Abstract
Versican/PG-M is a large chondroitin sulfate proteoglycan in the extracellular matrix, which is transiently expressed in mesenchymal condensation areas during tissue morphogenesis. Here, we generated versican conditional knock-out mice Prx1-Cre/Vcanflox/flox, in which Vcan is pruned out by site-specific Cre recombinase driven by the Prx1 promoter. Although Prx1-Cre/Vcanflox/flox mice are viable and fertile, they develop distorted digits. Histological analysis of newborn mice reveals hypertrophic chondrocytic nodules in cartilage, tilting of the joint, and a slight delay of chondrocyte differentiation in digits. By immunostaining, whereas the joint interzone of Prx1-Cre/Vcan+/+ shows an accumulation of TGF-β, concomitant with versican, that of Prx1-Cre/Vcanflox/flox without versican expression exhibits a decreased incorporation of TGF-β. In a micromass culture system of mesenchymal cells from limb bud, whereas TGF-β and versican are co-localized in the perinodular regions of developing cartilage in Prx1-Cre/Vcan+/+, TGF-β is widely distributed in Prx1-Cre/Vcanflox/flox. These results suggest that versican facilitates chondrogenesis and joint morphogenesis, by localizing TGF-β in the extracellular matrix and regulating its signaling.
Keywords: Development, Glycosaminoglycan, Proteoglycan Structure, Proteoglycan Synthesis, Transforming Growth Factor Beta (TGFbeta), Cartilage Development, Chondrocyte Differentiation, Joint Morphogenesis, Versican
Introduction
Versican/PG-M (1) consists of a core protein and chondroitin sulfate (CS)3 chains attached to the core protein. The core protein is composed of an N-terminal G1 and a C-terminal G3 globular domain and two CS-attachment domains CS-α and CS-β between the G1 and G3 domains. The N-terminal G1 domain comprises the A, B, and B′ subdomains and binds to both hyaluronan (HA) and link protein (2). The C-terminal G3 domain binds fibrillin-1 (3), fibulin-1 and -2 (4, 5), tenascins (4, 6), and heparan sulfate proteoglycans (7). Versican exhibits four spliced variants: V0, V1, V2, and V3 (8–11), with different CS domains. The V0 variant contains all of the domains G1, CS-α, CS-β, and G3; V1 contains G1, CS-β, and G3; V2 contains G1, CS-α, and G3; and V3 contains only G1 and G3. Interestingly, V0 and V1 are expressed widely, V2 is restricted to the nervous systems, and V3 has not been detected as a protein, although mRNA is detected. Thus, the number of CS chains required for the function of versican may vary among tissues. Versican is also characterized by two distinct expression patterns. In some adult tissues, such as heart, blood vessels, and brain, it is constitutively expressed, serving as a structural macromolecule of the ECM. In embryonic stages, it is transiently expressed in various developing tissues (12), including brain, hair follicles, developing heart, and mesenchymal condensation areas of cartilage primordium. Previous in vitro studies have revealed various effects of versican on cell behavior (13). For example, it inhibits cell adhesion of MG63 osteosarcoma cells and aggravates their malignant phenotype (14). It inhibits migration of neural crest cells and the outgrowth of motor and sensory axons (15). It is expressed in dermal papilla, a dense aggregate of dermis-derived stromal cells, suggesting its involvement in hair follicle formation (16). The fact that versican is transiently expressed at high levels in areas where cells are aggregating to a high density suggests that this proteoglycan plays a crucial role in cell aggregation that leads to tissue morphogenesis.
Cartilage development initiates from mesenchymal condensations where mesenchymal cells aggregate and further differentiate into chondrocytes. During mesenchymal condensation, there is an increased expression of ECM and cell surface molecules, such as versican, tenascin, syndecans, and N-CAM (17). With synchronized collaboration of these molecules, transforming growth factor-β (TGF-β) (18), bone morphogenetic protein (19), and GDF-5 (growth differentiation factor-5) (20) lead mesenchymal cells to commitment into the chondrocytic lineage and differentiation toward chondrocytes, which synthesize ECM molecules specific to cartilage, including type II collagen and aggrecan. During these processes, synovial joints are generated with packing and lining of mesenchymal cells at the future joint location termed the interzone (21–23). These mesenchymal cells are cells that are still undifferentiated (24) or derived from dedifferentiation of chondrocytes (25). Several signaling pathways are implicated in the molecular specification of the joint interzone. Wnt pathways mediated by Wnt-4, Wnt-14, and Wnt-16 (26, 27) have been shown to participate in joint formation, and another study indicates that the Wnt/β-catenin canonical signaling pathway is necessary and sufficient for the induction of synovial joints in the limb (28). Other studies reported that CD44-HA signaling is involved in synovial joint cavitation (29). Analysis of conditional null mice of TGF-β receptor II (TβRII) has demonstrated an essential role of TGF-β signaling in joint morphogenesis (30). During the process of cartilage development and synovial joint formation, versican exhibits dynamic expression patterns. Its expression initiates at a high level in mesenchymal condensation areas. While the cells differentiate into chondrocytes, it remains in pericondensation areas. When the joint is formed by accumulation and lining up of mesenchymal cells, versican is expressed in the joint interzone. After the formation of the joint cavity, versican is present in the articular cartilage and synovial tissue, lining the inside margin of the cavity (31, 32). These characteristic expression patterns of versican and its effects on cell behavior observed in cell culture systems strongly suggest that versican regulates, in the ECM, the function of ligands that mediate signaling toward cartilage development and synovial joint formation. An in vitro analysis of limb mesenchyme from gene-trapped versican-deficient mouse hdf (heart defect) by micromass indicated that versican is necessary for chondrocyte differentiation of mesenchymal cells (33). Our previous studies using N1511 chondrocytic cells demonstrated that versican is required for the mesenchymal matrix formation toward chondrocyte differentiation during chondrogenesis (34). However, the signaling pathways affected by versican have not been identified, and the mechanisms by which versican regulates their signal transduction toward cartilage development and joint formation remain to be elucidated.
To investigate the in vivo role of versican in cartilage development and joint formation, we generated versican conditional knock-out mice Prx1-Cre/Vcanflox/flox, in which Vcan is pruned out by site-specific Cre recombinase driven by the Prx1 promoter. Because Prx1 is expressed in limb mesenchyme, Prx1-Cre mice are widely used for the generation of conditional knock-out mice that enable in vivo analysis of molecules during limb development (35). Although Prx1-Cre/Vcanflox/flox mice are viable and fertile, they develop distorted digits. Histological analysis of newborn mice reveals 1) hypertrophic chondrocytic nodules in cartilage, 2) tilting of the joint, and 3) a slight delay of chondrocyte differentiation in limbs. We further observed decreased incorporation of TGF-β in the joint interzone and its altered distribution in micromass of Prx1-Cre/Vcanflox/flox. Our study provides evidence that versican regulates TGF-β-mediated signaling by localizing TGF-β in the ECM.
EXPERIMENTAL PROCEDURES
Generation of Conditional Vcan Knock-out Mice
To generate mice in which the Vcan gene is deleted in early limb mesenchyme, the conditional knock-out mice were generated by combining Cre/loxP with a Flp/FRT system as follows. A targeting vector harboring the Vcanflox allele was constructed by flanking exon 2 of the mouse Vcan gene with loxP sites in the combination of a PGK-neoR cassette flanked by the FRT sequence. Then mouse ES cells were electroporated with linearized targeting construct and cultured with G418 to positively select for clones that had integrated the targeting construct. Clones were screened by genomic PCR, and positive clones were confirmed by Southern blotting. After blastocyst injection and homologous recombination, chimeric mice whose genomic DNA contained the Vcanflox allele were obtained. Germ line transmission was attained by crossing these chimeric mice with C57BL/6 mice. Then, by crossing with CAG-flippase transgenic (Tg) mice, Vcan+/flox mice whose genomic DNA lacks the PGK-neoR fragment were obtained. We crossed these mice with C57BL/6 to segregate the CAG-flippase transgene and back-crossed to C57BL/6 at least four times (n ≥ 4). Then by crossing Vcan+/flox mice with Prx1-Cre Tg mice with a background of C57BL/6, Prx1-Cre/Vcan+/flox mice were obtained. Prx1-Cre/Vcan+/flox male and female mice were crossed to obtain Prx1-Cre/Vcanflox/flox mice whose Vcan gene was removed by Cre-mediated excision. The genotyping of transgenic mice was performed by PCR using DNA template from proteinase K digestion of a tail biopsy. WT and floxed alleles were identified by PCR using a forward primer of Int1–1 sequence 5′-TGAGCTCTAGCCATATAGGAAGGC- 3′ and a reverse primer of Kpn-1 sequence 5′-CGCATGCAGACGACATGAAGCAGGAGC-3′, generating a PCR product of 3 kb in wild-type and 2 kb in floxed alleles. Genomic DNA was amplified for 25 cycles of denaturation at 94 °C for 30 s, annealing at 62 °C for 30 s, and elongation for 5 min at 72 °C in reaction buffer containing 2 mm MgCl2, 1× Ex Taq buffer (10× Ex Taq buffer, Takara), 0.25 mm dNTPs (dNTP mixture, Takara), 0.2 μm each primer. PCR for Cre transgene was performed using the primer set of Prx1-Cre forward (5′-CCTGGAAAATGCTTCTGTCCGTTTGCC-3′) and Prx1-Cre reverse (5′-GAGTTGATAGCTGGCTGGTGGCAGATG-3′), which generated a PCR product of 620 bp.
Analysis of Prx1-Cre Activity
ROSA26 (Jackson Laboratory, Bar Harbor, ME) reporter mice were first crossed with the Prx1-Cre transgenic line to obtain the Prx1-Cre/R26R founders. Then Prx1-Cre/R26R were bred together to generate Prx1-Cre/R26R embryos at several stages. These embryos were then used for analysis.
X-ray Analysis and Histological Analysis
X-ray analysis was performed using a soft x-ray apparatus (Softex, Tokyo, Japan). Newborn mice or embryos from several embryonic stages were fixed in 10% neutral buffered formalin. Hind limbs were dissected and embedded in paraffin, 4- or 5-μm-thick paraffin sections were cut and mounted on Superfrost Mascot slides (Matsunami Glass Inc., Osaka, Japan), sections were put in a 37 °C incubator overnight and stored at room temperature, and then hematoxylin and eosin staining was performed.
Cryosectioning and X-gal Staining
Hind limbs from various embryonic stages were dissected and put into cold 1× phosphate-buffered saline (PBS) containing 2 mm MgCl2, and then fixed in 4% paraformaldehyde at 4 °C for 2–3 h. They were then washed with PBS twice, soaked in the gradient of sucrose solution at concentrations of 15 and 30% at 4 °C overnight or until they sank. They were briefly rinsed with PBS and then embedded in optimal cutting temperature compound and kept at −80 °C. Cryosections were cut at a thickness of 15 μm. The slides were fixed in 4% paraformaldehyde at 4 °C for 10–15 min and then washed twice with PBS at 4 °C. Slides were stained in X-gal solution (1 mg/ml X-gal, 5 mm K3Fe(CN)6, 5 mm K4Fe(CN)6, 2 mm MgCl2, 0.01% deoxycholate, 0.02% Nonidet P-40) at 37 °C overnight. After staining, they were washed in PBS and counterstained with eosin; dehydrated in ethanol, xylene; and mounted.
After 3 days of micromass culture (as described below), cultures were washed three times with PBS, fixed with 2% formaldehyde for 5 min, washed twice with PBS, and stained in staining solution (0.1 m citric acid, 0.2 m sodium phosphate, pH 6.0, 5 mm K3Fe(CN)6, 5 mm K4Fe(CN)6, 150 mm NaCl, 2 mm MgCl2) at 37 °C overnight.
Immunostaining and Hyaluronan Detection
Sections were deparaffinized in xylene, rehydrated in a gradient of ethanol, and briefly washed with PBS. Then endogenous peroxidase was inactivated through incubation in 3% H2O2 in methanol for 20 min. Pretreatment with chondroitinase ABC (5 milliunits/μl; Seikagaku, Tokyo, Japan) for 30–45 min was required for versican and aggrecan. Antigen retrieval with citrate buffer, pH 6.0, by autoclave for 20 min was also essential for CD44, β-catenin, and Cre immunostaining. Then they were treated with blocking solution (Dako) at room temperature for 1 h and incubated with primary antibodies, including anti-versican glycosaminoglycan (GAG) β-domain (Chemicon) at 1:1000, anti-aggrecan clone 1C6 at 1:20, anti-link protein clone 8A4 at 1:100, anti-CD44 (Chemicon) at 1:250, anti-β-catenin (Epitomics) at 1:500, anti-Cre (Novagen) at 1:2000, anti-TGF-β (R&D Systems) at 1:20, anti-TβRII (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) at 1:250, and anti-phospho-Smad2/3 (Cell Signaling) at 1:50. After washing, they were incubated with secondary antibodies consisting of LSAB2 kits (Dako) for colorimetric reaction or Alexa Fluor 594 streptavidin and Alexa Fluor 594 anti-rabbit IgG (Invitrogen) at 1:1000 for fluorescent detection. Images were photographed using an Olympus BX50 microscope and Olympus DP71 digital camera. For detection of HA, sections were incubated with biotinylated hyaluronan-binding protein (HABP; Seikagaku Biobusiness) at 1:500 at 4 °C overnight, followed by incubation with biotinylated streptavidin (Dako) for colorimetric reaction or Alexa Fluor 594 streptavidin (Invitrogen) at 1:1000 for fluorescent detection. HABP is used to detect HA. Because HA is masked by a large amount of chondroitin sulfate of aggrecan, pretreatment with chondroitinase ABC is usually necessary to detect HA in cartilage. Without the pretreatment, HABP mainly binds to HA, which is less incorporated or not incorporated into the matrix. To detect HA in the joint interzone, chondroitinase ABC pretreatment was not performed.
Micromass Culture
To generate cultures missing Vcan, embryos (n = 13) at E10.5 or E11.5 obtained from crossing Prx1-Cre/Vcanflox/flox × Vcanflox/flox were used, and to investigate Prx1 activity, embryos obtained from crossing Prx1-Cre/R26R × Prx1-Cre/R26R were used. Similarly, embryos (n ≥ 33) at E10.5 or E11.5 obtained from crossing Prx1-Cre/Vcan+/+ × Prx1-Cre/Vcan+/+ were used as a control. Limb micromass culture was carried out as described previously by Daniels et al. (47) with some modifications. Briefly, limb buds were removed and digested with 0.1% trypsin, 0.1% collagenase at 37 °C for 20 min to dissociate mesenchymal cells into a single cell suspension. Then their cell density was adjusted to 2 × 107 cells/ml, and 10 μl of the cell suspension was plated onto LabTek-II chamber slides (Nalge Nunc International, Tokyo, Japan) or 35-mm culture dishes. After cells were allowed to attach at 37 °C for 1 h, cultures were then flooded with Dulbecco's modified Eagle's medium/Ham's F-12 medium (Sigma) containing 10% fetal bovine serum. Cultures were incubated at 37 °C in a CO2 incubator for 6 days with fresh conditioned medium added daily. Following incubation, cultures were processed for Alcian blue staining, HA staining, or immunostaining. For double immunofluorescent staining, the sample was co-incubated with rabbit anti-versican glycosaminoglycan (GAG) β-domain (1:1000; Chemicon) and mouse anti-TGF-β (1:20; R&D Systems) for 4 °C overnight and, after washing with PBS, with a mixture of Alexa Fluor 594 anti-rabbit IgG (1:400) and Alexa Fluor 488 anti-mouse IgG (1:400). The photos of micromass culture were taken by a confocal laser-scanning microscope (LSM 710 Carl Zeiss MicroImaging, Tokyo, Japan). To precisely evaluate the localizations of molecules, the Z-stack program was applied. Immunostaining for CS chains was performed using anti-CS (LY111, 1:200; Seikagaku) and Alexa Fluor 488 anti-mouse IgM (1:400) as primary and secondary antibody, respectively.
Western Blot Analysis
Three micromasses at day 6 were collected, and the sample was subjected to 15% SDS-PAGE under a non-reducing condition. The proteins were electrotransferred to a polyvinylidene difluoride membrane, and the membrane was soaked in 5% skim milk in TBS containing 0.1% Tween 20 (TBS-T) for blocking. The membrane was treated with mouse anti-TGF-β (R&D Systems, 1:250) for 1 h at room temperature. After washing three times with TBS-T, the membrane was treated with peroxidase-conjugated goat anti-mouse IgG (1:1000) for 1 h at room temperature. After washing three times as above, the signal was detected with Western LightningTM Plus-ECL (PerkinElmer Life Sciences). The data were analyzed using an LAS 4000 mini, luminescent image analyzer (Fujifilm). For dilution of antibodies, Can Get SignalTM (Toyobo Life Science, Osaka) was used. The membrane was treated with RestoreTM Western blot stripping buffer (Pierce) and used for immunoblot analysis of actin. The band density was analyzed using ImageJ.
Alcian Blue Staining
Whole limbs from embryos at E13.5 and E15.5 days of gestation were fixed in 96% ethanol overnight and then placed in Alcian blue staining solution (0.1% Alcian blue, 70% ethanol, 1% HCl) for 2–3 days and dehydrated in 100% ethanol for 5 days, followed by maceration in 1% KOH for 3–4 days for close observation. After that, they were cleared in 25, 50, and 80% glycerol in distilled water for 1 day for each step and stored in 100% glycerol.
Alcian blue staining was carried out on day 6 of micromass culture as follows. Cultures were washed twice with cold PBS, fixed in 100% ethanol for 5 min, and incubated with Alcian blue solution overnight. Excess stain was washed off with deionized distilled water, and photos were taken using a Zeiss Stemi SV11 microscope with a Nikon CoolPix 995 digital camera or an Olympus SZX12 microscope with an Olympus DP12 camera. The staining density was analyzed using ImageJ.
RESULTS
Generation of Prx1-Cre/Vcanflox/flox Mice
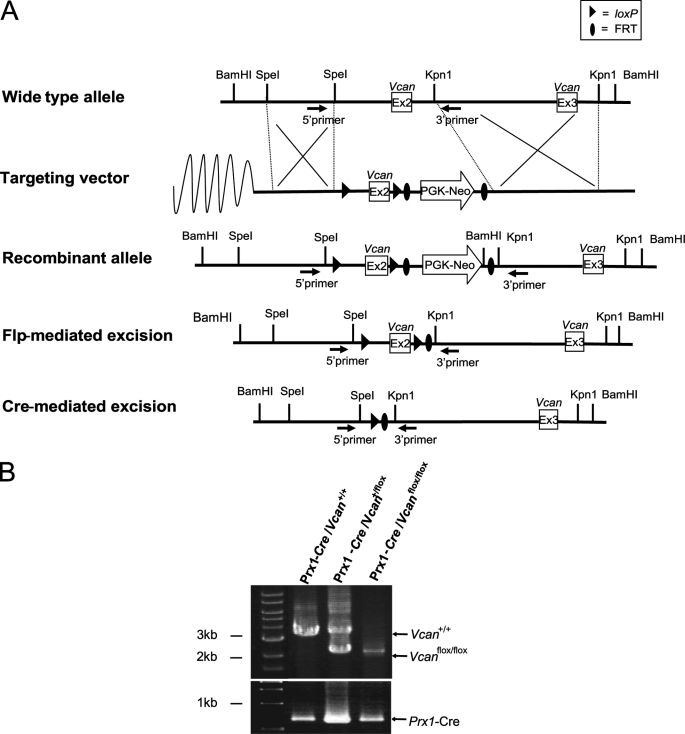
We designed a conditional targeting vector for the versican gene Vcan, in which exon 2 was flanked by the loxP sequence (Fig. 1A). The targeting vector contained a PGK-neoR cassette flanked by the FRT sequence. We transfected ES cells with the vector. After selection with G418 and screening by genomic PCR and Southern blot analyses, we obtained five ES cell clones with homologous recombination. After generation of chimeric mice by blastocyst injection, they were crossed with wild-type mice, and offspring mice with germ line transmission were obtained. Then we crossed them with CAG-flippase Tg mice and obtained Vcan+/flox mice whose genomic DNA lacks the PGK-neoR cassette. These mice were back-crossed with C57BL/6 mice to segregate the CAG-flippase transgene and further back-crossed to C57BL/6 mice at least four times (n ≥ 4). Then, by crossing Vcan+/flox male and female mice, we obtained Vcanflox/flox mice. Both Vcan+/flox and Vcanflox/flox mice were healthy and fertile.
FIGURE 1.
Genomic construct for conditional deletion of the Vcan mice. A, the wild-type Vcan locus is depicted on the top line. A Vcan targeting vector was constructed by flanking exon 2 with loxP sites, and flanking a PGK-neoR cassette with FRT sites (Targeting vector). ES cell clones with homologous recombination of the vector segment (Recombinant allele) were obtained by positive selection and used for generation of chimera mice, followed by germ line transmission. The FRT-flanked PGK-neoR cassette was subsequently deleted from the recombinant allele by crossing with CAG-Flp Tg mice. These mice, termed Vcanflox/flox, were crossed with Prx1-Cre Tg mice. These mice have cells with the null allele of Vcan, in regions where Prx1 promoter is and was active. B, genotyping was performed by PCR analysis on DNA prepared from a tail biopsy using the primers shown by black arrows. The WT (Vcan+/+) PCR product is 3 kb, and the flox (Vcanflox/flox) product without PGK-neoR cassette is 2 kb. The Prx1-Cre transgene was detected by PCR using another primer set. The reaction product (∼620 bp) is shown.
Versican is transiently expressed at a high level in the mesenchymal condensation areas of cartilage primordium. There, a transcription factor, Prx-1, is also expressed at a high level (35). We crossed Vcan+/flox with Prx1-Cre Tg mice with the background of C57BL6 and obtained Prx1-Cre/Vcan+/flox mice. Then, by crossing these mice, we obtained Prx1-Cre/ Vcanflox/flox mice, which were supposed to lack versican expression in the mesenchymal condensation areas, where Prx1 promoter activity is present.
Genotyping of the mice was performed by genomic PCR, as described under “Experimental Procedures.” The bands at 3 and 2 kb represent the WT (Vcan+/+) and Vcanflox/flox allele, respectively (Fig. 1B).
Prx1-Cre/Vcanflox/flox Mice Exhibit Distortion of Digits
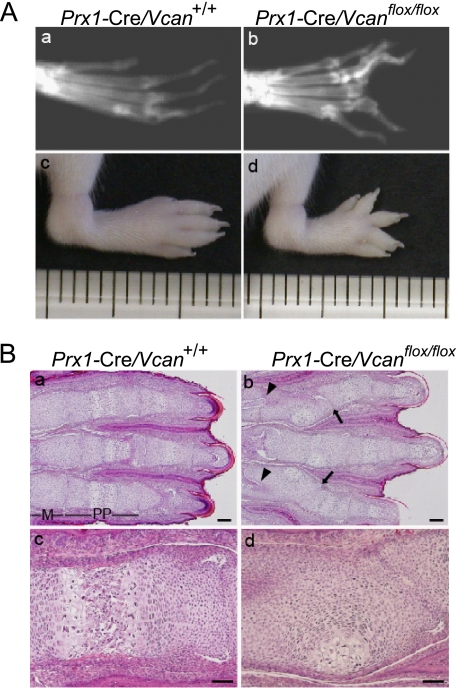
Prx1-Cre/Vcanflox/flox mice grew apparently normal by gross appearance and were fertile, but all of the Prx1-Cre/Vcanflox/flox mice displayed distorted digits in the hind limbs. No craniofacial anomalies were detected in Prx1-Cre/Vcanflox/flox mice, although Prx1 is expressed in craniofacial tissues and central nervous systems (35, 36). X-ray examination of adult mice revealed distortion of bones, especially in the proximal phalanges (Fig. 2, A (b)). By gross observation, the distortion became obvious as early as 1 week of age and was accompanied with limb shortening (Fig. 2A (d)). In contrast, distortion was not obvious in the forelimbs by gross appearance.
FIGURE 2.
Conditional removal of Vcan in limb bud mesenchyme leads to limb deformities. A, x-ray visualization of 8-month-old mice (a and b) and gross observations of 1-week-old mouse hind limbs (c and d). Prx1-Cre/ Vcanflox/flox mice (b and d) show distortion and shortening of digits, compared with Prx1-Cre/Vcan+/+ mice (a and c). B, histological analysis of newborn Prx1-Cre/Vcan+/+ (a and c) and Prx1-Cre/Vcanflox/flox (b and d) hind limbs by hematoxylin and eosin staining. Prx1-Cre/Vcanflox/flox digits display joint tilting (arrows in b) and clefting (arrowheads in b), formation of hypertrophic chondrocyte nodules in proximal phalanges, and delayed endochondral ossification (d). M, metatarsus; pp, proximal phalanges. Scale bars, 200 μm (a and b) and 40 μm (c and d). All of the histological sections (n ≥ 5) of Prx1-Cre/Vcanflox/flox digits show similar abnormalities.
The distortion might have derived from abnormality of bone, cartilage, ligament, and tendon. To determine which region was responsible for the phenotype, we performed histological analyses. Newborn hind limbs of WT showed good alignment of metatarsus and phalanges, which contain a well organized structure of differentiating chondrocytes (Fig. 2B, a and c). In contrast, Prx1-Cre/Vcanflox/flox mice showed distorted digits, which contained nodules of hypertrophic chondrocytes surrounded by proliferative and prehypertrophic chondrocytes (Fig. 2B, b and d). The joint surface between phalanges was tilted (arrow), presumably due to altered orientation of chondrocyte columns. In addition, the proximal region of proximal phalanges, adjacent to the metatarsophalangeal (MP) joint, exhibited cleft formation (arrowhead). These abnormalities were observed with complete penetrance in more than 48 mutant mice examined. Although gross observations did not disclose abnormalities in forelimbs, histological analyses revealed aberrant nodules of hypertrophic chondrocytes in digits and tilting and clefting of the metacarpophalangeal joint, similar to that in hind limbs (supplemental Fig. S1). Because the deformity was more obvious in hind limbs, we decided to investigate hind limbs.
Prx1-Cre/Vcanflox/flox Digits Exhibit Tilted Joint Surface and Delayed Cartilage Development
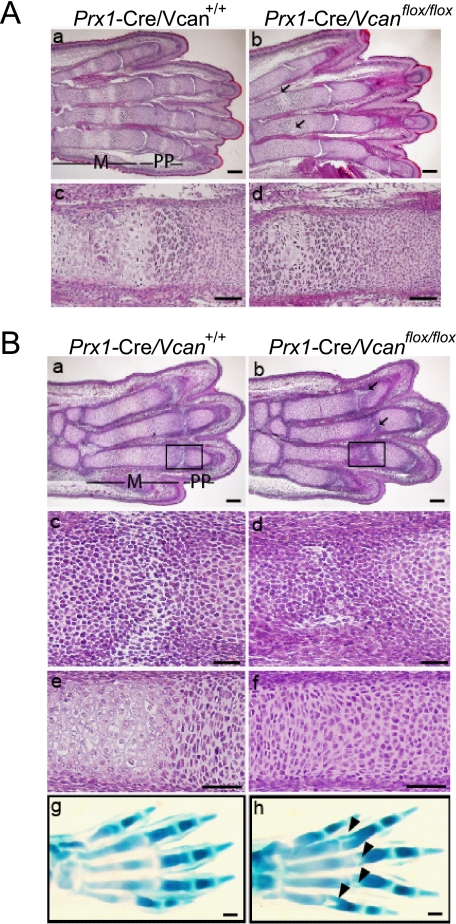
To determine the initiation of the deformities, we performed histological analysis on embryonic stages. At E18.5, metatarsus of Prx1-Cre/Vcan+/+ showed well organized chondrocyte columns in the growth plate and the primary ossification center with vascular invasion (Fig. 3A, a and c). In contrast, that of Prx1-Cre/Vcanflox/flox had an aberrant nodule of hypertrophic chondrocytes surrounded by prehypertrophic and proliferative chondrocytes in a concentric pattern, and it had not yet exhibited vascular invasion. At this stage, clefting of some metatarsophalangeal joints was seen (Fig. 3A, b and d).
FIGURE 3.
Prx1-Cre/Vcanflox/flox digits exhibit tilted joint and delayed cartilage development. A, histological analysis of E18.5 Prx1-Cre/Vcan+/+ (a and c) and Prx1-Cre/Vcanflox/flox (b and d) hind limbs by hematoxylin and eosin staining. Prx1-Cre/Vcan+/+ digits demonstrate a well organized columnar structure of prehypertrophic chondrocytes with vascular invasion in metatarsus (m) and proximal phalanges (pp), whereas Prx1-Cre/Vcanflox/flox digits exhibit formation of hypertrophic chondrocyte nodules in a concentric pattern in proximal phalanges (pp) (arrows in b) and delayed endochondral ossification of metatarsus (d). In addition, clefting of some metatarsophalangeal joints is observed (b). Scale bars, 100 μm (a and b) and 40 μm (c and d). B, histological analysis of E15.5 Prx1-Cre/Vcan+/+ and Prx1-Cre/Vcanflox/flox hind limbs by hematoxylin/eosin and Alcian blue staining. Prx1-Cre/Vcan+/+ digits display horizontal stripes of the metatarsophalangeal joint interzones (a and c (as an enlarged image of the boxed area in a)). Prx1-Cre/Vcan+/+ metatarsus shows prehypertrophic and hypertrophic chondrocyte layers (e). In contrast, Prx1-Cre/Vcanflox/flox digits display tilted joint interzones (b and d (as an enlarged image of the boxed area in b)). Prx1-Cre/Vcanflox/flox metatarsus shows prehypertrophic chondrocytes but not hypertrophic chondrocytes (f). Alcian blue staining of Prx1-Cre/Vcan+/+ hind limb shows well aligned stripes of the interzone (g), whereas that of Prx1-Cre/Vcanflox/flox digits reveals tilting of the joint interzone (indicated by arrowheads in h). Scale bars, 150 μm (a and b), 30 μm (c and d), 50 μm (e and f), and 70 μm (g and h). The number of embryos analyzed was as follows: n = 2 each at E18.5; n = 3 and 5 for Prx1-Cre/Vcanflox/flox and Prx1-Cre/Vcan+/+ at E15.5, respectively). At least two paraffin-embedded blocks were obtained, and their tissue sections were used for histological analysis.
At E15.5, Prx1-Cre/Vcan+/+ digits exhibited a horizontally oriented interzone, forming a joint cavity (Fig. 3B, a and c), whereas Prx1-Cre/Vcanflox/flox digits exhibited a broader interzone of small mesenchymal cells, giving rise to a wedge-shaped or vertical/longitudinal cavity (Fig. 3B, b and d). In addition, their metatarsus contained no hypertrophic chondrocytes yet (Fig. 3B (f)), contrasting with Prx1-Cre/Vcan+/+, which already contained a hypertrophic cell mass (Fig. 3B (e)). At this stage, cleft formation was obvious in the proximal region of proximal phalanges (Fig. 3B, b and h (arrowhead)).
At E14.5, we observed the joint interzone, defined as the area that consists of compact and closely associated mesenchymal cells located along the presumptive joint location (24, 25), but not a clear joint cavity yet. Hematoxylin and eosin staining exhibited no obvious differences between Prx1-Cre/Vcan+/+ and Prx1-Cre/Vcanflox/flox in the joint interzone, although there were no clear joint interzone stripes in Prx1-Cre/Vcanflox/flox, compared with Prx1-Cre/Vcan+/+ embryos (supplemental Fig. S2A). Immunostaining for Ki67, a protein expressed in the growing phase, showed a similar level of proliferation in cartilage between Prx1-Cre/Vcan+/+ and Prx1-Cre/Vcanflox/flox. Taken together, Prx1-Cre/Vcanflox/flox digits exhibited two abnormalities, delayed chondrocyte differentiation, as observed by a decrease in hypertrophic cells, and tilting of joint surface, which initially appear at E15.5. Formation of hypertrophic chondrocyte nests at E18.5 is probably due to a shift of the columnar axis by tilting of the joint surface.
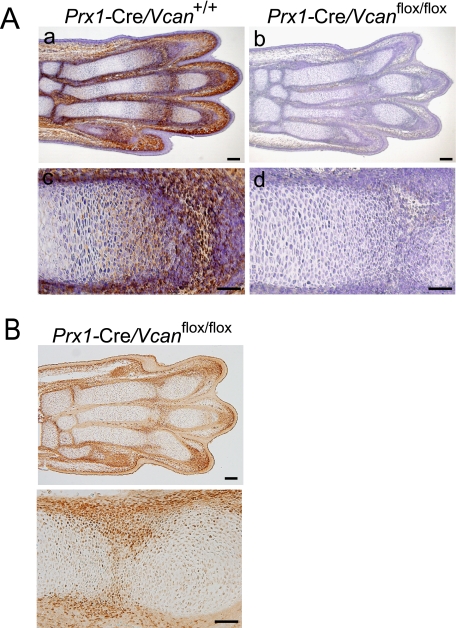
Versican Distribution in Mice Digits
Next, we examined expression patterns of versican at E15.5, when abnormalities initiated. Whereas versican was strongly immunostained in the joint interzone and perichondrium and moderately immunostained in the proliferative zone of cartilage in Prx1-Cre/Vcan+/+ (Fig. 4A, a and c), it was not immunostained in the interzone, perichondrium, and cartilage of Prx1-Cre/ Vcanflox/flox (Fig. 4A, b and d). The regions lacking versican specifically in Prx1-Cre/Vcanflox/flox were supposed to be the regions where Cre enzyme was or had been expressed. When immunostained, Cre enzyme was observed in the perichondrium and the interzone in Prx1-Cre/Vcanflox/flox (Fig. 4B). By X-gal staining at E14.5 of Prx1-Cre/R26R, β-galactosidase activity was found in chondrocytes as well as in perichondrium (supplemental Fig. S2B). These observations support the lack of versican expression in Prx1-Cre/Vcanflox/flox.
FIGURE 4.
Versican is absent in Prx1-Cre/Vcanflox/flox digits. A, immunostaining for versican of E15.5 Prx1-Cre/Vcan+/+ and Prx1-Cre/Vcanflox/flox hind limbs. In Prx1-Cre/Vcan+/+ digits, versican is strongly immunostained in the interzone and perichondrium and moderately immunostained in the proliferative zone of cartilage (a and c), whereas in Prx1-Cre/Vcanflox/flox digits, versican is immunostained neither in the interzone, the perichondrium, nor the proliferative zone of cartilage (b and d). Scale bars, 120 μm (a and b) and 70 μm (c and d). B, immunostaining for Cre enzyme of E15.5 Prx1-Cre/Vcanflox/flox hind limbs at low (top) and high (bottom) magnifications. Cre enzyme is immunostained in the perichondrium and in the tilted interzone. Scale bars, 100 and 50 μm, respectively. Two individual Prx1-Cre/Vcanflox/flox digits showed the same abnormalities.
Distribution of Hyaluronan and Its Binding Molecules in the Joint
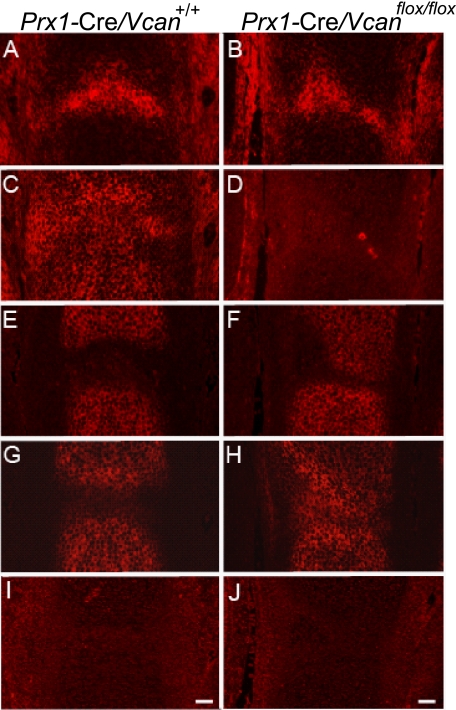
Whereas HA is bound to both aggrecan and link protein and profoundly incorporated as the proteoglycan aggregate in cartilage, it is also accumulated in the joint interzone, which contributes to joint formation. Recent studies suggested that the accumulated HA mediates signals via CD44 toward joint cavity formation (29). Because our immunostaining demonstrated the presence of versican in the interzone at E15.5 of Prx1-Cre/Vcan+/+ and its absence in Prx1-Cre/Vcanflox/flox, we speculated that versican is necessary for adequate levels of HA accumulation and HA-mediated signaling toward joint formation and that its absence caused the joint abnormality. By the detection method using biotinylated HABP, accumulation of HA was observed at a similar level in the interzone of both Prx1-Cre/Vcan+/+ and Prx1-Cre/Vcanflox/flox (Fig. 5, A and B). Immunofluorescent staining displayed the presence of versican in the interzone, future articular surface, and perichondrium in Prx1-Cre/Vcan+/+ and its absence in Prx1-Cre/ Vcanflox/flox (Fig. 5, C and D). Immunofluorescent staining for aggrecan that clearly demarcates cartilage tissue from the interzone revealed clefting of the future joint surface in Prx1-Cre/Vcanflox/flox (Fig. 5, E and F). Whereas link protein was colocalized with aggrecan in Prx1-Cre/Vcan+/+, it was also present in the interzone in Prx1-Cre/Vcanflox/flox (Fig. 5, G and H). These observations suggest that whereas versican retains HA in the interzone, link protein does so in the absence of versican. CD44 was not detected at E15.5, eliminating involvement of CD44 in the joint formation at this embryonic stage (Fig. 5, I and J).
FIGURE 5.
Distribution of hyaluronan in the joint interzone is unaffected by the absence of versican. Distributions of HA and HA-binding molecules in the metatarsophalangeal joint interzone at E15.5 of Prx1-Cre/Vcan+/+ (A, C, E, G, and I) and Prx1-Cre/Vcanflox/flox (B, D, F, H, and J) are shown. By detection of HA using b-HABP, unincorporated HA is accumulated at a similar level in the joint interzone of both Prx1-Cre/Vcan+/+ and Prx1-Cre/Vcanflox/flox (A and B). Immunofluorescent staining confirms the presence of versican in the interzone, future articular surface, and perichondrium in Prx1-Cre/Vcan+/+ and its absence in Prx1-Cre/Vcanflox/flox (C and D). Immunofluorescent staining for aggrecan, demarcating the cartilage tissue, demonstrates tilting of the joint surface in Prx1-Cre/Vcanflox/flox (E and F). Link protein (LP) is immunostained in cartilage of Prx1-Cre/Vcan+/+ (G), whereas it is immunostained in both cartilage and the joint interzone of Prx1-Cre/Vcanflox/flox (H). CD44 is not detected in both Prx1-Cre/Vcan+/+ and Prx1-Cre/Vcanflox/flox at this stage (I, J), eliminating the possibility of its participation in joint cavitation. Scale bars: 30 μm. Immunostaining was performed on at least two individual digits for Prx1-Cre/Vcan+/+ and Prx1-Cre/Vcanflox/flox, with the same immunostaining patterns.
TGF-β Signaling Is Attenuated in Prx1-Cre/Vcanflox/flox Interzone
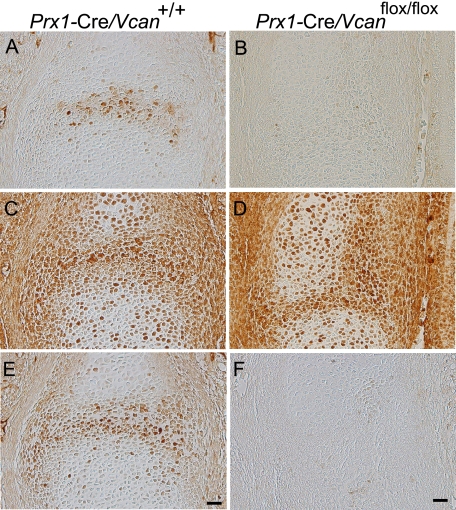
When compared with animals that exhibit joint malformation, the abnormalities found in Prx1-Cre/ Vcanflox/flox embryos resembled conditional knock-out mice of TβRII generated by crossing Prx1-Cre transgenic and Tgfbr2flox/flox mice, which exhibit the failure of joint interzone development (30). We speculated that TGF-β signaling is altered in Prx1-Cre/Vcanflox/flox. When the expression patterns of TGF-β and its related molecules were investigated by immunostaining, whereas TGF-β was localized in Prx1-Cre/Vcan+/+ interzone, it was not detected in Prx1-Cre/Vcanflox/flox (Fig. 6, A and B). TβRII was broadly immunostained in interzone, perichondrium, and chondrocytes of both Prx1-Cre/Vcan+/+ and Prx1-Cre/Vcanflox/flox digits (Fig. 6, C and D). Phospho-Smad2/3 was detected in the nuclei of cells in the Prx1-Cre/Vcan+/+ interzone (Fig. 6E). In contrast, it was not detected in Prx1-Cre/Vcanflox/flox (Fig. 6F). These results indicate that TGF-β signaling is substantially diminished in Prx1-Cre/Vcanflox/flox, although the expression of its receptor is unaffected.
FIGURE 6.
TGF-β signaling is attenuated in Prx1-Cre/Vcanflox/flox joint interzone. Immunostaining patterns of TGF-β (A and B), TβRII (C and D), and phospho-Smad2/3 (E and F) in the metatarsophalangeal joint interzone at E15.5 of Prx1-Cre/Vcan+/+ (A, C, and E) and Prx1-Cre/Vcanflox/flox (B, D, and F) are shown. Whereas TGF-β is localized in the Prx1-Cre/Vcan+/+ joint interzone (A), it is not detected in Prx1-Cre/Vcanflox/flox (B). TβRII is broadly immunostained in the joint interzone, perichondrium, and chondrocytes of both Prx1-Cre/Vcan+/+ (C) and Prx1-Cre/Vcanflox/flox (D). Whereas phospho-Smad2/3 is found in the nuclei of cells in the Prx1-Cre/Vcan+/+ interzone, it is not detected in Prx1-Cre/Vcanflox/flox. Scale bars, 20 μm. Immunostaining was performed on at least two individual digits for Prx1-Cre/Vcan+/+ and Prx1-Cre/Vcanflox/flox, with the same immunostaining patterns.
Impaired Mesenchymal Condensations and Altered TGF-β Signaling in Prx1-Cre/Vcanflox/flox Mice Lead to Delayed Chondrocyte Differentiation
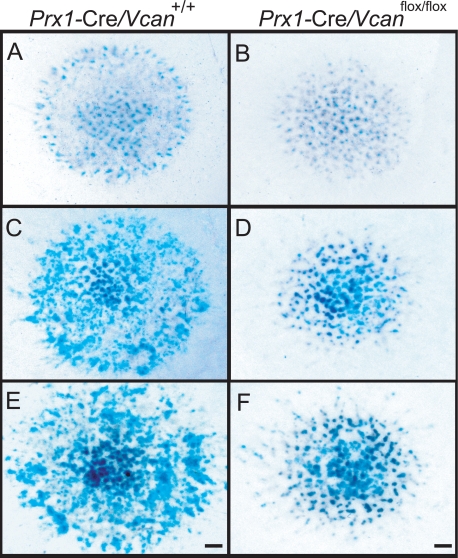
Prx1-Cre/Vcanflox/flox digits showed a delay in chondrocyte differentiation at E15.5 (Fig. 3B). Interestingly, whole mount Alcian blue staining demonstrated a decrease in areas positive for Alcian blue in Prx1-Cre/ Vcanflox/flox hind limbs compared with Prx1-Cre/Vcan+/+ (supplemental Fig. S3), suggesting that delay of chondrocyte differentiation took place prior to E13.5. To obtain insight into the mechanisms of delayed chondrocyte differentiation in Prx1-Cre/Vcanflox/flox digits, we applied a high density micromass culture system of limb bud mesenchymal cells. At day 3 of micromass culture, Prx1-Cre/Vcan+/+ contained some cartilaginous nodules stained with Alcian blue (Fig. 7A). At day 6, the number of the nodules positive for Alcian blue increased considerably (Fig. 7C). At day 9, their number further increased, and the nodules in the center were strongly stained with Alcian blue (Fig. 7E). When compared with Prx1-Cre/Vcan+/+, Prx1-Cre/Vcanflox/flox micromass at day 3 contained a small number of cartilaginous nodules (Fig. 7B). At day 6, the number of the nodules positive for Alcian blue increased, but it remained smaller than Prx1-Cre/Vcan+/+ (Fig. 7D). At day 9, their number increased, but the nodules remained smaller than Prx1-Cre/Vcan+/+ micromass (Fig. 7F). Quantitatively, the percentage of area positive for Alcian blue was 21 ± 2 and 18 ± 4.2% at day 3, and 35 ± 2.6 and 32 ± 5.6% at day 6 in Prx1-Cre/Vcan+/+ and Prx1-Cre/Vcanflox/flox micromass (from two different littermates), respectively. These observations indicate that Prx1-Cre/Vcanflox/flox cells in the mesenchymal condensation showed a delay in chondrocyte differentiation.
FIGURE 7.
Prx1-Cre/Vcanflox/flox exhibits delayed chondrocyte differentiation in micromass culture. Patterns of micromass stained by Alcian blue at day 3 (A and B), 6 (C and D), and 9 (E and F) of Prx1-Cre/Vcan+/+ (A, C, and E) and Prx1-Cre/Vcanflox/flox (B, D, and F) are shown. At day 3, micromass of Prx1-Cre/Vcan+/+ contains some cartilaginous nodules stained with Alcian blue (A), whereas that of Prx1-Cre/Vcanflox/flox contains a smaller number of them (B). At day 6, the number of the nodules positive for Alcian blue in micromass of Prx1-Cre/Vcanflox/flox increases, but it remains smaller than that of Prx1-Cre/Vcan+/+ (C and D). At day 9, the number of cartilaginous nodules in micromass of Prx1-Cre/Vcan+/+ further increases, and the nodules in the center become strongly stained with Alcian blue (E). Although the number of cartilaginous nodules in micromass of Prx1-Cre/Vcanflox/flox increases during culture (F), it remains smaller than Prx1-Cre/Vcan+/+. Scale bars, 300 μm.
Then we investigated expression of versican in the micromass. At day 3 in Prx1-Cre/Vcan+/+, versican was localized in the center of the condensation areas of future cartilaginous nodules (supplemental Fig. S4A), confirming its transient high expression in mesenchymal condensation areas. At day 6, it was found in areas surrounding the cartilaginous nodules, designated perinodular regions (Fig. 8A). In contrast, versican was not observed in micromass of Prx1-Cre/Vcanflox/flox, which suggests that the Cre enzyme driven by Prx1 promoter successfully abrogated versican expression in all of the mesenchymal cells in the micromass (Fig. 8B). This was further confirmed by X-gal staining of micromass obtained from Prx1-Cre/R26R limb buds (supplemental Fig. S4B).
FIGURE 8.
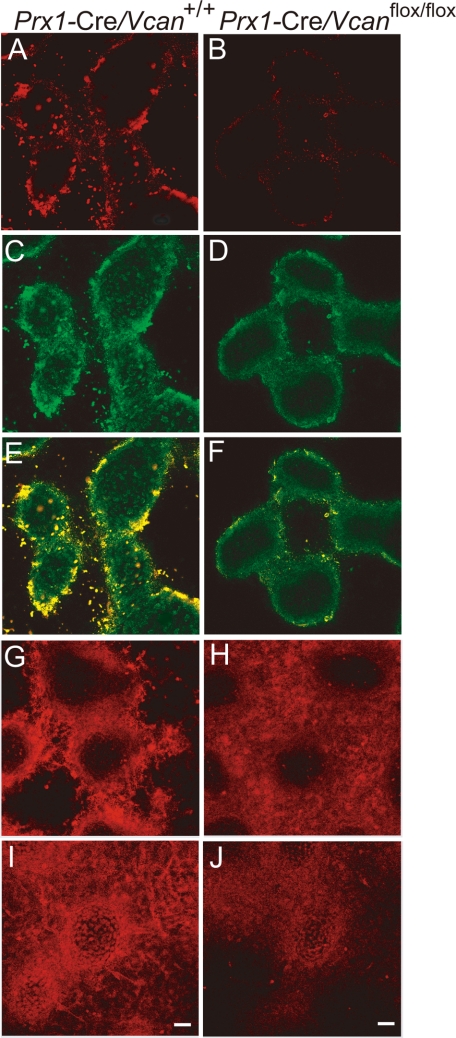
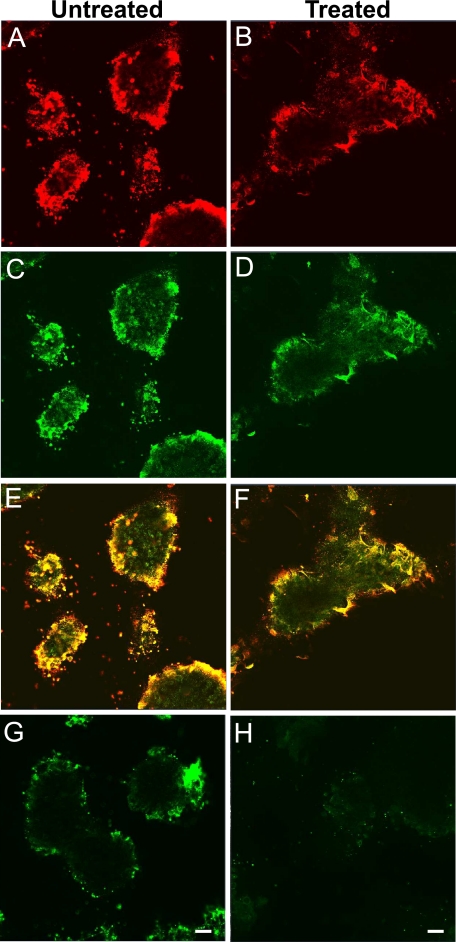
TGF-β signaling is altered in the micromass of Prx1-Cre/ Vcanflox/flox. Immunofluorescent staining at day 6 of culture, for versican (A and B), TGF-β (C and D), both merged (E and F), TβRII (G and H), and phospho-Smad2/3 (I and J) of Prx1-Cre/Vcan+/+ (A, C, E, G, and I) and Prx1-Cre/ Vcanflox/flox (B, D, F, H, and J) are shown. Versican is immunostained strongly in the perinodular region and moderately in the internodular region in Prx1-Cre/Vcan+/+ (A), whereas it is immunostained very faintly in Prx1-Cre/Vcanflox/flox (B). TGF-β is mainly localized in the perinodular regions in Prx1-Cre/Vcan+/+ similar to versican (C), whereas it is immunostained diffusely at a substantially lower intensity in Prx1-Cre/Vcan+/+ (D). When merged, both verican and TGF-β are well co-localized in the perinodular region (E). TβRII is immunostained mainly in the perinodular region in both Prx1-Cre/Vcan+/+ (G) and Prx1-Cre/Vcanflox/flox (H) micromass. In addition, it is immunostained rather diffusely in Prx1-Cre/Vcanflox/flox (H). Phospho-Smad2/3 is immunostained in the nuclei of perinodular cells strongly and in chondrocytes in the nodules moderately in Prx1-Cre/Vcan+/+ micromass (I). In contrast, it is immunostained moderately in the nuclei of chondrocytes in the nodules and weakly in the perinodular cells in the Prx1-Cre/Vcanflox/flox micromass (J). Immunofluorescent staining was visualized by Alexa Fluor 594. Scale bars, 13 μm. Immunofluorescent staining was performed at least twice, with essentially the same results. Immunostaining was performed at least three times from individual micromass for Prx1-Cre/Vcan+/+ and Prx1-Cre/Vcanflox/flox, with essentially the same immunostaining patterns.
TGF-β signaling is known to facilitate chondrocyte differentiation because the chondrocyte differentiation medium of bone marrow mesenchymal stem cells contains TGF-β (37). As shown in Fig. 6, TGF-β was substantially decreased in the Prx1-Cre/Vcanflox/flox interzone at E15.5. We speculated that TGF-β signaling is similarly down-regulated in Prx1-Cre/Vcanflox/flox micromass culture, which caused the delay in chondrocyte differentiation. By immunofluorescent staining, TGF-β was localized in the perinodular regions in Prx1-Cre/Vcan+/+ micromass (Fig. 8C), similar to versican. In contrast, it was observed diffusely in the extracellular matrix at a substantially lower intensity in Prx1-Cre/Vcanflox/flox micromass (Fig. 8D). When merged, both versican and TGF-β were well co-localized in Prx1-Cre/Vcan+/+ micromass (Fig. 8E). TβRII was immunostained mainly in the perinodular regions in both Prx1-Cre/Vcan+/+ (Fig. 8G) and Prx1-Cre/Vcanflox/flox micromass. In addition, it was immunostained rather diffusely in Prx1-Cre/Vcanflox/flox (Fig. 8H). Phospho-Smad2/3 was immunostained strongly in the nuclei of perinodular cells and moderately in the nuclei of chondrocytes within the nodules in the Prx1-Cre/Vcan+/+ micromass (Fig. 8I). In contrast, it was immunostained in the nuclei of chondrocytes in the nodules moderately and in those of the cells weakly in the perinodular regions of the Prx1-Cre/Vcanflox/flox micromass (Fig. 8J). These results strongly suggest that versican accumulates TGF-β in the extracellular matrix of the perinodular regions and facilitates its signaling. The diffuse patterns of TGF-β in the extracellular matrix and the positive immunostaining for phospho-Smad2/3 in chondrocytes indicate that TGF-β signaling functions toward the differentiation even in the absence of versican. By Western blot analysis, Prx1-Cre/Vcanflox/flox micromass exhibited comparable expression levels of TGF-β (supplemental Fig. S6).
Co-localization of versican and TGF-β in Prx1-Cre/Vcan+/+ micromass and diffuse patterns of TGF-β in the absence of versican in the Prx1-Cre/Vcanflox/flox micromass strongly suggest a direct binding of versican to TGF-β or its complex. Versican contains at least three functional domains: the G1 domain that binds hyaluronan, the G3 domain that binds various ECM molecules, and CS chains. To investigate whether CS chains are necessary for localizing TGF-β in the ECM, we treated Prx1-Cre/Vcan+/+ micromass with chondroitinase ABC for 48 h before immunostaining. In the micromass treated with chondroitinase ABC, versican remained in the areas surrounding the nodules (Fig. 9, A and B), confirming that CS chains are not essential for incorporation of versican in the ECM. TGF-β also similarly remained in the areas surrounding the nodules (Fig. 9, C and D). When merged, these molecules were well co-localized in the perinodular region (Fig. 9, E and F), even after ablation of CS chains (Fig. 9H). These observations indicate that versican without CS chains may retain the function of localizing TGF-β.
FIGURE 9.
Versican localizes TGF-β in the extracellular matrix. Immunofluorescent staining of versican (A and B) and TGF-β (C and D) of Prx1-Cre/Vcan+/+ micromass untreated (A, C, and E) and treated with chondroitinase ABC for 48 h before fixation (B, D, and F). The areas immunostained for versican become diffuse by the treatment, although versican essentially remains in the perinodular region (B). The areas immunostained for TGF-β become diffuse by the treatment, concomitantly with versican. Merged images are shown (E and F). Immunostaining for CS chains that confirm ablation of CS is shown (untreated (G) and treated (H)). Scale bars, 15 μm. Immunofluorescent staining was performed at least three times, with essentially the same results.
DISCUSSION
In this study, we have generated the conditional versican-null mice, in which the versican gene Vcan is conditionally inactivated in limb buds and a subset of mesenchyme tissues starting at very early embryonic limb development. These mice, termed Prx1-Cre/Vcanflox/flox, grow normally and are fertile, although they grossly show distorted digits. Histologically, their digits display hypertrophic chondrocyte nodules, tilting and clefting of the joint surface, and a slight delay of cartilage development. The joint interzone at E15.5 of these mice exhibited a decrease in TGF-β in the extracellular matrix, concomitant with a substantially decreased number of nuclei positive for phospho-Smad2/3. Our micromass culture system of limb bud mesenchymal cells demonstrated diffuse TGF-β localization in the mutant micromass, contrasting to its restrictive localization to regions surrounding developing cartilaginous nodules in Prx1-Cre/Vcan+/+. These results strongly suggest that versican localizes TGF-β in the extracellular matrix and regulates its signaling. Our study uncovers, for the first time, the in vivo role of versican in skeletogenesis.
Versican Is Required for TGF-β Signaling during Joint Morphogenesis
Joint formation begins with the condensations of uninterrupted mesenchyme differentiating to chondrocytes. In the sites of joint formation, the mesenchymal cells remaining undifferentiated or conversely dedifferentiated from chondrocytes become densely packed to form a region termed the joint interzone, followed by joint cavity formation (24, 25). In this study, we have shown that whereas the metatarsophalangeal joint interzone at E15.5 of Prx1-Cre/Vcan+/+ digits gives rise to a well defined space representing early joint cavities, that of Prx1-Cre/Vcanflox/flox forms a tilted joint interzone filled with closely packed mesenchymal cells. Interestingly, the deposition of TGF-β in the ECM and the number of nuclei positive for phospho-Smad2/3 were markedly diminished. Recently, TGF-β type II receptor gene (Tgfbr2) conditional knock-out mice under the control of the Prx1 promoter have demonstrated a failure of interphalangeal joint interzone development, resulting from an aberrant persistence of differentiated chondrocytes and a failure of Jagged-1 expression, thereby indicating an essential role of TGF-β signaling in joint formation (30). The absence of versican in the joint interzone of Prx1-Cre/Vcanflox/flox digits may disturb levels and spatially ordered distribution of TGF-β signaling, leading to altered localization of mesenchymal cells in the joint interzone. Despite marked inhibition of TGF-β signaling in the joint interzone of Prx1-Cre/Vcanflox/flox digits, accumulation of mesenchymal cells was observed in the joint interzone. Therefore, TGF-β signaling necessary for maintaining a mesenchymal phenotype appears viable. It has also been proposed that mesenchymal cells in the interzone are derived from dedifferentiation of chondrocytes. Because chondrocytes were absent in the interzone of Prx1-Cre/Vcanflox/flox as evaluated by immunostaining for aggrecan, the process of their dedifferentiation to mesenchymal cells is probably intact, even with considerable reduction of TGF-β signaling.
HA and its receptor CD44 have been proposed to play key roles in joint cavitation (38–40). We have shown that unincorporated HA in the joint interzone of both Prx1-Cre/Vcan+/+ and Prx1-Cre/Vcanflox/flox is accumulated at similar levels in the joint interzone. In addition, the expression of CD44 in the joint was not detected until E18.5 (supplemental Fig. S5A). These observations may exclude direct involvement of HA- and CD44-mediated signaling in the joint abnormalities of Prx1-Cre/Vcanflox/flox digits.
The Wnt/β-catenin canonical signaling pathway has been reported to be necessary and sufficient for the induction of synovial joints in the limb (28). By immunostaining of the joint interzone at E15.5, β-catenin was expressed in closely associated mesenchymal cells at similar levels between Prx1-Cre/Vcan+/+ and Prx1-Cre/Vcanflox/flox (supplemental Fig. S5B), although more mesenchymal cells were observed in the interzone. Together with the fact that the joints themselves are formed in Prx1-Cre/Vcanflox/flox, versican is unlikely to have direct effects on Wnt signaling.
It is intriguing that the abnormalities of Prx1-Cre/ Vcanflox/flox appeared only in autopods, although versican is expressed in both proximal and distal joint interzone, articular cartilage, and synovia of limbs (28). Our immunostaining confirmed the absence of versican in joints where no abnormalities were observed, suggesting that the role of versican in joint formation is confined to digits. It is of note that TβRII conditional knock-out mice exhibit impaired joint formation only in digits, although TβRII is also expressed in proximal joints. These observations support the notion that versican contributes to joint formation by regulating TGF-β signaling and that versican does not play a major role in formation of the joints where TGF-β is unnecessary.
Versican Accumulates TGF-β to Perinodular Regions in Mesenchymal Condensation
Because versican was identified in mesenchymal condensation areas of chick limb bud, this proteoglycan has been assumed to play an important role in cartilage development. Our previous study using N1511 chondrogenic cells clearly demonstrated that inhibition of versican expression at an early stage substantially decelerates following chondrocyte differentiation (34). High density micromass culture systems of limb bud obtained from versican-null hdf homozygote embryos demonstrated that versican is necessary for mesenchymal cell aggregation but that these mesenchymal cells undergo chondrocyte differentiation in three-dimensional type I collagen gel culture (33). These results suggest that versican interacts with other ECM molecules and cell membrane molecules and mechanically ties up individual cells to form aggregates. In contrast to these results, the fact that Prx1-Cre/Vcanflox/flox mice undergo almost normal cartilage development suggests that versican does not have a profound effect on mesenchymal cell aggregation in vivo, although a slight delay of cartilage development occurs.
It has not been understood why mesenchymal condensation is required for proper chondrocyte differentiation. It may facilitate signal transduction by increasing local concentrations of growth factors and cytokines secreted by the cells and by gathering target cells close with each other. We have shown that both versican and TGF-β are restricted to the area surrounding the cartilaginous nodules in Prx1-Cre/Vcan+/+, whereas TGF-β is widely distributed in the Prx1-Cre/Vcanflox/flox micromass. This suggests that versican accumulates TGF-β in the ECM and elicits a strong TGF-β-mediated signal to the target cells. Indeed, phospho-Smad2/3 was strongly immunostained in the nuclei of cells in the perinodular regions in Prx1-Cre/Vcan+/+ micromass, whereas it was immunostained weakly in Prx1-Cre/Vcanflox/flox micromass.
Functional Domain of Versican
The functional domain of versican that participates in mesenchymal condensation has not yet been determined. By chondroitinase ABC treatment of Prx1-Cre/Vcan+/+ micromass for 48 h before fixation, both versican and TGF-β retained essentially the same localization patterns as the untreated micromass, eliminating a direct function of CS chains in accumulating TGF-β in the ECM. In addition, CS chains attached to versican in cartilage are undersulfated compared with those of aggrecan (41). Furthermore, gene-trapped mice of C4st1 (chondroitin-4-sulfotranferase 1), C4st1gt/gt, with a substantial decrease in CS amounts, show no abnormalities at E11.5 and E13.5 when mesenchymal condensation occurs in the limb bud (42). Thus, it is unlikely that CS chains of versican directly bind TGF-β and regulate its signaling, although they may affect joint formation because C4st1gt/gt mice display an impaired segmentation of cartilage in digits (42).
Another candidate is the G1 domain, which specifically binds to HA. Conditional knock-out mice of the Has2 (hyaluronan synthase 2) gene, Prx1-Cre/Has2flox/flox, display dwarfism, abnormal digit patterning, impaired chondrocyte maturation, and a defective synovial joint cavity (43). Their dwarfism and impaired chondrocyte maturation in the growth plate are probably due to the reduction of cartilage extracellular matrix containing HA and aggrecan. Although data regarding the mesenchymal condensation process of limb bud in these mice have not been presented, the result demonstrating that these mice undergo cartilage development even in the absence of HA in mesenchymal condensation areas suggests that HA is not essential for the initial stage of cartilage development. Because of a rather minor perturbation of mesenchymal condensation by the absence of HA, it is unlikely that the function of versican is via interaction with HA.
The C-terminal G3 domain interacts with fibulin-1 and -2 (4, 5) and fibrillin-1 (3). Fibrillin-1 binds LTBP-1 and -4 (latent TGF-β-binding proteins 1 and 4), which bind and store TGF-β in the ECM. Recently, fibrillin-1 has been shown to be required for the appropriate matrix assembly of LTBPs, which is affected by fibulins (44). Versican may regulate balance deposition of fibrillin-1 and fibulins and regulate the incorporation of LTBP into the ECM. Fibrillin-1 and -2 double knock-out mice (45) and LTBP-1-null mice (46) exhibit no clear defects in cartilage development. Our Western blot analysis showed similar levels of TGF-β deposition in the ECM of Prx1-Cre/Vcan+/+ and Prx1-Cre/Vcanflox/flox micromass, which suggests that other ECM molecules contribute to its deposition even in the absence of versican. The mechanisms by which versican accumulates TGF-β in the perinodular region of the ECM should be studied, in addition to LTBPs, fibulins, and fibrillin-1.
So far, we have not determined the functional domain of versican in regulation of TGF-β. Several ECM molecules have been shown to interact with each other and with TGF-β and its superfamily members, and the sequestration may involve the supramolecular structure built up via interactions of various ECM molecules. Storage and action of signaling molecules may be regulated by alteration of the tertiary structure of the complex. Further studies to connect the structure of the ECM and its regulation on cell signaling remain to be performed.
Supplementary Material
Acknowledgments
We thank Dr. Y. Ito for technical assistance and Dr. N. Sugiura for technical consultation.
This work was supported by Grants-in-aid for Scientific Research (B) (KAKENHI) (to Hideto Watanabe) and grants-in-aid for priority areas (to Hideto Watanabe).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
- CS
- chondroitin sulfate
- HA
- hyaluronan
- ECM
- extracellular matrix
- TGF
- transforming growth factor
- ES
- embryonic stem
- Tg
- transgenic
- WT
- wild type
- X-gal
- 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside
- PBS
- phosphate-buffered saline
- HABP
- hyaluronan-binding protein
- TβRII
- TGF-β receptor II
- LTBP
- latent TGF-β-binding protein
- En
- embryonic day n.
REFERENCES
- 1.Kimata K., Oike Y., Tani K., Shinomura T., Yamagata M., Uritani M., Suzuki S. (1986) J. Biol. Chem. 261, 13517–13525 [PubMed] [Google Scholar]
- 2.Matsumoto K., Shionyu M., Go M., Shimizu K., Shinomura T., Kimata K., Watanabe H. (2003) J. Biol. Chem. 278, 41205–41212 [DOI] [PubMed] [Google Scholar]
- 3.Isogai Z., Aspberg A., Keene D. R., Ono R. N., Reinhardt D. P., Sakai L. Y. (2002) J. Biol. Chem. 277, 4565–4572 [DOI] [PubMed] [Google Scholar]
- 4.Aspberg A., Miura R., Bourdoulous S., Shimonaka M., Heinegârd D., Schachner M., Ruoslahti E., Yamaguchi Y. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 10116–10121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olin A. I., Mörgelin M., Sasaki T., Timpl R., Heinegård D., Aspberg A. (2001) J. Biol. Chem. 276, 1253–1261 [DOI] [PubMed] [Google Scholar]
- 6.Day J. M., Olin A. I., Murdoch A. D., Canfield A., Sasaki T., Timpl R., Hardingham T. E., Aspberg A. (2004) J. Biol. Chem. 279, 12511–12518 [DOI] [PubMed] [Google Scholar]
- 7.Ujita M., Shinomura T., Ito K., Kitagawa Y., Kimata K. (1994) J. Biol. Chem. 269, 27603–27609 [PubMed] [Google Scholar]
- 8.Shinomura T., Nishida Y., Ito K., Kimata K. (1993) J. Biol. Chem. 268, 14461–14469 [PubMed] [Google Scholar]
- 9.Dours-Zimmermann M. T., Zimmermann D. R. (1994) J. Biol. Chem. 269, 32992–32998 [PubMed] [Google Scholar]
- 10.Ito K., Shinomura T., Zako M., Ujita M., Kimata K. (1995) J. Biol. Chem. 270, 958–965 [DOI] [PubMed] [Google Scholar]
- 11.Zako M., Shinomura T., Ujita M., Ito K., Kimata K. (1995) J. Biol. Chem. 270, 3914–3918 [DOI] [PubMed] [Google Scholar]
- 12.Bode-Lesniewska B., Dours-Zimmermann M. T., Odermatt B. F., Briner J., Heitz P. U., Zimmermann D. R. (1996) J. Histochem. Cytochem. 44, 303–312 [DOI] [PubMed] [Google Scholar]
- 13.Wu Y. J., La Pierre D. P., Wu J., Yee A. J., Yang B. B. (2005) Cell Res. 15, 483–494 [DOI] [PubMed] [Google Scholar]
- 14.Yamagata M., Saga S., Kato M., Bernfield M., Kimata K. (1993) J. Cell Sci. 106, 55–65 [DOI] [PubMed] [Google Scholar]
- 15.Landolt R. M., Vaughan L., Winterhalter K. H., Zimmermann D. R. (1995) Development 121, 2303–2312 [DOI] [PubMed] [Google Scholar]
- 16.Kishimoto J., Ehama R., Wu L., Jiang S., Jiang N., Burgeson R. E. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 7336–7341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall B. K., Miyake T. (1992) Anat. Embryol. 186, 107–124 [DOI] [PubMed] [Google Scholar]
- 18.Leonard C. M., Fuld H. M., Frenz D. A., Downie S. A., Massagué J., Newman S. A. (1991) Dev. Biol. 145, 99–109 [DOI] [PubMed] [Google Scholar]
- 19.Blessing M., Nanney L. B., King L. E., Jones C. M., Hogan B. L. (1993) Genes Dev. 7, 204–215 [DOI] [PubMed] [Google Scholar]
- 20.Storm E. E., Huynh T. V., Copeland N. G., Jenkins N. A., Kingsley D. M., Lee S. J. (1994) Nature 368, 639–643 [DOI] [PubMed] [Google Scholar]
- 21.Holder N. (1977) J. Embryol. Exp. Morphol. 39, 115–127 [PubMed] [Google Scholar]
- 22.Mitrovic D. (1978) Am. J. Anat. 151, 475–485 [DOI] [PubMed] [Google Scholar]
- 23.Hamrick M. W. (2001) Evol. Dev. 3, 355–363 [DOI] [PubMed] [Google Scholar]
- 24.Pacifici M., Koyama E., Iwamoto M. (2005) Birth Defects Res. C Embryo Today 75, 237–248 [DOI] [PubMed] [Google Scholar]
- 25.Archer C. W., Dowthwaite G. P., Francis-West P. (2003) Birth Defects Res. C Embryo Today 69, 144–155 [DOI] [PubMed] [Google Scholar]
- 26.Hartmann C., Tabin C. J. (2001) Cell 104, 341–351 [DOI] [PubMed] [Google Scholar]
- 27.Khan Z., Vijayakumar S., de la Torre T. V., Rotolo S., Bafico A. (2007) Mol. Cell. Biol. 27, 7291–7301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo X., Day T. F., Jiang X., Garrett-Beal L., Topol L., Yang Y. (2004) Genes Dev. 18, 2404–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pitsillides A. A. (2003) Cell Biochem. Funct. 21, 235–240 [DOI] [PubMed] [Google Scholar]
- 30.Spagnoli A., O'Rear L., Chandler R. L., Granero-Molto F., Mortlock D. P., Gorska A. E., Weis J. A., Longobardi L., Chytil A., Shimer K., Moses H. L. (2007) J. Cell Biol. 177, 1105–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Snow H. E., Riccio L. M., Mjaatvedt C. H., Hoffman S., Capehart A. A. (2005) Anat. Rec. A Discov. Mol. Cell. Evol. Biol. 282, 95–105 [DOI] [PubMed] [Google Scholar]
- 32.Shepard J. B., Krug H. A., LaFoon B. A., Hoffman S., Capehart A. A. (2007) Int. J. Biol. Sci. 3, 380–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Williams D. R., Jr., Presar A. R., Richmond A. T., Mjaatvedt C. H., Hoffman S., Capehart A. A. (2005) Biochem. Biophys. Res. Commun. 334, 960–966 [DOI] [PubMed] [Google Scholar]
- 34.Kamiya N., Watanabe H., Habuchi H., Takagi H., Shinomura T., Shimizu K., Kimata K. (2006) J. Biol. Chem. 281, 2390–2400 [DOI] [PubMed] [Google Scholar]
- 35.Logan M., Martin J. F., Nagy A., Lobe C., Olson E. N., Tabin C. J. (2002) Genesis 33, 77–80 [DOI] [PubMed] [Google Scholar]
- 36.Leussink B., Brouwer A., el Khattabi M., Poelmann R. E., Gittenberger-de Groot A. C., Meijlink F. (1995) Mech. Dev. 52, 51–64 [DOI] [PubMed] [Google Scholar]
- 37.Johnstone B., Hering T. M., Caplan A. I., Goldberg V. M., Yoo J. U. (1998) Exp. Cell Res. 238, 265–272 [DOI] [PubMed] [Google Scholar]
- 38.Craig F. M., Bayliss M. T., Bentley G., Archer C. W. (1990) J. Anat. 171, 17–23 [PMC free article] [PubMed] [Google Scholar]
- 39.Edwards J. C., Wilkinson L. S., Jones H. M., Soothill P., Henderson K. J., Worrall J. G., Pitsillides A. A. (1994) J. Anat. 185, 355–367 [PMC free article] [PubMed] [Google Scholar]
- 40.Pitsillides A. A., Archer C. W., Prehm P., Bayliss M. T., Edwards J. C. (1995) J. Histochem. Cytochem. 43, 263–273 [DOI] [PubMed] [Google Scholar]
- 41.Matsumoto K., Kamiya N., Suwan K., Atsumi F., Shimizu K., Shinomura T., Yamada Y., Kimata K., Watanabe H. (2006) J. Biol. Chem. 281, 18257–18263 [DOI] [PubMed] [Google Scholar]
- 42.Klüppel M., Wight T. N., Chan C., Hinek A., Wrana J. L. (2005) Development 132, 3989–4003 [DOI] [PubMed] [Google Scholar]
- 43.Matsumoto K., Li Y., Jakuba C., Sugiyama Y., Sayo T., Okuno M., Dealy C. N., Toole B. P., Takeda J., Yamaguchi Y., Kosher R. A. (2009) Development 136, 2825–2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ono R. N., Sengle G., Charbonneau N. L., Carlberg V., Bächinger H. P., Sasaki T., Lee-Arteaga S., Zilberberg L., Rifkin D. B., Ramirez F., Chu M. L., Sakai L. Y. (2009) J. Biol. Chem. 284, 16872–16881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carta L., Pereira L., Arteaga-Solis E., Lee-Arteaga S. Y., Lenart B., Starcher B., Merkel C. A., Sukoyan M., Kerkis A., Hazeki N., Keene D. R., Sakai L. Y., Ramirez F. (2006) J. Biol. Chem. 281, 8016–8023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Todorovic V., Frendewey D., Gutstein D. E., Chen Y., Freyer L., Finnegan E., Liu F., Murphy A., Valenzuela D., Yancopoulos G., Rifkin D. B. (2007) Development 134, 3723–3732 [DOI] [PubMed] [Google Scholar]
- 47.Daniels K., Reiter R., Solursh M. (1996) Methods Cell Biol. 51, 237–247 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.