Abstract
A major mechanism of antibiotic resistance in bacteria is the active extrusion of toxic compounds through membrane-bound efflux pumps. The TtgR protein represses transcription of ttgABC, a key efflux pump in Pseudomonas putida DOT-T1E capable of extruding antibiotics, solvents, and flavonoids. TtgR contains two distinct and overlapping ligand binding sites, one is broad and contains mainly hydrophobic residues, whereas the second is deep and contains polar residues. Mutants in the ligand binding pockets were generated and characterized using electrophoretic mobility shift assays, isothermal titration calorimetry, and promoter expression. Several mutants were affected in their response to effectors in vitro: mutants H70A, H72A, and R75A did not dissociate from promoter DNA in the presence of chloramphenicol. Other mutants exhibited altered binding to the operator: L66A and L66AV96A mutants bound 3- and 15-fold better than the native protein, whereas the H67A mutant bound with 3-fold lower affinity. In vivo expression assays using a fusion of the promoter of ttgA to lacZ and antibiotic tolerance correlated with the in vitro observations, namely that mutant H67A leads to increased basal expression levels and enhances antibiotic tolerance, whereas mutants L66A and L66AV96A exhibit lower basal expression levels and decreased resistance to antibiotics. The crystal structure of TtgR H67A was resolved. The data provide evidence for the inter-domain communication that is predicted to be required for the transmission of the effector binding signal to the DNA binding domain and provide important information to understand TtgR/DNA/effector interactions.
Keywords: Antibiotics, Bacterial Genetics, DNA Operators, Promoters, Repressor Protein, RND Efflux Pumps
Introduction
Microorganisms are continuously exposed to naturally occurring deleterious chemicals, for example, the antibiotics produced by members of microbial communities, fungi and plants, or detergents such as bile salts present in the intestinal tract of higher animals. Human activity has also led to the presence of a great diversity of noxious organic and inorganic chemicals (xenobiotics) in the environment; some chemicals, such as semi-synthetic antibiotics or biocides, have been specifically developed to act as antimicrobial agents. Toxic compounds usually affect the structure of biological membranes or impair biosynthetic pathways essential for microbial growth. Nevertheless, bacteria display resistance to the action of these compounds due to intrinsic long-standing mechanisms that protect the cells from the continuous exposure to harmful chemicals. Among the mechanisms underlying the resistance to toxic chemicals, active exclusion catalyzed by efflux pumps is considered to be the most effective and widespread (1, 2). Efflux pumps can be either specific for one substrate or can transport a range of structurally dissimilar compounds, including antibiotics of different chemical classes, biocides, dyes, detergents, metabolic inhibitors, plant secondary metabolites, and organic solvents (2, 3–5). Pumps that transport several compounds are grouped under the term of multidrug resistance. The phenomenon of multidrug recognition is not exclusively confined to multidrug transporters, but also to their transcriptional regulators; often the pattern of these systems is that the regulators controlling the expression of the transporters respond to the same range of compounds that the transporter extrudes (5–10).
An example of a microbe that is multidrug resistant is Pseudomonas putida DOT-T1E, a strain that can grow in liquid medium with >10% (v/v) toluene, is resistant to multiple antibiotics and capable of surviving in the presence of plant secondary metabolites (11–14). A key efflux pump responsible for these phenotypes is TtgABC, which is a member of the RND family of pumps. Expression of the ttgABC operon is controlled by a transcriptional repressor known as TtgR (9, 15). TtgR is a member of the TetR family of transcriptional repressors, which typically comprise two functional domains, a highly conserved N-terminal DNA binding domain, and a less conserved C-terminal domain involved in both dimerization and effector binding. The structures of the crystallized members of the family show that they are all α-helical proteins that bind to DNA utilizing a helix-turn-helix motif. Previously we showed that a DOT-T1E ttgR mutant overproduced the efflux pump proteins and was more resistant than the wild-type to carbenicillin, chloramphenicol, nalidixic acid, and tetracycline (15).
In vivo analysis of expression of the ttgABC efflux pump operon and its regulatory gene ttgR in response to many structurally different antibiotics and natural products demonstrated that TtgR from P. putida DOT-T1E binds a wide range of antibiotics and plant secondary metabolites (9, 10). These ligands were subsequently used in crystallization experiments for structural studies. The three-dimensional structure of TtgR complexed with five different effectors was resolved as a joint effort between our laboratories in Granada and London (UK) (16). TtgR was shown to be composed of 9 α-helices. Helices 1 to 3 constitute the DNA binding domain, with helix 3 being predicted as the one that makes most of the contacts with the operator DNA. Helix 4 serves as a link to the rest of the protein, which folds independently of the N-terminal domain, and constitutes the effector binding pocket. Most of the ligands that have been characterized bind at a similar location. They bind vertically in a hydrophobic binding pocket (general binding pocket) with few specific interactions, which most likely contributes to the versatility of the ligand binding and micromolar affinity of TtgR. Interestingly, we also showed that phloretin, a plant antimicrobial, is capable of binding in a second binding pocket of TtgR, known as the high affinity/specific binding pocket. The ligand binding sites consist of hydrophobic residues lining the side walls including Leu-66, Leu-92, Leu-93, Val-96, Phe-168, and Val-171, whereas the bottom of the binding site consists of polar residues Asn-110, His-114, and Asp-172. It was also shown that a mutation within the binding pocket (R176G) reduced the binding affinity to phloretin (16).
We have also investigated the DNA binding characteristics of TtgR and found that it binds to a pseudo-palindromic site that overlaps the ttgR/ttgA promoters (13). The minimal DNA fragment for TtgR binding was a 30-mer and analysis of its sequence revealed two partially overlapping inverted repeats. Using analytical ultracentrifugation it was also shown that TtgR forms stable dimers in solution, and that two dimers bind to the operator. Dimethyl sulfate DNA-footprint assays revealed a close interaction between TtgR and the central region of the operator. The binding of the two TtgR dimers to the operator was characterized and the results indicated positive cooperativity (13). A series of oligonucleotides were generated in which the imperfect palindrome of the TtgR operator was empirically optimized. Optimization of the palindrome did not significantly alter the binding of the initial TtgR dimer to the operator, but increased the cooperativity of binding and consequently the overall affinity (13).
In this study we describe the results obtained from a focused effort to determine the residues of importance for TtgR effector binding specificity. EMSA3 and isothermal titration calorimetry (ITC) assays of the mutant proteins indicated that several had altered ability to associate with effector molecules and that some of the mutants in the effector binding domain had either increased or decreased affinities for the DNA operator region. The in vitro results were shown to correlate with in vivo promoter expression and minimal inhibitory concentration assays. Complementing this data, we resolved the three-dimensional structure of one of the mutant proteins, providing a visual explanation for the altered operator affinity. Together the data suggest that residues within the effector binding pocket are not only required for effector binding but are also involved in cross-talk between the two functional domains of TtgR.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, and Culture Medium
The P. putida and Escherichia coli strains and the plasmids used in this study are listed in Table 1. P. putida strains were routinely grown in LB medium at 30 °C and E. coli in 2xYT medium for the production of the TtgR protein. Antibiotics used were kanamycin (Km), 30 μg/ml; rifampicin (Rif), 10 μg/ml; and tetracycline (Tc), 20 μg/ml. Plasmids pWtttgR-A::lacZ, pH67AttgR-A::lacZ, and pL66AttgR-A::lacZ are derivatives of pMP220 and carry a transcriptional fusion of the ttgA promoter to a promoterless lacZ gene; ttgR under the control of its own promoter is also cloned divergently to the ttgA promoter.
TABLE 1.
Pseudomonas putida and Escherichia coli strains and plasmids used in this study
| Strain or plasmid | Characteristicsa | Source/Ref. |
|---|---|---|
| Strains | ||
| P. putida DOT-T1E | TolR, RifR | 12 |
| P. putida DOT-T1E13 | RifR, KmR, ttgR::&OHgr;Km | 15 |
| E. coli (BL21 DE3) | Novagen | |
| Plasmids | ||
| pMP220 | TcR, lacZ promoter fusion vector | 18 |
| pET29▵+) | KmR, T7 protein expression vector | Novagen |
| pWilR2 | KmR, pET29▵+) with wild-type ttgR | 10 |
| pETttgRL66A | KmR, pET29▵+) with L66A mutant ttgR | This study |
| pETttgRH67A | KmR, pET29▵+) with H67A mutant ttgR | This study |
| pETttgRH70A | KmR, pET29▵+) with H70A mutant ttgR | This study |
| pETttgRH72A | KmR, pET29▵+) with H72A mutant ttgR | This study |
| pETttgRR75A | KmR, pET29▵+) with R75A mutant ttgR | This study |
| pETttgRV96A | KmR, pET29▵+) with V96A mutant ttgR | This study |
| pETttgRR130A | KmR, pET29▵+) with R130A mutant ttgR | This study |
| pETttgRF168A | KmR, pET29▵+) with F168A mutant ttgR | This study |
| pETttgRR176G | KmR, pET29▵+) with R176G mutant ttgR | 16 |
| pETttgRL66AV96A | KmR, pET29▵+) with L66AV96A mutant ttgR | This study |
| pWtttgR-A::lacZ | TcR, wild-type ttgR-ttgAp::lacZ promoter fusion | This study |
| pL66AttgR-A::lacZ | TcR, L66A ttgR-ttgAp::lacZ promoter fusion | This study |
| pL66AV96AttgR-A::lacZ | TcR, L66A ttgR-ttgAp::lacZ promoter fusion | This study |
| pH67AttgR-A::lacZ | TcR, H67A ttgR-ttgAp::lacZ promoter fusion | This study |
a TolR, RifR, KmR, and TcR stand for resistance to toluene, rifampicin, kanamycin, and tetracycline, respectively. ttgAP refers to the promoter in front of the ttgA that is fused to the promoterless lacZ gene.
Minimal Inhibitory Concentration Assays
Antimicrobial susceptibility was determined in LB medium by the microtiter broth dilution method (17). Microtiter plate wells each containing 100 μl of LB and appropriate antibiotic were inoculated with 105 colony forming units/ml. The microtiter plates were then incubated with shaking for 16 h at 30 °C. The growth was analyzed and the minimal inhibitory concentration (MIC) corresponded to the minimal concentration at which growth was inhibited by at least 90%.
β-Galactosidase Assays
Fresh bacterial colonies from LB agar plates were inoculated into LB liquid medium supplemented with tetracycline and grown overnight at 30 °C with shaking (200 rpm). The overnight cultures were diluted to an A660 ∼ 0.1 in 20 ml of fresh LB and incubated at 30 °C with shaking until the culture reached an A660 of ∼0.7. The cultures were then split into fresh tubes, and inducers at appropriate concentrations were added to one tube while an equivalent volume of DMSO was added to the control. The cultures were returned to the incubator for 1 h at which point β-galactosidase activity was determined according to Miller (19). All chemical inducers were used at concentrations below the MIC and therefore did not affect growth.
Overexpression and Purification of Native and Mutant TtgR Proteins
The native ttgR expression plasmid (pWilR2) has been previously described (10). Alterations of specific TtgR amino acids encoded in this plasmid were performed using QuikChange mutagenesis (Stratagene). For the purification of TtgR proteins, the pET29a(+)-based expression plasmids were transformed into E. coli BL21(DE3). The cells were grown at 30 °C in 2-liter Erlenmeyer flasks containing 1 liter of 2xYT culture medium supplemented with 30 μg/ml of Km. Protein expression was induced at an A660 of 0.5–0.6 by adding 1 mm isopropyl β-d-1-thiogalactopyranoside. Cells were grown for another 3 h at 18 °C and subsequently harvested by centrifugation (6,000 × g for 10 min). The pellet resulting from a 1-liter culture was resuspended in 30 ml of buffer A (20 mm Tris-HCl, pH 6.4, 20 mm NaCl, 5% (v/v) glycerol, 0.1 mm EDTA), containing CompleteTM EDTA-free protease inhibitor mixture (Roche) and benzonase. Cells were lysed by treatment with 20 μg/ml of lysozyme and two passes through a French Press at a pressure of 1000 p.s.i. Following centrifugation at 16,000 × g for 60 min, the TtgR protein was predominantly present (more than 80%) in the soluble fraction. The supernatant was loaded onto a Hitrap Heparin HP column (1 or 5 ml, Amersham Biosciences) previously equilibrated with buffer A and subsequently eluted with a gradient of 0.02–0.6 m NaCl. Fractions containing TtgR were pooled, concentrated to <5 ml by ultrafiltration using an Amicon 8050 apparatus (Amicon-Millipore), and dialyzed against buffer B (20 mm Tris-HCl, pH 7.5, NaCl 250 mm, 10% (v/v) glycerol, 0.1 mm EDTA, and 5 mm dithiothreitol). The sample was then submitted to size exclusion chromatography using a Sephacryl HR-200 preparative column (Amersham Biosciences). Eluted fractions of TtgR were pooled, concentrated using the Amicon 8050, and dialyzed against buffer C (10 mm Tris-HCl, pH 7.5, 250 mm NaCl, 50% glycerol (v/v), 0.1 mm EDTA, and 5 mm dithiothreitol) for protein storage at −70 °C. Protein concentrations were determined using either the Bio-Rad Protein Assay kit or Nanodrop spectrophotometer set with the parameters: Mr 23,854 and ϵ, 21,151.
Electrophoretic Mobility Shift Assays
Electrophoretic mobility shift assays were carried out as previously described (9). Briefly, the DNA probe (189-bp fragment containing the ttgABC-ttgR intergenic region) was obtained from P. putida DOT-T1E chromosomal DNA by PCR. Radiolabeled probe (1 nm, ∼10,000 cpm) was incubated with defined concentrations of either purified native or mutant TtgR in 10 μl of DNA binding buffer (10 mm Tris-HCl, pH 7.0, 250 mm NaCl, 10 mm magnesium acetate, 10 mm KCl, 5% glycerol (v/v), 0.1 mm EDTA, and 5 mm dithiothreitol) supplemented with 20 μg/ml of poly d(I-C) and 200 μg/ml of bovine serum albumin. Effectors (prepared in DMSO) were added to the binding reaction at a final concentration of 1 mm. Reactions were incubated for 10 min at 30 °C and samples were electrophoresed on 4.5% (w/v) non-denaturing polyacrylamide gels (Bio-Rad Mini-Protean II) for 2 h at 50 volts in Tris glycine buffer (25 mm Tris-HCl, pH 8.0, and 200 mm glycine). Gels were subsequently dried and analyzed using a Personal FX reader and QuantityOne software (Bio-Rad).
ITC
For effector binding studies, all ITC measurements were performed on a VP microcalorimeter (MicroCal, Northampton, MA) at 30 °C. The protein was thoroughly dialyzed against effector binding buffer (25 mm Pipes, pH 7.0, 250 mm NaCl, 5% (v/v) glycerol, 10 mm magnesium acetate, 10 mm KCl, 0.1 mm EDTA, and 1 mm dithiothreitol). The protein concentration was determined using the Nanodrop spectrophotometer. Stock solutions of naringenin, phloretin, and chloramphenicol were freshly prepared at 0.5 m in DMSO, and subsequently diluted in dialysis buffer. Note that tetracycline was not tested in ITC as it is extremely insoluble and when tried was precipitated during titration. The appropriate amount of DMSO (0.1–0.3% (v/v)) was added to the protein sample in each assay. To avoid evaporation and nonspecific binding all effectors were manipulated in glass vessels and were neither degassed nor filtered. Typically, an experiment involved a single 1.6 μl and a series of 4.8-μl injections of effector molecule (250 μm) into the protein solution (8–12 μm) until saturation. Mean enthalpy changes measured from the injection of the ligand into the buffer were routinely subtracted from raw titration data before data fitting with ORIGIN software (MicroCal).
Crystallization, Crystallographic Data Collection, Structural Determination, and Modeling
TtgR containing the H67A alteration was crystallized using the same protocol and in similar conditions to those previously reported (16). Briefly, H67A TtgR was concentrated to 9.1 mg/ml and crystals were grown using the sitting-drop vapor diffusion method in 0.1 m BisTris (pH 6.5), 0.35 m MgCl2, 20% (v/v) PEG3350. Crystals were soaked in crystallization buffer previously supplemented with 20% (v/v) glycerol as cryoprotectant before being frozen in liquid nitrogen. The data sets were collected under cryogenic conditions at beamline I02 at the Diamond Light Source, Chilton, Didcot, UK. Data were processed using Mosflm. The space group is P1 with unit cell dimensions a = 43.11Å, b = 42.17Å, c = 114.80Å, α = 96.9°, β = 99.3°, γ = 96.0°. The structure was solved by the molecular replacement method implemented in Phaser using the native TtgR structure (PDB code 2UXH) as a search model. Subsequent building/rebuilding of models was performed using program COOT. Model refinement was carried out using Phenix by setting aside 5% of the observed reflection data for cross-validation. The structure was refined to 2.2-Å resolution with final Rfree of 28.8% and Rwork of 21.8%. Modeling of the L66A TtgR structure was performed via in silico mutation of the residue in the interactive graphics system program “O,” followed by analysis of the structural alterations using superimposition over the wild-type TtgR.
RESULTS
In Vitro Effects of TtgR Mutation on DNA Operator Binding and Dissociation
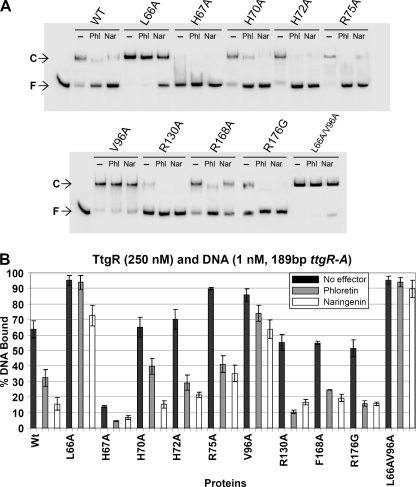
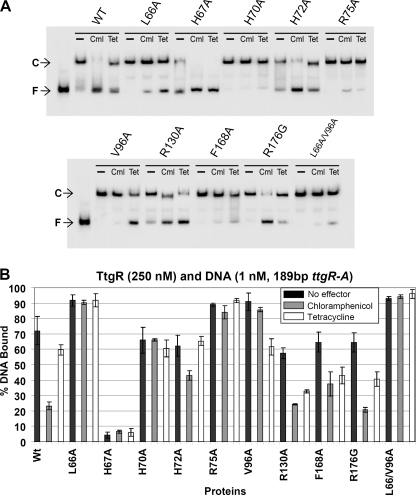
The ttgABC operon is expressed at a basal level that increases in the presence of antibiotics (e.g. tetracycline and chloramphenicol) and plant secondary metabolites (e.g. naringenin and phloretin) (9, 10). These molecules, commonly termed effectors cause the increase in expression of the TtgABC efflux pump by permitting the release of TtgR from the ttgR-ttgA operator region. In an attempt to identify the amino acids essential for effector binding and functional activation of TtgR we targeted amino acids within the putative effector binding pockets and effector portal of TtgR. The residues were rationally selected on the basis of the previously published TtgR apo and effector bound structures (16). Specifically, Leu-66 was targeted because it is indicated to associate with the phenolic ring of quercetin and naringenin. Residue Val-96 was chosen because it may form an aromatic ring sandwich with Leu-66 and thus shape a hydrophobic pocket in which chloramphenicol can bind. His-70 forms a bend in the middle of α4, which is a helix that forms one side of the effector portal; thus this residue is likely important for entry and or binding of effector molecules to TtgR. Three other residues were selected based on their proximity to the effector portal, the likelihood that they may be involved in forming a portal gate and that in TtgR from other Pseudomonas strains are replaced by other amino acids; His-67, which is Arg or Gln in Pseudomonas syringae and Pseudomonas fluorescens, and P. putida F1, His-72, which is Glu in P. syringae and P. fluorescens, and Arg-75, which has a large hydrophilic and flexible side chain and may form a gate across the portal of α7 and α4; this amino acid is substituted to Gln in P. syringae and Ala in P. fluorescens. We also targeted Arg-130 because it interacts with OH groups of phloretin and Phe-168 as it is an α8 side wall hydrophobic residue that is an His in P. fluorescens. Arg-176 is located in the high affinity binding site for phloretin (Gly or Tyr in other Pseudomonads) and is a residue that our group previously showed to be important in binding of phloretin (16). To assess the effects of alteration of these amino acids located in the effector binding pocket and portal of TtgR on its ability to bind to and be released from the operator DNA we performed electrophoretic mobility shift assays as previously described (9). A quantitative comparison of the effector-mediated release was performed using 250 nm TtgR (wild type or mutant) and 1 mm effector (Figs. 1 and 2). Most of the mutants were able to bind to and retard the DNA to a similar level as the native protein when tested under these conditions. However, whereas the native protein retarded ∼65% of the DNA when used at 250 nm, mutants L66A (93%), V96A (85%), and L66A/V96A (95%) retarded a much higher percentage of the DNA in the absence of effector, whereas mutant H67A bound less than 15% (Figs. 1A and 2A). The majority of mutants were also released from the DNA in the presence of both phloretin and naringenin; exceptions were again L66A and L66A/V96A, although mutant L66A did show some dissociation with naringenin. We then performed identical experiments using antibiotics as the effectors and found similar retardation profiles for the proteins in the absence of the effectors (Fig. 2, A and B). However, these effectors showed different capacities to dissociate the protein·DNA complexes (Fig. 2, A and B). Interestingly three of the mutants in the proposed effector portal, H70A, H72A, and R75A, were not noticeably dissociated from the DNA in the presence of tetracycline and both H70A and R75A did not respond to chloramphenicol. This data suggests that these mutations affect the ability of the antibiotics to access the general binding pocket or are preventing the signal transduction of the binding response to the DNA binding domain of the TtgR protein.
FIGURE 1.
EMSA of TtgR variants using the 189-bp ttgR-ttgA operator region. TtgR variants were tested in EMSA at 250 nm with 1 nm labeled operator DNA as described under “Experimental Procedures”; a representative assay is shown. All mutants were also tested with the addition of 1 mm of either phloretin (Phl) or naringenin (Nar). C and F followed by an arrow indicate the DNA·TtgR complex and free DNA, respectively. B, graphical analysis showing the results of three duplicate assays of each of the mutants in the above described EMSA and the standard error represented by bars. WT, wild-type.
FIGURE 2.
EMSA of TtgR variants using the 189-bp ttgR-A operator region in the presence of antibiotics. A, conditions as in the legend for Fig. 1 except that all mutants were also tested with the addition of 1 mm of either chloramphenicol (Cml) or tetracyline (Tet). B, graphical analysis showing the results of three duplicate assays of each of the mutants in the above described EMSA. WT, wild-type.
Determination of KD of Wild-type and Mutant TtgR for the ttgR-ttgA Promoter DNA
In the initial EMSAs it was noted that the mutant proteins had diverse affinities for the ttgR-ttgA operator DNA; we therefore determined the dissociation constants (KD) of the specific proteins for the operator DNA. We tested all of the mutant proteins and the wild-type over a range from 2 to 750 nm, the outcome of which are presented in Table 2. The results allowed the mutants to be divided into three different groups depending on their ability in both DNA and effector binding. Many of the mutant proteins showed a very similar affinity for the operator DNA as the wild-type protein (170 ± 5 nm); for example, H70A (170 ± 20 nm), H72A (140 ± 3 nm), R75A (150 ± 5 nm), F168A (125 ± 10 nm), R176G (130 ± 15 nm), and R130A (150 ± 3 nm) with KD values 1 to 1.3 times the wild-type value. The second group consisted of mutants that bound with much greater affinity than the wild-type protein and these included L66A (33 ± 1 nm, 5.1 times wild-type), L66A/V96A (11 nm, 15 times wild-type), and V96A (58 nm, 2.9 times wild-type). A third and final type of mutant protein, H67A, showed a marked decrease in affinity for the operator DNA (550 ± 30 nm). These titration results are interesting in that one would expect that mutations in the effector binding pocket of the TtgR protein would normally only influence effector binding and not necessarily the DNA binding ability of the protein. These data suggest that several of the effector binding pocket mutants that we generated and analyzed may be in amino acids involved in the signaling or putative conformational change required for effector-mediated DNA release. We reasoned that the changes in affinity for DNA of TtgR proteins with mutations in the effector binding pocket may also alter the specific DNA residues bound by TtgR, i.e. the footprint. To test this hypothesis we compared wild-type, L66A, V96A, L66A/V96A, and H67A mutants for their abilities to protect dimethyl sulfate-treated DNA in a footprint assay. The results indicated that all mutants were able to protect the same bases of the DNA as the wild-type (data not shown). However, as expected, mutants L66A, V96A, and L66A/V96A all protected the DNA more effectively than the wild-type protein. This together with the KD results suggests that the proteins are not altered in the location of DNA binding but that they differ in their affinity for the promoter region. These observations also suggest that some of the amino acids in the effector binding domain are important in the predicted cross-talk between this domain and the helix-turn-helix DNA binding domain.
TABLE 2.
Apparent dissociation constants for TtgR wild-type and mutant proteins for the ttgR-ttgA operator
Data were obtained from the densitometric analyses of EMSA performed using protein concentrations ranging from 2 to 750 nm. The DNA probe was 1 nm of a 189-bp fragment comprising the entire ttgR-ttgA intergenic region. Results shown are the mean of two independent assays each done in duplicate.
| Protein | KD | KD,wild-type/KD |
|---|---|---|
| nm | ||
| TtgR (wild-type) | 170 ± 5 | 1 |
| TtgR (L66A) | 33 ± 1 | 5.1 |
| TtgR (H67A) | 550 ± 30 | 0.3 |
| TtgR (H70A) | 170 ± 20 | 1 |
| TtgR (H72A) | 140 ± 3 | 1.2 |
| TtgR (R75A) | 150 ± 6 | 1.1 |
| TtgR (V96A) | 58 ± 4 | 2.9 |
| TtgR (R130A) | 150 ± 3 | 1.1 |
| TtgR (F168A) | 125 ± 10 | 1.3 |
| TtgR (R176G) | 130 ± 15 | 1.3 |
| TtgR (L66A/V96A) | 11 ± 1 | 15.2 |
In Vivo Expression of ttgABC in Response to Known TtgR Effector Molecules
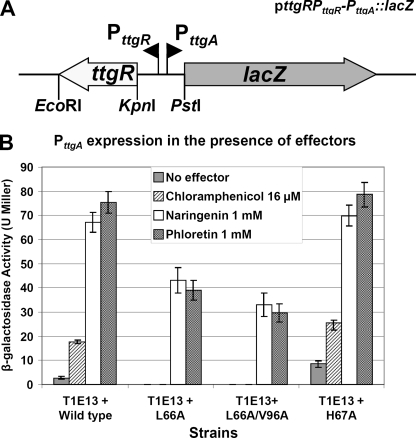
As a complement to the in vitro assays we decided to test the effects of several of the relevant effector binding domain mutants on the in vivo expression of the TtgABC efflux pump. For this, we used a PttgA::lacZ fusion and β-galactosidase assays to measure the in vivo effects of TtgR effector pocket mutations on the regulation of ttgABC expression in the presence of various effectors. To achieve this aim we first produced a construct that contained the wild-type ttgR gene and the full intergenic region between ttgR and ttgA fused to lacZ in the reporter plasmid pMP220 (Fig. 3A). The construction was designed in such a way that the wild-type ttgR gene could be easily replaced by mutated versions of the gene. Using this method we produced four constructs, containing wild-type, L66A, H67A, or L66A/V96A ttgR. These four plasmids along with the control plasmid pMP220 without insert were transformed into strain T1E13 (DOT-T1E ΔttgR) and the resulting transformants were tested for β-galactosidase activity in the presence and absence of effector molecules (chloramphenicol, naringenin, and phloretin) (Fig. 3B). Assays with the wild-type construct indicated that a low level of expression from the ttgA promoter was detectable in the absence of effector molecules. Interestingly, no such activity was detected with the L66A or L66A/V96A TtgR and a slightly higher basal level of expression was noted for the H67A construct. In the presence of chloramphenicol the wild-type showed a modest but reproducible increase in ttgA expression, whereas the L66A and L66A/V96A containing strains were not induced and the H67A strain gave higher β-galactosidase activities than the wild-type (Fig. 3B). Assays performed in the presence of the plant secondary metabolites naringenin and phloretin resulted in the highest activities; both the wild-type and H67A gave between 60 and 80 β-galactosidase units, whereas the L66A strain gave only 40 Miller units and the double mutant L66A/V96A only 30 Miller units. These data agree with the data obtained in the EMSA, which showed that mutants L66A and L66A/V96A bound with greater affinity to the operator region of ttgR-ttgA and that H67A bound with less affinity than the wild-type protein. These results also indicate that the altered DNA affinities noted during the in vitro EMSA can be directly translated to an in vivo effect on the expression of the ttgABC efflux pump operon and its cognate repressor ttgR.
FIGURE 3.
Construction used and graphical illustration of β-galactosidase assays. A, schematic representation of the ttgR-PttgR-PttgA::lacZ fusion clone used in the β-galactosidase and minimal inhibitory concentration assays. Restriction sites used in the cloning process are indicated below the figure and the divergent ttgR and ttgA promoters are shown as black triangles. B, graphical analysis showing the levels of β-galactosidase expression determined when strain T1E13 (ΔttgR) harboring the indicated plasmids were grown in the presence of the various effector molecules. Data represent the average of three duplicate assays including standard deviation.
In Vivo Susceptibility of Complemented DOT-T1E13 to Antimicrobials
To further analyze the effects of TtgR effector binding pocket mutation on ttgABC expression, and thus TtgABC production, we tested T1E13 (ΔttgR) cells that had been complemented with wild-type, L66A, L66A/V96A, and H67A ttgR for their ability to grow in the presence of a variety of antibiotics. The averaged results from three MIC assays are presented in Table 3. T1E13 cells bearing the ttgR wild-type allele or the ttgR H67A allele showed a very similar susceptibility profile to the antibiotics tested; only the sensitivity of H67A to ethidium bromide and nalidixic acid was decreased. However, the profile of T1E13 cells bearing L66A and L66A/V96A was different to the wild-type containing strain; the susceptibilities to chloramphenicol, cefalotoxin, nalidixic acid, norfloxacin, and ethidium bromide were all increased. The increased susceptibility of T1E13 cells expressing TtgR variants L66A and L66A/V96A complemented strains suggests that the higher affinity observed for the binding of these proteins to the promoter DNA of ttgR-ttgA is able to suppress efflux pump expression to such a level that it increases the sensitivity of the bacteria to various antibiotics. These data complement those already obtained from the EMSA and β-galactosidase assays and show that increasing or decreasing the binding affinity of the repressor TtgR for its promoter region have direct in vivo consequences.
TABLE 3.
Susceptibility of P. putida DOT-T1E and complemented ttgR mutant strains
MICs were tested using the microdilution assay as described under “Experimental Procedures”; results presented are the average of three experiments. Antibiotics used were: Ap, ampicillin; Cb, carbenicillin; Cm, chloramphenicol; Ctx, cefalotoxin; Nal, nalidixic acid; Nor, norfloxacin; Sm, streptomycin; Tc, tetracycline; Gm, gentamycin; EtBr, ethidium bromide.
| Strain/antimicrobial | MIC |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ap | Cb | Cm | Ctx | Nal | Nor | Sm | Tc | Gm | EtBr | |
| μg/ml | ||||||||||
| DOT-T1E (pMP220) | 250 | 512 | 64 | 6.25 | 64 | 3.9 | 12 | 64 | NDa | 2500 |
| T1E13 (pMP220) | 500 | 1024 | 256 | 12.5 | 64 | 7.8 | 16 | 64 | 0.5 | 5000 |
| T1E13 (Wt) | 250 | 512 | 128 | 6.25 | 64 | 3.9 | 16 | 64 | 0.5 | 2500 |
| T1E13 (L66A) | 250 | 512 | 64 | 3.125 | 32 | 1.95 | 16 | 64 | 0.5 | 1250 |
| T1E13 (L66A/V96A) | 250 | 512 | 64 | 3.125 | 32 | 1.95 | 16 | 64 | 0.5 | 1250 |
| T1E13 (H67A) | 250 | 512 | 128 | 6.25 | 128 | 3.9 | 16 | 64 | 0.5 | 5000 |
a ND, not determined.
Calorimetric Analysis of Wild-type and Mutated TtgR with Known Effector Molecules
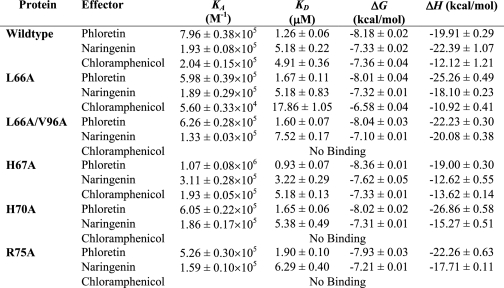
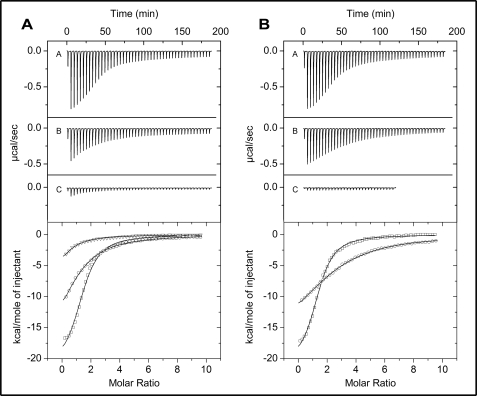
To quantitate the effects of mutating amino acid residues within the effector binding pocket of TtgR on effector binding, we assayed the interactions of the wild-type and a subset of the mutants using ITC. Titration of the wild-type protein with the three effectors showed that the thermodynamic modes of binding were enthalpy-driven giving ΔH of −12.12 ± 1.21 to −22.39 ± 1.07 kcal/mol (Table 4). The KD values for the wild-type TtgR protein were 1.26 ± 0.06 μm for phloretin, 5.18 ± 0.22 μm for naringenin, and 4.91 ± 0.36 μm for chloramphenicol (Fig. 4A and Table 4). Interestingly, the L66A mutant gave similar KD values for both phloretin and naringenin, however, the L66A affinity for chloramphenicol was decreased by a factor of 3.5-fold (KD = 18 ± 1 μm). This result showed that this mutation reduces binding to chloramphenicol, without affecting binding to the others. Identical results were obtained with the L66A/V96A double mutant, except that this mutant showed no detectable binding to chloramphenicol, because no enthalpy changes were observed upon its injection into the protein solution (Fig. 4B and Table 4). The previous EMSA studies had confirmed that the L66A/V96A protein could not be released from the ttgR-ttgA operator DNA in the presence of chloramphenicol; the ITC results substantiate this observation and suggest that its lack of release from the DNA is due to the inability of the protein to bind chloramphenicol. Two other proteins gave similar results to the L66A/V96A mutant; H70A and R75A TtgR proteins could bind both phloretin and naringenin with affinities equal to the wild-type protein, whereas neither of these proteins showed any detectable affinity for chloramphenicol (Table 4). The H67A mutant, which shows a 3-fold decrease in affinity for the DNA than the wild-type protein interestingly showed slightly higher affinities for the effectors phloretin and naringenin; 1.35- and 1.6-fold, respectively (Table 4). It is difficult to know if this increased affinity has any influence on the overall release of the H67A from the operator DNA because this protein binds poorly in both the presence and absence of effector. Overall the ITC data complement the results obtained for the mutant proteins from EMSA, β-galactosidase promoter expression assays, and MIC assays, and imply that several of the amino acids targeted are indeed required for effector binding to TtgR.
TABLE 4.
Thermodynamic parameters derived from calorimetric titration of TtgR proteins with different effectors
Data were obtained from TTC experiments carried out as described under “Experimental Procedures.” The assays were performed by titrating 8–12 μm TtgR protein with 250 μm effector in 25 mm Pipes, 250 mm NaCl, 5% (v/v) glycerol, 10 mm magnesium acetate, 10 mm KCl, 0.1 mm EDTA, and 1 mm dithiothreitol, pH 7.0.
FIGURE 4.
ITC of the binding of different effectors to wild-type and L66A/V96A TtgR. Experiments were performed as described under “Experimental Procedures” at 30 °C in effector binding buffer. Heat changes (upper panels) and integrated and fitted peak areas (lower panels) for the injection of a 1.6-μl and a series of 4.8-μl aliquots of 250 μm for: A, phloretin; B, naringenin; or C, chloramphenicol, into a solution of 8–12 μm of either wild-type TtgR (panel A) or L66A/V96A (panel B) are presented. Graphs in the lower panels are: □, phloretin; ○, naringenin; and ▵, chloramphenicol. All data were fitted with ORIGIN using the one set of sites algorithm.
Crystal Structure of TtgR H67A and Structural Basis of Mutant Data
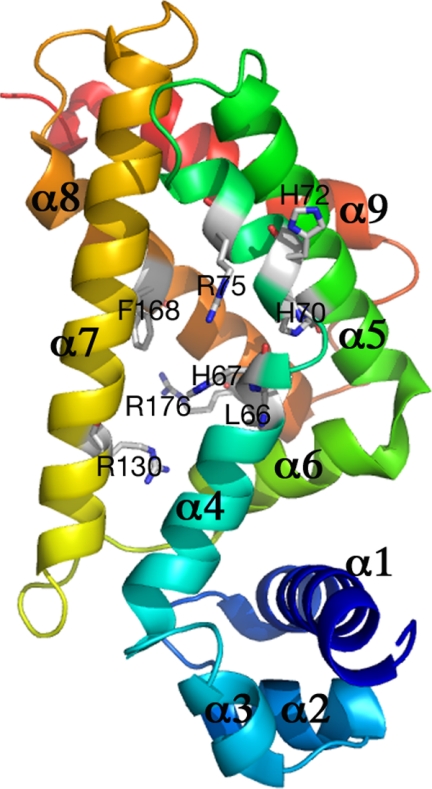
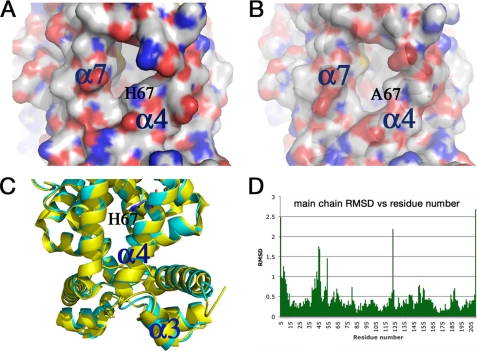
It is an interesting and potentially important finding that the H67A and L66A alterations located in the ligand binding pocket can affect the ability of TtgR to bind the operator DNA either positively or negatively. To investigate this further, we carried out structural characterization of the mutant protein, H67A. The structure was solved to 2.2 Å and refined to Rfree of 28.8% and Rwork of 21.8% (Table 5 and Fig. 5). The three-dimensional structure superimposes well with that of wild-type TtgR (16). However, detailed analysis reveals that H67A located at the end of helix α4 and the entrance of the effector portal, alters the conformation of α4 slightly, which is then transmitted to α3, the DNA recognition helix (Figs. 5–7). His-67 packs tightly between α4 and α7 in the wild-type structure (Fig. 6A). This packing is loosened in the H67A mutant structure causing α4 to rotate and hence this change is then amplified to α3 (Fig. 6, B and C). Indeed, detailed analysis of the structural differences reveals that α3 (residues 44 to 51) contains the largest deviations, with r.m.s. deviation of Cα being >1 Å (Fig. 6, C and D) between the H67A mutant and wild-type structures. The H67A mutation therefore results in far reaching conformational changes, leading to alterations in the orientation of the DNA recognition helix and subsequently reduced DNA binding affinity. Attempts to crystallize L66A or L66A/V96A mutants failed to yield high quality diffracting crystals; instead, we carried out in silico analysis using the interactive graphics system program O. The analysis revealed that the L66A alteration, which is adjacent to His-67 but instead faces α5 would indeed induce the opposite conformational changes in α4 to those observed for the H67A mutant; hence TtgR L66A might be expected to have enhanced DNA affinity, as observed. These results suggest that α4 is a key structural component in transmitting the conformational changes induced in the ligand binding pocket to the DNA binding domain, thereby modulating the ability of TtgR to be released from the DNA upon ligand binding. Interestingly, residues in the upper part of α4 do not seem to be involved in transmitting signals from the ligand binding pocket to the DNA binding domain, as mutations H70A, H72A, and R75A, all located in the upper part of α4, have no effect on DNA binding.
TABLE 5.
Data collection, phasing, and refinement statistics for H67A TtgR
Values in parentheses are for the highest-resolution shell.
| Data processing | |
|---|---|
| Wavelength (Å) | 0.9745 |
| Space group | P1 |
| Cell parameters | |
| a (Å) | 43.1 |
| b (Å) | 43.2 |
| c (Å) | 114.8 |
| α | 96.9° |
| β | 99.3° |
| γ | 96.0° |
| Resolution range (Å) | 56.1–2.2 |
| Rsym (%) | 4.9 (24.4) |
| I/s〈I〉 | 7.8 (2.9) |
| Completeness (%) | 90.9 (85.8%) |
| Multiplicity | 1.8 |
| Refinement | |
| Rwork/Rfree (%) | 22.65/27.68 |
| R.m.s. deviation bond length | 0.009 Å |
| R.m.s. deviation bond angle | 1.729° |
FIGURE 5.
Overall crystal structure of the TtgR H67A protein. Ribbon representation of the H67A monomer with α-helices labeled numerically from the N-terminal to the C-terminal. N-terminal is colored in blue and C-terminal in red. Side chains of residues analyzed in this study are indicated along with the residue labels. The portal to the effector binding domain is located toward the center of the illustrated structure between α4 and α7.
FIGURE 6.
Partial view of the crystal structure of TtgR H67A. A, His-67 packs closely against α7 in the wild-type structure, which is not the case in the mutant structure. B, representation of TtgR H67A structure. C, the rotation in α4 due to the H67A mutation is amplified to α3. D, this is supported by the largest root mean square deviations (RMSD) between wild-type and H67A mutant structures around α3 (residues 44 to 51).
FIGURE 7.
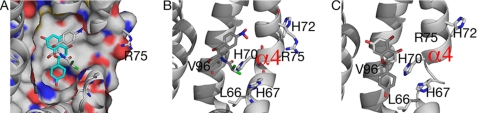
Comparison of ligand binding between chloramphenicol and naringenin. A, chloramphenicol (white) binds in the pocket toward the front entrance compared with naringenin (cyan). Most mutations are located at the front entry of the pocket, which interact with chloramphenicol (B) but not naringenin (C).
To understand the reduced binding ability to chloramphenicol for a number of mutants, we analyzed the structure of TtgR in complex with chloramphenicol and compared it with that of naringenin as the mutants are not affected in naringenin binding. Chloramphenicol differs from other ligands in the charge distribution due to the presence of Cl− ions. Consequently, chloramphenicol binds closer to the edge of the largely hydrophobic binding pocket compared with naringenin (Fig. 7A). Most of the mutated residues are located at the edge of the binding pocket, and hence could affect chloramphenicol binding, whereas having little effect on the inner walls of the pocket, where naringenin binds. In fact, Val-96 and His-70 interact directly with chloramphenicol (Fig. 7B). Mutating these residues therefore affects chloramphenicol binding. Arg-75 does not interact directly with chloramphenicol in the pocket. However, it is located in the opening of the binding pocket (Fig. 7A); therefore it is likely important for the entry of chloramphenicol due to the charge complementarity. Mutating Arg-75 to Ala would, as a result, be predicted to affect the entry of chloramphenicol, and subsequently its binding. His-67 does not interact with chloramphenicol directly nor is it located at the opening of the binding pocket and thus as expected, mutating His-67 to Ala has no effect on chloramphenicol binding.
DISCUSSION
The TtgR protein controls both its own expression and the expression of the ttgABC operon by binding to the ttgR-ttgA operator region. In this study we have shown that residues within the TtgR effector binding pocket are required for both effector and DNA binding. The amino acids Leu-66 and Val-96 are predicted to interact with the phenolic ring of naringenin and Leu-66 may form an aromatic ring sandwich with residue Val-96 as part of the hydrophobic pocket for chloramphenicol. Most interestingly, we have shown that alterations in these amino acid residues, namely L66A, V96A, and L66A/V96A not only alter the effector binding and release of TtgR from DNA but also influence the initial ability of TtgR to bind to its operator sequence. EMSA allowed us to define the KD of these mutated proteins for the ttgR-ttgA operator region and the results indicate that they have affinities 3–15-fold higher than the native TtgR protein. ITC showed that the L66A/V96A protein is in fact able to bind naringenin and phloretin with similar affinities to that of the native protein, indicating that the reduced release of the mutant proteins from the DNA in the EMSA is due to the higher overall affinity of the protein for the DNA. However, the L66A/V96A mutant protein was unable to bind to chloramphenicol when tested using ITC, a finding that agrees with the EMSA results. The in vivo results obtained with the mutant TtgR proteins strongly support both the EMSA and ITC data. Mutants L66A and L66A/V96A bound and repressed the expression from the ttgA promoter to a greater level than the wild-type TtgR protein, they also both showed drastically reduced expression of the promoter in the presence of all effector molecules tested (chloramphenicol, phloretin, and naringenin; Fig. 3B). Relative to these mutants, the wild-type TtgR has a moderately low affinity for the ttgR-ttgA operator; a phenomenon that is likely an evolutionary advantage allowing equilibrium to be established between the repression and expression of the TtgABC efflux pump. Evidence regarding the importance of this fine balance was provided by our minimal inhibitory concentration assay results (Table 3). Strains containing mutated TtgR, which showed higher affinity for the ttgR-ttgA promoter region (L66A and L66A/V96A), showed increased susceptibility to antibiotics when compared with native TtgR containing cells. Clearly, the evolutionary process has led to the selection of a native TtgR protein, which has an optimized balance between repression and basal level expression of the ttgABC operon.
The previously published three-dimensional structure of TtgR suggests that residue His-70 forms a bend in the middle of α4 and it has been hypothesized that it may be part of an effector portal (16). When tested in EMSA the TtgR H70A protein bound with a similar affinity to the wild-type protein and could be released from the DNA in the presence of phloretin, naringenin, and tetracycline. Interestingly the TtgR H70A was unable to bind chloramphenicol and was not released from the DNA in the presence of this effector. This is somewhat surprising as it would be expected that if the alteration was in an effector portal that the access and binding of most effectors would be altered. It is possible that the His-70 residue is important for guiding only molecules that harbor a negative charge, such as chloramphenicol, into the general binding pocket and that the alteration of the His to an Ala residue does not affect docking of non-charged effectors such as naringenin, phloretin, and tetracycline.
Two other residues that are predicted to be involved in forming a portal gate across the entrance between α7 and α4 were investigated in this study; His-72 (a residue with a large hydrophilic and flexible side chain), which is Glu in P. syringae and P. fluorescens, and Arg-75, which is Gln in P. syringae and Ala in P. fluorescens. When tested in EMSA both H72A and R75A could bind with equal affinity to the ttgR-ttgA operator DNA as the wild-type protein. Both proteins were released from the DNA in the presence of naringenin and phloretin, however, only mutant H72A responded to chloramphenicol. Data were confirmed with ITC as the R75A protein was able to bind naringenin and phloretin with similar affinity to the wild-type protein, whereas it did not show significant heats when titrated with chloramphenicol.
The investigation of residues that form part of the hydrophobic wall of the binding pockets and/or have been shown to have specific interactions with phloretin (Arg-130, Phe-168, and Arg-176) provided some insight into their importance in TtgR function. More specifically, the Arg-130 residue was shown from the previous structural analysis to interact with hydroxyl groups of the second phloretin located in the specific binding pocket (16). In our current assays the R130A mutant bound to operator DNA with a similar affinity as the wild-type protein and could be released in the presence of all effectors tested. The data suggest that although the Arg residue is involved in an interaction with phloretin during binding in the second effector binding pocket this interaction is not imperative for the function of TtgR or in modulating the effect of phloretin on the TtgR/DNA complex. Residue Phe-168 is an α8 side wall hydrophobic residue, and is a His in P. fluorescens. Mutants in this residue showed a modest increase in affinity for the ttgR-ttgA operator DNA in EMSA and were released from the DNA in the presence of all effectors tested. Again these results suggest that Phe-168 is not crucial for correct TtgR function. Arg-176 is in the high affinity binding site for phloretin and is Gly or Tyr in the TtgR of other Pseudomonads. In a previous publication (16) from our group it was shown that the R176G change causes a difference in the binding affinity for phloretin when tested in ITC. The EMSA with R176G in this current study showed that although the binding affinity for phloretin may be altered the effector could still cause dissociation of the complex.
The results for the H67A mutant afford much insight; this histidine residue is an Arg or Gln in other TtgR proteins from different Pseudomonads (P. syringae, P. fluorescens, and P. putida F1) and is predicted to form part of the portal through which effectors enter the general binding pocket. Surprisingly, rather than dramatic effects on substrate binding, the H67A protein showed a 3.3-fold decrease in affinity for the ttgR-ttgA operator DNA when compared with the native protein. This result seems independent of effector binding, because, using ITC the H67A TtgR protein was shown to bind naringenin, phloretin, and chloramphenicol with affinities close to the wild-type protein. Importantly, the lower DNA-binding affinity of H67A TtgR for the ttgR-ttgA operator led to clearly observable in vivo effects. As such, in β-galactosidase assays, H67A TtgR could not repress the expression of the ttgA promoter to the same degree as the native protein. Additionally, MIC assays showed that the higher basal level of expression of the ttgABC leads to higher resistance to nalidixic acid and ethidium bromide. These results make one wonder why the TtgR protein did not evolve to contain this mutation; it allows greater expression of the efflux pump and therefore a superior level of antibiotic resistance. The likely answer to this is that a lower basal level of TtgABC production is all that is required to allow the bacteria to survive until the effector (antibiotic or plant secondary metabolite) binds TtgR and forces the dissociation from the operator region. A H67A mutation results in a TtgR protein that is not a good repressor, the consequence of which is wasteful overproduction of TtgABC in non-challenging growth environments. The three-dimensional structure, combined with the ITC, EMSA, promoter expression, and MIC experiments highlight the peculiar conformational requirements for DNA binding affinity (not specificity, as shown). Together, emphasizing that the degree of rotation of the α3 helix acts as a stick shift, the precise angle of which has been chosen evolutionarily to optimize the balance between high and low DNA-binding affinities.
In summary the thorough in vitro and in vivo analyses of TtgR effector binding pocket mutants in this study, combined with structural analysis, has shown that amino acids within the effector binding pockets of TtgR are important for not only effector binding but also for DNA binding. The data provide evidence for the inter-domain communication that is predicted to be required for the transmission of the effector binding signal to the DNA binding domain and provide important insight into the mechanics of multidrug resistance.
Acknowledgments
We thank Benjamin J. Pakuts for critical reading of the manuscript and M. Mar Fandila and Carmen D. Lorente for secretarial assistance.
This work was supported in part by Junta de Andalucia FEDER Proyecto de Excelencia Grant CVI1912, FEDER Consolider C Grant BIO2006-05668, and Grant PSYSMO GEN2006-27750-C5-5-E/SYS from the Spanish Ministry of Science and Innovation.
- EMSA
- electrophoretic mobility shift assay
- ITC
- isothermal titration calorimetry
- MIC
- minimal inhibitory concentration
- DMSO
- dimethyl sulfoxide
- Pipes
- 1,4-piperazinediethanesulfonic acid
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- r.m.s.
- root mean square.
REFERENCES
- 1.Nikaido H. (2001) Semin. Cell Dev. Biol. 12, 215–223 [DOI] [PubMed] [Google Scholar]
- 2.Ramos J. L., Duque E., Gallegos M. T., Godoy P., Ramos-Gonzalez M. I., Rojas A., Teran W., Segura A. (2002) Annu. Rev. Microbiol. 56, 743–768 [DOI] [PubMed] [Google Scholar]
- 3.Kieboom J., de Bont J. (2001) Microbiology 147, 43–51 [DOI] [PubMed] [Google Scholar]
- 4.Nikaido H., Zgurskaya H. I. (2001) J. Mol. Microbiol. Biotechnol. 3, 215–218 [PubMed] [Google Scholar]
- 5.Poole K. (2003) Pseudomonas, Kluwer Academic/Plenum Publishers, New York [Google Scholar]
- 6.Chuanchuen R., Narasaki C. T., Schweizer H. P. (2002) J. Bacteriol. 184, 5036–5044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grkovic S., Brown M. H., Roberts N. J., Paulsen I. T., Skurray R. A. (1998) J. Biol. Chem. 273, 18665–18673 [DOI] [PubMed] [Google Scholar]
- 8.Lomovskaya O., Lewis K., Matin A. (1995) J. Bacteriol. 177, 2328–2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Terán W., Felipe A., Segura A., Rojas A., Ramos J. L., Gallegos M. T. (2003) Antimicrob. Agents Chemother. 47, 3067–3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Terán W., Krell T., Ramos J. L., Gallegos M. T. (2006) J. Biol. Chem. 281, 7102–7109 [DOI] [PubMed] [Google Scholar]
- 11.Ramos J. L., Duque E., Huertas M. J., Haïdour A. (1995) J. Bacteriol. 177, 3911–3916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramos J. L., Duque E., Godoy P., Segura A. (1998) J. Bacteriol. 180, 3323–3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krell T., Terán W., Mayorga O. L., Rivas G., Jiménez M., Daniels C., Molina-Henares A. J., Martínez-Bueno M., Gallegos M. T., Ramos J. L. (2007) J. Mol. Biol. 369, 1188–1199 [DOI] [PubMed] [Google Scholar]
- 14.Bernal P., Segura A., Ramos J. L. (2007) Environ. Microbiol. 9, 1658–1664 [DOI] [PubMed] [Google Scholar]
- 15.Duque E., Segura A., Mosqueda G., Ramos J. L. (2001) Mol. Microbiol. 39, 1100–1106 [DOI] [PubMed] [Google Scholar]
- 16.Alguel Y., Meng C., Terán W., Krell T., Ramos J. L., Gallegos M. T., Zhang X. (2007) J. Mol. Biol. 369, 829–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amsterdam D. (1991) in Antibiotics in Laboratory Medicine (Lorian V. ed) pp. 72–78, Williams & Wilkins, Baltimore, MD [Google Scholar]
- 18.Spaink H. P., Okker R. J., Wijffeman C. A., Pees E., Lugtenberg B. J. J. (1987) Plant Mol. Biol. 9, 27–39 [DOI] [PubMed] [Google Scholar]
- 19.Miller J. H. (1972) Experiments in Molecular Genetics, Chapter 48, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]