Abstract
Intraflagellar transport (IFT) particles of Chlamydomonas reinhardtii contain two distinct protein complexes, A and B, composed of at least 6 and 15 protein subunits, respectively. As isolated from C. reinhardtii flagella, IFT complex B can be further reduced to a ∼500-kDa core that contains IFT88, 2× IFT81, 2× IFT74/72, IFT52, IFT46, IFT27, IFT25, and IFT22. In this study, yeast-based two-hybrid analysis was combined with bacterial coexpression to show that three of the core B subunits, IFT88, IFT52, and IFT46, interact directly with each other and, together, are capable of forming a ternary complex. Chemical cross-linking results support the IFT52-IFT88 interaction and provide additional evidence of an association between IFT27 and IFT81. With previous studies showing that IFT81 and IFT74/72 interact to form a (IFT81)2(IFT74/72)2 heterotetramer and that IFT27 and IFT25 form a heterodimer, the architecture of complex B is revealing itself. Last, electroporation of recombinant IFT46 was used to rescue flagellar assembly of a newly identified ift46 mutant and to monitor in vivo localization and movement of the IFT particles.
Keywords: Kinesin, Molecular Motors, Protein Assembly, Protein Cross-linking, Protein-Protein Interactions, Chlamydomonas, Intraflagellar Transport, Cilia, Flagella
Introduction
Found on the surface of many eukaryotic cells, cilia and flagella (redundant terms) are organelles consisting of membrane-bounded microtubular projections that emanate from basal body templates. Either motile or nonmotile, cilia have been adapted for a variety of functions, including cellular motility, directional fluid movement, sensory reception, and cellular signaling (reviewed in Refs. 1–4). Ciliary-based sensory reception includes vision and olfaction, whereas ciliary-mediated receptor-dependent signaling includes sonic hedgehog, noncanonical Wnt, and platelet-derived growth factor pathways (reviewed in Refs. 5–7). Defects in the assembly and function of these organelles have been associated with ciliopathies, an expanding list of human diseases that include immotile cilia and Bardet-Biedl syndromes and cystic kidney disorders, such as polycystic kidney disease and nephronophthisis (reviewed in Refs. 8–12). Many of these ciliopathies have been linked to intraflagellar transport (IFT),3 a conserved process required for the assembly and maintenance of eukaryotic cilia (reviewed in Refs. 13–15).
IFT is characterized by the robust bidirectional movement of large proteinaceous particles along the length of the axonemal microtubules (16, 17). Kinesin-2 is responsible for driving the anterograde or outward movement (17–21), whereas the retrograde return to the cell body is powered by cytoplasmic dynein-1b/2 (22, 23). Formerly known as rafts, the long IFT trains contain multiple copies of two distinct protein complexes, A and B (20, 24, 25). As isolated from the flagella of the green alga, Chlamydomonas reinhardtii, complex A contains at least six distinct proteins (IFT144, IFT140, IFT139, IFT122, IFT121, and IFT43), whereas complex B contains at least 13 proteins (IFT172, IFT88, IFT81, IFT80, IFT74/72, CrDYF-1, IFT57, IFT52, IFT46, IFT27, IFT25, IFT22, and IFT20); the subunit names reflect the relative mobilities of each when separated by SDS-PAGE and, thus, represent the approximate mass (kDa) of each protein (reviewed in Ref. 26).
As observed in other ciliated organisms, mutations affecting Chlamydomonas IFT B genes often result in severe disruptions of flagellar assembly. Mutations of either the IFT52 or IFT88 genes resulting in loss-of-function produce “bald” cells that are unable to construct flagella past the transition zone (27, 28). A slightly less severe phenotype occurs in the ift46-1 mutant, where a fraction of the cells are able to assemble short (∼3-μm) paralyzed flagella (29). Although the exact mechanism underlying the specific role of each IFT protein is largely unknown, mutant analysis has revealed important clues. IFT46, for example, is required for the assembly of outer dynein arms onto axonemal microtubules and also appears to be important in the stabilization of complex B (29, 30). Knockdown of IFT27 results in reduced expression of other IFT genes and significantly slows down cell cycle progression and cytokinesis, indicating that this Rab-like G protein has important regulatory roles (31). Last, Chlamydomonas IFT172 is associated with the distal localization of EB1 and plays a role in the turnaround of IFT particles at the flagellar tip (32).
As isolated from Chlamydomonas flagella, complex B is much less stable than complex A (21, 33). Sucrose density gradient centrifugation of IFT proteins under conditions of increasing ionic strength, for example, results in the partial dissociation of complex B, whereas A stays intact. This partial dissociation has revealed a stable subset of B proteins termed the B core that contains IFT88, IFT81, IFT74/72, IFT52, IFT46, IFT27, IFT25, and IFT22 (33, 34). Within this core, IFT27 and IFT25 are thought to form a heterodimer (34), whereas IFT81, IFT74, and IFT72 are thought to form a heterotetrameric complex in a ratio of 2:1:1, respectively, although the nature of the difference between the algal IFT74 and IFT72 remains unknown (33). The only other published interaction between B subunits is that of the mammalian IFT20 with IFT57, as reported by Baker et al. (35). The studies presented here result from our efforts to identify additional interactions within the B complex.
Initially, we employed an exhaustive yeast-based two-hybrid screen and found three interactions that included the previously reported IFT81 homodimerization and IFT81-IFT74/72 association (33). As described here, we also found that IFT46 interacted directly with IFT52. Surprisingly, however, our screen was unable to identify additional interactions of B subunits. Hypothesizing that the assembly of the B complex is, at least partially, ordered, we exploited a heterologous coexpression system that facilitated tandem affinity purification of multiple proteins. After confirming the IFT46-IFT52 interaction, we found that IFT46 and IFT52 were also capable of interacting independently with IFT88; the IFT52-IFT88 interaction was further supported by separate cross-linking studies of the native complex B core. Coexpression of all three proteins followed by tandem affinity chromatography showed that they were able to form a ternary complex, suggesting that an in vivo association of these three subunits could occur in the absence of additional B proteins. Last, we show that, within hours of electroporation, recombinant IFT46 protein can rescue the Chlamydomonas ift46-2 flagellar assembly mutant phenotype. Furthermore, the N-terminal 25 amino acids of IFT46 were not essential to rescue the mutant phenotype. We conclude that the use of protein electroporation with Chlamydomonas IFT mutants could be used to dissect the function of additional IFT proteins as well as to provide a rapid means of introducing fluorescently tagged IFT protein into live cells.
EXPERIMENTAL PROCEDURES
Strains and Media
The Chlamydomonas cell wall-deficient strain CC-503 (cw92) was obtained from the Chlamydomonas Center. Chlamydomonas stains were grown on solid TAP medium (36). The Escherichia coli strain Rosetta BL21 DE3 (Novagen) was used for all protein expression. All bacterial cloning was performed using the E. coli TOP10 (Invitrogen) strain. Liquid or solid Luria-Bertani (LB) media with appropriate antibiotic were used for all bacterial growth.
Insertional Mutagenesis
Chlamydomonas motility mutants were generated by insertional mutagenesis of the cell wall-deficient strain, CC-503, using the pHyg3 plasmid carrying an aminoglycoside phosphotransferase (aph7″) gene that confers resistance to hygromycin B (37–39). In brief, CC-503 cells were spread liberally onto fresh TAP plates and placed under constant illumination (2300 lux average) 24–48 h prior to flooding with 14 ml of TAP medium for 1 h. Cells were resuspended in a total volume of ∼40 ml and shaken at 150 rpm for 1–2 h under constant light (1200 lux average); the density of cells at this stage was typically 1–2 × 107/ml. Cells were concentrated to ∼1.3 × 108/ml by centrifugation at 1200 × g for 2 min, followed by resuspension in TAP medium. Cell aliquots (330 μl; 0.5 × 108) were mixed with 10 μl of KpnI-digested pHyg3 plasmid (0.5 μg) and 112 μl 20% polyethylene glycol 8000 (Sigma) and vortexed on high for 15 s with 300 mg of sterile 0.5-mm glass beads (Propper Manufacturing Co.) according to Kindle (40). The cell mixture was immediately diluted with 10 ml of TAP medium, transferred to a 250-ml flask, further diluted to ∼20 ml with TAP medium, and allowed to recover for 4 h under light (1200 lux average) prior to plating on solid TAP medium (1.7% agar) containing 10 μg/ml hygromycin B. For plating, cells were concentrated, and ∼107 cells in 0.5 ml of TAP were quickly, albeit gently, mixed with 4.5 ml of TAP containing melted 0.45% agar (42 °C) with 10 μg/ml hygromycin B, before layering on top of solid TAP plates. After 10–12 days of constant illumination (2300 lux average), transformed colonies were visually screened for motility defects using a 96-well plate and an inverted phase microscope. Motility mutants displaying gross flagellar assembly defects were screened for disruptions in IFT genes using gene-specific PCR amplification with genomic DNA as the template; PCR primer sequences and amplification conditions are available upon request. After a strain was identified that failed to yield IFT46 PCR products, the genomic region containing the IFT46 gene was exhaustively screened using a variety of PCR primer sets to better define the disrupted region.
Bacterial Plasmid Construction
The pMAL-c2x plasmid (New England BioLabs) was used for expression of all maltose-binding fusion proteins, whereas the pRSF-Duet plasmid (Novagen) was used for expression of His6-tagged and untagged proteins. Multicloning site 1 (MCS-1) of pRSF-Duet was used for all His6 fusions, and multicloning site 2 (MCS-2) was used for production of untagged, full-length IFT88. All IFT52 and IFT88 constructs used in this study were PCR-amplified and subcloned using a nearly full-length IFT52 cDNA clone (LCL098a07, accession number AV631675) and a full length IFT88 cDNA clone (LCL045a10, accession number AV628646); both clones were obtained from Kazusa DNA Research Institute (Kisarazu, Chiba, Japan) (41, 42). Attempts to clone a full-length IFT52 cDNA from total cDNA and cDNA libraries were unsuccessful. A full-length IFT46 cDNA clone was generated using a cDNA clone obtained as a generous gift from Hongmin Qin and Joel Rosenbaum. The IFT52 constructs used for coexpression experiments include 52ΔN24 (amino acids 25–454), 52ΔN90 (amino acids 90–454; minus the N terminus), 52ΔC122 (amino acids 25–332), 52M (amino acids 90–332; mid domain), and 52CT (amino acids 258–454; C-terminal 197 amino acids). 52ΔN24 was subcloned into the pMAL-c2x and pRSF-Duet vectors using the EcoRI and SalI restriction sites in MCS-1. 52ΔC122, 52ΔN90, 52M, and 52CT were cloned into the pMAL-c2x EcoRI and SalI sites of MCS-1. The full-length IFT88 cDNA encoding amino acids 1–782 as reported by Pazour et al. (28) was cloned into the pRSF-Duet NdeI and BglII sites within MCS-2. IFT46 constructs used for coexpression experiments include 46F (amino acids 1–344; full length), 46ΔN100 (amino acids 101–344; minus the N terminus), 46ΔC89 (amino acids 1–255; minus the C terminus), 46M (amino acids 101–255; mid domain), and 46CT (amino acids 198–344; C-terminal 147 amino acids). The full coding sequence of IFT46 (46F) was subcloned into pMAL-c2x and pRSF-Duet using the EcoRI and SalI sites of MCS-1. DNA encoding 46ΔN100, 46ΔC89, 46M, and 46CT were cloned into pRSF-Duet using the EcoRI and SalI sites of MCS-1. Primer sequences for all amplifications are available upon request.
Yeast-based Two-hybrid Analysis
Yeast-based two-hybrid analysis was performed as described by Lucker et al. (33) using the Hybrizap 2.1 two-hybrid system based on the GAL4 transcriptional activator and YRG-2 host cells (Stratagene). IFT46, IFT52, and IFT88 GAL4 activation domain (AD) or DNA binding domain (BD) constructs were generated using the same cDNA sources utilized and described for the bacterial coexpression. Negative control plasmids expressed only the activation domain (AD-MCS) or DNA binding domain (BD-MCS). As supplied by the manufacturer, the two-hybrid strong interaction control consisted of amino acids 132–236 of wild-type λ cI, fragment C, fused to both the AD and BD domains (Stratagene). The weak interaction control consisted of a mutant form of the λ cI protein fused to both the AD and BD domains. The negative interaction control combined the AD-λ cI vector with the human lamin C (amino acids 67–230) in the BD vector.
Electroporation
Protein electroporation was performed as described by Hayashi et al. (43). Briefly, Chlamydomonas CC-503 cells were grown on solid TAP medium (1.7% agar) for 3–5 days and then suspended in liquid TAP+S (TAP medium containing 60 mm sucrose). Suspended cells were placed below a 60-watt incandescent plant grow light (1000–1400 lux) for 1–2 h and agitated by shaking. Cells were gently pelleted in a microcentrifuge at 250 × g for 2 min and washed three times with HMDKCaS buffer (30 mm HEPES, 5 mm MgSO4, 1 mm dithiothreitol, 50 mm potassium acetate, 1 mm calcium acetate, 60 mm sucrose, pH 7.4). After the third wash, cells were brought to a density of 108 cells/ml in HMDKCaS and incubated at 15 °C for 15 min. Cells were then mixed with recombinant protein in a 1:1 ratio with a final volume and protein concentration of 125 μl and 0.5 mg/ml, respectively, in a 2.0-mm electroporation cuvette (BTX, Harvard Bioscience). The cuvette was placed at a ∼135° angle for 1 min immediately prior to electroporation. Electroporation was performed using an ECM630 electroporator (BTX) set at a voltage of 1.6 kV/cm, with a resistance of 25 ohms and conductance of 500 microfarads. Immediately following electroporation, cells were diluted into 300 μl of TAP+S and gently agitated every few min by hand at room temperature (∼23–25 °C) for 1 h. Prior to fluorescent imaging, excess labeled protein was removed from the medium by gently washing cells three times in 1.5 ml of TAP+S medium with 2 min of centrifugation at 250 × g; cells were resuspended in a final volume of 300 μl in TAP+S.
Protein Expression and Affinity Purification
For protein coexpression, 100 ml of LB liquid medium containing both ampicillin (100 μg/ml) and kanamycin (30 μg/ml) was inoculated with overnight cultures of E. coli BL21 DE3 Rosetta cells (Novagen) containing both pMAL-c2x (New England BioLabs) and pRSF-Duet (Novagen) expression vectors. Shaken at 37 °C until the A600 was ∼0.6, cells were induced with 1 mm isopropyl-β-d-thiogalactopyranoside for a period of 2 h before harvest. Harvested cells were cooled on ice while protease inhibitors (0.2 mm phenylmethylsulfonyl fluoride, 3.4 μg/ml aprotinin, 20 μg/ml leupeptin, 0.2 μg/ml pepstatin A, and 10 μg/ml soybean trypsin inhibitor) were added. Cells were immediately centrifuged for 10 min at 3500 × g at 4 °C and then resuspended in 10 ml of amylose chromatography buffer (ACB; 20 mm Tris, 200 mm NaCl, and 1.0 mm EDTA, pH 7.4) prior to storage at −28 °C. For preparation of soluble lysates, cells were thawed to 4 °C while 1.0 mm dithiothreitol and the same protease inhibitors described above were added. Cells were lysed via eight 15-s sonication pulses with 30-s breaks on ice between pulses. Insoluble material was removed by centrifugation at 17,500 × g for 15 min at 4 °C. Soluble cell lysate was diluted to a final volume of 50 ml with ACB and loaded onto a 2-ml amylose resin column equilibrated in ACB. After washing with 20 bed volumes of ACB, protein was eluted with 3 ml of ACB supplemented with 10 mm maltose and collected in 8–10 fractions (∼300 μl/fraction). For consistency in comparative analyses, 5-μl samples were fractionated on SDS-polyacrylamide gels (44). For Western blots, proteins were transferred to nitrocellulose membranes and probed with primary antibodies prior to incubation with alkaline phosphatase-conjugated secondary antibodies and colorimetric development as described by Harlow and Lane (45). His6-tagged proteins were purified using nickel ion affinity chromatography following standard protocols, including imidazole elution, as recommended by the manufacturer (Novagen).
Protein Labeling and Microscopy
Recombinant IFT46 (His6-46F) and a control protein, BSA (Fraction V, Fisher), were fluorescently labeled with Alexa-fluor 488-activated succinimidyl ester (Invitrogen) following the recommended procedures. Briefly, bacterially expressed His6-46F was purified on a Ni2+ column following protocols recommended by Novagen. Purified His6-46F was concentrated to ∼10 mg/ml using an Amicon Ultra centrifugal concentration column with a 5000 Da limit (Millipore) and exchanged into conjugation buffer (0.1 m sodium bicarbonate, pH 8.3) using a 5-ml Sephadex G-25 (GE Healthcare) centrifugation column as described by Penefsky (46). After the reactive Alexa-fluor ester was solubilized in DMSO at a final concentration of 10 mg/ml, 50 μl was added slowly to 500 μl of His6-46F or BSA at ∼10 mg/ml with continuous agitation using a micro-stir bar for 1 h at room temperature. Free dye was removed, and labeled protein was exchanged into electroporation buffer using a 5-ml Sephadex G-25 centrifugation column. The ratio of fluorophore to protein following gel filtration was calculated to be ∼1.5 and ∼1.2 for His6-46F and BSA, respectively. Fluorescent protein was visualized within the ift46-2 mutant using an LSM 510 META confocal microscope (Carl Zeiss Microimaging, Inc.).
Chemical Cross-linking
Chemical cross-linking and isolation of cross-linked products were performed as described previously (33). In brief, soluble flagellar proteins were fractionated by high salt (300 mm NaCl in HMDEK buffer containing 10 mm Hepes, pH 7.2, 5 mm MgSO4, 1 mm DTT, 0.5 mm EDTA, and 25 mm KCI) sucrose density gradient centrifugation. The IFT complex B core fractions were pooled, divided into equal aliquots, and treated with 1,5-diflouro-2,4-dinitrobenzene (DFDNB; Pierce) at final concentrations of 0.0, 0.1, 0.3, 1.0, and 3.0 mm for 10 min on ice before being quenched with 10 mm Tris-HCl, pH 8.5. The complex B core was then selectively immunoprecipitated from each aliquot with ∼25 μl of an anti-IFT81 antibody resin (33). The resin was batch-washed three times in a 0.5-ml microcentrifuge tube using 15 bed volumes of HMEK-300 prior to a 95 °C 3-min incubation and elution with ∼25 μl of 2× SDS sample buffer. Eluted proteins were separated on 4.0% SDS-polyacrylamide gels and visualized with Coomassie Blue. Protein bands containing cross-linked products were excised from the gels, digested with trypsin, and analyzed by matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry. Resulting peptide masses were compared with predicted tryptic IFT particle protein peptides using Protein Prospector (47).
RESULTS
IFT52 and IFT46 Interact
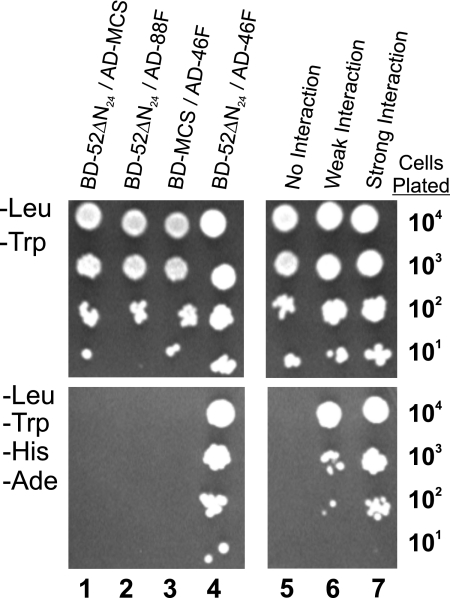
The protein subunits of the Chlamydomonas intraflagellar transport complex B are encoded by at least 13 distinct genes, known as IFT172, IFT88, IFT81, IFT80, IFT74/72, CrDYF-1, IFT57, IFT52, IFT46, IFT27, IFT25, IFT22, and IFT20 (reviewed in Ref. 26). In an attempt to identify which of the B subunits interact with one another, we tested every pairwise combination of B proteins, with the exception of CrDYF-1, using an exhaustive yeast-based two-hybrid screen. A direct interaction between IFT81 and IFT74/72 and a homodimeric interaction between two IFT81 proteins were described previously (33). In the extensive two-hybrid screen described here, the only additional interaction identified was between IFT52 and IFT46 (Fig. 1). In this assay, the GAL4 transcriptional activation domain was fused to the amino terminus of full-length IFT46 (AD-46F), whereas the GAL4 DNA binding domain was fused to the N-terminal end of a nearly full-length IFT52 (amino acids 25–454; BD-52ΔN24); cloning issues prevented use of a full-length IFT52 fusion protein. The combination of these two plasmids allowed yeast colonies to grow on selective medium missing histidine and adenine (−His, −Ade). The reciprocal experiment where the IFT46 and IFT52 fusion proteins are switched was not possible due to false positive results with the BD-46F construct (data not shown). As a verification of the IFT52-IFT46 interaction, we used the BD-52ΔN24 plasmid to screen a Volvox carteri two-hybrid library generously provided by Stephen Miller (University of Maryland). Using the Matchmaker mating protocol suggested by the manufacturer (Clontech), the initial library screen generated 18 positive clones, eight of which encoded for portions of the Volvox IFT46 protein. Besides verifying the IFT52-IFT46 interaction, this screen showed that Chlamydomonas proteins could be successful baits to screen a Volvox two-hybrid library.
FIGURE 1.
Direct interaction of IFT46 and IFT52 using yeast-based two-hybrid analysis. Serial dilutions of YRG-2 yeast containing both AD and BD plasmid constructs were grown on selective (−Leu, −Trp) medium (top panels) to verify the presence of both AD and BD plasmids and a more restrictive (−Leu, −Trp, −His, −Ade) medium (bottom panels) to test for protein interactions. Column 1, the lack of growth in the absence of histidine and adenine revealed that IFT52 did not interact with the control AD protein. Column 2, the lack of cell growth on the more restrictive medium (−His, −Ade) revealed that IFT52 and IFT88 failed to interact in this assay. Column 3, IFT46 (AD-46F) showed no interaction with the control BD protein. Column 4, the combination of the BD-52ΔN24 and AD-46F plasmids conferred growth in the absence of histidine and adenine, which was consistent with a direct interaction between IFT52 and IFT46. Columns 5–7, negative, weak, and strong interaction control plasmids are described under “Experimental Procedures.”
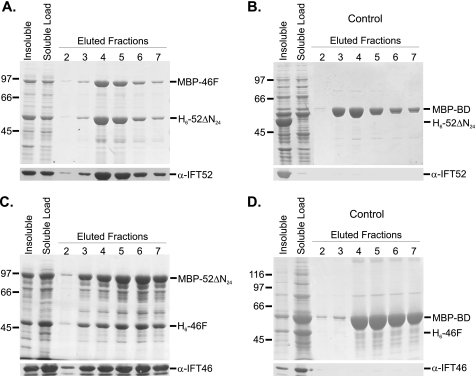
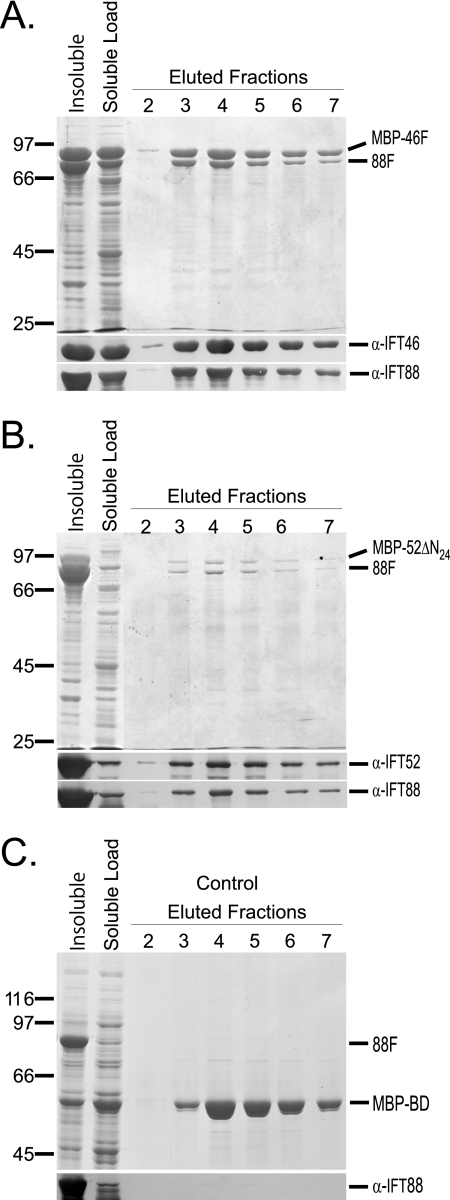
To confirm the two-hybrid results, Chlamydomonas IFT46 and IFT52 were coexpressed in a single bacterial host using hexahistidine (His6 or H6) and maltose-binding protein (MBP) purification tags. First, MBP-46F and H6-52ΔN24 were coexpressed, followed by amylose affinity chromatography to purify the MBP-tagged IFT46. As shown in Fig. 2A, the H6-52ΔN24 coeluted with IFT46, demonstrating that the bacterially expressed proteins interacted; control expressions using an MBP-BD fusion demonstrated that IFT52 did not interact with the MBP affinity tag (Fig. 2B). Interestingly, densitometry indicated that twice as many (1.9-fold) of the H6-52ΔN24 proteins co-purified with each MBP-46F. Because the amylose chromatography was specific for the maltose-binding protein, this result suggested that either IFT46 has two separate IFT52 binding sites or that the IFT52 was able to homodimerize. Our in vitro interaction analyses, however, have provided no evidence that IFT52 is capable of homodimerization (data not shown). When IFT52 was fused to the maltose-binding protein (MBP-52ΔN24) and subsequently coexpressed with His-tagged IFT46 (H6-46F), the two proteins coeluted from an amylose column at a stoichiometry of nearly 1:1 (Fig. 2C). Because the native B complexes isolated from flagella contain stoichiometric levels of both IFT46 and IFT52, we chose to pursue deletion analysis using the MBP-tagged IFT52 and His6-tagged IFT46 protein constructs, and, importantly, the His6-IFT46 deletion constructs were significantly more soluble relative to IFT52 constructs.
FIGURE 2.
Coexpression and copurification of recombinant IFT46 and IFT52 using amylose affinity chromatography. The first two lanes of each Coomassie Blue-stained gel contain insoluble and soluble fractions of bacterial cell lysates. The soluble fraction was loaded onto amylose MBP affinity resin, washed with column buffer (ACB), and eluted using 10 mm maltose in ACB; fractions 2–7 are shown here. Western blots were probed with either anti-IFT52 or anti-IFT46, as indicated. A, following coexpression, H6-52ΔN24 coeluted with MBP-46F. B, H6-52ΔN24 did not copurify with the MBP-BD control. C, following coexpression, H6-46F coeluted with MBP-52ΔN24. D, H6-46F did not copurify with the MBP-BD control.
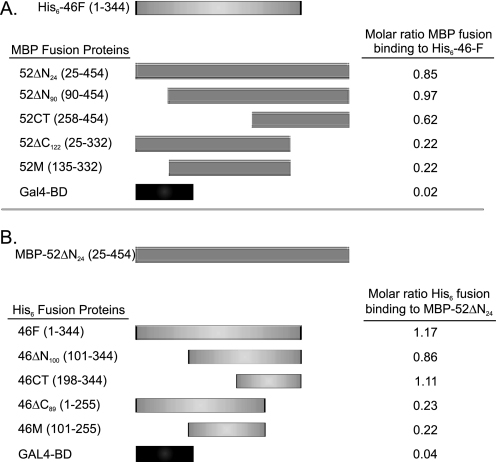
For higher resolution mapping of the IFT52- and IFT46-interacting domains, deletion analysis using bacterial coexpression was performed. As summarized in Fig. 3A, full-length IFT46 (H6-46F) was coexpressed with various MBP-IFT52 constructs, followed by amylose affinity chromatography. Deletion of the N-terminal 90 amino acids of IFT52 (MBP-52ΔN90) did not affect the ability of the two proteins to copurify at nearly equimolar stoichiometries. Removal of the N-terminal 258 amino acids (MBP-52CT) did reduce the ratio of IFT46 to IFT52 to 0.62. However, removing the C-terminal 122 amino acids resulted in an 80% decrease of the IFT46/IFT52 ratio, suggesting the IFT52 C terminus plays a pivotal role mediating the interaction between these two proteins. A similar analysis was performed by screening for copurification of MBP-52ΔN24 with deletion constructs of His-tagged IFT46 (Fig. 3B). In this case, over 50% of the amino terminus of IFT46 (197 amino acids) could be removed before stoichiometric copurification was compromised. Removing the C-terminal 89 amino acids from IFT46, however, was sufficient to drop the ratio of IFT46 to IFT52 to ∼0.2. In summary, the C termini of both IFT52 and IFT46 were necessary to generate stoichiometric interactions between the two proteins when coexpressed in the same host.
FIGURE 3.
Interactions between IFT46 and IFT52 are mediated by C-terminal domains. Pairwise combinations of proteins were coexpressed. Interactions were monitored using MBP affinity chromatography. Molar ratios of copurified proteins were determined using densitometric scanning of Coomassie Blue-stained gels. A, full-length IFT46 (His6-46F) was coexpressed with various MBP-tagged IFT52 deletion and control proteins. B, nearly full-length IFT52 (MBP-52ΔN24) was coexpressed with various His6-46 deletion and control proteins.
Recombinant IFT46 Rescues Ciliogenesis
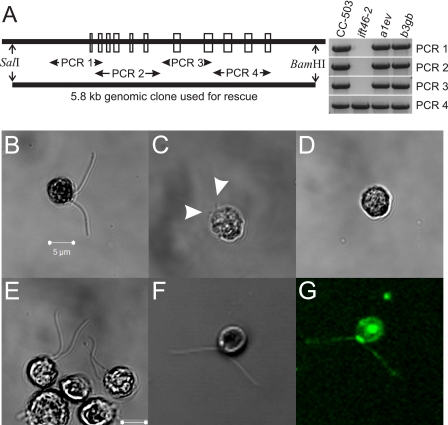
Biological activity of recombinant His6-tagged IFT46F protein was tested via electrical insertion of purified H6-46F into a newly identified ift46 mutant (Fig. 4). The ift46-2 mutant strain was identified by screening Chlamydomonas motility mutants generated by random insertional mutagenesis with the hygromycin B resistance gene, aph7″ (37–39). Like the parental CC-503 strain, the ift46-2 cells were conveniently cell wall-deficient, facilitating subsequent DNA transformation and protein electroporation. Similar to the recently described ift46-1 strain that assembles short, stumpy flagella (29), a minor fraction (∼6%) of the ift46-2 cells displayed short flagella of 1–5 μm (∼3 μm average). Extremely short flagella of less than 1 μm were not reliably visible and could not be counted. To confirm that the assembly phenotype resulted specifically from the loss of IFT46, ciliogenesis was rescued by transforming the mutant strain using a 5.8-kb SalI/BamHI genomic fragment that contained the intact IFT46 gene (Fig. 4A). PCR analysis showed that most of the IFT46 gene was absent in the ift46-2 strain but had been replaced in two transformed strains, a1ev and b3gb, that had recovered flagellar assembly (Fig. 4A).
FIGURE 4.
Electroporation of recombinant His6-46F rescues the ift46-2 flagellar assembly phenotype. A, a screen of random insertional motility mutants revealed a C. reinhardtii strain, ift46-2, that carries a deletion of most of the IFT46 gene; the exons of the wild-type IFT46 are depicted by open boxes. The lack of specific PCR amplification products (PCR 1–3) revealed that most of the IFT46 gene was disrupted in the ift46-2 strain. A 5809 bp SalI/BamHI genomic fragment was used to rescue the ift46-2 flagellar assembly phenotype. PCR analysis of two rescued strains, a1ev and b3gb, revealed that the IFT46 gene had been successfully reintroduced. B, the cell wall-deficient parental CC-503 strain assembles flagella of normal length and function. C, although most ift46-2 cells were bald, ∼6% were able to assemble short flagella (average ∼3 μm) as indicated by arrowheads. D, electroporation of BSA into ift46-2 resulted in no change in the bald phenotype; the image shown was taken 4 h postelectroporation. E, at 4 h postelectroporation of recombinant His6-IFT46, many cells displayed partial or full assembly of motile flagella. F and G, electroporation of Alexa-fluor 488-labeled His6-46F resulted in similar rates of rescue; 4-h postelectroporation images shown are white light (F) and emission at 518 nm (G). Many flagellated cells displayed concentrated pools of fluorescent IFT46 near the basal bodies with punctate staining throughout the flagella. Intraflagellar transport of the labeled IFT46 could also be observed in some cells.
To further verify the phenotype of the ift46-2 strain, purified H6-46F was used to rescue flagellar assembly. The recombinant protein was introduced into ift46-2 cells, as described previously for protein electroporation into Chlamydomonas (43, 48), except that it was unnecessary to treat the cell wall-deficient strain with autolysin to remove cell walls. Following electroporation and dilution with TAP medium, the cells were undisturbed with the exception of occasional gentle mixing prior to microscopic analysis. Within 2 h, a small percentage of cells displayed irregular non-Brownian movement, whereas at 4 h, an even greater number of cells displayed normal swimming behavior, which included normal photophobic and phototactic responses to varying illumination. The phototrophic responses allowed easy enrichment of swimming cells to record their movement, which was similar to that of the parental CC-503 cells (supplemental Movies 1 and 2). In contrast, ift46-2 cells receiving either no protein or the BSA control never recovered flagellar assembly or the ability to swim (supplemental Movie 3). For quantification of flagellar assembly, cells were fixed with Lugol's iodine stain, and flagellar lengths were measured at various times following electroporation using a final concentration of 0.1 mg/ml purified H6-46F. At 8 h postelectroporation, 22% of the cells had assembled two flagella that were 50–100% as long as the parental CC-503 strain, as shown in Fig. 4, E–G. As expected, the BSA control failed to rescue any flagellar assembly (Fig. 4D).
In order to visualize recombinant proteins in live cells, H6-46F and control BSA were conjugated with Alexa-fluor 488 (Alexa-H6-46F and Alexa-BSA) prior to electroporation. At 4 h postelectroporation, Alexa-H6-46F was often found concentrated near basal bodies of rescued cells (Fig. 4G), resembling patterns of previous IFT localization studies (20, 29, 31, 49). In some cells, punctate staining of Alexa-H6-46F could be observed within flagella, a phenomenon commonly observed with IFT localizations. Furthermore, movement of Alexa-H6-46F via IFT could also be monitored in select cells, making this the first time that intraflagellar movement of a chemically labeled IFT protein has been visualized in any organism (supplemental Movie 4). In control experiments, Alexa-BSA could sometimes be seen in pools near the basal bodies, but flagellar assembly was never rescued (data not shown).
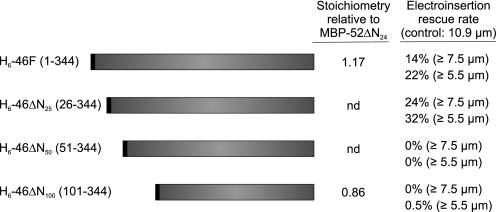
Next we exploited the recombinant rescue approach to examine the biological effect of N-terminal deletions of the IFT46 protein. The amino terminus was chosen because deletions removing up to 198 N-terminal amino acids of IFT46 did not significantly reduce interaction with IFT52. Electroporation of ift46-2 was performed in the presence of 0.1 mg/ml BSA or recombinant IFT46 protein. It was observed that removal of the first 25 amino acids (H6-46ΔN25) had no deleterious effect on the ability of the protein to rescue flagellar assembly (Fig. 5). Removal of 50 or 100 amino acids, however, completely inhibited the ability of the IFT46 protein to rescue ciliogenesis (supplemental Fig. 1). Thus, although H6-46ΔN100 was capable of binding directly to IFT52 (Fig. 3), this association was not sufficient to restore full biological activity.
FIGURE 5.
The N-terminal 25 amino acids of IFT46 are not required for recombinant protein rescue. Recombinant His6-46 proteins with the indicated N-terminal deletions were purified by metal chelate chromatography and introduced into ift46-2 cells using electroporation with a final concentration of recombinant protein at 0.1 mg/ml. For comparison, the stoichiometry of the full-length IFT46 (H6-46F) and the N-terminal 100-amino acid deletion (H6-46ΔN100) relative to the MBP-tagged IFT52 are shown (see Fig. 3). Both the full-length and N-terminal 25-amino acid deletion IFT46 proteins were capable of similar rates of flagellar assembly rescue 8 h postelectroporation. nd, not determined.
IFT88 Can Be Cross-linked to IFT52
To identify candidate IFT46 or IFT52 interactors, we chemically cross-linked neighboring proteins within the complex B core. Consisting of a subset of the complex B subunits, the Chlamydomonas B core was first isolated from flagella using moderately high salt (HMEK buffer plus 300 mm NaCl) sucrose density gradient centrifugation (supplemental Fig. 2) (33). The gradient fractions enriched in the B core (∼11 S) were pooled and treated with the hydrophobic 3.0-Å cross-linker, DFDNB. After blocking the unreacted DFDNB, the B core was immunoprecipitated with an anti-IFT81 resin and fractionated by low percentage acrylamide (4%) SDS-PAGE as shown in supplemental Fig. 2B. Prominent cross-linked species were excised from the gel, digested with trypsin, and then analyzed by MALDI-TOF mass spectrometry.
Cross-linked band 1 (supplemental Fig. 2B) contained only IFT81 and IFT74/72, as determined previously in similar DFDNB experiments (33). Cross-linked band 2 contained 16 tryptic masses falling between 900 and 2500 Da (Table 1). Seven of the masses were uniquely attributed to predicted tryptic peptides derived from IFT52 (∼23% coverage), whereas seven were attributed to IFT88 peptides (∼9% coverage). Two peptide masses matched more than one peptide sequence predicted for IFT52 and IFT88 and one mass could not be attributed to either IFT52 or IFT88. Similar MALDI-TOF analysis of cross-linked band 3 yielded a small number of peptides from IFT81 and IFT27, but the percentage of coverage for both proteins was correspondingly small (∼5% coverage each), so further analysis would be necessary to verify an association between these two subunits. Finally, the relative SDS-PAGE mobilities for the 88-52 and 81-27 cross-linked products, are within 10% of the predicted mobilities for the total masses of the respective products. Together, these results indicate that IFT88 and IFT52 are close enough in the complex B core to be linked together with a 3-Å cross-linker. Thus, IFT88 became a prime candidate to test for direct interactions with recombinant IFT52 and its binding partner, IFT46.
TABLE 1.
MALDI-TOF mass spectrometry analysis of tryptic peptides from cross-linked band 2
A total of 16 tryptic peptide masses were identified from band 2; 15 of those are listed below. One mass, 986.5200 Da, matched predicted sequences from both IFT52 and IFT88. Another mass, 1316.6300, matched two unique sequences from IFT88. One mass, 1357.73 Da, could not be attributed to any IFT protein.
| m/z submitted | MH+ matched | IFT52 peptide sequence | Modifications |
|---|---|---|---|
| Da | Da | ||
| 947.4700 | 947.4701 | IMDFFFK | |
| 986.5200 | 986.5423 | HVLSEVFR | |
| 1228.7100 | 1228.6901 | EVLISDGILNR | |
| 1473.7100 | 1473.6902 | DWTSLFDDSLFK | |
| 1681.8000 | 1681.8689 | FDTGLIPEAVSLYEK | |
| 2203.0500 | 2203.1235 | IAYPMNRPVGAVWAQPGYGR | |
| 2285.1100 | 2285.1414 | STYRPDKVDKDDFTLDTLR | |
| 2474.1300 | 2474.1727 | EPPPPALELFDLDESFASETNR |
| m/z submitted | MH+ matched | IFT88 peptide sequence | Modifications |
|---|---|---|---|
| 905.5400 | 905.5209 | NIGLSFVR | |
| 986.5200 | 986.5522 | LANEVFLAK | |
| 1104.5800 | 1104.5591 | NFPQSGWLR | |
| 1303.6600 | 1303.6184 | ALHYYQESHR | |
| 1316.6300 | 1316.6421 | VNMGNIHFEQK | |
| 1316.6300 | 1316.6455 | MLVKEHMGGGGGK | Met-ox |
| 1349.7400 | 1349.7469 | LNELPYALAAFK | |
| 1626.7900 | 1626.8491 | NYSEALNLYTAIVR | |
| 2016.9600 | 2017.0759 | WFELLTASLVSNDPGVLAR |
IFT88 Can Bind Directly to either IFT52 or IFT46
Expression of recombinant IFT88 did not include any affinity tags, such as MBP or His6; the full-length IFT88 cDNA, including the native stop codon, was inserted into the second multicloning site (MCSII) of the RSF Duet vector. Untagged IFT88 was then coexpressed with either MBP-46F or MBP-52ΔN24, followed by amylose affinity chromatography of the MBP fusion proteins (Fig. 6). IFT88 copurified with MBP-46F and MBP-52ΔN24 but did not copurify with the control MBP-Gal4 DNA binding domain (MBP-BD). It should be noted that very little of the recombinant IFT88 was soluble when expressed alone or when coexpressed with either the control protein or MBP-52ΔN24. More of the IFT88 was soluble when coexpressed with MBP-46F, suggesting that binding to IFT46 stabilized IFT88. An even more dramatic IFT46-dependent increase in the solubility of recombinant IFT52 can be seen by comparing A and B of Fig. 2. Very little of the H6-52ΔN24 was soluble when coexpressed with the control fusion, whereas ∼40% of the recombinant H6-52ΔN24 was soluble when coexpressed with MBP-46F. Combined, these results suggest that IFT46 plays a stabilizing role for both IFT52 and IFT88.
FIGURE 6.
IFT88 interacts separately with either IFT52 or IFT46. Untagged IFT88 was coexpressed separately with MBP fusions of IFT52, IFT46, and BD (negative control) proteins. The first two lanes of each gel contain the insoluble and soluble fractions of bacterial cell lysates. The soluble fraction was loaded onto amylose MBP-affinity resin and washed with ACB buffer prior to 10 mm maltose elution. Elution fractions 2–7 are shown on the Coomassie Blue-stained SDS-polyacrylamide gels. Corresponding Western blots are shown below each gel. A and B, the untagged IFT88 copurifies with MBP-46F and MBP-52ΔN24, respectively. C, IFT88 does not copurify with the MBP-BD control protein.
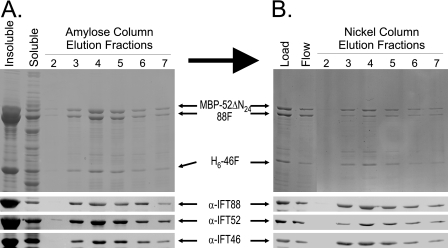
To test for simultaneous interactions, all three proteins were coexpressed in a single host using the MBP-52ΔN24, H6-46F, and the untagged 88F constructs. Soluble cell extract was first fractionated using amylose affinity purification (Fig. 7A). Not surprisingly, both H6-46F and 88F copurified with the MBP-52ΔN24. To show that all three proteins were together in a single complex, the peak MBP fraction was further fractionated using nickel chelate affinity chromatography to specifically purify the H6-46F (Fig. 7B). Even with the high salt conditions required for this chromatography, significant portions of both the MBP-52ΔN24 and 88F coeluted with the His-tagged IFT46. These results indicate that IFT46, IFT52, and IFT88 are capable of forming an independent complex in the absence of other complex A or B subunits. Subsequent sucrose density gradient centrifugation of the peak fractions (not shown) resulted in broad distributions of all three proteins, which is consistent with the presence of a heterotrimeric complex in addition to some higher order complexes.
FIGURE 7.
Copurification of H6-46F, MBP-52ΔN24, and IFT88 using tandem affinity chromatography. Upper panels, Coomassie Blue-stained SDS-polyacrylamide gels; lower panels, transfer membranes probed with antibodies directed against IFT88, IFT52, or IFT46. A, MBP affinity purification of coexpressed recombinant H6-46F, MBP-52ΔN24, and untagged IFT88. Soluble bacterial lysate was loaded onto an amylose column. After washing the resin, MBP-52ΔN24 and associated proteins were eluted using 10 mm maltose; fractions 2–7 are shown here. Both H6-46F and the untagged IFT88 coeluted with the MBP-52ΔN24. B, the peak fractions from the MBP affinity chromatography (A) were pooled and further purified using metal (Ni2+) chelate chromatography. Elution of the H6-46F from the Ni2+ resin using imidazole resulted in the coelution of both MBP-52ΔN24 and IFT88, indicating that the three proteins were capable of forming a stable ternary complex.
DISCUSSION
IFT46, IFT52, and IFT88 Form a Ternary Complex within the Complex B Core
As isolated from Chlamydomonas flagella, IFT complex B was initially reported to contain at least 12 polypeptide subunits (20, 24, 25), which are believed to include two copies each of IFT81 and IFT74/72 (33). Recently, three additional B polypeptides, IFT25, IFT22, and CrDYF-1, have been reported (34, 50), bringing the total number of algal complex B subunits to at least 15. The Chlamydomonas B complex is labile to higher ionic strength and can be partially dissociated to reveal a semistable core containing IFT88, 2× IFT81, 2× IFT74/72, IFT52, IFT46, IFT27, IFT25, and IFT22 (33, 34).4 Our previous analysis of the complex B core combined chemical cross-linking with yeast-based two- and three-hybrid analyses to show that IFT81 and IFT74/72 are able to form a heterotetrameric subcomplex consistent with the experimentally observed ratio of IFT81/IFT74/IFT72 of 2:1:1 in isolated IFT complexes. Initially, we employed these same techniques to identify additional interactions between complex B subunits. As reported here, however, exhaustive two-hybrid analysis only identified one additional B interaction, that between IFT52 and IFT46 (Fig. 1). A yeast-based two-hybrid interaction between mammalian IFT20 and IFT57/Hippi has been reported (35), but we were unable to confirm a similar interaction between the algal orthologs using either two-hybrid analysis or bacterial coexpression. Chemical cross-linking did, however, identify two additional B associations between IFT88 and IFT52 and between IFT81 and IFT27.
The small number of interactions identified using the yeast-based assay was disappointing because deduction implies that a minimum of 14 distinct interactions would occur for a complex containing 15 subunits. Because some complex B subunits are likely to interact with multiple neighboring subunits, the total number of protein-protein interactions within complex B could be much greater than 14. The inability to identify many of these expected interactions by two-hybrid analysis led to the hypothesis that assembly of the B complex might follow a stepwise process.
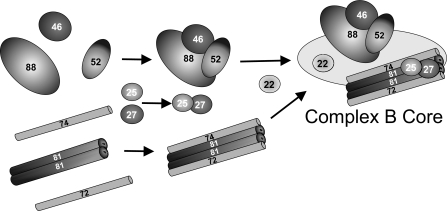
To test the ordered assembly hypothesis, we utilized a heterologous bacterial expression system (Duet vectors, Novagen), which allows simultaneous coexpression of up to eight proteins. Each protein can be expressed with an optional tag, such as a His6 or S·TagTM that can be used as an epitope for Western blot analysis or for affinity chromatography and/or batch-wise pull-downs. These studies were initiated with Chlamydomonas IFT88, IFT52, and IFT46 based on the two-hybrid IFT52-IFT46 interaction (Fig. 1) and the chemical cross-linking of IFT88 and IFT52 (supplemental Fig. 2). We show here that all three bacterially expressed proteins, IFT88, IFT52,, and IFT46, are able to interact independently with one another (Figs. 2 and 6). Furthermore, tandem affinity chromatography showed that when all three proteins were expressed simultaneously, the three can form a ternary complex (Fig. 7). Because any two of these three proteins are capable of sustaining an interaction, there may not be a specific order in which the three must interact for in vivo assembly. These results, however, demonstrate that these three proteins are capable of forming a subcomplex in the absence of additional B subunits. This allows us to suggest a working model of complex B assembly where IFT88, -52, and -46 subunits form an independent trimer, whereas IFT81 and IFT74/72 form an independent tetramer, and IFT27 and IFT25 form an independent dimer (Fig. 8). These three subcomplexes would then associate with one another along with the remaining core subunit, IFT22. Wang et al. (34) has recently shown that a portion of an IFT27-IFT25 subcomplex fractionates independently of complex B, indicating that these two proteins can also interact in the absence of any other B proteins. The more peripheral B subunits (e.g. IFT172, IFT80, etc.) could add on at any point in the assembly of the complete complex but should not be required to assemble the core.
FIGURE 8.
Hypothetical model of in vivo assembly of the IFT complex B core. IFT46, IFT52, and IFT88 form a ternary complex prior to assembly with the IFT81, IFT74/72 tetramer. Previous studies have shown that the IFT81 and IFT74/72 subunits and the IFT27 and IFT25 subunits are capable of forming stable associations in the absence of other complex B proteins (33). The actual order in which these and additional subunits assemble onto the core is unknown.
The results described here do provide evidence that IFT46 serves to stabilize IFT52 and IFT88. Repeated attempts to generate appreciable amounts of soluble IFT52 or soluble IFT88 failed when either protein was expressed by itself. Coexpression with IFT46, however, greatly increased the total fraction of soluble recombinant protein, suggesting that IFT46 helps to stabilize IFT52 and IFT88 (Figs. 2 and 6). When IFT52 (bld1) or IFT88 (ift88) is absent from Chlamydomonas, cells are completely bald and fail to assemble any flagellar structures past the transition zone. When IFT46 is absent (ift46-1, ift46-2), however, a fraction of the Chlamydomonas cells are able to assemble short flagella (Fig. 4) (29). These combined observations suggest that an important role for IFT46 is to provide stabilization for the IFT52 and IFT88 subunits. It has already been documented, however, that IFT46 also has a specific role in the assembly of outer dynein arms through a direct association with ODA16 (30). Thus, IFT46 may well serve multiple roles associated with IFT and flagellar assembly and function.
The importance of these three complex B proteins (IFT88, IFT52, and IFT46) in the assembly of cilia and flagella has been documented in diverse organisms. The Caenorhabditis elegans IFT52 gene, OSM-6, was the first IFT particle gene to be established as ciliogenic (20, 51). In addition to ciliogenesis, the mammalian IFT52 homologue, Ngd5, also has an essential role in sonic hedgehog (Shh) signaling (52). The mouse homologue to IFT88, TG737/Polaris, was the first IFT particle protein to be associated with a major mammalian ciliopathy, polycystic kidney disease (28, 53). In addition to ciliogenesis (28, 54), the mammalian IFT88 is also essential for Shh signaling (52, 55). Like IFT52 (OSM-6), IFT88 (OSM-5) and IFT46 (DYF-6) are each required for ciliogenesis in the nematode (56, 57). In summary, the common ciliogenic functions uniting these three IFT proteins reflect their importance to IFT and a conserved role in ciliary assembly and function.
Electroporation of Recombinant IFT Proteins Can Screen for Functional Domains
The heterologous expression assays described above are useful in identifying protein-protein interactions and dissecting the subunit architecture of the IFT complexes. These assays, however, do little to address the biological functions of structural proteins, such as these. In order to address specific functions, one can transform appropriate mutant strains with a variety of gene constructs to see what effect, if any, specific modifications have on the restoration of wild-type behavior. Transformation with the intact gene generally confers stable rescue of the phenotype resulting from the loss of said gene. For example, transformation of ift46-2 with a 5.8-kb genomic fragment containing the intact IFT46 gene was sufficient to rescue the flagellar assembly defect. One caveat to this very popular approach, however, is that it can take considerable time to isolate and characterize stable cell lines following transformation. A much quicker approach is to deliver recombinant protein directly into the appropriate mutant cells. Electroporation of bacterially expressed protein into Chlamydomonas was first used by Kamiya and co-workers (43, 48) to replace axonemal components required for normal flagellar beating. Although the rescue is temporary (≤24 h), we have adopted this approach to show that recombinant IFT46 can rescue flagellar assembly of treated ift46-2 cells. This sort of assay can test the biological activity of a modified protein in less than a day. As an example, we used this approach to show that the N-terminal 25 amino acids of IFT46 are not required to rescue ciliogenesis. In contrast, IFT46 does appear to require at least a portion of amino acids 26–50 in order to confer flagellar assembly. In order to address how the loss of amino acids 26–50 might affect complex B formation, future biochemical analysis could be directed at cell lines stably transformed with DNA constructs expressing either IFT46ΔN25 or IFT46ΔN50. Additionally, it will be interesting to see if the N-terminal domain of IFT46 is responsible for interaction with other proteins. ODA16, for example, is a flagellar protein that was recently found to interact with IFT46 (30); loss of the N-terminal 50 amino acids of IFT46 might interfere with the flagellar function of ODA16 or other proteins.
Direct electroporation of recombinant protein also raises the possibility that chemically labeled IFT proteins can be monitored in live cells. In our experiments, Alexa-fluor 488-labeled IFT46 was able to rescue ift46-2 cells with similar efficiency as the unlabeled IFT46 (Figs. 4 and 5). As expected for IFT proteins, the labeled IFT46 was concentrated near the base of the organelle with some protein distributed within the organelle. Active intraflagellar transport of the fluorescently tagged 46F could also be monitored, which raises the possibility that future experiments could include the electroporation of two or more IFT proteins containing distinct labels to analyze proximities and transport properties.
In conclusion, cilia and flagella are incredibly diverse organelles that are important for cellular motility and signaling. Due, in part, to these critical functions, cilia and flagella have essential roles during early development and sensory transduction. Defects that disrupt the important functions of these organelles have been linked to an expanding group of human diseases known collectively as ciliopathies. Because IFT is responsible for the assembly and function of these organelles, understanding the molecular mechanisms by which IFT operates is especially important. Although the identities of most IFT proteins are known, the specific functions of each subunit are poorly understood. This study has specifically addressed the architecture of complex B by exploring specific interactions between selected subunits. In summary, IFT88, IFT52, and IFT46 interact with one another and are able to form a ternary complex, which we believe to exist within the complex B core. Furthermore, the conserved ciliogenic nature of these three proteins leads us to believe that the IFT88-IFT52-IFT46 ternary complex may act as a scaffold to bind multiple B subunits and IFT-associated cargos.
Supplementary Material
Acknowledgments
We are grateful to Dr. Stephen M. Miller (University of Maryland) for the generous donation of a V. carteri two-hybrid library. We thank Drs. Hongmin Qin (Texas A&M University) and Joel L. Rosenbaum (Yale University) for sharing unpublished sequences and IFT antibodies. We are grateful to Dr. John Leszyk (University of Massachusetts Medical School) for expert analysis using MALDI-TOF mass spectrometry. We thank Drs. Allan Caplan, Elizabeth Fortunato, and Gustavo Arrizabalaga (University of Idaho) for valuable advice and generous donation of microscope time and facilities. We gratefully acknowledge the Washington State University Franceschi Microscopy and Imaging Center (Pullman, WA) for assistance with confocal microscopy. Last, we thank all members of the Cole laboratory who have provided critical feedback and technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants R01-GM61920 (to D. G. C.) and P20-RR016454 from the Idea Networks of Biomedical Research Excellence (INBRE) Program of the National Center for Research Resources.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and Movies S1–S4.
B. F. Lucker, M. S. Miller, S. A. Dziedzic, P. T. Blackmarr, and D. G. Cole, unpublished results.
- IFT
- intraflagellar transport
- AD
- GAL4 activation domain
- BD
- GAL4 DNA binding domain
- MBP
- maltose-binding protein
- H6
- His6
- DFDNB
- 1,5-diflouro-2,4-dinitrobenzene
- ACB
- amylose chromatography buffer
- TAP
- Tris-acetate-phosphate
- MCS-1 and -2
- multicloning site 1 and 2, respectively
- MALDI-TOF
- matrix-assisted laser desorption ionization time-of-flight.
REFERENCES
- 1.Wheatley D. N. (1995) Pathobiology 63, 222–238 [DOI] [PubMed] [Google Scholar]
- 2.Bray D. (2001) Cell Movements: From Molecules to Motility, pp. 3–16, 225–241, Garland Publishing, New York, NY [Google Scholar]
- 3.Praetorius H. A., Spring K. R. (2005) Annu. Rev. Physiol. 67, 515–529 [DOI] [PubMed] [Google Scholar]
- 4.Davis E. E., Brueckner M., Katsanis N. (2006) Dev. Cell 11, 9–19 [DOI] [PubMed] [Google Scholar]
- 5.Pazour G. J., Witman G. B. (2003) Curr. Opin. Cell Biol. 15, 105–110 [DOI] [PubMed] [Google Scholar]
- 6.Bisgrove B. W., Yost H. J. (2006) Development 133, 4131–4143 [DOI] [PubMed] [Google Scholar]
- 7.Eggenschwiler J. T., Anderson K. V. (2007) Annu. Rev. Cell Dev. Biol. 23, 345–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Afzelius B. A. (2004) J. Pathol. 204, 470–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pan J., Wang Q., Snell W. J. (2005) Lab. Invest. 85, 452–463 [DOI] [PubMed] [Google Scholar]
- 10.Badano J. L., Mitsuma N., Beales P. L., Katsanis N. (2006) Annu. Rev. Genomics Hum. Genet. 7, 125–148 [DOI] [PubMed] [Google Scholar]
- 11.Fliegauf M., Benzing T., Omran H. (2007) Nat. Rev. Mol. Cell Biol. 8, 880–893 [DOI] [PubMed] [Google Scholar]
- 12.Marshall W. F. (2008) J. Cell Biol. 180, 17–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosenbaum J. L., Witman G. B. (2002) Nat. Rev. Mol. Cell Biol. 3, 813–825 [DOI] [PubMed] [Google Scholar]
- 14.Scholey J. M. (2003) Annu. Rev. Cell Dev. Biol. 19, 423–443 [DOI] [PubMed] [Google Scholar]
- 15.Cole D. G. (2003) Traffic 4, 435–442 [DOI] [PubMed] [Google Scholar]
- 16.Kozminski K. G., Johnson K. A., Forscher P., Rosenbaum J. L. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 5519–5523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kozminski K. G., Beech P. L., Rosenbaum J. L. (1995) J. Cell Biol. 131, 1517–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shakir M. A., Fukushige T., Yasuda H., Miwa J., Siddiqui S. S. (1993) Neuroreport 4, 891–894 [DOI] [PubMed] [Google Scholar]
- 19.Walther Z., Vashishtha M., Hall J. L. (1994) J. Cell Biol. 126, 175–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cole D. G., Diener D. R., Himelblau A. L., Beech P. L., Fuster J. C., Rosenbaum J. L. (1998) J. Cell Biol. 141, 993–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Signor D., Wedaman K. P., Rose L. S., Scholey J. M. (1999) Mol. Biol. Cell 10, 345–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pazour G. J., Dickert B. L., Witman G. B. (1999) J. Cell Biol. 144, 473–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Porter M. E., Bower R., Knott J. A., Byrd P., Dentler W. (1999) Mol. Biol. Cell 10, 693–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piperno G., Mead K. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 4457–4462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piperno G., Siuda E., Henderson S., Segil M., Vaananen H., Sassaroli M. (1998) J. Cell Biol. 143, 1591–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cole D. G. (2009) The Chlamydomonas Sourcebook: Cell Motility and Behavior, pp. 71–113, 2nd Ed., Vol. 3, Academic Press, Inc., San Diego, CA [Google Scholar]
- 27.Brazelton W. J., Amundsen C. D., Silflow C. D., Lefebvre P. A. (2001) Curr. Biol. 11, 1591–1594 [DOI] [PubMed] [Google Scholar]
- 28.Pazour G. J., Dickert B. L., Vucica Y., Seeley E. S., Rosenbaum J. L., Witman G. B., Cole D. G. (2000) J. Cell Biol. 151, 709–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hou Y., Qin H., Follit J. A., Pazour G. J., Rosenbaum J. L., Witman G. B. (2007) J. Cell Biol. 176, 653–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahmed N. T., Gao C., Lucker B. F., Cole D. G., Mitchell D. R. (2008) J. Cell Biol. 183, 313–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qin H., Wang Z., Diener D., Rosenbaum J. (2007) Curr. Biol. 17, 193–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pedersen L. B., Miller M. S., Geimer S., Leitch J. M., Rosenbaum J. L., Cole D. G. (2005) Curr. Biol. 15, 262–266 [DOI] [PubMed] [Google Scholar]
- 33.Lucker B. F., Behal R. H., Qin H., Siron L. C., Taggart W. D., Rosenbaum J. L., Cole D. G. (2005) J. Biol. Chem. 280, 27688–27696 [DOI] [PubMed] [Google Scholar]
- 34.Wang Z., Fan Z. C., Williamson S. M., Qin H. (2009) PLoS ONE 4, e5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baker S. A., Freeman K., Luby-Phelps K., Pazour G. J., Besharse J. C. (2003) J. Biol. Chem. 278, 34211–34218 [DOI] [PubMed] [Google Scholar]
- 36.Gorman D. S., Levine R. P. (1965) Proc. Natl. Acad. Sci. U.S.A. 54, 1665–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pazour G. J., Witman G. B. (2000) Methods 22, 285–298 [DOI] [PubMed] [Google Scholar]
- 38.Sizova I., Fuhrmann M., Hegemann P. (2001) Gene 277, 221–229 [DOI] [PubMed] [Google Scholar]
- 39.Berthold P., Schmitt R., Mages W. (2002) Protist 153, 401–412 [DOI] [PubMed] [Google Scholar]
- 40.Kindle K. L. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 1228–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Asamizu E., Nakamura Y., Sato S., Fukuzawa H., Tabata S. (1999) DNA Res. 6, 369–373 [DOI] [PubMed] [Google Scholar]
- 42.Asamizu E., Miura K., Kucho K., Inoue Y., Fukuzawa H., Ohyama K., Nakamura Y., Tabata S. (2000) DNA Res. 7, 305–307 [DOI] [PubMed] [Google Scholar]
- 43.Hayashi M., Yanagisawa H. A., Hirono M., Kamiya R. (2002) Cell Motil. Cytoskeleton 53, 273–280 [DOI] [PubMed] [Google Scholar]
- 44.Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 45.Harlow E., Lane D. (1999) Using Antibodies: A Laboratory Manual, pp. 276–304, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 46.Penefsky H. S. (1977) J. Biol. Chem. 252, 2891–2899 [PubMed] [Google Scholar]
- 47.Clauser K. R., Baker P., Burlingame A. L. (1999) Anal. Chem. 71, 2871–2882 [DOI] [PubMed] [Google Scholar]
- 48.Watanabe Y., Hayashi M., Yagi T., Kamiya R. (2004) Cell Struct. Funct. 29, 67–72 [DOI] [PubMed] [Google Scholar]
- 49.Deane J. A., Cole D. G., Seeley E. S., Diener D. R., Rosenbaum J. L. (2001) Curr. Biol. 11, 1586–1590 [DOI] [PubMed] [Google Scholar]
- 50.Lechtreck K. F., Luro S., Awata J., Witman G. B. (2009) Cell Motil. Cytoskeleton 66, 469–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Collet J., Spike C. A., Lundquist E. A., Shaw J. E., Herman R. K. (1998) Genetics 148, 187–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu A., Wang B., Niswander L. A. (2005) Development 132, 3103–3111 [DOI] [PubMed] [Google Scholar]
- 53.Taulman P. D., Haycraft C. J., Balkovetz D. F., Yoder B. K. (2001) Mol. Biol. Cell 12, 589–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoder B. K., Tousson A., Millican L., Wu J. H., Bugg C. E., Jr., Schafer J. A., Balkovetz D. F. (2002) Am. J. Physiol. Renal Physiol. 282, F541–F552 [DOI] [PubMed] [Google Scholar]
- 55.Huangfu D., Liu A., Rakeman A. S., Murcia N. S., Niswander L., Anderson K. V. (2003) Nature 426, 83–87 [DOI] [PubMed] [Google Scholar]
- 56.Qin H., Rosenbaum J. L., Barr M. M. (2001) Curr. Biol. 11, 457–461 [DOI] [PubMed] [Google Scholar]
- 57.Bell L. R., Stone S., Yochem J., Shaw J. E., Herman R. K. (2006) Genetics 173, 1275–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.