Abstract
The human immunodeficiency virus, type 1 (HIV-1)-encoded Rev protein is essential for the expression of late viral mRNAs. Rev forms a large organized multimeric protein-protein complex on the Rev response element of these viral mRNA species and transports them from the nucleus to the cytoplasm, exploiting the CRM1-mediated cellular machinery. Here we report the selection of a nanobody, derived from a llama heavy-chain only antibody, that efficiently blocks the assembly of Rev multimers. The nanobody inhibits HIV-1 replication in cells and specifically suppresses the Rev-dependent expression of partially spliced and unspliced HIV-1 RNA. In HIV-susceptible cells, this nanobody thus has potential as an effective anti-HIV agent using genetic immunization strategies. Its binding site was mapped to Rev residues Lys-20 and Tyr-23 located in the N-terminal α-helical multimerization domain. In the presence of this nanobody, we observed an accumulation of dimeric Rev species, supporting a head-to-head/tail-to-tail molecular model for Rev assembly. The results indicate that the oligomeric assembly of Rev follows an ordered stepwise process and identify a new epitope within Rev that could guide strategies for the development of novel HIV inhibitors.
Keywords: Fluorescence Resonance Energy Transfer (FRET), HIV, Protein-Protein Interactions, RNA-binding Protein, RNA Transport, Nanobody, Rev Multimerization, Single-domain Antibody
Introduction
Nuclear export of viral mRNA is critical for the life cycle of the human immunodeficiency virus (HIV).2 Fully spliced viral mRNA is exported to the cytoplasm of the infected cell through cellular mechanisms. However, in contrast to cellular systems, HIV uses a sophisticated molecular machinery to transport unspliced and partially spliced forms of its viral mRNA. The nuclear export of these late viral messengers is required for both the expression of late viral genes and packaging of genomic RNA and is mediated by the HIV-encoded regulatory protein Rev (1). The viral Rev protein forms a multimeric complex on a secondary structured RNA element (the Rev response element (RRE)) present in all unspliced and partially spliced mRNAs (2–4) and exploits the CRM1-mediated cellular machinery to transport these RNAs from the nucleus to the cytoplasm (5–7).
Rev is a small protein of 116 amino acid residues. Under steady state conditions, Rev localizes mainly to the nucleoli of cells (8). However, different functional elements cause Rev to continuously shuttle between the nucleus and the cytoplasm (9–11). A short stretch of basic amino acids characterized by 10 arginine residues serves both as a nuclear localization signal for the nuclear import of Rev and as an RNA-binding domain during the export of RNA-Rev complexes (4). This arginine-rich region is flanked on both sides by sequences that contribute to the oligomerization of Rev on the RRE. Its strong tendency to oligomerize has hampered the structure determination of Rev by x-ray crystallography or NMR spectroscopy. However, circular dichroism, nuclear magnetic resonance, and Raman spectroscopy studies strongly suggest that the oligomerization domains and the nuclear localization signal are components of a helix-turn-helix motif (12–14). A leucine-rich nuclear export signal (15) binds the cellular export receptor CRM1 and mediates nuclear export (5–7, 16, 56). Disruption of the nuclear export signal results in a dominant negative mutant (e.g. RevM10) that retains nucleolar localization and RRE binding but is defective in nuclear export because it does not engage with CRM1 (17). On the other hand, compounds disrupting the Rev-CRM1 interaction and hence nuclear export of Rev have been described (18–20).
One important aspect of the Rev function is its requirement for multimerization (21, 22). Oligomerization of Rev has been shown in vitro and in cell culture (21, 23–25). Initial binding to the high affinity Rev binding site of the RRE (stem-loop IIB) is followed by multimerization of Rev along the RRE template via a combination of cooperative hydrophobic protein-protein interactions and electrostatic protein-RNA interactions leading to the further coating of stem IIA and stem I of the RRE (3, 22, 26). Compared with the monomer, the Rev multimer forms a higher affinity complex with the RRE, indicating that the oligomer Rev molecules can expose their RNA-binding domain to alternative binding sites on the RRE (26, 28). According to the current model for the intermolecular interactions between Rev monomers on the RRE, Rev cooperatively assembles one molecule at a time via a series of symmetrical tail-to-tail and head-to-head protein-protein interactions (24, 27). However, although it is essential for Rev function, the mechanistic role of multimerization in the HIV replication has remained uncertain. The progress of Rev assembly on the RRE may determine the threshold to achieve a functional Rev response. Indeed, there is a strong correlation between the affinity of the Rev multimer for the RRE and efficiency of RNA export (26). Upon reaching threshold levels of Rev, its multimerization on RRE thus acts as a “molecular rheostat” that triggers the export and expression of viral mRNAs encoding late gene products (21, 22, 29).
In this study, we selected a molecule that specifically targets the N-terminal multimerization domain of Rev. We took advantage of the distinguishing feature of Camelidae that in addition to conventional antibodies also produce smaller fully functional antibodies solely composed of homodimeric heavy chains (30). Nanobodies derived from these heavy-chain only antibodies are minimally sized, highly soluble single domain antibody fragments (31). Therefore, we have immunized a llama with recombinant HIV-1 Rev protein and performed a phage display screen for the variable domain repertoire of the heavy-chain only antibodies that bind to Rev. From a dozen nanomolar affinity Rev binders, we identified one nanobody that blocks the multimerization of Rev and inhibits HIV-1 replication. It is the first known molecule directed against the Rev N-terminal α-helical multimerization domain. The use of this nanobody also allowed us to support the molecular model for concerted assembly of Rev multimers to a stepwise head-to-head/tail-to-tail mechanism.
EXPERIMENTAL PROCEDURES
Nanobody Generation, Selection, Expression, and Purification
Llama immunizations, phage display construction, and nanobody purifications were performed according to earlier described protocols (32). One llama (Lama glama) was immunized with 2 mg of purified recombinant HIV-Rev over a period of 6 weeks. The immunization, library construction, and nanobody selection have been performed following standard procedures according to Conrath et al. (33), and a nanobody library in pHen4 (32) of 3.7 × 107 independent clones was established.
Recombinant HIV-Rev-specific phages were recovered by incubating recombinant HIV-Rev-coated wells with 100 mm triethylamine, pH 11.0, for 10 min. The eluate containing the phages was neutralized with Tris-HCl, pH 6.8, and added to freshly grown TG1 cells. The HIV-Rev-coated wells were then washed once with Tris-HCl, pH 6.8, and several times with phosphate-buffered saline. Finally, freshly grown TG1 cells were added to the wells to recover the non-eluted phages. After two and three rounds of selection, individual colonies were screened for the expression of HIV-specific nanobodies: Maxisorb 96-well plates were coated overnight at 4 °C with 1 μg/ml purified recombinant HIV-Rev in sodium bicarbonate buffer, pH 8.2. Residual protein-binding sites in the wells were blocked for 2 h at room temperature with 2% milk in phosphate-buffered saline. Detection of antigen-bound nanobodies was performed with a mouse anti-hemagglutinin-decapeptide tag (clone 16B12, Babco). Subsequent detection of the mouse anti-tag antibodies was done with an alkaline phosphatase anti-mouse IgG conjugate (Sigma). The absorption at 405 nm was measured 15–30 min after adding the enzyme substrate p-nitrophenyl phosphate. Finally, all selected nanobody genes were cloned in a pHEN6 vector for expression with a His tag in Escherichia coli (33).
Cell Culture, Transfections, and Plasmids
Prokaryotic and eukaryotic expression vectors were constructed using standard molecular cloning techniques. pRev-GFP expresses the Rev protein fused to the green fluorescent protein, and pNb-mKO produces fusion proteins of nanobody with monomeric Kusabira Orange. HeLa, human epithelial cells, and human embryonal kidney 293T cells were maintained using standard procedures. For transfection of plasmid DNA in HeLa, cells were plated onto glass bottom microwell dishes (MatTek corporation) at 0.1 × 106 cells/plate and cultured until 50% confluent. Then cells were washed with phosphate-buffered saline and transfected with plasmid DNA using SuperFect® transfection reagent (Qiagen, Valencia, CA) according to the manufacturer's manual and incubated overnight. 293T cells were cultured in microwell dishes until 50% confluence and transfected by the calcium phosphate coprecipitation technique. For the virus expression experiments, typically 0.5 μg of pNL4-3 and different amounts of pcDNA3.1-Nb- or pcDNA3.1-Dronpa-expressing plasmids were used. pBKS DNA was added to each transfection mixture so that in every transfection equal amounts of DNA were transfected. Virus expression was analyzed by measuring the virus-associated core antigen (p24) in the supernatants of transfected cells by an enzyme-linked immunosorbent assay (GE Healthcare).
The NL4-3 molecular clone has been described previously (34). NL-Nb190 and NL-Nb163 were generated by subcloning the nanobody sequence into the BlpI-XhoI site of p83-10 (35). The resulting plasmids were obtained by replacing the EcoRI-XhoI fragment from NL4-3 with the EcoRI-XhoI fragment from p83-10 containing the nanobody sequences. NLRev(−)RRE(−)CTE(+) was from B. Felber and G. Pavlakis.
Microscopy and Fluorescence Recovery after Photobleaching
Transfected HeLa cells were imaged with an ArrayScan HCS Reader (Thermo Scientific Cellomics) or a laser-scanning SP5 confocal microscope (Leica Microsystems) equipped with an AF 6000 microscope and an Acousto optical beam splitter, using an HCX plan apochromat ×63 (numerical aperture 1.2) water immersion objective magnification. Enhanced green fluorescent protein was monitored with the argon laser using the 488-nm line for excitation, and emission was detected between 492 and 558 nm. Monomeric Kusabira Orange fluorescent protein (mKO) was imaged using the DPSS 561-nm laser for excitation, and emission was detected between 570 and 670 nm.
Fluorescence recovery after photobleaching studies were performed by obtaining a series of 20 prebleach images of a nucleolus at 5% Acousto optical tunable filter laser setting, followed by a single 0.39 s bleach iteration at full laser power of a 1 μm wide circle in the nucleolus. Subsequent post-bleach images were acquired at 5% Acousto optical tunable filter with the following time scheme: 20 images every 0.39 s, 20 images every 1 s, and 15–55 images every 10 s. Data were background-subtracted, bleach-corrected, and normalized according to the method described (36). Half-times of recovery were obtained by fitting the recovery curves employing GraphPad Prism (GraphPad Software, Inc.).
Protein Expression and Purification
pET29b(+) and pET21b(+) constructs encoding, respectively, the Rev protein and fusion proteins ECFP-Rev, EYFP-Rev, ECFP-EYFP, free ECFP, and free EYFP were transformed in E. coli BL21(DE3) and expressed by a 3.5-h induction with 1 mm isopropyl 1-thio-β-d-galactopyranoside. Cells were lysed using the SLM Amino French pressure cell press (Beun-DeRonde), and the proteins were purified via Ni2+-nitrilotriacetic acid affinity chromatography. For immunization of the llama, the Rev protein was further purified by anion exchange chromatography (GE Healthcare).
Rev Multimerization Assay
To evaluate the effect of the nanobodies on the Rev multimerization by fluorescence resonance energy transfer (FRET), nanobody concentrations were mixed (in a 96-well plate in 100 μl of phosphate-buffered saline) with 0.1 μm ECFP-Rev and 0.2 μm EYFP-Rev either prior to or following the assembly of Rev multimers. A sample with only ECFP-Rev and EYFP-Rev was used as positive control for FRET, whereas the combination of free ECFP plus EYFP-Rev was used as negative control. After a 30-min incubation, the FRET percentage was determined using a spectrofluorometer (Safire2, Tecan). Hence, emission was measured at 480 ± 5 nm (C) and 530 ± 5 nm (F) after excitation with 430 ± 5 nm and at 530 ± 5 nm (Y) after excitation with 490 ± 5 nm. The relative FRET efficiency was then calculated as follows. The total FRET signal (F) was first corrected for the “bleed-through” of the donor ECFP (i.e. 44% of C) and the “direct excitation” of the acceptor EYFP (i.e. 5% of Y), resulting in the “real FRET” W,
The FRET efficiency (E) is then given by the following,
 |
where γ is the ratio between the quantum yields of acceptor and donor (i.e. 1.53). The relative FRET percentage was obtained by expressing E relative to the FRET efficiency of both the positive control (100%) and the negative control (0%).
Nothern Blot Analysis
mRNA was extracted using the Oligotex Direct mRNA kit (Qiagen). After contaminating DNA was digested using RNase-free DNase I (Invitrogen), the mRNA was denatured by heating to 50 °C in glyoxal load dye and separated by agarose electrophoresis under denaturing conditions. mRNA was blotted using the NorthernMax-Gly system (Ambion) according to the manufacturer's manual. The RNA probe spanning exon 7 from the BamHI to the BglII site (nucleotides 8475–9056) of the NL4-3 genome was produced by in vitro transcription from T7 primer PCR products.
Quantitative Reverse Transcription-PCR
Two days after transfection, total mRNA from cells was extracted using the Oligotex Direct mRNA kit (Qiagen) followed by DNA digestion using RNase-free DNase I (Invitrogen). DNase I-treated mRNA was used to generate cDNA along with Thermoscript reverse transcriptase (Invitrogen) and oligo(dT)20. Quantitative reverse transcription-PCR for unspliced and multiply spliced HIV mRNA was performed according to a protocol described earlier (37) that we adapted slightly. For unspliced mRNA, 0.2 mm primers GAG1 and SK431 and 0.2 mm FAM-BHQ1 fluorescent probe GAG3 was used. For multiply spliced mRNA, 0.2 mm primers mf83 and mf84 and 0.2 mm FAM-BHQ1 fluorescent probe ks2-tq was used. Control reactions omitted reverse transcriptase, and the number of cDNA copies was determined using the HIV-1NL4-3 molecular clone or the pTat/Rev DNA as standards. pTat/Rev represents partial sequence (exons 4 and 7) of HIV-1 multiply spliced RNA encoding Tat and Rev proteins. Results are expressed as the number of copies of unspliced HIV mRNA per copy of multiply spliced RNA.
Mobility Shift Assays
High affinity IIB hairpin RRE labeled with Alexa Fluor 633 was purchased from Sigma. RRE RNA transcripts were prepared by a T7 in vitro transcription (DuraScribe T7 Transcription Kit, Epicenter) and purified by electrophoresis on a 6% polyacylamide gel. These RREs contain 2′-fluorine-modified UTP and CTP to make them resistant to degradation. The transcripts where then labeled with Alexa Fluor 647 by an Ulysis Nucleic Acid Labeling Kit (Molecular Probes). Binding reactions of Rev to RNA were performed in buffer containing 20 mm Tris-HCl (pH 8.0), 100 mm NaCl, 10 mm dithiothreitol with 0.1 mg/ml bakers' yeast tRNA. Typically, either 5 nm IIB RRE and 100 nm Rev protein or 3 nm RRE, 60 nm Rev, and 300 nm Nb190 or Nb163 were used. The samples were incubated for 20 min at room temperature and then run on a 6% polyacrylamide gel for 1 h. The bands were visualized and quantified using an Ettan DIGE Imager (GE Healthcare).
RESULTS
Identification of Nanobodies That Bind Rev and Inhibit Multimerization
A llama (L. glama) was immunized with 2 mg of purified recombinant HIV-1 Rev protein over a period of 6 weeks. The variable domain repertoire of the heavy-chain only antibodies from this llama was then expressed in a phage display library containing 3.7 × 107 independent clones. In order to select Rev-specific phages, the phage display library was panned on immobilized Rev protein. After three rounds of selection, individual colonies were expressed in E. coli and confirmed for binding to recombinant Rev. Twelve different nanobodies interacting with recombinant Rev were selected. To identify those nanobodies that selectively block the multimerization of Rev, we designed an in vitro multimerization assay based on FRET. In this assay, Rev fused to cyan fluorescent protein (ECFP-Rev) was incubated with a Rev fusion of the yellow fluorescent protein (EYFP-Rev). Multimerization of Rev brings the fused ECFP and EYFP labels in close proximity, resulting in an effective FRET efficiency of about 20% (Fig. 1A). The addition of excess unlabeled Rev protein to the ECFP-Rev/EYFP-Rev mix reduced the FRET signal below 5% because it intercalates between the labeled Rev molecules, thus increasing the distance between ECFP and EYFP. One nanobody (Nb190) drastically reduced the FRET signal from the ECFP-Rev/EYFP-Rev mix (Fig. 1, B and C), indicating inhibition of Rev protein-protein interaction.
FIGURE 1.
Selection of Rev multimerization-inhibiting nanobodies. A, FRET for ECFP-Rev incubated with EYFP-Rev and suitable negative controls. As a positive control, the FRET efficiency of an ECFP-EYFP fusion protein was measured. The FRET efficiency from the ECFP-Rev/EYFP-Rev mix was inhibited by recombinant unlabeled wild-type Rev protein. Results are mean ± S.E. (error bars), with n = 3. B, effect of the selected Rev-binding nanobodies on the Rev multimerization as measured by FRET. The graph shows the percentage of FRET relative to the ECFP-Rev/EYFP-Rev mixture without nanobody added. The control nanobody Nb163, cloned from a llama that was not immunized with Rev, was included as a negative control. C, amino acid sequence of Nb190. The hypervariable regions are shown in red, green, and blue.
Nb190 Disassembles Multimers of HIV-1 Rev
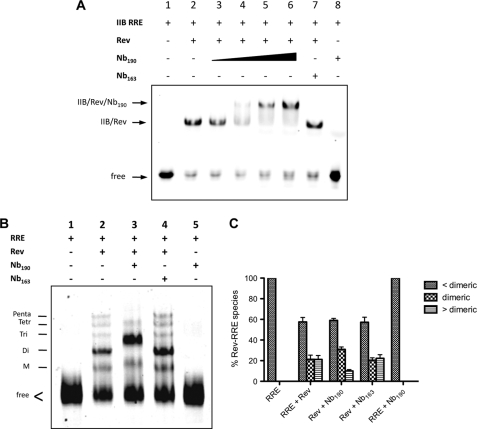
In our multimerization assay, Nb190 specifically reduced the FRET. It remained unclear, however, if the nanobody, besides disrupting the Rev-Rev interaction, also interferes with the Rev-RRE interaction. Therefore, gel shift experiments of Rev binding to the high affinity IIB hairpin of RRE were performed (Fig. 2A). The IIB stem-loop of RRE binds one single Rev monomer only (38). In contrast to the control nanobody Nb163 that was cloned from a llama not immunized with Rev (Fig. 2A, lane 7), the addition of increasing amounts of Nb190 caused a reduction of the mobility of the IIB-Rev complex (Fig. 2A, lanes 3–6). This result confirms the dose-dependent binding of the Nb190 to this Rev-RRE complex without disrupting the protein-RNA interaction. Nb190 per se does not interact with the IIB stem-loop of the RRE (Fig. 2A, lane 8).
FIGURE 2.
Nb190 blocks the HIV Rev multimerization in vitro. A, effect of Nb190 on the Rev-RRE interaction. Gel mobility shifts of labeled high affinity stem IIB RRE RNA in complex with Rev were performed in the presence of increasing amounts of Nb190 (lanes 3–6; 0.02, 0.1, 0.5, and 2.5 μm, respectively). Bands corresponding to free high affinity binding stem IIB RRE (lane 1), IIB-Rev (lane 2), and IIB-Rev-Nb190 complexes (lanes 4–6) are indicated. Nb163 (lane 7) was included as negative control. B, effect of Nb190 on the organization of the Rev-RRE assembly. Gel mobility shift analysis of multimeric Rev binding to fluorescently labeled full-length RRE, containing 2′-fluorine-modified UTP and CTP, in the presence or absence of Nb190. The supershift of the Rev-RRE complex in the presence of Nb190 is indicative of a ternary complex. M, monomeric; Di, dimeric; Tri, trimeric; Tetr, tetrameric; Penta, pentameric. C, quantification of the ribonucleoprotein Rev-RRE species in gel mobility shift experiments from B. Bands corresponding to the sum of free RRE plus monomeric Rev-RRE (<dimeric), dimeric Rev-RRE (dimeric), and the sum of tri-, tetra-, and pentameric Rev-RRE complexes (>dimeric) were quantified and expressed as a percentage of total RRE. Results are mean ± S.E. (error bars) from four independent experiments, p < 0.01 for >dimeric, and p < 0.005 for dimeric.
Nb190 Accumulates Dimers of Rev on the RRE
Efficient Rev function requires the formation of a large organized Rev-RRE ribonucleoprotein complex in which a Rev homomultimer assembles on the RRE RNA (21, 26). To verify whether Nb190 also blocks the multimerization of Rev on the RRE, we assessed the effect of Nb190 on the organization of the Rev-RRE assembly by gel retardation experiments. Fluorescently labeled full-length RRE, containing 2′-fluorine-modified UTP and CTP to make it resistant to degradation, was produced by in vitro transcription. Wild-type Rev forms higher order multimeric complexes on this RRE (Fig. 2B, lane 2) (3). In the presence of Nb190, we consistently observed reduced amounts of higher order Rev-RRE complexes while the species containing dimeric Rev accumulated (Fig. 2B, lane 3; quantified in Fig. 2C). It thus appears that Nb190 disrupts higher order multimeric Rev complexes along this RRE, resulting in a dimeric Rev phenotype on the RNA.
Nb190 Is an Intrabody That Induces a Multimerization-impaired Rev Phenotype in Cells
Next, we investigated whether Nb190 interacts with Rev in living cells using confocal fluorescence microscopy (Fig. 3). For these experiments, we fused Rev to GFP, and the nanobodies were fused to the orange-colored mKO. In agreement with earlier observations, transiently expressed Rev-GFP primarily localized to the nucleoli of HeLa cells (10, 11). In these cells, Nb190-mKO was found both in the nucleus and in the cytoplasm, and it distributed evenly over both cellular compartments, as did Nb163-mKO. However, when co-expressed, Rev-GFP and Nb190-mKO both redistributed and co-localized in the cytoplasm, in contrast to control experiments with Nb163-mKO. Upon inhibition of the nuclear export by leptomycin B, a redistribution of both Rev-GFP and Nb190-mKO to the nucleoli was observed (supplemental Fig. S1), suggesting active shuttling of the complex. Similarly, Nb190-mKO and RevM10-GFP demonstrated nucleolar co-localization (Fig. 3). Taken together, these data indicate that Nb190 is an intrabody that binds Rev within the different relevant cellular compartments and that it does not drastically prohibit the shuttling of Rev between the nucleus and the cytoplasm of the cell. Most importantly, our nanobody provokes a cellular redistribution of Rev that is very similar to the distribution caused by mutations that impair the multimerization of Rev. Specifically, the RevV16D (24) and RevM4 (Y23D/S25D/N26L) (17, 39) multimerization-deficient mutants display a comparable diffuse cytoplasmic distribution in the absence of Nb190. Interestingly, Nb190-mKO co-localized with the RevV16D mutant but not with the multimerization-deficient RevM4 mutant, pointing toward a possible role of the Tyr23–Asn26 segment of Rev in the recognition by Nb190.
FIGURE 3.
Nb190 interacts with Rev in cells. HeLa cells were transfected separately or co-transfected with expression plasmids as indicated. Subcellular localization of the proteins was visualized by confocal fluorescence microscopy in both GFP and mKO channels. The right column shows overlay images. DIC, differential interference contrast. Scale bar, 25 μm.
Nb190 Interacts with Lys-20 and Tyr-23 in the N-terminal α-Helix of Rev
To further map the epitope in more detail, we constructed Y23D, S25D, and N26L single Rev mutants and investigated their interaction with Nb190 in living cells (Fig. 4A). The Y23D and S25D mutations triggered a cytoplasmic Rev localization, whereas RevN26L-GFP remained nucleolar. The Y23D mutation per se was sufficient to inhibit the Rev-GFP/Nb190-mKO co-localization in cells, suggesting a significant role of Tyr-23 in the interaction with Nb190. We next constructed a series of alanine scan mutants in this region and investigated the co-localization of Nb190 with these Rev mutants in living cells (Fig. 4B). Per se, all of these Rev alanine mutants localized to the cytoplasm of the cell, in contrast to wild-type Rev, which is found in the nucleoli. Nb190-mKO still co-localized with both the K20A and Y23A single mutants; however, it did not interact any longer with the K20A/Y23A double mutant. To confirm the contribution of each of these single point mutations to the loss of interaction by the double mutant, fluorescence recovery after photobleaching experiments were performed (Fig. 4, C and D). For these interaction studies, we used the nuclear export-deficient RevM10, which remains nucleolar upon binding with Nb190 (Fig. 3). Cells were transfected with RevM10-GFP, RevM10K20A-GFP, or RevM10Y23A-GFP together with an Nb190-mKO-expressing construct (Fig. 4C). Photobleaching of Nb190-mKO when bound to RevM10-GFP in the nucleolus is followed by a slow recovery with a half-recovery time of 102 s, which indicates strong interaction between the two proteins. In contrast, the recovery of Nb190-mKO when bound to RevM10K20A or RevM10Y23A is much faster, with half-recoveries of 12 and 20 s, respectively. These results show that both K20A and Y23A mutations have a drastic effect on the binding with Nb190, with Lys-20 being more crucial than Tyr-23 for the Rev-Nb190 interaction.
FIGURE 4.
Rev residues Lys-20 and Tyr-23 form the binding epitope for Nb190. A and B, HeLa cells were co-transfected with expression plasmids as indicated. Subcellular localization of the proteins was visualized by confocal fluorescence microscopy in both GFP and mKO channels. The right column shows overlay images. Scale bar, 25 μm. C, fluorescence recovery after photobleaching of Nb190-mKO when bound to RevM10-BFP or RevM10 alanine mutants in the nucleoli of cells is shown. The dashed circles indicate the photobleached area. Bleaching and recovery of Nb190-mKO in the nucleolus were monitored for 540 s. D, quantitative fluorescence recovery after photobleaching analysis of Nb190-mKO bound to RevM10 (blue) or RevM10 containing the single K20A (green) or Y23A (red) mutation in the putative binding site. Values are averages from at least eight cells. E, the K20A and Y23A mutations disrupt the interaction between Rev and Nb190 in vitro. Gel mobility shifts of labeled high affinity stem IIB RRE RNA in complex with Rev or Rev mutants were performed in the absence or presence of Nb190, as indicated. wt, wild type.
These results were confirmed in vitro by gel shift analysis for Nb190 binding to the Rev mutants containing mutations in the putative binding site. Gel shift experiments of the respective alanine mutant Rev binding to the high affinity IIB hairpin of RRE were performed in the presence of Nb190 (Fig. 4E). In vitro, the Nb190-Rev interaction was completely abolished by the K20A and Y23A mutations. Indeed, the addition of Nb190 to IIB-RevK20A, IIB-RevY23A, or IIB-RevK20AY23A complexes did not cause a reduction of the mobility, further confirming that both Lys-20 and Tyr-23 of Rev are critical residues for the binding of Nb190.
Nb190 Specifically Inhibits the Rev-dependent Expression of RRE-containing RNA
To evaluate whether Nb190 influences the Rev function in cells, we first analyzed its effect on the Rev-dependent nucleocytoplasmic RNA export using the system developed by Hope et al. (40). The pDM128CATRevRE plasmid produces an RRE-containing transcript that requires Rev for expression of the CAT reporter. 293T cells were co-transfected with pDM128CATRevRE and a Rev-producing plasmid, resulting in the Rev-dependent expression of the CAT reporter (Fig. 5A, left). Co-expression of Nb190 resulted in dose-dependent reduction of Rev-dependent CAT production (Fig. 5A, right).
FIGURE 5.
Nb190 specifically inhibits the Rev-dependent expression of RNA. A, Nb190 decreases the expression of RRE-containing RNAs in cells. Left, 293T cells were transfected with pDM128CATRevRE in the absence or presence of 1 μg of Rev-expressing plasmid. As a control, the co-expression of RevM10 decreases the Rev-dependent CAT expression. The transdominant negative RevM10 mutant disrupts export and expression of Rev-dependent viral mRNAs (55). Right, Rev-dependent CAT expression in the presence of different amounts (μg) of nanobody-expressing plasmid as indicated. The graphs show the levels of the CAT reporter expression as a percentage relative to the condition where Rev but no nanobody was added. Results are mean ± S.E. (error bars) from four independent experiments. B and C, Nb190 inhibits the Rev-RRE- but not the CTE-dependent Gag expression. Left, 293T cells were transfected with pGag-RRE or pGag-CTE in the absence or presence of Rev and RevM10 encoding plasmid as indicated. Right, Rev-dependent Gag expression in the presence of different amounts (μg) of nanobody-expressing plasmid, as indicated. Results are mean ± S.E. (error bars), with n = 4.
To further determine whether the nanobody is specific for the Rev-RRE-dependent RNA export, we compared the effect of Nb190 on the RRE-dependent Gag expression with the expression from the same vector in which the RRE was substituted by a constitutive transport element (Fig. 5, B and C). For this experiment, we used the constitutive transport element (CTE) from the Mason-Pfizer monkey virus (41). CTE-dependent expression is Rev-independent (42, 43). Gag production from pGag-RRE is greatly receptive to Rev (44) and inhibited by Nb190 but not by the Nb163 control, whereas Nb190 had no effect on Rev-independent CTE-driven expression of Gag.
Inhibition of the Rev Protein-Protein Interaction in Cells
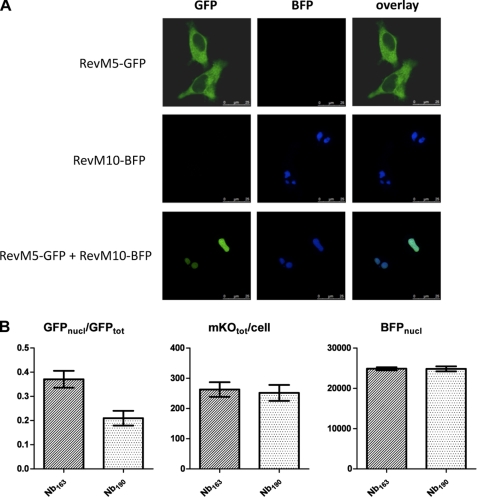
Next, the ability of Nb190 to inhibit Rev-Rev protein-protein interactions in cells was investigated by testing the potency of the nanobody to disrupt the RevM5-GFP/RevM10-BFP interaction in the nucleoli of HeLa cells. RevM5 is a mutant that is unable to bind RNA (21) and localizes to the cytoplasm (Fig. 6A). RevM10 is a trans-dominant negative mutant that is not exported to the cytoplasm and is found in the nucleoli of cells. When RevM5-GFP and RevM10-BFP were co-expressed, RevM5-GFP co-localized with RevM10-BFP in the nucleoli, demonstrating an interaction between both proteins that holds RevM5 in the nucleolus (Fig. 6A). We investigated whether Nb190 can elicit the redistribution of M10-bound RevM5 from the nucleolus to the cytoplasm. Therefore, we measured the relative amount of RevM5-GFP found in the nucleoli of cells that co-expressed RevM5-GFP, RevM10-BFP, and Nb190-mKO (Fig. 6B). This ratio was compared with that of cells co-expressing RevM5-GFP, RevM10-BFP, and the control Nb163-mKO. Indeed, in cells expressing equal amounts of RevM10-BFP and equal amounts of Nb-mKO, significantly less RevM5-GFP was present in the nucleoli of the cells expressing Nb190-mKO compared with the cells expressing Nb163-mKO (Fig. 6B), suggesting that Nb190-mKO disrupts the RevM5/RevM10 interaction, allowing RevM5 to redistribute to the cytoplasm.
FIGURE 6.
Nb190 inhibits the interaction between RevM5-GFP and RevM10-BFP in the nucleoli of cells. A, RevM5-GFP co-localizes with RevM10-BFP in cells. HeLa cells were transfected with expression plasmids, as indicated, and localization of the proteins was visualized by confocal fluorescence microscopy. B, quantification of the colocalization of RevM5-GFP with RevM10-BFP in the presence of Nb190-mKO or the control Nb163-mKO. HeLa cells were co-transfected with RevM5-GFP, RevM10-BFP, and Nb190-mKO or Nb163-mKO and imaged. Cells expressing equal amounts of RevM10-BFP in the nucleoli (BFPnucl) and equal amounts of mKO (BFPtot), respectively, were analyzed for their amount of nucleolar RevM5-GFP (GFPnucl) versus total ReM5-GFP over the cell (GFPtot). Results are mean ± S.E. (error bars), with n = 50, p < 0.0005.
Rev-specific Inhibition of the HIV-1 Production
To assess the potential of Nb190 for anti-HIV therapy, we investigated the effects of Nb190 on HIV-1 expression. First, an Nb190 expression plasmid was co-transfected together with the viral HIV-1NL4-3 molecular clone in 239T cells. Transfection of cells with this molecular clone recapitulates the late events in the viral life cycle, including Rev function, production of all viral mRNAs and proteins, and virus maturation. Virus production was measured by analyzing the p24 Gag amount in the supernatants of transfected cells. In contrast to cells transfected with an expression plasmid for the control nanobody (Nb163), virus production was markedly suppressed in a dose-dependent manner in cells expressing Nb190 (Fig. 7A). To confirm the inhibitory effect of this nanobody on viral production, we constructed chimeric HIV-1 molecular clones by inserting the nanobody coding sequence in the nef open reading frame (Fig. 7B). Such inserted unrelated genes are expressed by HIV-1 in place of Nef during virus replication. This set-up takes into account the equal ratio of Rev versus nanobody in the cells because they are both expressed from multiply spliced RNA in the same molecular clone. These pNL-Nb chimeric molecules were transfected in 293T cells. Virus production from the NL-Nb190 clone was reduced by 300-fold compared with the expression of wild-type NL4-3 or a control virus expressing the control nanobody Nb163 (Fig. 7B).
FIGURE 7.
Nb190 efficiently inhibits the HIV-1 production. A, dose-dependent inhibition of the HIV-1 production by the intracellular expression of Nb190. 293T cells were co-transfected with the pNL4-3 molecular clone (0.5 μg) and increasing amounts of the expression vector for Nb190 (red circles) or the control nanobody Nb163 (green squares). A vector expressing the GFP variant Dronpa was included as negative control (blue triangles). Production of virus was analyzed by quantifying the virus-associated core antigen (Gag p24) in the supernatants of the transfected cells. Values are expressed as percentages relative to the p24 expression levels of cells transfected with pNL4-3 only. Results are mean ± S.E. (error bars), with n = 3. B, comparison of NL4-3 and chimeric NL-Nb expression in cells. Nb190 and Nb163 were cloned in the nef open reading frame of pNL4-3 (top), and these chimeric molecular clones were transfected in 293T cells. The amount of p24 in the supernatants of transfected cells was monitored as a measure for HIV-1 expression (bottom). Results are mean ± S.E. (error bars), with n = 4, p < 0.0005. C, Nb190 fails to inhibit the viral production of a Rev-independent molecular clone. 293T cells were transfected with pNLRev(−)RRE(−)CTE(+) (0.5 μg) and different amounts of Nb190, Nb163, or Dronpa expression plasmids. Expression of pNLRev(−)RRE(−)CTE(+) was analyzed by measuring the virus-associated core p24 antigen in the supernatants of the transfected cells. p24 production levels are presented relative to control cells expressing pNL4-3Rev(−)RRE(−)CTE(+) only. Results are mean ± S.E., with n = 3.
To confirm our inhibitor is inhibiting the HIV-1 production by specifically targeting the Rev function, we assessed the effect of Nb190 on the expression of a Rev-independent HIV-1 molecular clone NLRev(−)RRE(−)CTE(+) (42). In this engineered infectious HIV-1 clone, Rev and RRE are substituted by the CTE from the Mason-Pfizer monkey virus. CTE-dependent expression is Rev-independent (43). Upon co-transfection, Nb190 was unable to reduce the expression of this Rev-independent virus, reinforcing our conclusion that the antiviral effect of this nanobody, targeting the N-terminal α-helical multimerization domain of Rev, closely correlates with inhibition of the Rev function (Fig. 7C).
Nb190 Inhibits the Expression of Late Viral RNAs
Compelling independent confirmation of the Rev-specific effects of Nb190 was obtained by analyzing its effect on the expression levels of the different viral mRNA species. Wild-type HIV-1 produces three predominant size classes of viral mRNAs with sizes of 9.3, 4, and 2 kb (45). The late unspliced 9.3-kb and partially spliced 4-kb species are expressed in a Rev-dependent manner, whereas the expression of the early multiply spliced 2-kb mRNAs is Rev-independent. In Northern blot (Fig. 8A) and real-time quantitative reverse transcription-PCR experiments (Fig. 8, B and C) on RNA extracted from cells transfected with the chimeric NL-Nb190 molecular clone or co-transfected with NL4-3 and Nb190, we found that the ratio of the late unspliced transcripts over the early fully spliced mRNA was greatly reduced compared with cells transfected with the wild-type NL4-3 or the NL-Nb163 virus. These data establish that Nb190 inhibits the HIV-1 production in cell culture by specifically disrupting Rev function.
FIGURE 8.
Nb190 inhibits the expression of partially spliced and unspliced viral RNA. A, Northern blot analysis of viral mRNA species in cells transfected with wild-type or chimeric pNL-Nb molecular clones. B and C, levels of late unspliced relative to early multiply spliced viral mRNA species, as measured by real-time quantitative reverse transcription-PCR. 293T cells were transfected with wild-type or chimeric pNL-Nb clones (B) or co-transfected with pNL4-3 and Dronpa control, Nb, or RevM10 vector plasmid, as indicated (C). The difference in RNA sizes observed for the NL4-3 versus the chimeric NL-Nb molecular clones is due to the insertion of the nanobody coding sequence in the nef open reading frame for the chimeric viruses. Results are mean ± S.E., with n = 3, p < 0.025.
DISCUSSION
In this study, we selected a nanobody, derived from a llama heavy-chain only antibody, that binds the N-terminal α-helical multimerization domain of Rev and potently inhibits its oligomerization. This nanobody specifically suppresses the Rev-dependent expression of late viral RNAs. Previous attempts to develop Rev antibodies have used a single-chain antibody approach by incorporating the variable regions of light and heavy chains from a conventional monoclonal antibody in a single gene (SFv). These anti-Rev SFv possess potent anti-HIV activity by sequestering Rev to the cytoplasm (46). The nanobody described here also triggers a cytoplasmic relocalization of Rev, but in contrast to the previously reported anti-Rev SFv, it does not retain Rev in the cytoplasm because Rev was still able to shuttle between the nucleus and cytoplasm in the presence of Nb190. We have demonstrated that this nanobody binds residues Lys-20 and Tyr-23 in the N-terminal multimerization domain of Rev, independently confirming its anti-multimerization activity. However, we cannot completely exclude the possibility that, in addition to its anti-multimerization effect, in cells the nanobody exerts an extra anti-Rev activity that involves the same or overlapping Lys-20 and Tyr-23 residues. The N terminus of Rev is essential for Rev function and plays a role in its multimerization (17, 24, 25, 47). The DEAD box helicase DDX1 is a co-factor that was also found to interact with the N terminus of Rev (48). Also, other DEAD box helicases like DDX3 have an important role in Rev-dependent HIV-1 gene expression (49). Future research will verify whether in cells the nanobody, in addition to its anti-multimerization activity, also affects other functions. Nevertheless, by identifying a new reactive site in the Rev protein, our findings set the stage for the development of a novel class of anti-HIV drugs.
The nanobody Nb190 identified here represents the first known molecule that destabilizes and prevents the formation of a large organized homoprotein complex required for efficient HIV-1 mRNA export from the nucleus. Drugs directed against this novel and highly specific viral target are not expected to show any cross-resistance to all currently available clinically used anti-HIV drugs. Given the vital and conserved role of Rev in the HIV replication process, Rev inhibitors are also expected to affect replication of a wide variety of HIV-1 strains. In HIV-susceptible cells, this nanobody by itself may be used directly as an effective anti-HIV agent in vivo using genetic immunization strategies. Although the therapeutic potential of antibodies aimed at intracellular targets may be questionable, our nanobody provides a strong proof of principle that HIV-1 Rev, and especially its residues Lys-20 and Tyr-23, is a potential target for the development of therapeutic anti-HIV-1 agents. Indeed, these residues are highly conserved: 92.7% for Lys-20 and 98.5% for Tyr-23. Moreover, the Rev-Nb190 binding may be of value as a tool for drug screening and as a starting point for drug design. Indeed, by resolving the epitope, this nanobody represents a suitable lead for antiviral drug development, and small molecules may be modeled to interact with these residues. Progress has been made in developing small molecules capable of disrupting protein-protein interactions; however, this remains a challenging but not impossible task (50). In particular, small molecule inhibitors dissociating trimeric TNF-α complexes or the p53-HDM2 interaction have been described (50, 51). The identification of this nanobody will also facilitate the establishment of highly specific and sophisticated drug discovery methods both in vitro and in cells for the discovery of a new class of inhibitors directed against this target.
Understanding the three-dimensional structure of Rev would be advantageous for the rational design of small molecules against this target. Although circular dichroism, nuclear magnetic resonance, and Raman spectroscopy studies strongly suggest that the oligomerization domains and the nuclear localization signal are components of a helix-turn-helix motif (12–14), the full molecular Rev structure is still unknown. This is mainly because in solution Rev is strongly prone to aggregation at concentrations above 1 μm, both in the presence and in the absence of RNA (52). Ultimately, blocking the aggregation of Rev in solution by nanobodies may be the key to unveil its structure by x-ray crystallography. Moreover, nanobodies have proven potential as a crystallization aid for structure determination of proteins that present particular experimental challenges and are not conducive to more conventional crystallization strategies (53).
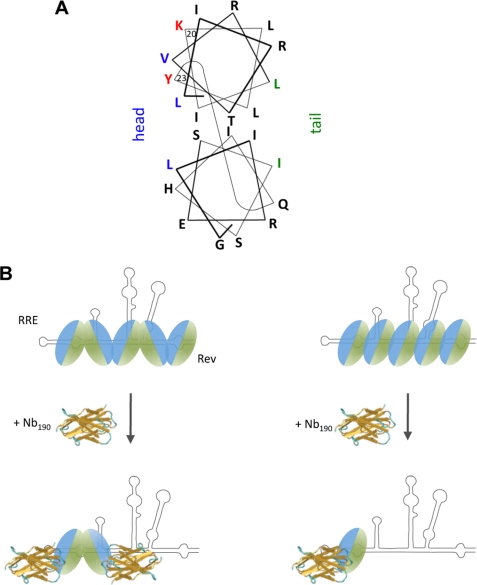
This nanobody also proved a powerful tool to study the mechanism of Rev oligomer assembly and allowed us to support a molecular model for Rev assembly. Rev includes two separate multimerization domains that are joined, yielding a head and tail protein surface (24) (Fig. 9A). Two structural models for the Rev assembly along the RRE are possible: a head-to-tail and a head-to-head/tail-to-tail configuration. Nanobodies that selectively bind the head or the tail of the monomer can distinguish these two scenarios as outlined in Fig. 9B. Accumulation of monomeric Rev in the presence of nanobody would indicate a head-to-tail configuration (Fig. 9B, right), whereas accumulation of Rev dimers would favor the model of symmetrical head-to-head and tail-to-tail multimer formation (Fig. 9B, left). We consistently observed that Nb190 fails to completely eliminate the FRET signal in the in vitro multimerization assay, whereas purified recombinant Rev completely reduces the FRET signal to background levels. We speculate that the residual FRET signal in the presence of Nb190 results from ECFP-Rev/EYFP-Rev dimers. Indeed, in gel shift experiments, the presence of Nb190 reduced the amounts of multimeric Rev-RRE complexes and concomitantly led to the accumulation of dimeric Rev species. The observed accumulation of dimeric Rev upon inhibition of multimerization provides independent support for a head-to-head/tail-to-tail model for Rev assembly (24) (Fig. 9B, left). In addition, alanine scan mutations revealed that Nb190 is binding to Rev residues Lys-20 and Tyr-23, which are, according to the latest structural models for Rev, located within the α-helical surface that contributes to the head multimerization facade of Rev (24) (Fig. 9A). This result obtained both in vitro and in physiological conditions, confirms the anti-multimerization effect of the nanobody and indicates that Nb190 selectively binds the head surface of Rev. Moreover, recent single molecule experiments have indicated that the assembly of Rev on RRE proceeds stepwise after nucleation of a single Rev monomer bound to a high affinity site in stem-loop IIB (27). These findings, together with our observation that Nb190 interacts with the head multimerization surface, suggests that the first Rev molecule is positioned on the RRE in such a way that the second monomer binds through a tail-to-tail interaction (Fig. 9B, left).
FIGURE 9.
Model for inhibition of Rev multimerization. A, helical wheel projection model of a Rev monomer molecule (adapted from Ref. 24). Protein residues implicated in multimerization are indicated in blue for the head surface and in green for the tail surface. Residues Lys-20 and Tyr-23, important for nanobody binding, are indicated in red. B, structural model for Rev multimeric assembly of Rev on the RRE and inhibition by Nb190. Possible models for Rev multimerization are shown: the head-to-head/tail-to-tail model for Rev multimerization, which is supported by our data (left), and head-to-tail model (right).
Assembly of a functional multimeric Rev-RRE ribonucleoprotein complex requires the multimerization of Rev onto multiple specific RNA sites. Rev shows a cooperative chameleon-like RNA binding behavior, implying that it employs different surfaces of its RNA-binding domain to recognize different low affinity binding sites on the RRE (26, 54). Our data show that inhibition of the oligomerization at the stage of the first dimer formed on the RRE prevents the further coating of the RNA (Fig. 2B). These data confirm that the oligomeric assembly of Rev follows an ordered stepwise process that starts at a single site on the RRE.
In conclusion, by selecting a nanobody that targets the Rev N-terminal α-helical multimerization domain and blocks the oligomerization of Rev, we have identified a reactive site in the Rev protein that will inspire future novel approaches for therapeutic intervention against HIV. The nanobody also proved to be a unique tool to refine the molecular model for concerted assembly of Rev multimers to a stepwise head-to-head/tail-to-tail mechanism. Furthermore, it is a valuable instrument to further unravel in cells the complex pathway of HIV mRNA nucleocytoplasmic transport and the interplay with cellular co-factors.
Supplementary Material
Acknowledgments
We thank L. Bral, L. Dedier, N. Buys, and B. Van Laer for excellent technical assistance; Dr. C. Pannecouque and members of the Rega Institute and the Structural Biology Laboratory for discussions; Drs. L Vandenberghe and P. Cherepanov for critical reading of the manuscript; and Drs. G. Pavlakis and B. Felber for plasmids. A number of reagents were obtained through the National Institutes of Health AIDS Reagent Program.
This work was supported by “Fonds voor Weteschappelijk Onderzoek-Vlaanderen” Grants 1.5.104.07 and 1.5.165.10, the Belgian Government under the framework of the Interuniversity Attraction Poles (I.A.P. P6/19), the “Instituut voor de Aanmoediging van Innovatie door Wetenschap en Technologie,” and the “K.U. Leuven Impulsfinanciering.”

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- HIV
- human immunodeficiency virus
- FRET
- fluorescence resonance energy transfer
- RRE
- Rev-responsive element
- ECFP
- enhanced cyan fluorescent protein
- EYFP
- enhanced yellow fluorescent protein
- Nb
- nanobody
- GFP
- green fluorescent protein
- CTE
- constitutive transport element
- mKO
- monomeric Kusabira Orange fluorescent protein.
REFERENCES
- 1.Pollard V. W., Malim M. H. (1998) Annu. Rev. Microbiol. 52, 491–532 [DOI] [PubMed] [Google Scholar]
- 2.Dayton E. T., Powell D. M., Dayton A. I. (1989) Science 246, 1625–1629 [DOI] [PubMed] [Google Scholar]
- 3.Daly T. J., Cook K. S., Gray G. S., Maione T. E., Rusche J. R. (1989) Nature 342, 816–819 [DOI] [PubMed] [Google Scholar]
- 4.Malim M. H., Hauber J., Le S. Y., Maizel J. V., Cullen B. R. (1989) Nature 338, 254–257 [DOI] [PubMed] [Google Scholar]
- 5.Fornerod M., Ohno M., Yoshida M., Mattaj I. W. (1997) Cell 90, 1051–1060 [DOI] [PubMed] [Google Scholar]
- 6.Fukuda M., Asano S., Nakamura T., Adachi M., Yoshida M., Yanagida M., Nishida E. (1997) Nature 390, 308–311 [DOI] [PubMed] [Google Scholar]
- 7.Neville M., Stutz F., Lee L., Davis L. I., Rosbash M. (1997) Curr. Biol. 7, 767–775 [DOI] [PubMed] [Google Scholar]
- 8.Cochrane A. W., Perkins A., Rosen C. A. (1990) J. Virol. 64, 881–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Felber B. K., Hadzopoulou-Cladaras M., Cladaras C., Copeland T., Pavlakis G. N. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 1495–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalland K. H., Szilvay A. M., Brokstad K. A., Saetrevik W., Haukenes G. (1994) Mol. Cell. Biol. 14, 7436–7444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meyer B. E., Malim M. H. (1994) Genes Dev. 8, 1538–1547 [DOI] [PubMed] [Google Scholar]
- 12.Auer M., Gremlich H. U., Seifert J. M., Daly T. J., Parslow T. G., Casari G., Gstach H. (1994) Biochemistry 33, 2988–2996 [DOI] [PubMed] [Google Scholar]
- 13.Blanco F. J., Hess S., Pannell L. K., Rizzo N. W., Tycko R. (2001) J. Mol. Biol. 313, 845–859 [DOI] [PubMed] [Google Scholar]
- 14.Tan R., Chen L., Buettner J. A., Hudson D., Frankel A. D. (1993) Cell 73, 1031–1040 [DOI] [PubMed] [Google Scholar]
- 15.Fischer U., Huber J., Boelens W. C., Mattaj I. W., Lührmann R. (1995) Cell 82, 475–483 [DOI] [PubMed] [Google Scholar]
- 16.Daelemans D., Costes S. V., Lockett S., Pavlakis G. N. (2005) Mol. Cell. Biol. 25, 728–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malim M. H., Böhnlein S., Hauber J., Cullen B. R. (1989) Cell 58, 205–214 [DOI] [PubMed] [Google Scholar]
- 18.Daelemans D., Afonina E., Nilsson J., Werner G., Kjems J., De Clercq E., Pavlakis G. N., Vandamme A. M. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 14440–14445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Neck T., Pannecouque C., Vanstreels E., Stevens M., Dehaen W., Daelemans D. (2008) Bioorg. Med. Chem. 16, 9487–9497 [DOI] [PubMed] [Google Scholar]
- 20.Wolff B., Sanglier J. J., Wang Y. (1997) Chem. Biol. 4, 139–147 [DOI] [PubMed] [Google Scholar]
- 21.Malim M. H., Cullen B. R. (1991) Cell 65, 241–248 [DOI] [PubMed] [Google Scholar]
- 22.Mann D. A., Mikaélian I., Zemmel R. W., Green S. M., Lowe A. D., Kimura T., Singh M., Butler P. J., Gait M. J., Karn J. (1994) J. Mol. Biol. 241, 193–207 [DOI] [PubMed] [Google Scholar]
- 23.Daelemans D., Costes S. V., Cho E. H., Erwin-Cohen R. A., Lockett S., Pavlakis G. N. (2004) J. Biol. Chem. 279, 50167–50175 [DOI] [PubMed] [Google Scholar]
- 24.Jain C., Belasco J. G. (2001) Mol. Cell 7, 603–614 [DOI] [PubMed] [Google Scholar]
- 25.Thomas S. L., Oft M., Jaksche H., Casari G., Heger P., Dobrovnik M., Bevec D., Hauber J. (1998) J. Virol. 72, 2935–2944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Daugherty M. D., D'Orso I., Frankel A. D. (2008) Mol. Cell 31, 824–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pond S. J., Ridgeway W. K., Robertson R., Wang J., Millar D. P. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 1404–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith C. A., Calabro V., Frankel A. D. (2000) Mol. Cell 6, 1067–1076 [DOI] [PubMed] [Google Scholar]
- 29.Pomerantz R. J., Seshamma T., Trono D. (1992) J. Virol. 66, 1809–1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hamers-Casterman C., Atarhouch T., Muyldermans S., Robinson G., Hamers C., Songa E. B., Bendahman N., Hamers R. (1993) Nature 363, 446–448 [DOI] [PubMed] [Google Scholar]
- 31.Muyldermans S., Cambillau C., Wyns L. (2001) Trends Biochem. Sci. 26, 230–235 [DOI] [PubMed] [Google Scholar]
- 32.Arbabi Ghahroudi M., Desmyter A., Wyns L., Hamers R., Muyldermans S. (1997) FEBS Lett. 414, 521–526 [DOI] [PubMed] [Google Scholar]
- 33.Conrath K. E., Lauwereys M., Galleni M., Matagne A., Frère J. M., Kinne J., Wyns L., Muyldermans S. (2001) Antimicrob. Agents Chemother. 45, 2807–2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fisher A. G., Collalti E., Ratner L., Gallo R. C., Wong-Staal F. (1985) Nature 316, 262–265 [DOI] [PubMed] [Google Scholar]
- 35.Gibbs J. S., Regier D. A., Desrosiers R. C. (1994) AIDS Res. Hum. Retroviruses 10, 343–350 [DOI] [PubMed] [Google Scholar]
- 36.McNally J. G. (2008) Methods Cell Biol. 85, 329–351 [DOI] [PubMed] [Google Scholar]
- 37.Pasternak A. O., Adema K. W., Bakker M., Jurriaans S., Berkhout B., Cornelissen M., Lukashov V. V. (2008) J. Clin. Microbiol. 46, 2206–2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tiley L. S., Malim M. H., Tewary H. K., Stockley P. G., Cullen B. R. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 758–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Madore S. J., Tiley L. S., Malim M. H., Cullen B. R. (1994) Virology 202, 186–194 [DOI] [PubMed] [Google Scholar]
- 40.Hope T. J., McDonald D., Huang X. J., Low J., Parslow T. G. (1990) J. Virol. 64, 5360–5366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sonigo P., Barker C., Hunter E., Wain-Hobson S. (1986) Cell 45, 375–385 [DOI] [PubMed] [Google Scholar]
- 42.Bray M., Prasad S., Dubay J. W., Hunter E., Jeang K. T., Rekosh D., Hammarskjöld M. L. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 1256–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zolotukhin A. S., Valentin A., Pavlakis G. N., Felber B. K. (1994) J. Virol. 68, 7944–7952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hadzopoulou-Cladaras M., Felber B. K., Cladaras C., Athanassopoulos A., Tse A., Pavlakis G. N. (1989) J. Virol. 63, 1265–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim S. Y., Byrn R., Groopman J., Baltimore D. (1989) J. Virol. 63, 3708–3713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Duan L., Bagasra O., Laughlin M. A., Oakes J. W., Pomerantz R. J. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 5075–5079 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 47.Kubota S., Pomerantz R. J. (1998) Oncogene 16, 1851–1861 [DOI] [PubMed] [Google Scholar]
- 48.Fang J., Kubota S., Yang B., Zhou N., Zhang H., Godbout R., Pomerantz R. J. (2004) Virology 330, 471–480 [DOI] [PubMed] [Google Scholar]
- 49.Yedavalli V. S., Neuveut C., Chi Y. H., Kleiman L., Jeang K. T. (2004) Cell 119, 381–392 [DOI] [PubMed] [Google Scholar]
- 50.He M. M., Smith A. S., Oslob J. D., Flanagan W. M., Braisted A. C., Whitty A., Cancilla M. T., Wang J., Lugovskoy A. A., Yoburn J. C., Fung A. D., Farrington G., Eldredge J. K., Day E. S., Cruz L. A., Cachero T. G., Miller S. K., Friedman J. E., Choong I. C., Cunningham B. C. (2005) Science 310, 1022–1025 [DOI] [PubMed] [Google Scholar]
- 51.Fischer P. M., Lane D. P. (2004) Trends Pharmacol. Sci. 25, 343–346 [DOI] [PubMed] [Google Scholar]
- 52.Watts N. R., Misra M., Wingfield P. T., Stahl S. J., Cheng N., Trus B. L., Steven A. C., Williams R. W. (1998) J. Struct. Biol. 121, 41–52 [DOI] [PubMed] [Google Scholar]
- 53.Lam A. Y., Pardon E., Korotkov K. V., Hol W. G., Steyaert J. (2008) J. Struct. Biol. 166, 8–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bayer T. S., Booth L. N., Knudsen S. M., Ellington A. D. (2005) RNA 11, 1848–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bevec D., Dobrovnik M., Hauber J., Böhnlein E. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 9870–9874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Costes S. V., Daelemans D., Cho E. H., Dobbin Z., Pavlakis G., Lockett S. (2004) Biophys. J. 86, 3993–4003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.