Abstract
Upon a stimulus of light, histamine is released from Drosophila photoreceptor axonal endings. It is taken up into glia where Ebony converts it into β-alanyl-histamine (carcinine). Carcinine moves into photoreceptor cells and is there cleaved into β-alanine and histamine by Tan activity. Tan thus provides a key function in the recycling pathway of the neurotransmitter histamine. It is also involved in the process of cuticle formation. There, it cleaves β-alanyl-dopamine, a major component in cuticle sclerotization. Active Tan enzyme is generated by a self-processing proteolytic cleavage from a pre-protein at a conserved Gly-Cys sequence motif. We confirmed the dependence on the Gly-Cys motif by in vitro mutagenesis. Processing time delays the rise to full Tan activity up to 3 h behind its putative circadian RNA expression in head. To investigate its pleiotropic functions, we have expressed Tan as a His6 fusion protein in Escherichia coli and have purified it to homogeneity. We found wild type and mutant His6-Tan protein co-migrating in size exclusion chromatography with a molecular weight compatible with homodimer formation. We conclude that dimer formation is preceding pre-protein processing. Drosophila tan1 null mutant analysis revealed that amino acid Arg217 is absolutely required for processing. Substitution of Met256 in tan5, on the contrary, does not affect processing extensively but renders it prone to degradation. This also leads to a strong tan phenotype although His6-Tan5 retains activity. Kinetic parameters of Tan reveal characteristic differences in Km and kcat values of carcinine and β-alanyl-dopamine cleavage, which conclusively illustrate the divergent tasks met by Tan.
Keywords: Cysteine Protease, Drosophila, Eye, Neurotransmitter Transport, Neurotransmitters, Photoreceptors, Histamine
Introduction
Photoreceptor synapses of Drosophila melanogaster release histamine neurotransmitter in a tonic fashion. It binds to histamine-gated chloride channels of postsynaptic neurons. Chloride influx hyperpolarizes the postsynaptic cell, which terminates spontaneous firing. To achieve this reverse signaling, two prerequisites have to be met at the synaptic cleft. The binding affinity of histamine to chloride channels must be low. In addition, opening times of histamine-gated channels must be short enough to facilitate transmission of tiny changes of photoreceptor depolarization into a postsynaptic signal (1). From studies on the giant barnacle Balanus nubilus it was concluded that histamine synthesis by decarboxylation of histidine is low in arthropod photoreceptor cells (2). Therefore, a continuous recycling of neurotransmitter is indispensable in visual signal transduction. This is facilitated by fast transmitter uptake into surrounding glia and inactivation of histamine by Ebony-mediated β-alanine conjugation to yield β-alanyl-histamine (carcinine). Various experimental evidence substantiate this concept of inactivation (3–5). It further requires carcinine uptake into photoreceptor cells where it is cleaved by Tan into β-alanine and histamine. Histamine is then available for transport into synaptic vesicles whereas β-alanine might shuttle back into glia (Fig. 1).
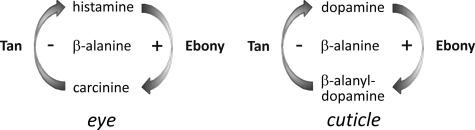
FIGURE 1.
Tan and Ebony regulate histamine and dopamine concentrations in eye and cuticle, respectively. In the eye histamine neurotransmitter is inactivated by β-alanyl conjugation to yield β-alanyl histamine (carcinine). Later Tan can hydrolyze carcinine to again supply histamine for neurotransmission. In the cucticle the interplay between Tan and Ebony activity adjusts the concentration of free dopamine.
We had previously shown that Tan expression in fly heads is confined to photoreceptor cells (5). Ebony is strongly expressed in optic ganglion glia cell types and in a large proportion of glia of the protocerebrum, although at lower concentrations (5–8). The broad expression might reflect the pleiotropic function of Ebony in biogenic amine inactivation (4). In the visual system, Ebony together with Tan is thought to sustain the recycling pathway required to replenish the histamine neurotransmitter for vesicular release (3, 4). Histamine injections into the head of the larger fly Sarcophaga bullata have shown that conversion into carcinine starts within seconds. Carcinine concentration accumulates to reach an equilibrium with histamine in a 1:2 ratio after about one-half hour (3). These results reveal that Tan and Ebony serve different roles within the recycling pathway: whereas Ebony in combination with a transporter has to inactivate the neurotransmitter quickly by converting it into a derivative after delocalization, Tan balances carcinine and histamine concentration, which guarantees a continuous flow of neurotransmitter.
Tan activity is also involved in cuticle sclerotization in the pupal stage. Cuticle sclerotization depends on the availability of catecholic substrates such as N-acetyldopamine and β-alanyl-dopamine (10), which are formed in epidermal cells by arylalkylamine N-acetyltransferases, and N-β-alanyl-biogenic amine synthase (Ebony), respectively. After secretion of β-alanyl-dopamine into the cuticle it is oxidized by phenoloxidases and serves as a cross-linking agent. Free dopamine is oxidized to melanin, which contributes to the formation of the brown cuticle pigment. Limiting the concentration of β-alanyl-dopamine by Tan activity seems to be critical for cuticle sclerotization and tanning (10–12). The details of spatial interaction between Ebony and Tan in the cuticle still wait to be determined. Because inactivation of Tan by gene mutation diminishes the amount of free dopamine, reduction of melanin pigment disclosed tan as a body color mutant, which was discovered as one of the first mutants in Drosophila genetics (13, 14). Among the various tan mutants, the functional null alleles tan1 and tan5 have recently been characterized by sequencing. They both contain single nucleotide changes that lead to amino acid substitutions within the protein sequence (15).
Tan is closely related to isopenicillin N-acyltransferase (IAT)2 of the filamentous fungus Penicillium chrysogenum (15), which catalyzes the last steps in penicillin-G biosynthesis (16, 17). IAT activates itself by self-processing into a 11-kDa α-subunit and a 29-kDa β-subunit at a Gly-Cys sequence motif, a process that identifies it as cysteine peptidase (18). Neither subunit alone shows activity; instead, it is only observed when both subunits can interact in a cooperative folding event (19). Sequence similarity of IAT with Tan extends over the whole amino acid sequence. We have previously shown that Tan is also cleaved from a pre-protein into two subunits at this motif (5). Contrary to IAT, which is transported into peroxisomes (20), Tan was shown to be a cytosolic protein (5). IAT cleaves the carboxy-amide bond of isopenicillin N to yield 6-aminopenicillanic acid and l-α-aminoadipic acid. In a second step, IAT acylates 6-aminopenicillanic acid using acyl-CoA. Altogether it gives rise to an exchange of acyl groups eventually yielding penicillin-G. Tan hydrolase activity coincides with the first step of IAT activity, cleavage of a carboxy-amide, or peptide bond. Despite its close relationship to IAT in size, sequence, and processing requirement, Tan activity has not been assigned with a biogenic amine N-acylation reaction although N-acylation of histamine would also be conceivable as mode of neurotransmitter inactivation.
Activation by timely self-processing at a Gly-Cys sequence motif is an inherent feature of members of the cysteine peptidase protein family (21). The affiliation of IAT to this group had been confirmed by in vitro mutagenesis, particularly at the site encoding the consensus amino acid motif. Self-processing was strictly dependent on the Gly-Cys motif. Also, enzymatic activity appeared to depend on cleavage into α,β-heterodimers. Because unprocessed IAT mutants were shown to exert a regulatory role on processing of wild type IAT, it was speculated that processing might require pre-protein dimer formation and self-cleavage might be an inter-molecular process (22). If Tan processing reveals the same stringent dependence on the Gly-Cys motif, activation of enzymatic activity might also depend on this process and maturation of activity might require a certain time period. In light of the reported circadian expression of tan this should shift the daily rise of activity (23).
To understand the role of Tan in the histamine recycling pathway of the eye as opposed to the sclerotization process in the cuticle, we have first investigated the dependence of Tan processing and activity on the availability of the Gly-Cys motif by in vitro mutagenesis. We purified His6-Tan to homogeneity and showed that native Tan associates to a dimer containing two α- and two β-subunits. We observed that prevention of processing into α,β subunits keeps the protein inactive and prone to degradation in the tan1 mutant. We further determined that the Tan5 mutant enzyme retained about one-third of the wild type activity. We conclude that instead of functional impairment, degradation of the mutant protein must give rise to the tan5 null mutant phenotype. We determined the kinetic parameters of carcinine as well as β-alanyl-dopamine cleavage and observed fundamental differences between both substrates in Km and kcat values. These differences provide a basis to explain how Tan serves its pleiotropic roles in unrelated biochemical processes.
EXPERIMENTAL PROCEDURES
Fly Stocks
D. melanogaster was cultured on standard corn meal medium. The wild type strain examined was Canton-S (C-S). tan1 and tan5 mutants examined by immunocytochemistry were kept in white1118 mutant background.
Mutation of Tan cDNA
Mutant tan cDNAs were constructed from the pQE82L (Qiagen, Hilden, Germany):tan cDNA plasmid (5) using the QuikChange® site-directed mutagenesis procedure (Novagen, La Jolla, CA). 42-mer primers comprising the altered codon sequence in the middle were employed. They were obtained from MWG-Biotech AG (Ebersberg, Germany). The identity of the resulting cDNAs was confirmed by re-sequencing.
RNA Isolation and RT-PCR
Whole RNA from 100 Canton-S, t1, and t5 heads was isolated using the ZR Tissue and Insect MicroPrepTM kit (Zymo Research Corp., Orange, CA). 1 μg of RNA was used to generate cDNA from an oligo(dT)16-Primer (MWG Biotech AG, Ebersberg, Germany) with the Transcriptor First Strand cDNA Synthesis Kit (Roche Applied Science). cDNA-specific primer pairs for tan and ebony, as an internal control, were designed using the NCBI/Primer BLAST tool. These spanned an exon-exon border, if possible, or resulted in a significantly bigger genomic PCR product. Fragments were amplified with native Taq DNA polymerase (Fermentas GmbH, St. Leon-Rot, Germany) with 35 cycles in a Mastercycler® personal (Eppendorf, Hamburg, Germany). Samples were analyzed on a 3% (w/v) agarose gel.
His6-Tan Expression in Escherichia coli
Wild type and mutated pQE82L:tan cDNA plasmids were transformed into BL21-RosettaTM cells (Novagen-Merck, Darmstadt, Germany). Single colonies were incubated for 16 h at 37 °C in lysogeny broth medium (24) with ampicillin (100 μg/ml) and chloramphenicol (25 μg/ml). The cultures were used to inoculate fresh medium at 28 °C until an A600 of 0.6 was reached. Expression of the His6-Tan fusion proteins was induced by addition of isopropyl β-d-thiogalactopyranoside (IPTG) to 0.1 mm. After incubation for an additional 16 h the cells were reaped and re-suspended in 0.3 m sodium chloride (NaCl), 50 mm HEPES/NaOH, pH 8.0.
Tan Self-processing Assessment in E. coli
Self-processing time was determined in growth-competent E. coli according to Garcia-Estrada et al. (22). Tan expression from pQE82L in BL21-RosettaTM cells was induced with IPTG as described above. After Tan expression for 3 h at 28 °C cells were washed twice with 0.9% NaCl to remove IPTG. They were taken up in lysogeny broth medium and incubated at 25 °C to allow self-processing. Aliquots were taken out at time intervals and equal amounts of cells were prepared for loading on a SDS-PAGE gel and subsequent Western blot (25) evaluation of Tan processing as described below.
His6-Tan Purification
Cells were lysed with 10 cycles of 750 bar pressure using an M-110L microfluidizer (Microfluidics, Newton, MA). The lysate was cleared by centrifugation. The first step in purification was affinity chromatography using Protino® Ni-TED Resin (Macherey and Nagel, Dueren, Germany). Fractions with the highest concentrations were pooled. They were dialyzed against 0.1 m NaCl, 50 mm HEPES/NaOH, pH 7.0, 1 mm dithiothreitol, and were applied to a SourceTM 15S column (GE Healthcare, Uppsala, Sweden). The column was washed with the same buffer and the enzyme was eluted by gradually increasing the NaCl concentration of the buffer to 1 m. Fractions with the highest concentrations were pooled. Protein solution was dialyzed against 0.3 m NaCl, 50 mm HEPES/NaOH, pH 7.0, 1 mm dithiothreitol.
His6-Tan Detection
Samples of the His6-Tan protein were separated by discontinuous 15% SDS-PAGE (26) using mini-gel chambers (Hoefer, Hollistom, MA). Whole protein was stained with a Coomassie Brilliant Blue G-250 solution. Alternatively, silver staining was performed according to Heukeshoven and Dernick (27). Electroblotting (28) was performed on nitrocellulose membrane (Whatman, Schleicher and Schuell, Dassel, Germany). Free binding sites were blocked with 1% milk powder. Affinity purified anti-Tan peptide antisera ap61 and ap63, each directed against a mixture of an amino-terminal and a carboxyl-terminal peptide, (5) in 25 mm Tris/HCl, pH 7.5, 144 mm NaCl, 0.2% Tween 20 (TBST) were applied at 1:10,000 dilution. Alkaline phosphatase-coupled goat anti-rabbit IgG (H+L Dianova, Hamburg, Germany) was used at 1:10,000 dilution. Protein bands were visualized by stain development with nitro blue tetrazolium chloride/5-bromo-4-chloro-3-indolyl phosphate.
Immunocytochemistry
Preparation of 10-μm thin sections of Drosophila heads and immunolabeling with anti-Tan antiserum ap63 was performed essentially as described previously (5).
Determination of Hydrolase Activity
Substrate solutions were prepared in 50 mm HEPES, 100 mm NaCl, pH 7.0 (HEPES/NaCl). Tan was applied in final concentrations between 4 and 0.04 nm in HEPES/NaCl, 0.2 mg/ml of bovine serum albumin. For each assay, the substrate solution was preincubated at the selected temperature. The enzyme solution was added rapidly to start the reaction. Aliquots were taken from the reaction mixture after each time interval and added to the same volume of ice-cold 0.56 m perchloric acid to stop the reaction. Insolubles were spun down at 20,800 × g for 15 min at 4 °C. The supernatant was mixed with an internal standard in a ratio of 5:1 for sample preparation. 20 μl of this mixture were injected into the HPLC system for dopamine determination. For histamine determination, pre-column derivatization was required: 40 μl of the assay plus internal standard sample was added to 520 μl of mobile phase plus 30 μl of orthophthaldialdehyde (1% v/v in methanol) and 30 μl of 2-mercaptoethanol (1% v/v in methanol). It was adjusted to pH 11.0 with 65 μl of 100 mm NaBO4/KOH buffer. 100 μl were injected into the HPLC system. The mobile phase was prepared from high purity Millipore water. It contained 150 mm chloroacetic acid, 117 mm NaOH, 0.86 mm 1-octanesulfonic acid, 0.67 mm Na2EDTA, and 5% acetonitrile, 1.8% tetrahydrofuran for dopamine determination and 18% acetonitrile, 7% methanol for histamine detection, respectively. The HPLC system consisted of an isocratic Elite LaChrome 2130 pump (Hitachi, Japan) and an amperometric EP30 electrochemical detector (Biometra, Gottingen, Germany) with a radial flow electrode MF-1091 (BAS, Stareton, UK). Chromatographic separation was performed with a 5-μm C18, 100 A Nucleosil column (150 × 4.6 mm) coupled with a 5-μm C18, 100 A Nucleosil guard column (10 × 4.6 mm) (Macherey Nagel, Duren, Germany) at a flow rate of 1 ml/min. Mobile phase and column were maintained at 26 °C. The working electrode potential was adjusted to 0.7 V against an Ag/AgCl reference. Pre-column derivatization and column injection was automatically performed employing a Triathlon autosampler (Spark-Holland B.V., Emmen, The Netherlands). Data were acquired using Clarity software (DataApex, Prague, The Czech Republic). Kinetic parameters were calculated and graphing was performed with Origin software (Additive GmbH, Friedrichsdorf, Germany).
Size Exclusion Chromatography
Separation of proteins under native conditions was performed with a Superdex 200 high resolution column 10/30 (GE Healthcare, Uppsala, Sweden) with HEPES/NaCl, pH 7.0, 1 mm dithiothreitol at a flow rate of 1 ml/min.
RESULTS
The Gly-Cys Motif Is Required for Tan Self-processing
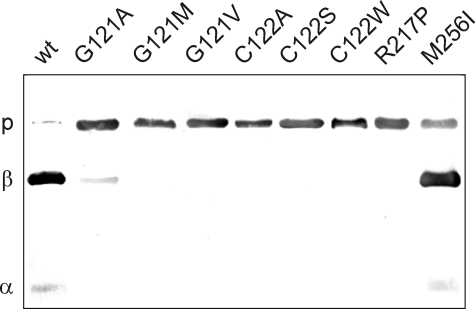
Tan exhibits sequence similarity with P. chrysogenum IAT, which is processed into an 11- and 29-kDa subunit. IAT cleavage depends on a Gly-Cys motif, the consensus sequence of cysteine peptidases (18). We have previously shown that His6-Tan also undergoes a post-translational cleavage reaction at a Gly-Cys motif into a 15-kDa α-subunit and a 30-kDa β-subunit (5). To experimentally prove dependence of Tan processing on this motif and its affiliation to the cysteine peptidase protein family, we mutagenized the two codons in vitro, expressed the cDNAs as His6-tagged fusion proteins in E. coli, and partially purified the resulting proteins by affinity chromatography. Processing was evaluated after SDS-PAGE and Western blot transfer by immunolabeling with antisera against both Tan subunits (5). First, Gly121 of the Gly-Cys motif was replaced by small amino acids Ala, Val, and Met, which carry uncharged side chains of growing size. Substitution with Val or Met abolished processing completely (Fig. 2). Only replacement of Gly by Ala, the amino acid with the smallest side chain, allowed a small degree of cleavage into both subunits, which discloses the necessity for unrestricted flexibility of the protein at this position. Cys122 mutation focused on the dispensability of the SH-group during processing. We either exchanged it with a hydroxyl group, which yields Ser, removed it by inserting Ala or introduced a bulky side chain by replacing Cys by Trp. Processing was completely lost in each case (Fig. 2). This demonstrates the strict requirement for Cys122 in His6-Tan, which for high efficiency processing should be preceded by an amino acid, preferably Gly that confers structural flexibility. These results unequivocally assign Tan as a new member of the cysteine peptidase protein family.
FIGURE 2.
Self-processing activity of His6-Tan. Western blot analysis of E. coli expressed, affinity purified wild type, and mutant His6-Tan with the anti-Tan antiserum ap61/ap63 (5) directed against the amino- and carboxy-terminal ends. Tan mutant variants applied in each lane are indicated on the top using 1 letter amino acid abbreviations. Tan pre-protein (P), β-subunit (β) and α-subunit (α) are running at 45, 30, and 15 kDa, respectively.
Mutations Outside the Gly-Cys Motif Affect Protein Maturation and Stability
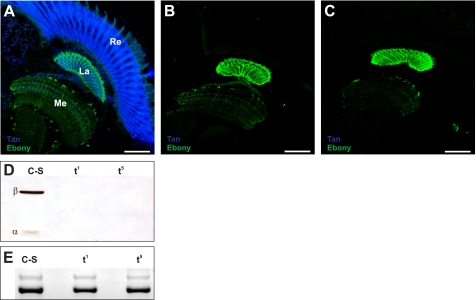
The Drosophila tan1 mutation has previously been shown to contain a point mutation within the translational reading frame (15). It replaces Arg by Pro at position 217, a region of pronounced sequence similarity to IAT (15). Mimicking the tan1 mutation, we expressed His6-TanR217P encoding cDNA in E. coli. Western blot analysis of Ni-TED affinity purified protein labeled a single band of pre-protein size indicating that His6-TanR217P is not able to process into α- and β-subunits (Fig. 2). This result demonstrates that Tan pre-protein processing depends also on structural constraints other than the Gly-Cys motif. In addition, we investigated processing of tan5 whose mutation is localized in a region of reduced sequence similarity with IAT by replacing Met256 with Ile (15). We also expressed His6-TanM256I encoding cDNA in E. coli. Western blot analysis revealed strong labeling of the 30-kDa β-subunit leaving only a minor portion of the pre-protein uncleaved (Fig. 2) reflecting only slightly reduced processing efficiency in tan5. Surprisingly, Western blot analysis of Drosophila head extracts as well as immunolabeling of head cryo-sections of tan1 and tan5 flies with anti-Tan antiserum failed to detect the mutant Tan proteins (Fig. 3). Therefore, transcriptional activity of both mutant tan genes was determined to exclude impairment of gene expression by additional mutations as the cause for their null phenotype. Indeed, RT-PCR amplification of a 456-nucleotide tan gene fragment from the head RNA of wild type C-S, tan5, and tan1 flies gave rise to DNA bands of comparable intensity (Fig. 3E).
FIGURE 3.
Expression of the tan gene in wild type C-S, tan1, and tan5 heads. A–C, immunolabeling of Tan in wild type (A), tan1 (B), and tan5 (C) 10 μm Drosophila head freeze sections with anti-Tan antiserum ap61/ap63 (5). The Tan label is in blue. Ebony labeled in green is used as control. Re, La, and Me indicate retina, lamina, and medulla, respectively. D, Western blot analysis of wild type Canton-S (C-S) and mutants tan1 (t1) and tan5 (t5) head extracts with anti-Tan antiserum. E, RT-PCR amplification of a 456-nucleotide tan-RNA fragment (strong lower band) from head total RNA preparations of wild type Canton-S (C-S) and mutants tan1 (t1) and tan5 (t5). RT-PCR amplification of a 581-nucleotide ebony-RNA fragment (weak upper band) served as control.
His6-Tan Purification for Kinetic Measurements
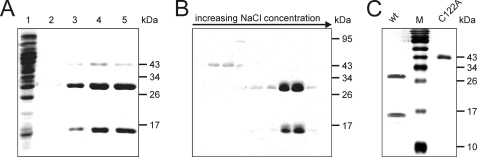
To evaluate Tan activity in carcinine as compared with N-β-alanyl-dopamine cleavage, we expressed it as His6-Tan fusion protein in E. coli. Expression in log phase bacteria was performed overnight at 28 °C to avoid inclusion body formation. The cleared bacterial lysate was applied to a Protino Ni-TED resin column. A broad peak of the Tan protein was eluted from the column by increasing imidazole to 250 mm (Fig. 4). The resulting staining pattern of SDS-PAGE showed the expected two polypeptides corresponding in size with the two subunits of mature enzyme and a weak band of pre-protein size. Additional Coomassie staining was observed at about 43 kDa, which was not labeled by anti-Tan antiserum and was later identified by mass spectrometry as the Lac-repressor (data not shown). Finally, essentially pure His6-Tan was obtained by subsequent cation exchange chromatography applying a Source 15S column (Fig. 4). Applying this purification scheme we also prepared the mutant protein Tan5 and the Gly-Cys motif processing mutant His6-TanC122A (Fig. 4).
FIGURE 4.
Purification of His6-Tan proteins. A, Protino Ni-TED affinity purification of E. coli cell lysate. 1, whole cell lysate; 2–5, peak fractions containing His6-Tan and a 43-kDa contaminant. B, elution profile of a Source 15S column loaded with the peak fraction of the Ni-TED step. C, peak fraction of a Source 15S column purification of wild type Tan and TanC122A (C122A). Coomassie staining was used in A and B, silver staining in C.
Native Tan Has a Protein Size Compatible with Dimer Formation
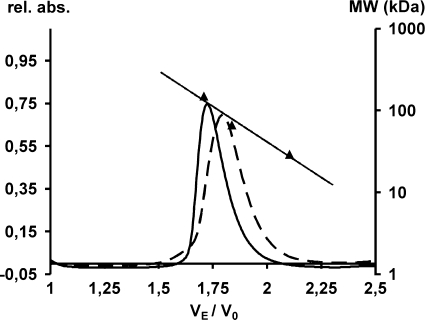
The proposal that IAT activation depends on dimer formation as a prerequisite for an intermolecular processing step (22) led us to investigate the molecular weight of native E. coli expressed wild type and of processing mutant His6-TanC122A fusion protein. Tan protein preparations were applied to a SuperdexTM 200 size exclusion chromatography column. The linear dependence of the logarithm of the molecular mass of the protein standards revealed a molecular mass of wild type His6-Tan at 120 kDa (Fig. 5). The elution profile of mutant His6-TanC122A matched the calculated dimer size of 90 kDa. Apparently, both Tan protein preparations show a higher molecular mass than 45 kDa of the monomer. Within variation of column velocity of flow, both elution positions are compatible with a dimer configuration of Tan.
FIGURE 5.
Size evaluation of native wild type His6-Tan compared with mutant His6-TanC122A. Superdex 200 high resolution chromatography of wild type Tan (solid line) and TanC122A (broken line) as a ratio of elution volume (VE) to void volume (V0). Triangles indicate the elution ratio for size marker proteins alcohol dehydrogenase (150 kDa), bovine serum albumin (66 kDa), and carbonic anhydrase (29 kDa). From their linear regression indicated as a straight line, the molecular weights of Tan have been calculated.
Impact of Tan Processing Time on Its Circadian Activity Profile
Pre-protein processing is a time consuming process that eventually yields active heterodimer. In light of the recently reported circadian expression of Tan in the head (23), timely processing would slow down the formation activity of the newly expressed Tan portion. To evaluate the extent of delay, we determined His6-Tan processing time in vivo in E. coli at 25 °C, a temperature that Drosophila stock is facing in the wild and is often used in stock maintenance in the laboratory. After cessation of Tan expression by removal of IPTG, aliquots were taken from the E. coli culture at time points over 3 h. Bacterial proteins were separated by SDS-PAGE, transferred to nitrocellulose, and labeled with anti-Tan antiserum (5). Reduction of pre-protein labeling with time starting at IPTG withdrawal indicates the processing progress. Completion of processing at 25 °C took about 3 h (Fig. 6), a time period that coincides roughly with the time required for recombinant P. chrysogenum IAT to be processed in E. coli (22).
FIGURE 6.
His6-Tan pre-protein processing time. Western blot of His6-Tan pre-protein expressed in E. coli cell extracts after removal of IPTG and additional growth periods at 25 °C as indicated in minutes. Anti-Tan antiserum labels diminishing proportions of pre-protein.
Determination of His6-Tan Activity
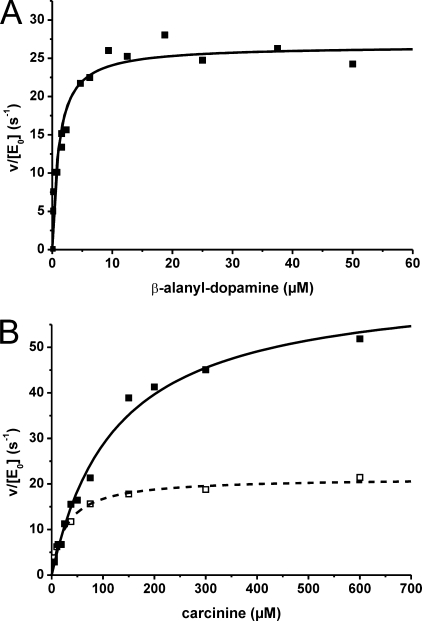
Purified His6-Tan was diluted in assay buffer supplemented with 0.2 mg/ml of bovine serum albumin to preserve protein stability. A series of control assays was performed beforehand with Tan concentrations between 0.04 and 4 nm at pH 7.0. At 37 °C, as well as at 25 °C, the product was formed linearly to the enzyme concentration. All further enzyme assays were performed at 25 °C, a temperature relevant for Drosophila life. Substrate stability was verified under enzyme assay and work up conditions for HPLC. Product formation at a fixed Tan concentration remained in linear correlation to reaction time for at least 40 min revealing velocity (v). Within a pH range of 6.0 to 8.0, enzymatic activity of Tan changed by less then 10%, indicating a broad pH optimum; supplementing Mg2+ in a concentration between 0.1 and 4.0 mm had no effect on product formation. Velocities were determined and plotted against the particular substrate concentration. Data points of two independent sets of experiments describe a hyperbolic function according to Michaelis-Menten (29) that was used to determine mean values of kcat and Km with their corresponding standard deviation. Hydrolysis of β-alanyl-dopamine revealed a very steep rise to maximal enzyme activity with a half-maximum velocity Km value at 0.9 ± 0.3 μm substrate concentration (Fig. 7). The turnover number kcat reached its maximum at 23.5 ± 4.9/s. Markedly different values were observed when carcinine was used as substrate (Fig. 7). The hyperbolic rise of kcat in the Michaelis-Menten plot revealed a Km value of 119 ± 8.5 μm. The observed maximum catalytic activity kcat at 68 ± 5.4/s is more than twice the value compared with β-alanyl-dopamine as substrate. Employing processing deficient TanC122A in assays with carcinine as substrate yielded no activity at all (data not shown). His6-Tan5 carrying the Met256 to Ile substitution showed with carcinine as substrate a reduced kcat value of 22.7 ± 1.8/s and a reduced Km of 30.3 ± 3.8 μm compared with wild type Tan (Fig. 7).
FIGURE 7.
Enzyme kinetics of carcinine and β-alanyl-dopamine hydrolysis. Representative Michaelis-Menten curves from each data set for each hydrolysis reaction are depicted. A, wild type His6-Tan with β-alanyl-dopamine as substrate. B, wild type His6-Tan and His6-TanM256I (broken line) with carcinine as substrate.
DISCUSSION
We have purified Tan to homogeneity to determine physical parameters of activation and activity. This is specifically interesting in light of the diverse sites of action of Tan in the fly, neurotransmitter inactivation and cuticle sclerotization. At both sites, Ebony is counteracting the activity of Tan by conjugating β-alanine with either histamine or dopamine. Like Ebony (4), Tan is related to a fungal protein. Interestingly, the Tan homologues IATs and the Ebony homologues non-ribosomal peptide synthetases interact as components of antibiotic synthesis. Penicillin-G synthesis of P. chrysogenum, for example, starts with an non-ribosomal peptide synthetase-catalyzed tripeptide formation and is completed with the exchange of an acyl group by IAT. Although the enzymatic activities of Tan and Ebony in Drosophila are unrelated to antibiotic synthesis, they also interact biochemically in cellular processes like neurotransmitter recycling or cuticle sclerotization. Analogous biochemical activities in vertebrates have not been discovered so far.
The amino acid sequence of Tan reveals remarkable similarity to IAT of P. chrysogenum (15), which belongs to the protein family of cysteine peptidases. These are proteolytic enzymes, which depend on a cysteine residue for activity. The large protein family contains at least seven subgroups, or clans, with distinctive structures and properties (21, 30). IAT belongs to clan PB comprising a special subgroup of enzymes, which are not peptidases in the classical sense, but instead, independently from an additional co-factor, cleave a single internal peptide bond in their precursor form (31). The resulting mature proteins acquire different features including hydrolase activity. Previous investigations on IAT processing had revealed a strong dependence of its autoproteolytic activity on the Gly-Cys sequence motif (18). We had shown previously that Tan processing also occurs at a Gly-Cys sequence (5). To unambiguously assign Tan to the cysteine peptidase family, dependence of processing on the Gly-Cys motif had to be proven experimentally. To address whether Gly121 and Cys122 are required for pro-enzyme cleavage, we expressed in vitro mutagenized his6-tan cDNA analogs in E. coli and evaluated the degree of processing after SDS-PAGE and Western blotting (Fig. 2). Only substitution of Gly121 by Ala revealed some heterodimer formation, a result that indicates a more stringent dependence on freedom of flexibility than observed previously for IAT. Previously, substitution of Gly with Ala in IAT yielded a high proportion of mature enzyme, whereas substitution with Val also abolished processing (18). This indicates that flexibility of the peptide chain to some extent must be guaranteed at this position to enable unrestricted processing of both, Tan and IAT. Cys122, however, is absolutely required for processing of Tan. Neither a change of the functional SH-group by an OH-group nor a switch of the size of the side chain was tolerated, a result also obtained in IAT processing (18). The results are reminiscent of reaction schemes proposed for N-S acyl rearrangements initiating protein processing (31). Here, the nucleophilic side chain of cysteine attacks the carbonyl carbon of the preceding amino acid to generate a cyclic hydroxythiazolidine intermediate. This intermediate converts to a thioester, which is then hydrolyzed into the α- and β-subunits. Taken together, both amino acid substitutions undoubtedly revealed the dependence of Tan processing on the Gly-Cys motif, which in turn classifies Tan as belonging to the family of cysteine peptidases. Like other enzymes of this family, Tan could operate as an N-terminal nucleophile hydrolase, because the N-terminal cysteine residue of the mature enzyme could function as a nucleophile in catalysis of peptide bond cleavage of β-alanyl conjugates. This activity has also been proposed for IAT (22). Experimental evidence, however, is lacking in both cases.
It is not surprising that amino acids other than the Gly-Cys motif are also indispensable for processing because the enzyme must fold into a certain structure to initiate the proposed rearrangements. Previous investigations had disclosed amino acid substitutions of IAT outside the Gly-Cys motif that are detrimental to pre-protein cleavage and activity (32) and others with intact processing that lead to loss of enzymatic activity (33) demonstrating a separation of pro-enzyme cleavage activity from enzymatic activity. We asked whether this also holds for Drosophila Tan. The Drosophila tan alleles give rise to amino acid substitutions separated in sequence from the Gly-Cys motif (15). Expression of the respective mutant proteins in E. coli disclosed His6-Tan1 as an example for a block in pre-protein processing caused by the amino acid substitution of Arg217 with Pro (Fig. 2). The observed sequence similarity with IAT in this area (15) indicates shared structural constraints likely to be required for processing to proceed. Therefore, it is not surprising that proline as an uncharged and inflexible amino acid with helix-breaking characteristics disturbs the local folding structure and thereby renders the Tan1 enzyme inactive as demonstrated by the strong loss of function phenotype of tan1. His6-Tan5, however, where substitution of Met256 by Ile also gives rise to a functional null phenotype, is only insignificantly impaired in processing (Fig. 2). One would therefore expect that Tan5 belongs to a type of mutant that affects the active center in a way that abolishes activity. This expectation, however, was not confirmed by our assays performed with E. coli expressed His6-Tan5. Cleavage of carcinine with His6-Tan5 revealed reduced Km and kcat values that still resembled catalytic efficiency of wild type enzyme activity. This activity is not compatible with the observed null mutant phenotype of tan5. Because transcriptional activity of the tan gene was confirmed in both mutants by RT-PCR (Fig. 3E), we looked for Tan protein expression in fly heads. To our surprise, we could trace Tan protein neither in Western blots of head extracts nor in freeze sections of tan5 specimens. We conclude that the amino acid substitution in TanM256I of tan5 flies leads to rapid degradation. We also observed loss of Tan protein in the tan1 mutant. There, the block in autoprocessing obviously gives rise to an unusual structure that must also be prone to degradation. Consistently, in both mutants the structural deviations from native Tan lead to degradation no matter whether the enzyme is locked in a pre-structure and inactive (Tan1) or the enzyme is active but contains an amino acid substitution that changes its structure (Tan5).
Recent evidence on IAT pre-protein processing in P. chrysogenum led to the hypothesis that processing might be an inter-molecular process between two IAT pro-enzymes (22). Upon co-expression of wild type IAT and processing incompetent IATC103S, Garcia-Estrada et al. (22) observed a delay in formation of mature IAT protein and hypothesized that maturation of IAT might be post-translationally regulated in a process that involves homodimer formation. Our observation that wild type Tan as well as maturation defective TanC122A both form dimers provides strong evidence that intermolecular interaction is required for processing into active α2β2 homodimers to occur. This could give rise to an inhibitory effect of processing incompetent Tan1 on processing of a simultaneously expressed wild type form in tan heterozygote flies that should become apparent as dominant phenotype. Because this has not been observed, the inhibitory effect is only weak or the misfolded Tan1 polypeptide is rapidly degraded, a possibility that is supported by the observed failure of Tan detection in homozygous tan1 mutant fly heads.
Pre-protein processing is an internal reaction process that has been shown for IAT in E. coli to require about 3–5 h (22). Tan processing time was also determined in E. coli. It is likely, but not proven, that processing time in vivo in Drosophila is comparable. Because Tan expression in Drosophila at least in heads is subject to circadian regulation (23), a delay of active Tan formation caused by a sustained maturation process should have an impact on the time of day at which Tan regains its full activity. This could affect the flow of histamine in the recycling process.
When one compares Tan with related IATs, it is puzzling that Tan only exhibits hydrolase activity, whereas IAT is not only capable of removing the α-amino-adipic acid group of isopenicillin N but can also replace it with acyl groups. Here, one has to keep in mind that CoA-activated acyl groups that are successfully transferred by IAT are carbonic acids with a hydrophobic side chain of at least five carbon residues (34). We have investigated the possibility of histamine neurotransmitter inactivation by employing acetyl-CoA as substrate. Because no N-acetylhistamine was detected (data not shown), its formation, if it occurs at all in Drosphila, is not dependent on Tan. Whether or not Tan is able to transfer long chain acyl groups perhaps in an unrelated physiological process remains an open question that requires additional investigation in the future. The recent evaluation of the three-dimensional structures of IATs (35) will provide clues to this question by comparing the respective active centers as soon as the Tan structure is also available.
Carcinine cleavage by Tan is in all photoreceptor cells controlling the histamine flow into synaptic vesicles, because histidine decarboxylase (36) activity is too low to supply the amount of neurotransmitter required to sustain basic visual function. Mechanisms of histamine transport into photoreceptor axonal endings and regulation of the flow of carcinine formed by Ebony in glia (5) back into the photoreceptor are still obscure. A participation of the Inebriated (Ine) transporter in this process has been proposed (37). However, as long as a demonstration of the carcinine transport function of Ine is still lacking, an indirect impact on carcinine transport is also feasible. Transporters together with Ebony and Tan constitute a chain of interacting partners that keep neurotransmitter recycling working. They are considered to react very fast and should thus not be a rate-limiting component of the chain. kcat values of Ebony activity are still lacking. We do, however, expect that Ebony, whenever a transporter removes a tiny increase of histamine concentration in the cleft, must immediately convert it into carcinine with comparable speed avoiding accumulation of neurotransmitter in glia next to a synaptic cleft. In contrast to Ebony, the primary role of Tan is not to react on tiny changes in carcinine concentration, but instead to ensure that a steady state concentration of histamine is maintained. This assures continuous synaptic vesicle filling. Adjustment of a fixed carcinine:histamine ratio of 1:2 in head after injection of histamine has been demonstrated. An enzyme revealing a steady rise of activity with increasing substrate concentration is ideally suited to balance this equilibrium. The determined Michaelis-Menten curve exactly concurs with the requirements of Tan activity. The Km value of 119 ± 8.5 μm carcinine reflects a rather weak substrate binding constant. Because according to the equation describing the kcat curve maximal Tan activity is only induced with about 0.5 mm carcinine, we consider it as exceptional that this value will be reached even when a flash of light induces a brief maximal amount of neurotransmitter release (9). Whereas experimental results revealed that Tan adjusts a continuous flow of histamine in photoreceptor cells, in the cuticle the contrary seems to be required. One can envisage that only a limited concentration of β-alanyl-dopamine is tolerated for incorporation into the cuticle to build up the appropriate degree of cross-linking. In this case, a certain concentration of β-alanyl-dopamine would be necessary but not be exceeded. If Tan activity is controlling an upper limit of β-alanyl-dopamine concentration, it should reveal very low activity up to a maximally tolerated substrate concentration. Above this concentration a switch to the highest cleavage activity would reduce β-alanyl-dopamine concentration below the putative limit. This is exactly what the Michaelis-Menten curve reflects when β-alanyl-dopamine is used as substrate: already at low concentration (Km = 0.9 ± 0.3 μm) Tan activity shoots toward full activity.
The presented kinetic parameters of carcinine and β-alanyl-dopamine cleavage exhibit profound differences. The Km of both substrates differs by 2 orders of magnitude. More importantly, concentration dependence of activity reflects the necessities required by the respective cellular pathway, in which Tan is participating. Our results show how the enzyme can manage different tasks in pleiotropic functions by simply varying substrate affinity and reaction speed and extend our understanding on cuticle sclerotization and histamine neurotransmitter recycling in Drosophila.
Acknowledgments
We gratefully acknowledge Irmi Sures and Christian Herrmann for valuable comments on the manuscript. We thank Wolfgang Schuhmann, Christian Herrmann, Jianyong Li, Janusz Borycz, Peter Wolff, and Petra Pennekamp for technical help. Alexander Neuhaus and Fabian Hertel provided tan mutant expression clones; Silke Wittlinger assisted in the kinetic measurements.
This work was supported by Deutsche Forschungsgemeinschaft Grant HO 714/13-2 (to B. T. H.) and a scholarship of the International Graduate School of Neuroscience (to S. A.).
- IAT
- isopenicillin N-acyltransferase
- RT
- reverse transcription
- IPTG
- isopropyl β-d-thiogalactopyranoside
- HPLC
- high pressure liquid chromatography.
REFERENCES
- 1.Stuart A. E., Borycz J., Meinertzhagen I. A. (2007) Prog. Neurobiol. 82, 202–227 [DOI] [PubMed] [Google Scholar]
- 2.Morgan J. R., Gebhardt K. A., Stuart A. E. (1999) J. Neurosci. 19, 1217–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borycz J., Borycz J. A., Loubani M., Meinertzhagen I. A. (2002) J. Neurosci. 22, 10549–10557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richardt A., Kemme T., Wagner S., Schwarzer D., Marahiel M. A., Hovemann B. T. (2003) J. Biol. Chem. 278, 41160–41166 [DOI] [PubMed] [Google Scholar]
- 5.Wagner S., Heseding C., Szlachta K., True J. R., Prinz H., Hovemann B. T. (2007) J. Comp. Neurol. 500, 601–611 [DOI] [PubMed] [Google Scholar]
- 6.Hovemann B. T., Ryseck R. P., Walldorf U., Störtkuhl K. F., Dietzel I. D., Dessen E. (1998) Gene 221, 1–9 [DOI] [PubMed] [Google Scholar]
- 7.Richardt A., Rybak J., Störtkuhl K. F., Meinertzhagen I. A., Hovemann B. T. (2002) J. Comp. Neurol. 452, 93–102 [DOI] [PubMed] [Google Scholar]
- 8.Suh J., Jackson F. R. (2007) Neuron 55, 435–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borycz J. A., Borycz J., Kubów A., Kostyleva R., Meinertzhagen I. A. (2005) J. Neurophysiol. 93, 1611–1619 [DOI] [PubMed] [Google Scholar]
- 10.Wright T. R. F. (1987) Adv. Genet. 24, 127–209 [PubMed] [Google Scholar]
- 11.True J. R. (2003) Trends Ecol. Evol. 18, 640–647 [Google Scholar]
- 12.Sugumaran M., Giglio L., Kundzicz H., Saul S., Semensi V. (1992) Arch. Insect Biochem. Physiol. 19, 271–283 [DOI] [PubMed] [Google Scholar]
- 13.Morgan C. B., Bridges T. H., Sturtevant A. H. (1925) Bibliog. Genet. 2, 237 [Google Scholar]
- 14.McEwen R. S. (1918) J. Exp. Zool. 25, 49–106 [Google Scholar]
- 15.True J. R., Yeh S. D., Hovemann B. T., Kemme T., Meinertzhagen I. A., Edwards T. N., Liou S. R., Han Q., Li J. (2005) PLoS Genet. 1, e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alvarez E., Cantoral J. M., Barredo J. L., Díez B., Martín J. F. (1987) Antimicrob. Agents Chemother. 31, 1675–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tobin M. B., Fleming M. D., Skatrud P. L., Miller J. R. (1990) J. Bacteriol. 172, 5908–5914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tobin M. B., Cole S. C., Miller J. R., Baldwin J. E., Sutherland J. D. (1995) Gene 162, 29–35 [DOI] [PubMed] [Google Scholar]
- 19.Tobin M. B., Baldwin J. E., Cole S. C., Miller J. R., Skatrud P. L., Sutherland J. D. (1993) Gene 132, 199–206 [DOI] [PubMed] [Google Scholar]
- 20.Müller W. H., Bovenberg R. A., Groothuis M. H., Kattevilder F., Smaal E. B., Van der Voort L. H., Verkleij A. J. (1992) Biochim. Biophys. Acta 1116, 210–213 [DOI] [PubMed] [Google Scholar]
- 21.Barrett A. J., Rawlings N. D. (2001) Biol. Chem. 382, 727–733 [DOI] [PubMed] [Google Scholar]
- 22.García-Estrada C., Vaca I., Fierro F., Sjollema K., Veenhuis M., Martín J. F. (2008) Fungal Genet. Biol. 45, 1043–1052 [DOI] [PubMed] [Google Scholar]
- 23.Keegan K. P., Pradhan S., Wang J. P., Allada R. (2007) PLoS Comput. Biol. 3, e208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bertani G. (1951) J. Bacteriol. 62, 293–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Southern E. M. (1975) J. Mol. Biol. 98, 503–517 [DOI] [PubMed] [Google Scholar]
- 26.Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 27.Heukeshoven J., Dernick R. (1988) Electrophoresis 9, 28–32 [DOI] [PubMed] [Google Scholar]
- 28.Kyhse-Andersen J. (1984) J. Biochem. Biophys. Methods 10, 203–209 [DOI] [PubMed] [Google Scholar]
- 29.Michaelis L., Menten M. L. (1913) Biochem. Z. 49, 333–369 [Google Scholar]
- 30.Rawlings N. D., Morton F. R., Kok C. Y., Kong J., Barrett A. J. (2008) Nucleic Acids Res. 36, D320–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perler F. B., Xu M. Q., Paulus H. (1997) Curr. Opin. Chem. Biol. 1, 292–299 [DOI] [PubMed] [Google Scholar]
- 32.Tobin M. B., Cole S. C., Kovacevic S., Miller J. R., Baldwin J. E., Sutherland J. D. (1994) FEMS Microbiol. Lett. 121, 39–46 [DOI] [PubMed] [Google Scholar]
- 33.Fernández F. J., Gutierrez S., Velasco J., Montenegro E., Marcos A. T., Martín J. F. (1994) J. Bacteriol. 176, 4941–4948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alvarez E., Meesschaert B., Montenegro E., Gutiérrez S., Díez B., Barredo J. L., Martín J. F. (1993) Eur. J. Biochem. 215, 323–332 [DOI] [PubMed] [Google Scholar]
- 35.Bokhove M., Yoshida H., Hensgens C. M., van der Laan J. M., Sutherland J. D., Dijkstra B. W. (2010) Structure 18, 301–308 [DOI] [PubMed] [Google Scholar]
- 36.Burg M. G., Sarthy P. V., Koliantz G., Pak W. L. (1993) EMBO J. 12, 911–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gavin B. A., Arruda S. E., Dolph P. J. (2007) PLoS Genet. 3, e206. [DOI] [PMC free article] [PubMed] [Google Scholar]