Abstract
Using x-ray crystallography we have determined the structure of the cytoplasmic fragment (residues 384–732) of the flagellum secretion system protein FlhA from Helicobacter pylori at 2.4-Å resolution (r = 0.224; Rfree = 0.263). FlhA proteins and their type III secretion homologues contain an N-terminal integral membrane domain (residues 1–350), a linker segment, and a globular C-terminal cytoplasmic region. The tertiary structure of the cytoplasmic fragment contains a thioredoxin-like domain, an RNA recognition motif-like domain inserted within the thioredoxin-fold, a helical domain, and a C-terminal β/α domain. Inter-domain contacts are extensive and the H. pylori FlhA structure appears to be in a closed conformation where the C-terminal domain closes against the RNA recognition motif-fold domain. Highly conserved surface residues in FlhA proteins are concentrated on a narrow surface strip comprising the thioredoxin-like and helical domains, possibly close to the export channel opening. The conformation of the FlhA N-terminal linker segment suggests a likely orientation for the FlhA cytoplasmic fragment relative to the membrane-embedded export pore. Comparison with the recently published structures of the Salmonella FlhA cytoplasmic fragment and its type III secretion counterpart InvA highlight a conformational change where the C-terminal β/α domain in H. pylori FlhA moves 15 Å relative to Salmonella FlhA. The conformational change is complex but primarily involves hinge-like movements of the helical and C-terminal domains. Interpretation of previous mutational screens suggest that the C-terminal domain of FlhAC plays a regulatory role in substrate class switching in flagellum export.
Keywords: Bacteria, Cell Motility, Crystal structure, Membrane Proteins, Protein Secretion, Flagellar Export Apparatus, Flagellum, FlhA, Helicobacter pylori, Type III Secretion
Introduction
Helicobacter pylori is a motile Gram-negative ϵ-proteobacterium that colonizes the human gastric mucosa. Infection with H. pylori causes inflammation typically resulting in gastritis, peptic and duodenal ulcer diseases, and in the most severe cases, gastric adenocarcinoma (reviewed in Refs. 1 and 2). H. pylori is motile by means of multiple flagella that are similar in many respects to the well studied flagella of enteric bacteria, with the exception that they are sheathed by an extension of the outer membrane and located at the cell poles (3, 4). Motility is also an essential factor for colonization and persistence of H. pylori in the gastric mucosa of the host organism (5, 6).
Bacterial flagella consist of four major substructures: a basal body containing the flagellar motor and the export apparatus, a hollow rod that spans the cell envelope, a hook that functions as the torque-generating universal joint, and the flagellar filament that protrudes into the aqueous medium and provides motility (reviewed in Refs. 7 and 8). The basal body is composed of two connected, but distinct structures, the C- or cytoplasmic ring containing FliG, FliM, and FliN, which interact with both the flagellar motor (MotA and MotB proteins) and the MS (motor-stator)-ring. The MS-ring houses the export apparatus, and is composed of 24–26 copies of the ∼570 residue FliF protein (9–11). The MS-ring is anchored to the cell membrane via two membrane spanning segments of FliF situated at approximately residues 23–42 and 449–460 (12). Associated with the MS-ring are the membrane-spanning proteins FlhA, FlhB, FliO, FliP, FliQ, and FliR, which are thought to assemble together in the cytoplasmic membrane to make an export pore (8). The pore proteins interact with FliI and FliH, two soluble cytoplasmic components of the export apparatus, and together direct substrates to and facilitate their passage through the export pore in a defined order to ensure correct flagellum assembly. Efficient export of flagellum components requires both ATP hydrolysis, by the flagellum specific ATPase FliI, and the transmembrane protonmotive force (13, 14).
Of the membrane-inserted components of the flagellar export apparatus, FlhA and FlhB are the most well characterized subunits (13, 15–19). FlhA encodes a ∼700 residue protein that is essential for flagellum biogenesis and which contains an N-terminal integral membrane domain (amino acids 1–350) predicted to contain either six or eight membrane-spanning helices (16). A globular C-terminal domain, FlhAC (residues 350–730 of Hp FlhA), resides in the cytoplasm and likely interacts with the soluble components of the export apparatus and/or export substrates as Salmonella strains harboring a plasmid carrying the FlhAC fragment in a wild-type background exhibit a dominant-negative phenotype, including impaired motility and impaired export of rod/hook substrates (16). A number of Salmonella FlhAC point mutants have been mapped that impair motility at restrictive temperatures (19). Genetic suppressor and biochemical studies indicate that the membrane-spanning segment of FlhA interacts directly with the MS-ring protein FliF (16, 18). The globular C-terminal cytoplasmic region of FliF is also known to interact with the C-ring protein FliG (12, 20) and has been reported to interact with the cytoplasmic C-terminal fragment of FlhA in Chlamydia pneumoniae (21). Other studies show that FlhA plays an important role in keeping the flagellar export gate closed in the absence of the export regulator FliH or the flagellar ATPase FliI (13, 17). The Salmonella FlhAC fragment has been well characterized (16) and crystallized (22), and a structure for Salmonella FlhAC has been published online (23). The structure of the C-terminal domain of Salmonella InvA, the type III secretion homolog of FlhA, was also published online (24).
In H. pylori, FlhA is a 733-amino acid polypeptide essential for motility (25). Deletion of FlhA or FlgM (the anti-σ factor for σ28) causes repression of the transcription of a broad range of flagellum genes (26). FlgM/FlhA double mutants have higher than normal flagellar gene expression levels for the genes down-regulated in the flhA mutant, suggesting an interaction between FlhA and FlgM (26–29). An interaction between H. pylori FlhAC and FlgM was recently demonstrated by a bacterial two-hybrid assay and also affinity pulldowns where purified FlgM is added to His-tagged FlhAC in Escherichia coli extracts, the interaction was, however, not confirmed using the two purified proteins, suggesting an additional interaction factor in E. coli extracts (29). In Salmonella, FlgM is a filament class export substrate, and is exported after hook completion. Unlike its Salmonella orthologue, H. pylori FlgM is not secreted and remains in the cytoplasm (29). Hence there are likely to be important differences in the function of FlhA in H. pylori, as compared with its role in Salmonella. Studies on FlhA from Campylobacter and other bacteria suggest functions in addition to flagellar protein export, where secretion of certain non-flagellar outer membrane proteins requires FlhA (30–32). Studies on LcrD, the Yersinia pestis type III secretion system homolog of FlhA, also demonstrate a role in the regulation of other secretion system components, suggesting this may be a general feature of the FlhA homolog function (33–35). Given the high level of sequence conservation between FlhA and its type III secretion orthologs, the important central role of FlhA in regulation of H. pylori flagellum assembly and the secretion of flagellum components, and the diverse role of FlhA in the secretion of non-flagellar outer membrane proteins, we chose to study the three-dimensional structure of the soluble cytoplasmic domain of FlhA (FlhAC) from H. pylori CCUG17874.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Plasmids
E. coli XL-1 Blue (Invitrogen) was used as the host strain for molecular cloning. Genomic DNA from H. pylori CCUG17874 (Culture Collection University of Gothenburg, Sweden) was used as a template for gene amplification. E. coli BL21(DE3) (Novagen) was used for protein overexpression.
Molecular Cloning
H. pylori CCUG 17874 genomic DNA was extracted as previously described (36). The gene fragment corresponding to the C-terminal cytoplasmic domain of the H. pylori flhA gene (designation hp1041 in strain 26695) was amplified by PCR from H. pylori CCUG 17874 gDNA. PCR was performed using Pfu polymerase according to the manufacturer's instructions (New England Biolabs). The fragment corresponding to amino acids 373–732 (the C-terminal Phe residue was deleted) of H. pylori FlhA was cloned into the glutathione S-transferase gene fusion vector pGEX-6P-3 (GE Healthcare) using EcoRI and BamHI restriction sites. Forward and reverse primer sequences are 5′-GAC CTG GGA TCC ACA AGG GCT AAA ACC CAA GAA GAG 3′, and 5′-GAC GAC GAA TTC TTA GTT AAT ATG GAT CGT GCC TAA GGC. Sequencing of the insert indicated that the predicted amino acid sequence of the hp1041 gene fragment from strain 17874 differed at five positions from the published sequence from the HP strain 26695. The mutations are conservative and likely have minimal impact on the tertiary structure or function of the protein. They are L381I, K482R, E502D, T503A, V670G, and I710V.
Protein Overexpression and Purification
E. coli BL21 cells harboring the pFlhAC plasmid were grown at 37 °C with shaking (225 rpm) in Luria-Bertani broth, containing 100 μg/ml of ampicillin. Cells were grown at 37 °C to an A600 of 0.7 and expression of the truncated recombinant FlhAC protein was induced with 0.1 mm isopropyl 1-thio-β-d-galactopyranoside. After induction, cells were grown at 25 °C for an additional 16–18 h. E. coli cells were harvested by centrifugation and lysed by passage twice through a French Press. The resultant supernatant was clarified by centrifugation two times at 16,000 × g for 30 min. The selenomethionine-labeled protein was prepared in a similar fashion, but once cells were grown to an A600 of 0.7 in LB, the E. coli cells were transferred to M9 minimial medium supplemented with selenomethionine (60 mg) and the following amino acids to inhibit the E. coli methionine biosynthetic pathway (Lys, Phe, Thr (100 mg); Val, Leu, Ile (50 mg)). After the cells recovered for 0.5 h at 37 °C, they were then induced with 0.1 mm isopropyl 1-thio-β-d-galactopyranoside and grown at 25 °C for an additional 16–18 h. For both wild-type and SeMet2 protein, the soluble fraction containing the recombinant glutathione S-transferase (GST) fusion protein was loaded onto an ÄKTA FPLC instrument, fitted with a column packed with 10 ml of glutathione-Sepharose 4B resin and equilibrated in lysis buffer. After extensive washing, the GST fusion protein was eluted using a gradient wash of reduced glutathione. The eluted GST fusion protein fractions were pooled, dialyzed overnight into cleavage buffer, and then incubated with PreScission Protease for 16 h at 4 °C (GE Healthcare). The GST tag and PreScission Protease were removed by a second passage of the sample over the glutathione-Sepharose column. The C-terminal domain of FlhA (FlhAC) was further purified by anion exchange chromatography (Source Q) on an ÄKTA FPLC (50 mm Tris-HCl buffer, pH 8.0, elution gradient of 0.0–0.5 m NaCl). FlhAC was further purified by gel filtration chromatography in 50 mm Tris buffer with 150 mm NaCl at pH 7.2 using a Superdex 200 HR 10/30 column. The purity of FlhAC was assessed after each purification step by SDS-PAGE. Protein samples were concentrated using 10-kDa molecular mass Amicon Ultra-15 Centrifugal Filter Units. Protein concentrations were measured using the Bradford dye binding assay, and/or absorbance of aromatic residues at 280 nm (ϵ280 = 14,400 m−1 cm−1; absorbance of 0.373 for 1 mg/ml soln).
Protein Crystallization
Highly purified FlhAC protein was concentrated to 10 mg/ml in 20 mm BisTris propane buffer, pH 6.5, and 150 mm NaCl. The FlhAC protein was crystallised at 20 °C in hanging-drop vapor diffusion experiments. Drops contained 1.0 μl of purified protein and an equivalent volume of reservoir solution and were sealed over a reservoir filled with 1 ml of crystallization solution. Small clusters of plate-like crystals were obtained from commercial sparse matrix screens containing ∼20% (w/v) polyethylene glycol 3350, 0.2 m Ca2+ or Mg2+ salts. Extensive optimization of the crystallization conditions yielded large, but thin plate crystals (0.5 × 0.3 × 0.03 mm) from 13.5 to 15% (w/v) monomethyl ether-polyethylene glycol 5000, 100 mm sodium cacodylate buffered at pH 6.2–6.7, 0.2 m (NH4)2SO4, and 6–8% (v/v) isopropyl alcohol.
Structure Determination
Crystals of selenometionine-incorporated FlhAC protein were grown as described above and incubated in mother liquor plus 18% (w/v) glycerol, mounted in 20-μm nylon loops (Hampton Research), and flash frozen in a stream of liquid nitrogen gas. A Selenium K-edge MAD experiment was performed on the CMCF-I 08ID-1 beamline at the Canadian Light Source Synchrotron (Saskatoon, Canada). Diffraction data at peak, inflection, and remote (low energy) wavelengths (Table 1) were then collected and processed with the HLK-2000 package (37). The crystals of the SeMet-labeled protein diffracted to a nominal resolution of 3.0 Å. The unit cell parameters fitted a C-centered orthorhombic lattice (space group C2221), with one molecule in the asymmetric unit (Table 1). Analysis of the anomalous scattering signal from the selenium atoms, using Solve, revealed a strong signal to ∼3.4-Å resolution (38–40). Eight of the nine Se atoms were located with Solve, the resultant Z-score was 31.5 with a figure of merit of 0.55 for all data between 50.0 and 3.00 Å resolution. A solvent-flattened electron density map (70% solvent in the unit cell) was calculated to 3.0-Å resolution with Resolve clearly revealed the overall trace of the polypeptide chain, and the positions of eight of the nine expected SeMet residues (supplemental Fig. S3). Subsequent model building was carried out with Coot (41). The initial model was refined against the SeMet peak data using CNS (42). Native data to 2.4-Å resolution were then obtained from crystals prepared from unlabeled protein (Table 2). These data were used for all further model refinement. Initially CNS (42) and then Refmac 5.2 were used in conjunction with Rfree (5% of the data) for refinement (43–45). A Ramachandran plot and representative electron density map for the final model can be found in the supplementary data. Crystallographic refinement statistics are provided in Table 2. The atomic coordinates have been deposited with the Protein Data Bank code 3MYD.
TABLE 1.
Data collection, phasing, and structure refinement statistics for H. pylori FlhAC
| Data collection statisticsa,b | Native (C2221) | SeMet |
||
|---|---|---|---|---|
| Peak | Inflection | Remote | ||
| Unit cell parameters (a, b, c) (Å) | 69.193, 136.20, 107.68 | 71.71, 132.8, 106.4 | 71.75, 132.9, 106.4 | 71.74, 132.9, 106.5 |
| Wavelength (Å) | 0.97934 | 0.97882 | 0.97907 | 0.98166 |
| Resolution (Å) | 50-2.4 (2.49-2.40) | 50-3.0 (3.11-3.00) | 50-3.0 (3.11-3.00) | 50-3.0 (3.11-3.00) |
| No. unique reflections | 20,380 (2005) | 19,692 (1935) | 19,715 (1946) | 19,662 (1952) |
| Redundancy | 7.3 (7.0) | 3.9 (3.8) | 3.9 (3.8) | 3.9 (3.9) |
| Completeness (%) | 99.9 (99.9) | 99.7 (98.0) | 99.7 (98.6) | 99.9 (99.9) |
| Mosaicity (°) | 0.540 | 0.62 | 0.49 | 0.63 |
| Average I/σ | 29.8 (3.7) | 25.2 (3.7) | 25.4 (3.6) | 22.0 (2.8) |
| Rmergec | 0.074 (.386) | 0.051 (0.34) | 0.050 (0.34) | 0.056 (0.46) |
a Values in parentheses correspond to the highest resolution shell.
b Data processing statistics calculated using Denzo/HKL2000 (37). The anomalous pairs F(+hkl) and F(−hkl) were not merged during data processing. Data were collected over 180° (180 frames). An identical orientation matrix was used for processing the three data sets.
c Rmerge = Σhkl Σi|〈I(hkl)obs〉 − I(hkl)obs,i|/Σhkl,i I(hkl)obs,I, where I(hkl)obs,i is the individual measurement of an hkl intensity and 〈I(hkl)obs〉 = Σi I(hkl)obs,i/N; where i = 1 to n individual reflections are measured.
TABLE 2.
Structure refinement statistics for H. pylori FlhAC
| Statistics | |
|---|---|
| Structure refinement | 30.0-2.40 Å (2.462-2.400)a |
| No. of reflections in working set | 19,217 (1,390)a |
| No. of reflections in test set | 1,037 (82)a |
| Rworkb | 0.224 (0.262)a |
| Rfreec | 0.263 (0.315)a |
| No. of amino acid residues | 361 |
| No. of water molecules | 70 |
| No. of ligands | 0 |
| Average B-factor (Å2)d | 26.2 |
| R.m.s. deviation B bonded MC atoms (Å2)d | 0.715 |
| R.m.s. deviation B bonded SC atoms (Å2)d | 1.36 |
| Rmsd B angle MC atoms (Å2)d | 2.16 |
| Rmsd B angle SC atoms (Å2)d | 3.40 |
| R.m.s. deviation bond lengths (Å)d | 0.011 |
| R.m.s. deviation angles (°)d | 1.24 |
| Residues in preferred Ramachandran regions (%)e | 91.7 |
| Residues in allowed regions (%)e | 8.3 |
a Values in parentheses correspond to the highest resolution shell.
b Rwork = Σhkl‖Fobs(hkl)‖−|Fcalc(hkl)‖/Σhkl|Fobs(hkl)|, where |Fobs(hkl)| and |Fcalc(hkl)| are the observed and calculated amplitudes, respectively, for the structure factor F(hkl).
c Rfree is the equivalent of Rwork for 5% of the reflections (randomly selected), which were not used in structure refinement.
e The Ramachandran plot was generated with Procheck in CCP4 (see supplemental Fig. S3) (45, 53).
RESULTS AND DISCUSSION
FlhAC Tertiary Structure
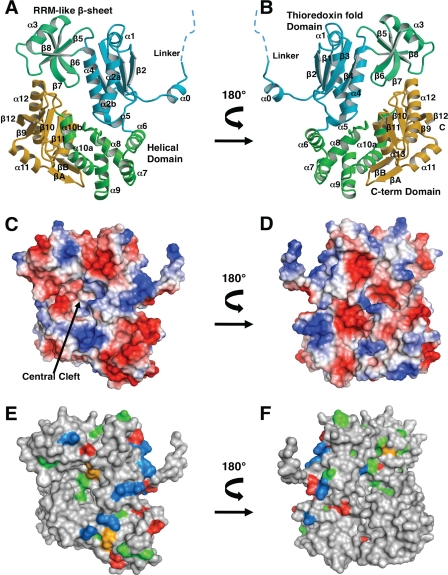
The structure of the H. pylori FlhA C-terminal domain (residues 384–732) has been fully refined against diffraction amplitudes from the native crystals to 2.4-Å resolution (supplemental Figs. S1–S3) (r = 22.4%, Rfree = 26.3%). The structure contains one molecule in the crystallographic asymmetric unit and is well ordered, with the exception that helices α6 and α7 have higher than average thermal displacement factors. H. pylori FlhAC is composed of four domains arranged roughly at the four corners of a rectangle, and begins with a short extended fragment (residues 384 to 394) that connects the N-terminal integral membrane segment of FlhA (residues 1–350) to FlhAC (Fig. 1). A Dali search (46) using the refined H. pylori FlhAC atomic coordinates revealed similarities to known protein domains (supplemental Fig. S4). The first domain resembles thioredoxin (residues 395–458 and 519–540 are similar to residues 16–81 of PDB 2P0J, Z-score = 6.5, r.m.s. deviation 2.2 Å). Inserted in the thioredoxin-like fold is a small antiparallel β-sheet domain with the same topology of RNA recognition motifs (RRM) (residues 470 to 518) (Figs. 1 and 2). The two segments connecting the thioredoxin and RRM domains each contain a proline residue (Pro-469 and Pro-519) that likely make this domain connection in H. pylori FlhA fairly rigid. The last helix of the thioredoxin-like domain is quite long and connects to an all α domain (residues 541 to 614) that remotely resembles the N-terminal helical domain of ribonucleotide reductase (PDB 2R1R, Z-score = 5.5, r.m.s. deviation 2.6 Å). The FlhA helical domain is in intimate contact with the FlhA C-terminal domain (residues 630–729). The C-terminal domain superficially resembles the β-strand arrangement and placement of two helices in the redox protein rhodanese (residues 6–95 of PDB 3G5J, Z-score = 4.6, r.m.s. deviation 3.3 Å) (Figs. 1 and 3). However, there is no sequence conservation between FlhA and any of these structurally similar domains.
FIGURE 1.
Tertiary structure of H. pylori FlhAC. Two orientations related by a 180° rotation about the vertical axis are shown for each rendering of the molecule drawn with Molscript/Raster3D (A and B) (49, 50) or PyMOL (parts C–F). A and B, ribbon diagram of FlhAC labeled with domain identifiers and labeled secondary structure elements. C and D, molecular surfaces depicting qualitative electrostatic potentials. E and F, highly conserved amino acids mapped onto the FlhAC molecular surface. Moderately conserved residues are not shown. Positively charged residues are colored blue, negatively charged residues red, polar residues orange, and hydrophobic residues green.
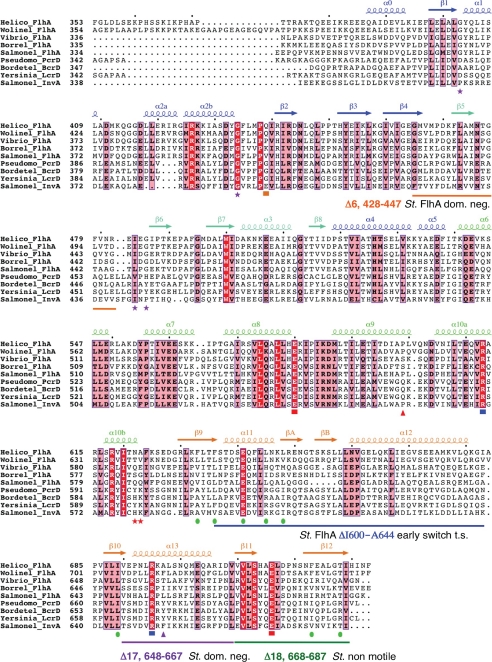
FIGURE 2.
Structurally based alignment of FlhAC and type III secretion homologue sequences. Calculated with T-Coffee (51) and rendered using Espript (52). Secondary structure assignments for the x-ray structure of H. pylori FlhAC are shown, and labeled according to the Salmonella FlhAC structure (23). Highly conserved residues are boxed in red, moderately conserved residues in pink. Salmonella temperature-sensitive secretion point mutations in FlhAC are marked with purple stars (19). Salmonella flhA nonsense mutations (Q588Stop, Q589Stop) that permit early secretion of FlgM are marked with red stars (48). The Salmonella flhA V404M mutation is marked with an orange square (13, 17). Y. pestis LcrD secretion-defective mutants are marked with triangles, red indicates a nonsense mutation at Gln-574 that deletes the C-terminal domain, pink depicts a Y670C missense mutation (34, 35). Conserved salt bridge interactions between the helical and C-domains of H. pylori FlhAC are marked with red (Glu) or blue (Arg) squares. Green circles mark residues that make up a predominantly hydrophobic surface on the C-domain. Salmonella FlhA deletion mutants mentioned in the text are marked by colored lines (16, 48).
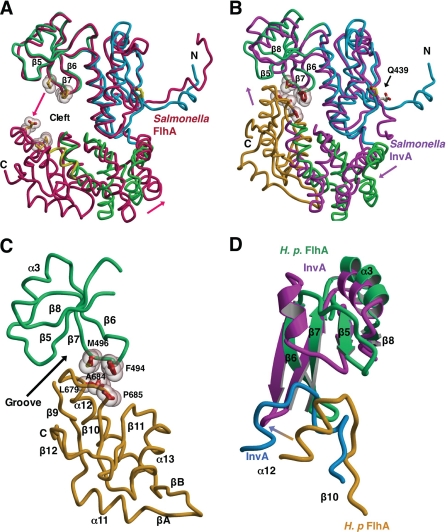
FIGURE 3.
Evidence for a FlhAC conformational change. Structural overlays calculated using lsqkab (45) and drawn with Molscript/Raster3D (49, 50). H. pylori FlhAC drawn as described in the legend to Fig. 1A. A, overlay of H. pylori and Salmonella FlhAC (3A5I, in red) (23) structures based on superposition of the thioredoxin-like and RRM-like domains. The C-domain of H. pylori FlhAC has been omitted for clarity. Arrows depict the relative movement of Salmonella FlhA relative to H. pylori FlhA. Residues in Salmonella FlhA likely to make contact in the presumed closed conformation of the structure are shown as ball-and-stick models with van der Waals surfaces. B, Salmonella InvA (2X49, in magenta) (24) is overlaid onto the structure of H. pylori FlhAC. The C-domain of Salmonella InvA is omitted for clarity. Side chains making van der Waals contact between the RRM-like and C-domains of H. pylori FlhA are drawn as in A and labeled. C, close up of the interface between the RRM-like and C-terminal domains of Hp FlhA. D, comparison of the interface between the RRM-like and C-terminal domains of H. pylori FlhA (in green and gold) and Salmonella InvA (in magenta and blue) (24).
At the juncture of the four domains of the H. pylori FlhAC molecule, there is a noticeable surface depression (Fig. 1). The bottom of this depression or cleft is occupied by the β6-β7 hairpin from the RRM-like domain and it contacts the α12-β10 loop on the C-terminal domain, making van der Waals contacts between residues Leu-679, Gly-682, Ala-684, and Pro-685 of the α12-β10 linker, and residues Phe-494 and Met-496 of the β6-β7 hairpin (Figs. 2 and 3). In addition, a cis-peptide bond connects residues Ala-684 and Pro-685 and contributes to this interaction surface (Fig. 3).
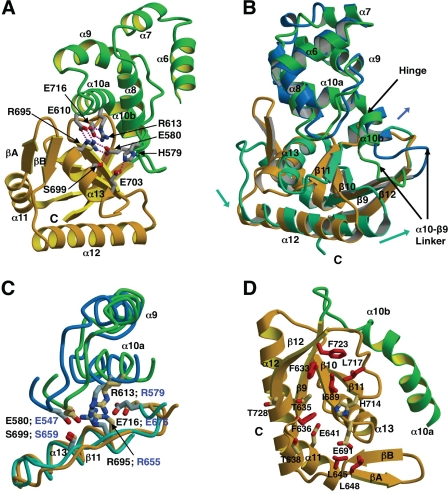
Interactions between the helical and C-terminal domains of H. pylori FlhA are mostly polar in nature, and are centered on four highly ordered salt bridge interactions (Fig. 4). Residues making two of these salt bridges are highly conserved in FlhA sequences (Fig. 2), they include Glu-580 (at the end of helix α8) that forms a salt bridge with Arg-695 (helix α13), and Glu-716 (β11-β12 loop) that forms a salt bridge with Arg-613 (helix α10a). Both Glu-580 and Glu-716 are known sites of temperature-sensitive point mutations in Salmonella FlhA (Glu-547 and Glu-676) that impair flagellum export function (19). In the H. pylori FlhAC structure, the guanidinium groups of Arg-613 and Arg-695 are highly ordered and in close contact, effectively making a π-π stacking interaction. Other less well conserved interactions between the helical and C-terminal domains include salt bridges between Glu-610 and Arg-695, His-579, and Glu-703 (Fig. 4).
FIGURE 4.
Inter-domain salt bridges and conformational rearrangement in the helical and C-terminal domains of FlhAC. Drawn with Molscript/Raster3D (49, 50). A, ribbon diagram depicting salt bridge interactions between the helical (green) and C-terminal (gold) domains of Hp FlhA. Side chains are shown as ball and stick models with hydrogen bonds drawn as dotted red lines. Salt bridges shown are: Glu-610 (helical) and Arg-695 (C-domain), Arg-613 (helical) and Glu-716 (C-domain), Glu-580 (helical) and Arg-695 (C-domain), His-579 (helical), and Glu-703 (C-domain). B, depiction of the helical and C-terminal domains in H. pylori (colored green and gold) and Salmonella (dark blue and aquamarine) FlhAC structures based on a least squares superposition of the helical domains. Movements of the Salmonella domains relative to H. pylori FlhA are indicated with arrows. C, structural superposition depicting conserved salt bridge residues at the interface of the helical and C-terminal domains of H. pylori and Salmonella FlhAC. Colored as in B. D, ribbon representation showing the side chains of mostly hydrophobic residues making up a conserved surface depression in the H. pylori FlhA C-domain. Side chains are drawn as red ball-and-stick models and labeled accordingly.
Sequence and Structural Conservation in the FlhA/LcrD Family
FlhA sequences and those of their type III secretion counterparts (InvA/LcrD etc.) are well conserved in bacteria, but are not related to any other protein families or domains. The FlhA cytoplasmic fragment sequences are less well conserved than the N-terminal transmembrane segment, and the membrane-cytoplasmic linker segment is the most poorly conserved region of the molecule (Fig. 2). For FlhAC, the highest sequence conservation is found in the thioredoxin and helical domains that contain a number of invariant amino acid residues (Fig. 2). In addition, conserved surface residues in these two domains tend to cluster on a fairly confined strip on one face of the FlhAC molecular surface (Fig. 1). A string of positively charged residues is part of this conserved surface patch (Fig. 1).
In contrast, sequence conservation in the RRM-like domain of FlhA homologues is less extensive, especially for the type III secretion system members (Fig. 2). However, residue Trp-500 in this domain is highly conserved and contacts residues in the thioredoxin-like domain. Importantly, the recently reported structure of Salmonella InvA (24) shows that the InvA RRM-like domain adopts a noticeably different conformation from the FlhA RRM-like domains in H. pylori and Salmonella FlhA structures, and contains an additional helix between strands β5 and β6 (Fig. 3).
The C-terminal β/α domain of FlhA is moderately well conserved both in sequence and structure and four residues making inter-domain salt bridges with the adjacent helical domain in H. pylori FlhAC are highly conserved (Figs. 2 and 4). In addition, a moderately hydrophobic surface patch on the C-terminal β/α domain of H. pylori FlhAC (secondary structure elements β9 and α11 plus β10 and β12) is conserved in hydrophobic character in other FlhA and type III secretion homologue sequences (Fig. 2). This surface also includes a highly conserved glutamate residue (Glu-641) (Figs. 2 and 4). The presence of this hydrophobic surface patch on the FlhA α/β C-domain is suggestive of a ligand binding site.
The recently published structures of Salmonella FlhAC (23) and Salmonella InvA (24) permit comparison with the structure of H. pylori FlhAC (Figs. 3 and 4). In all three molecules, the thioredoxin-like domain, the helical domain, and the C-terminal domain are very similar in tertiary structure and only differ significantly in conformation at a few insertions or deletions at surface loops (Figs. 2–4). Although the RRM-like domain structure and orientation are very similar in Salmonella and H. pylori FlhA, the RRM-like domain differs significantly in structure and orientation in Salmonella InvA (24) (Fig. 3). The orientation of the RRM-like domain of InvA is twisted away from the C-domain relative to FlhA when the thioredoxin-like domains are superimposed, and the InvA RRM-like domain also contains an extra helix (Fig. 3).
The highly curved N-terminal linker segment connecting the FlhA transmembrane domain to the cytoplasmic domain is also very similar in structure in H. pylori and Salmonella FlhA. This similarity in conformation of the linker segment was unexpected, as sequence conservation is minimal in the linker region and modeled thermal displacement factors are high (Figs. 2–3). Hence, this suggested to us the conformation of the linker segment may help to precisely position the cytoplasmic domain relative to the FlhA membrane-spanning segment. If the linker conformations observed in the two FlhA crystal structures are truly reflective of the in vivo conformation of FlhA, then we can tentatively assume that the plane of the cytoplasmic membrane would be situated at the top of Fig. 3. However, the linker fragment is quite short in the crystallized InvA cytoplasmic fragment and noticeably different in conformation (24).
FlhAC Undergoes a Dramatic Conformational Change
The relative positioning of the four domains in each of the three independent FlhA family member structures highlights overall similarities, but also exposes significant differences in the positions of the helical and C-terminal domains relative to the N-terminal half of the molecule. In H. pylori FlhA, Salmonella FlhA, and Salmonella InvA, the thioredoxin-like and RRM-like domains superimpose reasonably well with only small differences in backbone atom positions, although the InvA RRM-like domain does not overlay as well (Fig. 3). Differences in the position and conformation of helical and C-terminal domains in the three FlhA family structures are more pronounced (Fig. 3). In Salmonella FlhAC, the helical and C-terminal domains are displaced in position relative to H. pylori FlhA and Salmonella InvA, creating a large open cleft between the RRM- and C-terminal domains (Fig. 3). For instance, when the N-terminal two domains of H. pylori and Salmonella FlhAC molecules are superimposed by least squares, the C-terminal end of helix α12 in Salmonella FlhA is displaced by at least 15 Å relative to H. pylori FlhA. In contrast, the conformations of both the helical and C-terminal domains of Salmonella InvA and H. pylori FlhA are quite similar, and each molecule can be described as being in a closed conformation relative to Salmonella FlhA. However, the InvA molecule is closed to a greater extent than H. pylori FlhA, as the helical and C-terminal domains have undergone rigid body movements that places the α12-β10 linker of the C-terminal domain closer to the RRM-like domain, facilitating interdomain interactions (Fig. 3).
Because H. pylori FlhA and Salmonella InvA adopt a similar arrangement of their four domains, the RRM-like and C-terminal domains are in close contact in each of the two structures (24), resembling a closed conformation of the molecule (Fig. 3). Although the details of this interaction differ significantly in H. pylori FlhA and Salmonella InvA (not shown), and are partly compounded by differences in the structure and conformation of the RRM-like domain, the overall mechanism of contact is similar as the α12-β10 linker of the C-terminal β/α domain fits into a cleft between strands β5 and β7 of the RRM-like domain (Fig. 3).
A comparison of the domain movements contributing to the open-closed structural transition suggested by the structures of the three FlhAC homologues reveals that the helical domain shifts relative to the thioredoxin domain by small structural perturbations in the vicinity of helix α5 that links the two halves of FlhAC, resulting in a hinge-like rigid movement of the helical domain (Fig. 3). The C-domain largely moves in conjunction with the helical domain. However, there appears to be a second hinge between helices α10a and α10b at the juncture of the helical and C-terminal domains. The movement of α10b relative to α10a is further amplified by a structural rearrangement of the loop connecting helix α10b to strand β9 at the start of the C-terminal domain in Salmonella FlhA (Figs. 3–4). The α10a-α10b hinge movement is most evident in Salmonella FlhA when compared with H. pylori FlhA (Fig. 4). The orientations of highly conserved salt bridge residues at the interface between the FlhAC helical and C-terminal domains are also different in Salmonella FlhA, as Arg-579 in Salmonella FlhA appears to be partially disordered and does not make stacking interactions with Arg-653 or form hydrogen bonds with Glu-676 (Fig. 4). In H. pylori FlhA (and InvA), the analogous residue, Arg-613, makes stacking interactions with Arg-695 and also makes two hydrogen bonds to Glu-716 (Fig. 4). Hence, the conformations of these highly conserved salt bridge residues bridging the helical and C-terminal domains are essentially identical in the two closed FlhA homologue structures, but different in Salmonella FlhA, which exhibits an open conformation, strongly suggesting that the precise orientations of the conserved salt bridge residues may contribute to the relative positioning of the helical and C-terminal domains and hence possibly to the maintenance of the open/closed conformation of the FlhA molecule.
Insights into FlhA/LcrD Mutational Data from the H. pylori FlhA Structure
A number of genetic and functional studies have been published probing the function of FlhA, and in particular the FlhAC fragment from Salmonella enterica (13, 16–19, 21). A V404M Salmonella flhA point mutation (Gln-439 in Hp FlhA) confers basal levels of motility in a normally non-motile fliH fliI null background (13, 17) and lies on a highly conserved surface patch of the thioredoxin-like domain of FlhAC (Figs. 2 and 3). The flhA V404M mutation is thought to increase the likelihood of the export pore being in an open conformation, based on its ability to restore moderate levels of motility in a ΔfliHΔfliI non-motile background (13). As strains carrying flhA (V404M) are non-motile in a ΔfliI background (FliH inhibits flagellum protein export in the absence of FliI) (13, 17), it is unlikely that this region of FlhA interacts with FliH. This surface of FlhA more likely interacts with an N-terminal cytoplasmic extension of the FlhB transmembrane domain as several FlhB N-terminal mutants have a very similar phenotype to the flhA V404M mutation and residues 1–33 of FlhB most likely extend into the cytoplasm (17). The proximity of Val-404 to the transmembrane linker segment in the three-dimensional structures of Salmonella and H. pylori FlhAC is consistent with this interpretation. Gln-439 (equivalent to Salmonella Val-404) is also near a modest dimer interface in the H. pylori FlhAC crystal structure (supplemental Fig. S5). The significance of the dimer interface in the H. pylori FlhAC crystal is not clear, as the protein elutes as a monomer on analytical gel filtration columns.
Several temperature-sensitive Salmonella FlhAC missense mutants that cannot regrow flagella at the restrictive temperature have been reported (19). These mutants have been extensively analyzed with the recently reported Salmonella FlhAC structure and will not be discussed further (23, 47). However, the positions of the mutations are indicated in Fig. 2, and two of the mutations (St. E547K and E676K) have already been mentioned in relation to the conserved salt bridges at the interface between the helical and C-terminal domains (Figs. 2 and 4).
The phenotypes of 18 20-residue deletions (Δ1:328–347 through to Δ18:668–687) spanning Salmonella FlhAC have also been described (16). All but one of these deletion mutants partially complemented the loss of motility when introduced into a flhA null background. However, the Δ18 deletion, corresponding to removal of the last two β-strands of FlhAC (Figs. 2 and 5) did not restore motility in a flhA null strain (16). The relative severity of the Δ18 deletion suggests the deleted region is critical for FlhA function. Two other Salmonella flhA deletion mutants identified in the same study (Δ6, 428–447, and Δ17, 648–667) were reported to be dominant-negative for motility when introduced into fla+ strains (Fig. 2) (16). Interpretation of the dominant flhA deletions, however, is difficult within the context of the many other non-dominant flhA deletions that would also likely severely disrupt the FlhAC structure.
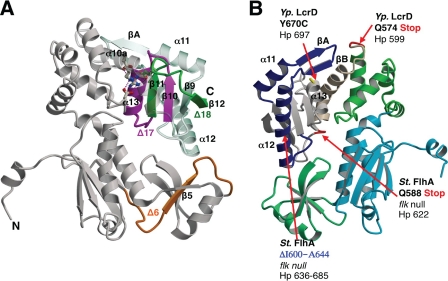
FIGURE 5.
Summary of Salmonella FlhA and Y. pestis LcrD mutants. A, the locations of Salmonella FlhAC Δ6, Δ17, and Δ18 deletions are mapped onto the H. pylori FlhA structure, depicted in a ribbon diagram, and colored according to Fig. 2 (16). B, the positions of nonsense mutations that result in truncation of the C-terminal domain in Salmonella FlhA or Y. pestis LcrD, respectively, are mapped onto the H. pylori FlhAC structure and drawn in red and labeled according to the position in the parent sequence (34, 35, 48). The position of an internal Salmonella FlhA deletion (Δ600–644) that permits early secretion of filament class substrates is also shown in blue (48).
Mutational screens of the Y. pestis type III secretion system have revealed the importance of the FlhA orthologue LcrD in controlling the transcription of other Type III export components, a feature that these proteins have in common with FlhA from H. pylori (27, 28, 33–35). An lcrD nonsense mutation (Q574Stop) truncates the corresponding region in FlhAC at the end of helix α9, hence deleting helices α10a and α10b and the entire C-terminal domain (Figs. 2 and 5) (35). The resultant truncated LcrD protein was defective in export substrate transcription and secretion, presumably through the cytoplasmic retention of the unidentified co-repressor of LcrH (35). An lcrD point mutant with a similar secretion defective phenotype (lcrD Y670C) (35) maps to position 697 on helix α13 of Hp FlhA (Fig. 5).
In the flagellum system, null mutations in the Salmonella flk gene permit premature switching from rod/hook substrates to filament type substrates, either in a fla+ background, or in a non-motile strain that lacks an outer rod and secretes flagellum components directly into the periplasm (48). Two nonsense mutations in FlhA at Gln-588 or Gln-589 (Asn-622 or Ala-623 in Hp FlhA) that cleanly delete the FlhAC C-terminal domain yielded a phenotype similar to the flk null in a temperature-sensitive screen. A third early switching mutant corresponded to an internal deletion of helices α11 through to the end of α12 (ΔI600–A644; ΔL634-I683 in Hp FlhA) (Figs. 2 and 5). The truncated Salmonella FlhA mutants permit early secretion of FlgM (a filament type substrate and anti-σ28 factor) at permissive temperatures, however, secretion of FlgM into the periplasm is not observed at the restrictive temperature (48). These FlhA truncation mutants also disrupt flagellar export when introduced to a fla+ background (48). A surprising feature of the early switching FlhA truncation mutations is that they overlap almost exactly with the Y. pestis LcrD truncation that is defective in type III effector secretion (Figs. 2 and 5) (34).
The phenotype of the flhA early secretion mutations suggests that the C-terminal domain of Salmonella FlhA not only prevents premature secretion of filament class substrates but also facilitates the secretion of rod/hook substrates, as the same mutants typically do not complete rod/hook structures in an otherwise wild type background. Although the flk gene is likely not present in H. pylori or other ϵ-proteobacteria, it is tempting to speculate that the C-terminal domain of H. pylori FlhA will function similarly to Salmonella FlhA in substrate switching and protein export due to the high level of sequence conservation for FlhA, FlhB, and FliK. Furthermore, the finding of similar mutants in Y. pestis LcrD with secretion-defective phenotypes suggest a common mechanism of FlhA homologue function in both flagellar and type III secretions systems. Nevertheless, there are likely important differences for FlhA function in H. pylori, as FlgM remains cytosolic during normal flagellum function, FlhA and FlgM appear to interact with each other, likely via an unknown third partner protein (29), and FlhA or FlgM null mutants have dramatic global effects on flagellum gene expression (27, 28).
Supplementary Material
Acknowledgments
Stacy McDonald carried out the cloning of the FlhAC domain. George Wong and Michael Lane contributed to early protein purification and crystallization efforts of a shorter FlhAC construct not presented in this study. Paul W. O'Toole generously provided H. pylori genomic DNA samples. We thank Michel Fojde, James Gorin, and Pawel Grochulski of the Canadian Macromolecular Crystallography (CMCF) beamline at the Canadian Light Source Synchrotron, Saskatoon, Canada, for timely access, advice, and assistance with x-ray data collection. The Canadian Light Source is funded by the Canadian Institutes for Health Research, the National Science and Engineering Research Council of Canada, the Canadian Foundation for Innovation, and the University of Saskatchewan. Many thanks to Dr. Paul W. O'Toole for providing support and encouragement, and for reading the manuscript.
This work was supported by a Health Sciences Utilization and Research Committee of Saskatchewan establishment grant and National Sciences and Engineering Research Council of Canada Discovery Grant RGPIN 262138-05.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
The atomic coordinates and structure factors (code 3MYD) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- SeMet
- selenomethionine
- GST
- glutathione S-transferase
- BisTris propane
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- r.m.s.
- root mean square
- RRM
- RNA recognition motif
- PDB
- Protein Data Bank.
REFERENCES
- 1.Cover T. L., Blaser M. J. (2009) Gastroenterology 136, 1863–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Veldhuyzen van Zanten S. J., Sherman P. M. (1994) Can. Med. Assoc. J. 150, 177–185 [PMC free article] [PubMed] [Google Scholar]
- 3.O'Toole P. W., Kostrzynska M., Trust T. J. (1994) Mol. Microbiol. 14, 691–703 [DOI] [PubMed] [Google Scholar]
- 4.O'Toole P. W., Lane M. C., Porwollik S. (2000) Microbes Infect. 2, 1207–1214 [DOI] [PubMed] [Google Scholar]
- 5.Eaton K. A., Morgan D. R., Krakowka S. (1992) J. Med. Microbiol. 37, 123–127 [DOI] [PubMed] [Google Scholar]
- 6.Kavermann H., Burns B. P., Angermuller K., Odenbreit S., Fischer W., Melchers K., Haas R. (2003) J. Exp. Med. 197, 813–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aizawa S. I. (1996) Mol. Microbiol. 19, 1–5 [DOI] [PubMed] [Google Scholar]
- 8.Macnab R. M. (2003) Annu. Rev. Microbiol. 57, 77–100 [DOI] [PubMed] [Google Scholar]
- 9.Suzuki H., Yonekura K., Murata K., Hirai T., Oosawa K., Namba K. (1998) J. Struct. Biol. 124, 104–114 [DOI] [PubMed] [Google Scholar]
- 10.Suzuki H., Yonekura K., Namba K. (2004) J. Mol. Biol. 337, 105–113 [DOI] [PubMed] [Google Scholar]
- 11.Thomas D. R., Francis N. R., Xu C., DeRosier D. J. (2006) J. Bacteriol. 188, 7039–7048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jenal U., Shapiro L. (1996) EMBO J. 15, 2393–2406 [PMC free article] [PubMed] [Google Scholar]
- 13.Minamino T., Namba K. (2008) Nature 451, 485–488 [DOI] [PubMed] [Google Scholar]
- 14.Paul K., Erhardt M., Hirano T., Blair D. F., Hughes K. T. (2008) Nature 451, 489–492 [DOI] [PubMed] [Google Scholar]
- 15.Minamino T., Yoshimura S. D., Morimoto Y. V., González-Pedrajo B., Kami-Ike N., Namba K. (2009) Mol. Microbiol. 74, 1471–1483 [DOI] [PubMed] [Google Scholar]
- 16.McMurry J. L., Van Arnam J. S., Kihara M., Macnab R. M. (2004) J. Bacteriol. 186, 7586–7592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Minamino T., González-Pedrajo B., Kihara M., Namba K., Macnab R. M. (2003) J. Bacteriol. 185, 3983–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kihara M., Minamino T., Yamaguchi S., Macnab R. M. (2001) J. Bacteriol. 183, 1655–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saijo-Hamano Y., Minamino T., Macnab R. M., Namba K. (2004) J. Mol. Biol. 343, 457–466 [DOI] [PubMed] [Google Scholar]
- 20.Grünenfelder B., Gehrig S., Jenal U. (2003) J. Bacteriol. 185, 1624–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stone C. B., Bulir D. C., Gilchrist J. D., Toor R. K., Mahony J. B. (2010) BMC Microbiol. 10, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saijo-Hamano Y., Imada K., Minamino T., Kihara M., Macnab R. M., Namba K. (2005) Acta Crystallogr. F Struct. Biol. Cryst. 61, 599–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saijo-Hamano Y., Imada K., Minamino T., Kihara M., Shimada M., Kitao A., Namba K. (2010) Mol. Microbiol. 76, 260–268 [DOI] [PubMed] [Google Scholar]
- 24.Worrall L. J., Vuckovic M., Strynadka N. C. (2010) Protein Sci. 19, 1091–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmitz A., Josenhans C., Suerbaum S. (1997) J. Bacteriol. 179, 987–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Colland F., Rain J. C., Gounon P., Labigne A., Legrain P., De Reuse H. (2001) Mol. Microbiol. 41, 477–487 [DOI] [PubMed] [Google Scholar]
- 27.Josenhans C., Niehus E., Amersbach S., Hörster A., Betz C., Drescher B., Hughes K. T., Suerbaum S. (2002) Mol. Microbiol. 43, 307–322 [DOI] [PubMed] [Google Scholar]
- 28.Niehus E., Gressmann H., Ye F., Schlapbach R., Dehio M., Dehio C., Stack A., Meyer T. F., Suerbaum S., Josenhans C. (2004) Mol. Microbiol. 52, 947–961 [DOI] [PubMed] [Google Scholar]
- 29.Rust M., Borchert S., Niehus E., Kuehne S. A., Gripp E., Bajceta A., McMurry J. L., Suerbaum S., Hughes K. T., Josenhans C. (2009) J. Bacteriol. 191, 4824–4834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ghelardi E., Celandroni F., Salvetti S., Beecher D. J., Gominet M., Lereclus D., Wong A. C., Senesi S. (2002) J. Bacteriol. 184, 6424–6433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carrillo C. D., Taboada E., Nash J. H., Lanthier P., Kelly J., Lau P. C., Verhulp R., Mykytczuk O., Sy J., Findlay W. A., Amoako K., Gomis S., Willson P., Austin J. W., Potter A., Babiuk L., Allan B., Szymanski C. M. (2004) J. Biol. Chem. 279, 20327–20338 [DOI] [PubMed] [Google Scholar]
- 32.Konkel M. E., Klena J. D., Rivera-Amill V., Monteville M. R., Biswas D., Raphael B., Mickelson J. (2004) J. Bacteriol. 186, 3296–3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hueck C. J. (1998) Microbiol. Mol. Biol. Rev. 62, 379–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Plano G. V., Straley S. C. (1993) J. Bacteriol. 175, 3536–3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Plano G. V., Straley S. C. (1995) J. Bacteriol. 177, 3843–3854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Douillard F. P., Ryan K. A., Caly D. L., Hinds J., Witney A. A., Husain S. E., O'Toole P. W. (2008) J. Bacteriol. 190, 7975–7984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 38.Terwilliger T. C., Berendzen J. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Terwilliger T. C. (2000) Acta Crystallogr. D Biol. Crystallogr. 56, 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Terwilliger T. C. (2003) Acta Crystallogr. D Biol. Crystallogr. 59, 38–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 42.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 43.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 44.Winn M. D., Isupov M. N., Murshudov G. N. (2001) Acta Crystallogr. D Biol. Crystallogr. 57, 122–133 [DOI] [PubMed] [Google Scholar]
- 45.Collaborative Computational Project, No. 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 46.Holm L., Kääriäinen S., Rosenström P., Schenkel A. (2008) Bioinformatics 24, 2780–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Minamino T., Shimada M., Okabe M., Saijo-Hamano Y., Imada K., Kihara M., Namba K. (2010) J. Bacteriol. 192, 1929–1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hirano T., Mizuno S., Aizawa S., Hughes K. T. (2009) J. Bacteriol. 191, 3938–3949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kraulis P. (1991) J. Appl. Crystallogr. 24, 946–950 [Google Scholar]
- 50.Merritt E. A., Bacon D. J. (1997) Methods Enzymol. 277, 505–524 [DOI] [PubMed] [Google Scholar]
- 51.Notredame C., Higgins D. G., Heringa J. (2000) J. Mol. Biol. 302, 205–217 [DOI] [PubMed] [Google Scholar]
- 52.Gouet P., Courcelle E., Stuart D. I., Métoz F. (1999) Bioinformatics 15, 305–308 [DOI] [PubMed] [Google Scholar]
- 53.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.