K. L., van Gemert G. J., Sauerwein R. W., Dautry F., Boucheix C., Mazier D., Rubinstein E. (2006) J. Cell Sci. 119, 1992–2002 [DOI] [PubMed] [Google Scholar]
- 44.Espenel C., Margeat E., Dosset P., Arduise C., Le Grimellec C., Royer C. A., Boucheix C., Rubinstein E., Milhiet P. E. (2008) J. Cell Biol. 182, 765–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kapadia S., Barth H., Baumert T., McKeating J. A., Chisari F. V. (2007) J. Virol. 81, 374–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yancey P. G., Rodrigueza W. V., Kilsdonk E. P., Stoudt G. W., Johnson W. J., Phillips M. C., Rothblat G. H. (1996) J. Biol. Chem. 271, 16026–16034 [DOI] [PubMed] [Google Scholar]
- 47.Timpe J. M., McKeating J. A. (2008) Gut 57, 1728–1737 [DOI] [PubMed] [Google Scholar]
- 48.Decaens C., Durand M., Grosse B., Cassio D. (2008) Biol. Cell 100, 387–398 [DOI] [PubMed] [Google Scholar]
- 49.Blanchard E., Belouzard S., Goueslain L., Wakita T., Dubuisson J., Wychowski C., Rouillé Y. (2006) J. Virol. 80, 6964–6972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meertens L., Bertaux C., Dragic T. (2006) J. Virol. 80, 11571–11578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Farquhar M. J., McKeating J. A. (2008) J. Viral Hepat. 15, 849–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Furuse M., Sasaki H., Tsukita S. (1999) J. Cell Biol. 147, 891–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Coyne C. B., Gambling T. M., Boucher R. C., Carson J. L., Johnson L. G. (2003) Am. J. Physiol. Lung Cell Mol. Physiol. 285, L1166–L1178 [DOI] [PubMed] [Google Scholar]
- 54.Daugherty B. L., Ward C., Smith T., Ritzenthaler J. D., Koval M. (2007) J. Biol. Chem. 282, 30005–30013 [DOI] [PubMed] [Google Scholar]
- 55.Wang L., Boyer J. L. (2004) Hepatology 39, 892–899 [DOI] [PubMed] [Google Scholar]
- 56.Paris L., Tonutti L., Vannini C., Bazzoni G. (2008) Biochim. Biophys. Acta 1778, 646–659 [DOI] [PubMed] [Google Scholar]
- 57.Reynolds G. M., Harris H. J., Jennings A., Hu K., Grove J., Lalor P. F., Adams D. H., Balfe P., Hübscher S. G., McKeating J. A. (2008) Hepatology 47, 418–427 [DOI] [PubMed] [Google Scholar]
- 58.Coller K. E., Berger K. L., Heaton N. S., Cooper J. D., Yoon R., Randall G. (2009) PLoS Pathog. 5, e1000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.