Abstract
Hydroxyurea (HU) is a well tolerated ribonucleotide reductase inhibitor effective in HIV, sickle cell disease, and blood cancer therapy. Despite a positive initial response, however, most treated cancers eventually progress due to development of HU resistance. Although RNR properties influence HU resistance in cell lines, the mechanisms underlying cancer HU resistance in vivo remain unclear. To address this issue, we screened for HU resistance in the plant Arabidopsis thaliana and identified seventeen unique catalase mutants, thereby establishing that HU toxicity depends on catalase in vivo. We further demonstrated that catalase is a direct HU target by showing that HU acts as a competitive inhibitor of catalase-mediated hydrogen peroxide decomposition. Considering also that catalase can accelerate HU decomposition in vitro and that co-treatment with another catalase inhibitor alleviates HU effects in vivo, our findings suggests that HU could act as a catalase-activated pro-drug. Clinically, we found high catalase activity in circulating cells from untreated chronic myeloid leukemia, offering a possible explanation for the efficacy of HU against this malignancy.
Keywords: Arabidopsis, Cancer Therapy, Drug Action, Drug Resistance, Enzyme Inhibitors, Enzyme Mutation, Genetics
Introduction
Since the early 1960s hydroxyurea (HU)2 has been used as a well tolerated antiproliferative drug in the treatment of chronic myeloproliferative disorders, in particular chronic myeloid leukemia (CML), and is now also used to treat non-neoplastic diseases (1). Currently, HU is the only FDA-approved drug for treatment of sickle cell disease, and it has also been used in HIV therapy (2, 3).
When treated with HU, most patients suffering from myeloproliferative disorders initially respond well with normalization of blood counts, but ultimately the malignant process progresses due to development of HU resistance through an unknown mechanism (4). Early on, it became clear that HU limits the cellular supply of deoxyribonucleotides by acting as an inhibitor of ribonucleotide reductase (RNR) (5). The importance of the HU-RNR interaction was bolstered by a number of cell line studies, where changes in RNR properties were shown to influence HU resistance (6–10). In this context, relatively little attention has been given to alternative potential HU targets, perhaps both due to lack of evidence for their in vivo importance and to the dominant position of the HU-RNR interaction in the field.
We hypothesized that plants, which depend on mitotically active stem cell populations for growth, could serve as a novel platform for investigation of HU resistance. By unbiased forward genetic screening, we identified the hydrogen peroxide-decomposing enzyme catalase (EC 1.11.1.6) as a major determinant of HU resistance in vivo and further found that HU directly targets and inhibits catalase.
MATERIALS AND METHODS
Plant Growth Conditions
Arabidopsis thaliana ecotype Columbia (Col-0) wild-type and mutant plants were grown on ½ Murashige and Skoog medium (Duchefa, M0222) solidified with 0.9% agar in growth chambers at 21 °C, 16/8 h light/dark cycles, and 50 μmol m−2s−1 photosynthetic photon flux. HU (US Biological, H9120) and 3-amino-1,2,4-triazole (3-AT) (Sigma-Aldrich, A8056) were dissolved in water to prepare 2.5 m and 1 m stock solutions, respectively, and added to the medium immediately before pouring plates.
RNR Activity Assay
RNR activity was quantified as described (11). Cytidine 5′-diphosphate, triammonium salt, 2-14C-labeled (Moravek), was used for labeling. 300 mg of plant tissue per sample was collected and ground in liquid nitrogen.
Quantification of HU Content
HU was quantified as described (12). Between 8 and 30 mg of leaf tissue per sample was collected and ground in phosphate-buffered saline. The concentration of HU in plant shoots was calculated assuming a water content of 85% (13).
Catalase Enzyme Assay and Materials
Catalase activity was quantified by monitoring the rate of H2O2 breakdown at 240 nm (14). GraphPad Prism 5 was used for statistical analysis and curve fitting. Plant extracts were prepared by grinding ∼60 mg of tissue in phosphate-buffered saline. Rat erythrocytes were isolated by centrifugation and lysed in distilled water. Blood samples were drawn into 10 ml of EDTA stabilized tubes. Less than 1 h after vein puncture samples were processed into mononuclear cells by Lymphoprep differential centrifugation at 2100 rpm for 10 min. The interface mononuclear cell suspension was harvested and washed twice with phosphate-buffered saline. Isolated mononuclear cells were counted and subsequently frozen in DMSO in liquid nitrogen. Cell lysates were prepared from 5 × 106 cells thawed at 37 °C, spun at 300 × g for 5 min, washed with phosphate-buffered saline, and finally lysed in distilled water. Catalase activity was normalized to the total protein content of each sample. For experiments with purified enzyme, a commercially available aqueous catalase solution was used (Sigma, C3155).
RESULTS
A. thaliana as a Model for Studying HU Toxicity
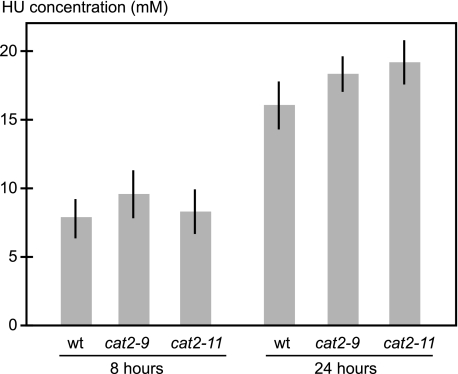
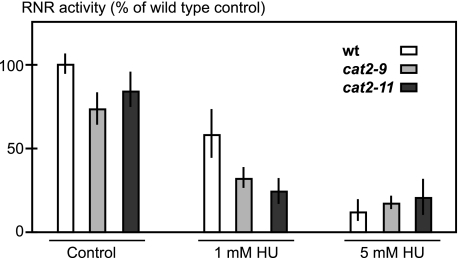
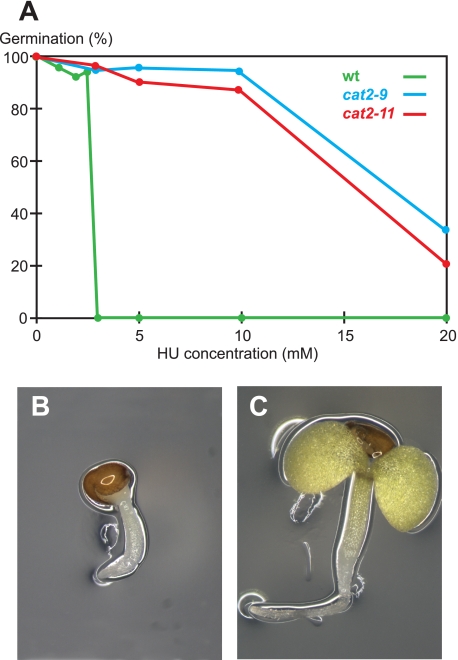
Plant growth depends on the activity of pluripotent stem cells within the root and shoot apical meristems, allowing for easy scoring of plant stem cell viability. This makes plants convenient for studying stem cell resistance to cytotoxic agents such as HU. Ideally, HU taken up by A. thaliana should inhibit its primary target RNR and kill plants at a concentration physiologically relevant to cancer treatment. To examine these criteria, we first investigated plant uptake of HU by measuring the HU concentration in plants grown for 8 or 24 h on 25 mm HU. Roots, not shoots, were in contact with the medium, and we measured HU content in leaves to ensure quantifying only internalized HU. After 24 h, the concentration in plant shoots approached that of the medium, showing that A. thaliana seedlings take up HU (Fig. 1). Second, we measured the effect of HU on A. thaliana RNR activity in protein extracts and found a clear inhibition (Fig. 2), supporting previously published results showing that HU can quench the tyrosyl radical of a purified subunit of A. thaliana RNR (15). Having established HU uptake and RNR inhibition, we then quantified HU toxicity in A. thaliana to establish screening conditions. On medium containing 2.5 mm HU the germination rate was >90% but dropped sharply to less than 1/20,000 on 3 mm HU, which demonstrated the feasibility of a tight and sensitive screen (Fig. 3A). A 3 mm HU concentration is comparable to the peak plasma levels of 1 mm HU detected after HU administration to blood cancer patients, indicating that screening conditions were within a physiologically relevant range (16).
FIGURE 1.
HU uptake by wild-type and HU-resistant mutants. 11-day-old wild-type and mutant seedlings grown on drug-free medium were moved to plates containing 25 mm HU and incubated for 8 or 24 h before shoots were harvested. After 24 h, the HU concentration in seedling shoots approached that of the medium for both wild-type and HU-resistant plants. HU could not be detected in plants grown on drug-free medium. Error bars indicate ±S.E.
FIGURE 2.
Inhibition of RNR activity by HU in wild-type and HU-resistant mutants. RNR activity in extracts from wild-type and HU-resistant mutants is shown. In the control, no HU was added to the RNR reaction. Where indicated, 1 or 5 mm HU was added to the RNR reaction. Error bars indicate ±S.E.
FIGURE 3.
HU response in wild-type and HU-resistant mutants. A, >90% of wild-type plants germinated on 2.5 mm HU, whereas <1/20,000 germinated on 3 mm HU. Two HU-resistant mutants germinating at significantly higher concentrations of HU than the wild type are shown. B and C, 7-day-old seedlings grown on 3 mm HU. B, wild type; C, cat2-9.
Loss of Catalase Function Causes HU Resistance
We screened 250,000 ethane methyl sulfonate mutagenized seeds from 20,000 parental lines on medium containing 3 mm HU and recovered more than 20 independent HU-resistant mutants. Resistant mutants were easily distinguished from the wild type by their elongated roots and expanded cotyledons (Fig. 3, B and C), and they tolerated HU concentrations more than five times higher than that lethal to the wild type (Fig. 3A and Table 1).
TABLE 1.
A. thaliana CAT2 mutant alleles
Accession number: AtCAT2: AT4G35090.1.
| Allele name | Amino acid position | Amino acid | Mutant amino acid | HU survivala | Catalase activityb | S.E. |
|---|---|---|---|---|---|---|
| % of wt | ||||||
| cat2-1c | 4th exond | 76 | 19.3 | 2.4 | ||
| cat2-2e | 3rd exond | 79 | 14.1 | 4.4 | ||
| cat2-3 | 61 | E | K | 81 | 29.9 | 3.9 |
| cat2-4 | 73 | G | D | NDf | ND | ND |
| cat2-5 | 104 | S | F | 57 | 29.6 | 2.1 |
| cat2-6 | 111 | G | E | 37 | 29.4 | 4.0 |
| cat2-7 | 114 | E | K | 82 | 59.0 | 7.1 |
| cat2-8 | 137 | G | R | 78 | 40.6 | 5.2 |
| cat2-9 | 182 | S | N | 69 | 19.0 | 4.2 |
| cat2-10 | 185 | M | I | 28 | 38.8 | 2.6 |
| cat2-11 | 251 | A | T | 73 | 15.7 | 0.7 |
| cat2-12 | 265 | P | L | 80 | 21.8 | 2.4 |
| cat2-13 | 267 | W | Stop | 70 | 49.4 | 7.1 |
| cat2-14 | 293 | W | Stop | 72 | 38.3 | 5.4 |
| cat2-15 | 332 | G | R | 99 | 18.6 | 3.3 |
| cat2-16 | 344 | R | Q | 97 | 30.2 | 5.2 |
| cat2-17 | 450 | W | Stop | 94 | 23.6 | 1.8 |
| cat2-18 | 473 | No splice | 93 | 31.3 | 3.4 | |
| cat2-19 | 480 | Q | Stop | 80 | 21.2 | 3.7 |
a Average germination rate for plants grown on 5, 10, and 25 mm HU. For wild-type plants, this value is zero.
b All measured mutant catalase activities were significantly lower than that of the wild type (95% CL, 1-way analysis of variance, Tukey post-test, GraphPad Prism 5.0).
c SALK_076998 T-DNA line.
d As described (20).
e SALK_057998 T-DNA line.
f ND, not determined.
To determine whether HU resistance was caused by an increase in RNR activity or by RNR activity resistant to HU inhibition, as previously described (9, 17–19), we compared RNR activity between wild-type plants and HU-resistant mutants. RNR enzyme activity in resistant mutants could be inhibited by HU and was not higher than in wild-type plants (Fig. 2), suggesting a novel RNR-independent resistance mechanism in the isolated mutants.
We therefore proceeded to identify the causative mutations by map-based cloning. One of the resistant mutants, cat2-9, in A. thaliana Columbia background, was crossed to the closely related ecotype A. thaliana Landsberg erecta, and the offspring were allowed to self-fertilize to produce a mapping population. We delimited a region containing five genes by genetic mapping and identified a point mutation in the CATALASE2 gene (AT4G35090). Upon sequencing the catalase open reading frame in all identified HU-resistant mutants, we found lesions in the catalase gene in the majority of mutants, confirming that the causative mutation had been identified. In total, 17 unique alleles were recovered (Table 1). All catalase mutants were then assayed for hydrogen peroxide decomposition activity. When compared with the wild type, all mutants displayed a significant decrease (95% confidence level) in catalase activity (Table 1). Two previously described catalase loss-of-function T-DNA insertion mutants also displayed HU resistance, reconfirming correct identification of the causal mutation (Table 1) (20, 21). The HU resistance of the T-DNA mutants also indicated that loss of catalase function was sufficient for inducing HU resistance and that no specific amino acid changes modifying enzyme function were required. Comparing the catalase activities of our newly identified mutants with those of the null-allele T-DNA mutants cat2-1 and cat2-2 indicated that we may have identified several additional CATALASE2 null alleles (Table 1) (20).
We scored mutants for their degree of HU resistance quantified as the germination rate in the presence of HU. No correlation between residual catalase activity and HU resistance was apparent, suggesting that other properties of catalase than catalysis of hydrogen peroxide decomposition may influence HU resistance (Table 1). However, because the catalase mutant lines analyzed have not been backcrossed, confounding influence of secondary mutations on catalase activity and HU resistance cannot be ruled out.
HU Inhibits Catalase Activity
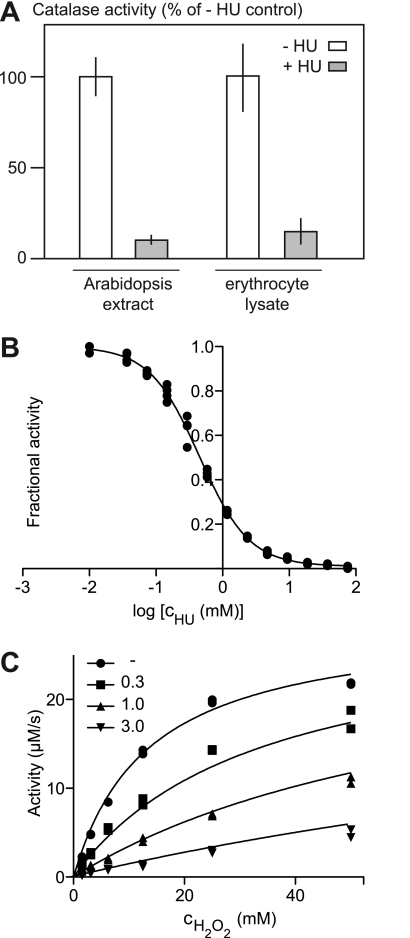
To study the HU resistance mechanism, we investigated the effect of HU on catalase-mediated hydrogen peroxide decomposition. In complex mixtures, we found that addition of HU inhibited hydrogen peroxide decomposition in both plant extracts and rat erythrocyte lysates (Fig. 4A), indicating that the interaction was conserved across species in line with the high similarity of plant and animal catalases, which are ∼40% identical at the amino acid level.
FIGURE 4.
HU inhibition of catalase-mediated decomposition of H2O2. A, catalase activity in plant and rat protein preparations in the absence (− HU) and presence of 1 mm HU (+ HU) is shown. Triplicate measurements were performed, and error bars indicate ±S.D. B, fractional activity, with respect to catalase activity under the same conditions in the absence of HU, is plotted versus log[cHU (mm)]. The black line indicates the best fit to a sigmoidal response curve. C, catalase activity plotted versus cH2O2 in the presence of varying concentrations of HU (millimolar). Black lines indicate the best fit to a competitive inhibition model.
To confirm that the HU-catalase interaction was direct and did not require additional components, we characterized the interaction quantitatively using commercially available purified bovine catalase, a representative mammalian catalase. Measuring catalase activity in the presence of 12 mm H2O2 and varying concentrations of inhibitor produced a sigmoidal response curve, from which we determined an IC50 value of 0.44 ± 0.02 mm (95% confidence level) using nonlinear regression (Fig. 4B). Next, both substrate (H2O2) and inhibitor (HU) concentrations were varied to allow determination of the mode of inhibition using curve fitting (Fig. 4C). Best fits of the collected data to the models “competitive inhibition,” “noncompetitive inhibition,” and “uncompetitive inhibition” were compared. Competitive inhibition was the preferred model with a probability of >99.99% according to Akaike's method, with an information criterion with second order correction for small sample size (AICc) scores of 26.80 (noncompetitive) and 78.54 (uncompetitive). Based on the best fit to the competitive inhibition model, we calculated the following kinetic characteristics: Km = 13.52 (11.22–15.82 mm), Ki = 0.23 (0.19–0.26 mm), and Vmax = 29 (27–31 μm/s) (95% confidence level indicated). If Ki was instead calculated from the IC50 value obtained from the data shown in Fig. 4B using the Cheng-Prusoff equation (22), a value of Ki = IC50/(1 + [S]/Km) = 0.23 mm was still obtained. This Ki value is comparable to those previously published for RNR inhibition by HU (0.07–0.13 mm) (23). Because peak plasma levels can reach concentrations of 1 mm after HU administration to blood cancer patients (16), HU treatment could have an impact on cellular catalase activity.
HU: A Catalase-activated Pro-drug?
Our results show that HU directly inhibits catalase-mediated hydrogen peroxide decomposition. Conversely, the decomposition of HU itself can be accelerated by catalase (24). This suggests two alternative ways by which HU could restrict plant growth in a catalase-dependent manner: by inhibiting catalase activity or by catalase-mediated conversion to cytotoxic compounds.
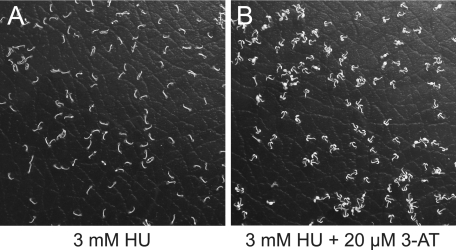
To distinguish between these two hypotheses we co-treated plants with HU and the known catalase inhibitor 3-AT. If catalase inhibition were the restricting factor, co-treatment with an additional inhibitor should exacerbate the effect of HU. Conversely, if cytotoxic HU-derived metabolites restricted growth, adding 3-AT should alleviate this restriction by obstructing catalase-mediated HU conversion. We found that co-treatment with 3-AT restored germination (Fig. 5), supporting the hypothesis that HU could act as a catalase-activated pro-drug.
FIGURE 5.
Co-treatment of A. thaliana seedlings with HU and 3-AT. A. thaliana seedlings germinated on medium containing 3 mm HU (A) and 3 mm HU and 20 μm 3-AT (B). Note that cotelydons are unfolded in B but not in A.
Increased Catalase Activity in Samples from CML Patients
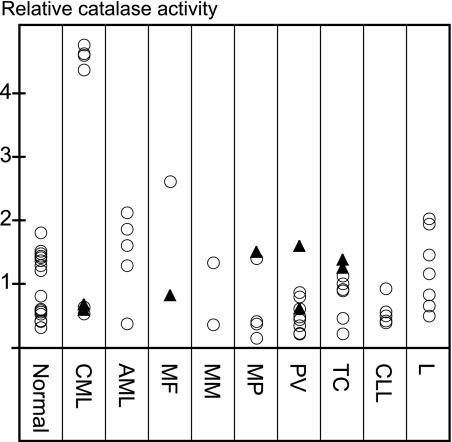
Because catalase and HU interacted directly, we proceeded to study the potential of catalase activity as a biomarker in circulating blood cells from patients with hematological malignancies. Most patient samples displayed catalase activity levels within the normal range with the exception of isolated mononuclear cells from four patients suffering from CML, where the catalase activity was four times increased compared with the normal average. Four other CML samples had activity levels within the normal range, and for two of these, patients were undergoing HU treatment at the time of sampling (Fig. 6 and supplemental Table S1). Considering the dependence of HU toxicity on catalase seen in A. thaliana, the elevated catalase activity in circulating cells from CML patients offers a possible explanation for HU efficacy against this malignancy and calls for further investigation.
FIGURE 6.
Catalase activity in patient blood samples. Catalase activity in blood samples from a control group (Normal), patients diagnosed with chronic myeloid leukemia (CML), acute myeloid leukemia (AML), myelofibrosis (MF), myelomatosis (MM), unspecified myeloproliferative disease (MP), polycytaemia vera (PV), thrombocytosis (TC), chronic lymphatic leukemia (CLL), or lymphoma (L). Values were normalized to the average activity of the Normal group. Closed triangles indicate that patients were undergoing HU treatment at the time of sampling. Standard errors for technical replicates and more detailed diagnoses, where available, are shown in supplemental Table S1.
DISCUSSION
In this study, we exploited the strong genetics and easily scorable stem cell activity of A. thaliana for the unbiased identification of catalase as a major determinant of HU toxicity in vivo and further showed that HU directly targets and inhibits catalase.
We find that in vivo conversion of HU to cytotoxic compounds by catalase is the simplest and most probable model compatible with our data. Still, although we consider it much less likely, it cannot be ruled out that the direct HU-catalase interaction is coincidental and not directly related to the resistance mechanism.
Separation of HU Effects Mediated by RNR and Catalase
Could the HU-catalase interaction be important for the treatment of diseases where HU is known to be effective? With respect to cancer treatment, CML cells with elevated levels of catalase activity could be HU-sensitive due to increased catalase-mediated conversion of HU to cytotoxic compounds. In sickle cell disease, the beneficial effect of HU results from an increased expression of fetal hemoglobin (2). This in turn is thought to depend on the cytotoxic effect of HU on bone marrow cells, which results in modification of hemoglobin gene expression following erythroid regeneration (25). RNR and/or catalase could be necessary for this cytotoxic effect, and catalase could further be involved in NO formation from HU, which may also contribute to induction of fetal hemoglobin expression (24, 26). For treatment of HIV, the potentiation of HU by the dideoxynucleotide didanosine indicates that RNR inhibition could be the main beneficial effect of HU, whereas bone marrow toxicity could be an unwanted side effect (27, 28).
In summary, ex vivo evidence from mammalian cell culture experiments and in vivo evidence from A. thaliana point to two direct HU targets, RNR and catalase. Between them, they have the potential to explain many of the pharmacological effects of HU, and the separation of HU effects mediated by these two targets now becomes an important task. This could be addressed pharmacologically by developing hydroxyurea analogs specifically targeting either RNR or catalase or genetically by using viable catalase null mice and by recruiting acatalasemic patients for clinical trials (29, 30). Using such methods, the hypothesis that catalase properties are clinically relevant for patient HU response becomes testable.
Supplementary Material
This work was supported by The Danish Research Council for Technology and Production (Grant 274-06-0096 to J.-E. J.), the K. E. Jensen Foundation (to H. E. J. and K. D.), and a Proof-of-Concept grant (to T. J., J.-E. J., and S. U. A.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
- HU
- hydroxyurea
- CML
- chronic myeloid leukemia
- RNR
- ribonucleotide reductase
- 3-AT
- 3-amino-1,2,4-triazole
- HIV
- human immunodeficiency virus.
REFERENCES
- 1.Navarra P., Preziosi P. (1999) Crit. Rev. Oncol. Hematol. 29, 249–255 [DOI] [PubMed] [Google Scholar]
- 2.Hankins J., Aygun B. (2009) Br. J. Haematol. 145, 296–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lori F., Foli A., Kelly L. M., Lisziewicz J. (2007) Curr. Med. Chem. 14, 233–241 [DOI] [PubMed] [Google Scholar]
- 4.Dalziel K., Round A., Stein K., Garside R., Price A. (2004) Health Technol. Assess. 8, iii, 1–120 [DOI] [PubMed] [Google Scholar]
- 5.Elford H. L. (1968) Biochem. Biophys. Res. Commun. 33, 129–135 [DOI] [PubMed] [Google Scholar]
- 6.Choy B. K., McClarty G. A., Chan A. K., Thelander L., Wright J. A. (1988) Cancer Res. 48, 2029–2035 [PubMed] [Google Scholar]
- 7.Hurta R. A., Wright J. A. (1990) Biochim. Biophys. Acta 1087, 165–172 [DOI] [PubMed] [Google Scholar]
- 8.McClarty G. A., Chan A. K., Wright J. A. (1986) Somat. Cell Mol. Genet. 12, 121–131 [DOI] [PubMed] [Google Scholar]
- 9.Tagger A. Y., Wright J. A. (1988) Int. J. Cancer 42, 760–766 [DOI] [PubMed] [Google Scholar]
- 10.Tonin P. N., Stallings R. L., Carman M. D., Bertino J. R., Wright J. A., Srinivasan P. R., Lewis W. H. (1987) Cytogenet. Cell Genet. 45, 102–108 [DOI] [PubMed] [Google Scholar]
- 11.Jong A. Y., Yu K., Zhou B., Frgala T., Reynolds C. P., Yen Y. (1998) J. Biomed. Sci. 5, 62–68 [DOI] [PubMed] [Google Scholar]
- 12.Bachir D., Hulin A., Huet E., Habibi A., Nzouakou R., El Mahrab M., Astier A., Galacteros F. (2007) Hemoglobin 31, 417–425 [DOI] [PubMed] [Google Scholar]
- 13.Gigon A., Matos A. R., Laffray D., Zuily-Fodil Y., Pham-Thi A. T. (2004) Ann Bot 94, 345–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beers R. F., Jr., Sizer I. W. (1952) J. Biol. Chem. 195, 133–140 [PubMed] [Google Scholar]
- 15.Elleingand E., Gerez C., Un S., Knüpling M., Lu G., Salem J., Rubin H., Sauge-Merle S., Laulhère J. P., Fontecave M. (1998) Eur. J. Biochem. 258, 485–490 [DOI] [PubMed] [Google Scholar]
- 16.Rodriguez G. I., Kuhn J. G., Weiss G. R., Hilsenbeck S. G., Eckardt J. R., Thurman A., Rinaldi D. A., Hodges S., Von Hoff D. D., Rowinsky E. K. (1998) Blood 91, 1533–1541 [PubMed] [Google Scholar]
- 17.Wright J. A., Cory J. G. (1983) Biosci. Rep. 3, 741–748 [DOI] [PubMed] [Google Scholar]
- 18.Lewis W. H., Srinivasan P. R. (1983) Mol. Cell. Biol. 3, 1053–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akerblom L., Ehrenberg A., Gräslund A., Lankinen H., Reichard P., Thelander L. (1981) Proc. Natl. Acad. Sci. U.S.A. 78, 2159–2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Queval G., Issakidis-Bourguet E., Hoeberichts F. A., Vandorpe M., Gakière B., Vanacker H., Miginiac-Maslow M., Van Breusegem F., Noctor G. (2007) Plant J. 52, 640–657 [DOI] [PubMed] [Google Scholar]
- 21.Bueso E., Alejandro S., Carbonell P., Perez-Amador M. A., Fayos J., Bellés J. M., Rodriguez P. L., Serrano R. (2007) Plant J. 52, 1052–1065 [DOI] [PubMed] [Google Scholar]
- 22.Cheng Y., Prusoff W. H. (1973) Biochem. Pharmacol. 22, 3099–3108 [DOI] [PubMed] [Google Scholar]
- 23.Lewis W. H., Wright J. A. (1978) J. Cell. Physiol. 97, 87–97 [DOI] [PubMed] [Google Scholar]
- 24.Huang J., Kim-Shapiro D. B., King S. B. (2004) J. Med. Chem. 47, 3495–3501 [DOI] [PubMed] [Google Scholar]
- 25.Stamatoyannopoulos G. (2005) Exp. Hematol. 33, 259–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cokic V. P., Smith R. D., Beleslin-Cokic B. B., Njoroge J. M., Miller J. L., Gladwin M. T., Schechter A. N. (2003) J. Clin. Invest. 111, 231–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paton N. I., Aboulhab J., Karim F. (2002) Lancet 359, 1667–1668 [DOI] [PubMed] [Google Scholar]
- 28.Gao W. Y., Johns D. G., Mitsuya H. (1994) Mol. Pharmacol. 46, 767–772 [PubMed] [Google Scholar]
- 29.Takahara S. (1952) Lancet 2, 1101–1104 [DOI] [PubMed] [Google Scholar]
- 30.Ho Y. S., Xiong Y., Ma W., Spector A., Ho D. S. (2004) J. Biol. Chem. 279, 32804–32812 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.