Abstract
Altered neurotrophic support as a result of reduced brain-derived neurotrophic factor (BDNF) expression and trafficking has been revealed as a key factor in Huntington disease (HD) pathology. BDNF binds to and activates the tyrosine kinase receptor TrkB, leading to activation of intracellular signaling pathways to promote differentiation and cell survival. In order to design new neuroprotective therapies based on BDNF delivery, it is important to define whether BDNF-mediated TrkB signaling is affected in HD. Here, we demonstrate reduced TrkB-mediated Ras/MAPK/ERK1/2 signaling but unchanged phosphatidylinositol 3-kinase/Akt and phospholipase Cγ activation in knock-in HD striatal cells. Altered BDNF-mediated ERK1/2 activation in mutant huntingtin cells is associated with reduced expression of p52/p46 Shc docking proteins. Notably, reduced BDNF-induced ERK1/2 activation increases the sensitivity of mutant huntingtin striatal cells to oxidative damage. Accordingly, pharmacological activation of the MAPK pathway with PMA prevents cell death induced by oxidative stress. Taken together, our results suggest that in addition to reduced BDNF, diminished Ras/MAPK/ERK1/2 activation is involved in neurotrophic deficits associated with HD pathology. Therefore, pharmacological approaches aimed to directly modulate the MAPK/ERK1/2 pathway may represent a valuable therapeutic strategy in HD.
Keywords: Huntington Disease, MAP Kinases (MAPKs), Neurodegeneration, Neurotrophic Factor, Oxidative Stress, Signal Transduction, TrkB Receptors
Introduction
Trophic factors have been suggested as therapeutic candidates for the treatment of several neurodegenerative diseases, given that they are essential contributors for maintenance of neuronal survival and differentiation. A number of studies have focused on the study of BDNF3 and its role in HD because striatal neurons, the most prominently neurons affected in HD, require BDNF for their normal function and survival (1–4). Thus, recent evidence demonstrates that wild type huntingtin facilitates BDNF transcription, whereas mutant huntingtin impairs it (5, 6). Accordingly, BDNF mRNA expression and protein levels are reduced in cortex and striatum of HD patients and in mouse and cellular models of HD (7–10). In order to respond to BDNF, striatal neurons must exhibit an appropriate expression and function of its high affinity receptor TrkB. Notably, we have previously described a significant reduction of total TrkB receptors in several HD mouse models, HD knock-in cells, and HD human brain (11). Moreover, reduced TrkB mRNA levels were also found in caudate but not in the cortex of HD patients (10). These results suggest that altered neurotrophic support as a result of BDNF deficit may contribute to the selective degeneration of striatal neurons in HD. However, no study has been reported on the functional consequences of TrkB down-regulation in HD pathology. The neuroprotective action of BDNF through TrkB receptors is associated with activation of MAPKs, the phosphatidylinositol 3-kinase (PI3K)/Akt, and the PLC-γ pathways (12–14). We therefore examined in a genetically precise HD cell model (immortalized STHdhQ111 striatal neuronal cells derived from HdhQ111 embryos (9, 15)) whether altered TrkB levels associated with mutant huntingtin expression involve altered BDNF signaling. We found in mutant huntingtin striatal cells a defective BDNF-mediated Ras-MAPK-ERK1/2 activation but unaffected BDNF-induced PI3K-Akt and PLC-γ signaling. We provide evidence that diminished levels of p52/p46 adapter proteins rather than decreased cell surface TrkB receptor per se may account for the reduced BDNF-mediated ERK1/2 activation. Finally, the functional relevance of altered BDNF-induced ERK1/2 activation in mutant cells was demonstrated by analysis of cell death and dysfunction induced by oxidative stress. Based on these data, we propose that pharmacological modulation of the ERK1/2 pathway may provide a new therapeutic target in HD.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
BDNF (Peprotech EC Ltd., London UK), EGF (R&D Systems), and phorbol 12-myristate 13-acetate (PMA) were obtained from Sigma. PD098059 was purchased from Calbiochem. Phospho-p44/42 ERK1/2 (Thr202/Tyr204), phospho-MEK1/2 (Ser217/221), phospho-c-Raf (Ser338), phospho-Akt (Ser473), phospho-PLC-γ (Tyr783), total ERK1/2, total MEK1/2, total c-Raf, total Akt, and total PLC-γ were obtained from Cell Signaling Technology (Beverly, MA). TrkB polyclonal antibody that only recognizes the full-length TrkB isoform (sc-7268; C-tal epitope) and phospho-Trk (which detects Trk phosphorylated at Tyr496/490) (sc-8058) were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). TrkB polyclonal antibody that recognizes the extracellular domain of TrkB, IRS-1 polyclonal antibody, and p52/p46Shc and p66Shc polyclonal antibodies were obtained from Millipore (Billerica, MA). Ras monoclonal antibody was purchased from BD Biosciences. Anti-HA (Sigma), TRITC-phalloidin, and α-tubulin were purchased from Sigma.
Cell Cultures
Conditionally immortalized wild type STHdhQ7 and mutant STHdhQ111 striatal neuronal progenitor cell lines expressing endogenous levels of normal and mutant full-length huntingtin with 7 and 111 glutamines, respectively, were generated from wild type HdhQ7 and homozygous HdhQ111 littermate embryos (9, 15). The knock-in striatal models faithfully represent the HD mutation carried by patients because elongated polyglutamine tracts are placed within the correct context of the murine Hdh gene. Thus, immortalized striatal cells accurately express normal and mutant huntingtin and do not exhibit amino-terminal inclusions, which allow us to study changes involved in early HD pathogenesis (15). Striatal cells were grown at 33 °C in Dulbecco's modified Eagle's medium (Sigma) supplemented with 10% fetal bovine serum, 1% streptomycin/penicillin, 2 mm l-glutamine, 1 mm sodium pyruvate, and 400 μg/ml G418 (Geneticin, Invitrogen).
Western Blotting Analysis
To analyze TrkB signaling, wild type and mutant huntingtin striatal cells were placed in serum-free Dulbecco's modified Eagle's medium for 3 h and then exposed to BDNF (100 ng/ml), EGF (25 ng/ml), or PMA (1 μm) for different time periods (0, 5, 15, and 30 min). To analyze the neuroprotective role of ERK1/2 activation against oxidative stress, wild type and mutant huntingtin striatal cells were treated with 200 μm H2O2 or pretreated with BDNF (100 ng/ml for 15 min) or BDNF in the presence of the MEK inhibitor PD98059 (10 μm for 15 min) before H2O2 treatment. Total cellular extracts were collected in lysis buffer containing 50 mm Tris base (pH 7.4), 150 mm NaCl, 2 mm EDTA, 0.1 mm phenylmethylsulfonyl fluoride, 1% Nonidet P-40, and supplemented with 1 mm sodium orthovanadate and protease inhibitor mixture (Sigma). Samples were centrifuged at 10,000 × g for 10 min, and the protein contents were determined by a (detergent-compatible) protein assay (Bio-Rad). Protein extracts (30 μg) were mixed with 5× SDS sample buffer, boiled for 5 min, resolved by 6–10% SDS-PAGE, and transferred to nitrocellulose membranes (Schleicher & Schuell). Blots were blocked in 10% nonfat powdered milk in TBS-T (50 mm Tris-HCl, 150 mm NaCl, pH 7.4, 0.05% Tween 20) for 30 min at room temperature. The membranes were then incubated overnight at 4 °C with primary antibodies (phospho-p44/42 ERK1/2 (1:1000), total ERK1/2 (1:2500), phospho-MEK1/2 (Ser217/221) (1:1000), total MEK1/2 (1:1000), phospho-c-Raf (Ser338) (1:1000), total c-Raf (1:1000), phospho-Akt (Ser473) (1:1000), total Akt (1:1000), phospho-PLC-γ (Tyr783) (1:1000), PLC-γ (1:1000), phospho-Trk (Tyr496/490) (1:1000), total TrkB (1:500), total Shc (1:1000), total IRS-1 (1:1000), total Ras (1:2500), anti-HA (1:1000), or α-tubulin (1:50,000)). The membranes were then rinsed three times with TBS-T and incubated with horseradish peroxidase-conjugated secondary antibody for 1 h at room temperature. After washing for 30 min with TBS-T, the membranes were developed using the enhanced chemiluminescence substrate kit (Santa Cruz Biotechnology). The Gel-Pro densitometry program (Gel-Pro Analyzer for Windows version 4.0.00.001) was used to quantify the different immunoreactive bands relative to the intensity of the α-tubulin band in the same membranes. Data are expressed as the mean ± S.D. of band density obtained in three independent experiments.
Surface Biotinylation and Western Blot Analysis of TrkB
Surface TrkB receptors were measured by biotinylation followed by Western blot using a specific antibody that only recognizes full-length TrkB isoforms (Santa Cruz Biotechnology, Inc.), as described elsewhere (16). Wild type and mutant huntingtin striatal cells were incubated in Sulfo-NHS-LC-Biotin (0.5 mg/ml; Pierce) in cold PBS with Ca2+ and Mg2+ for 30 min. The surface biotinylation was stopped by removing the above solution and incubating the cells in 10 mm ice-cold glycine in PBS for 20 min. Cells were washed three times with cold PBS with Ca2+ and Mg2+ and an additional once with cold PBS and then harvested and lysed in lysis buffer containing 50 mm Tris base (pH 7.4), 150 mm NaCl, 2 mm EDTA, 0.1 mm phenylmethylsulfonyl fluoride, 1% Nonidet P-40, supplemented with 1 mm sodium orthovanadate and protease inhibitor mixture (Sigma). Samples were centrifuged at 10,000 × g for 10 min, and the protein contents were determined by (detergent-compatible) protein assay (Bio-Rad). Surface-biotinylated TrkB receptors were immunoprecipitated from total extracts (500 μg) by Immunopure immobilized streptavidin (Pierce), separated by 6% SDS-PAGE, and subjected to Western blot using a polyclonal anti-TrkB antibody (Santa Cruz Biotechnology, Inc.).
Immunocytochemistry and Fluorescence Microscopy Analysis
Wild type and mutant huntingtin striatal cells were plated at a density of 6 × 105/well (triplicate wells), fixed in 4% (w/w) paraformaldehyde for 15 min to detect surface TrkB immunostaining, or fixed and then permeabilized in PBS containing 0.1% saponin (5 min) to visualize total TrkB and Ras protein. Blocking was done in 1% bovine serum albumin in PBS for 1 h, and cells were then incubated for 2 h in blocking solution containing anti-TrkB antibody (extracellular domain, 1:100; Millipore), anti-TrkB (total TrkB, 1:500; Santa Cruz) or anti-Ras antibody (1:200; BD Bioscience). After several washes in PBS (three times for 5 min each), cells were then incubated for 1 h in blocking solution containing Cy2-conjugated anti-mouse (1:200) or Cy3-conjugated anti-rabbit (1:300) (Invitrogen). For TrkB staining, cells were examined by confocal microscopy using a TCS SL laser-scanning confocal spectral microscope (Leica Microsystems Heidelberg, Mannheim, Germany) with argon and helium-neon lasers attached to a DMIRE2 inverted microscope (Leica Microsystems, Heidelberg). Images were taken using a ×63 numerical aperture objective with a ×4 digital zoom and standard (1 Airy disk) pinhole. For each cell, the entire three-dimensional stack of images from the ventral surface to the top of the cell was obtained by using the Z drive in the TCS SL microscope. For Ras staining, cells were observed with a BX60 epifluorescence microscope (Olympus, Tokyo, Japan) with an Orca-ER cooled CCD camera (Hamamatsu Photonics, Japan). TrkB and Ras immunofluorescence was quantified using the ImageJ version 1.33 by Wayne Rasband (National Institutes of Health). In all cases, 20–25 cells were randomly selected from at least three independent cultures.
To analyze the sensitivity of wild type and mutant huntingtin striatal cells to oxidative stress, striatal cells were treated for 30 min in Dulbecco's modified Eagle's medium containing 200 μm H2O2. Cells were then fixed in 4% formaldehyde in PBS for 15 min, washed in PBS (three times for 5 min each), and permeabilized for 10 min with PBS containing 0.1% saponin and 1% bovine serum albumin. Actin cytoskeleton was visualized by incubating cells for 30 min with 0.1 mg/ml of TRITC-labeled phalloidin. Stained cells were observed with a BX60 epifluorescence microscope (Olympus, Tokyo, Japan) with an Orca-ER cooled CCD camera (Hamamatsu Photonics).
To analyze whether PMA-mediated neuroprotection from H2O2 involved protein kinase C activation, MAPK signaling was inhibited in wild type and mutant huntingtin striatal cells by incubation with the MEK inhibitor PD098059 (10 μm for 15 min) prior to PMA treatment (1 μm for 15 min), and H2O2-induced cell death was determined by analysis of actin cytoskeleton.
Cell Survival
Cell survival was quantified by counting the total number of cells in treated conditions versus vehicle-treated conditions (100%). Forty fields were counted per condition and experiment, comprising at least 20–25 cells. Data are given as mean ± S.D. of values obtained in three independent experiments performed in triplicate.
Ras Activation Assay
Wild type and mutant huntingtin striatal cells were stimulated with BDNF (100 ng/ml) or EGF (25 ng/ml) for 15 min, harvested with the lysis buffer (the same as that for Western blot), and cleared by centrifugation at 10,000 × g for 10 min at 4 °C. Aliquots of lysates were set aside to allow quantification of total Ras by immunoblotting. Equal amounts of the remainder lysates were incubated for 90 min at 4 °C with beads coated with fusion protein (GST-Raf1-RBD) consisting of GST fused to the Ras binding domain of Raf-1 (Millipore). Beads were washed three times with cold PBS containing 0.1% Nonidet P-40 and an additional once with cold PBS. Bound protein was eluted for 5 min with 5× Laemmli sample buffer at 95 °C and analyzed by immunoblotting for Ras.
Cell Transfection
For the study of Ras-dependent induction of the ERK1/2 pathway, wild type and mutant huntingtin striatal cells were transfected using Lipofectamine 2000 as described by the manufacturer (Invitrogen). Wild type and mutant huntingtin cells at 70% confluence were transfected with HA-RasV12, a constitutively active form of Ras (generously provided by Dr. N. Agell, University of Barcelona), and the protein extracts were prepared 24 h post-transfection.
Luciferase Assay
The luciferase vector pGL3 Basic vector containing the promoter 2 (P2) of the trkB gene (1600trkBP2-Luc) (17) was kindly provided by Dr. Rodriguez-Peña (Consejo Superior de Investigaciones Científicas-Universidad Autonoma de Madrid, Madrid, Spain). Briefly, wild type and mutant huntingtin striatal cells were seeded into 24-well plates and co-transfected (Lipofectamine 2000, Invitrogen) with P2-CRE-TrkB-Luc (0.3 μg) and the pRL-TK construct (0.25 μg) (Promega, Madison, WI), which produces Renilla luciferase as a control for transfection efficiency. At 24 h after transfection, cells were lysed with Glo Lysis buffer, and firefly and Renilla luciferase activities in cell lysates were sequentially measured using the dual-luciferase reporter assay system (Promega) and luminometer (BIO-TEK Clarity). Firefly luciferase luminescence was normalized by Renilla luciferase luminescence. The mean values for each construct were obtained from three independent experiments performed in triplicate.
Quantitative PCR (Q-PCR) Assays
Total RNA from wild type and mutant huntingtin striatal cells was extracted using the Total RNA Isolation Nucleospin® RNA II Kit (Macherey-Nagel, Düren, Germany). Total RNA (500 ng) was used to synthesize cDNA using random primers with the StrataScript® first strand cDNA synthesis system (Stratagene, La Jolla, CA). The cDNA synthesis was performed at 42 °C for 60 min in a final volume of 20 μl according to manufacturer's instructions. The cDNA was then analyzed by Q-PCR using the following TaqMan® gene expression assays (Applied Biosystems, Foster City, CA): 18 S (Hs99999901_s1) and p52/p46 Shc (Mm00468940_m1). Reverse transcription-PCR was performed in 25-μl volumes on 96-well plates, in a reaction buffer containing 12.5 μl of Brilliant® Q-PCR Master Mix (Stratagene), 1.25 μl of TaqMan® gene expression assays, and 1 ng of cDNA. Reactions included 40 cycles of a two-step PCR, 95 °C for 30 s and 60 °C for 1 min, after initial denaturation at 95 °C for 10 min. All Q-PCR assays were performed in duplicate and repeated for at least three independent experiments. To provide negative controls and exclude contamination by genomic DNA, the reverse transcription was omitted in the cDNA synthesis step, and the samples were subjected to the PCR in the same manner with each TaqMan® gene expression assay. The Q-PCR data were analyzed using the MxProTM Q-PCR analysis software version 3.0 (Stratagene). Quantification was performed with the Comparative Quantitation Analysis program of the mentioned software and using the 18 S gene expression as internal loading control.
Statistical Analysis
All of the data were analyzed with the program GraphPad Prism version 4.0 (GraphPad Software, San Diego, CA). Results are expressed as mean ± S.D. Experimental data were analyzed by Student's t test. A value of p ≤ 0.05 was accepted as denoting statistical significance.
RESULTS
Mutant STHdhQ111 Striatal Cells Express Reduced Levels of Cell Surface TrkB Receptors
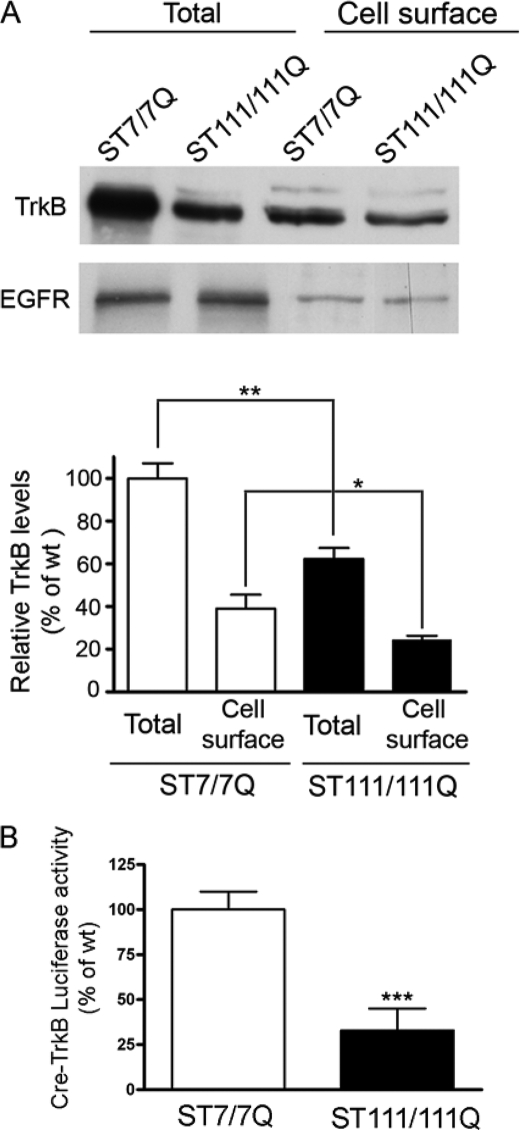
Decreased mRNA and protein levels of TrkB receptors have previously been described by our group in several HD models (11). Given the importance of cell surface TrkB receptors for activation of BDNF signaling, we first investigated by biotinylation and Western blotting the expression of cell surface TrkB in immortalized wild type STHdhQ7 and mutant STHdhQ111 striatal cells. These cells express endogenous levels of normal and mutant huntingtin with 7 and 111 glutamines, respectively, and provide an accurate genetic HD cell model (15). In agreement with our previous results (11), we found a significant reduction (48% ± 15%, p ≤ 0.01) of total TrkB levels in mutant huntingtin striatal cells compared with wild type cells (Fig. 1A). Notably, surface TrkB levels were reduced by 35 ± 10% (p ≤ 0.05) in the biotinylated fraction of mutant huntingtin cells compared with wild type cells, whereas no differences were found on total and cell membrane EGFR levels.
FIGURE 1.
Reduced levels of cell surface TrkB receptors in STHdhQ111 striatal cells. A, representative Western blot showing the levels of total and cell surface TrkB and EGFR receptors from wild type (ST7/7Q) and mutant (ST111/111Q) huntingtin striatal cells. Surface TrkB receptors and EGFR were detected by Western blotting after surface protein biotinylation and immunoprecipitation of biotinylated proteins by streptavidin-agarose beads. Total TrkB and EGFR were determined by Western blot of whole cell lysates. The histogram represents the percentage of TrkB expression relative to wild type values. Values are given as mean ± S.D. (error bars) of three independent experiments. **, p ≤ 0.01; *, p ≤ 0.05 as determined by Student's t test. B, histogram showing the transcription activity of the P2-CRE-TrkB promoter in transiently transfected wild type and mutant huntingtin striatal cells. The histogram represents the percentage of luciferase activity relative to wild type values. Values are given as mean ± S.D. of three independent experiments. ***, p ≤ 0.001 as determined by Student's t test.
We next examined whether reduced TrkB expression was associated with transcriptional deficits. Given that the trkB gene is a target of CREB transcription (18) and an altered CREB/CREB-binding protein pathway has previously been shown in mutant STHdhQ111 striatal cells (9), we have analyzed CRE-TrkB transcriptional activity in wild type and mutant huntingtin striatal cells by a luciferase reporter gene assay. As shown in Fig. 1B, a significant decrease of CRE-TrkB luciferase activity was detected in mutant compared with wild type striatal cells, suggesting that TrkB transcriptional deregulation may be involved in reduced TrkB expression in mutant huntingtin striatal cells.
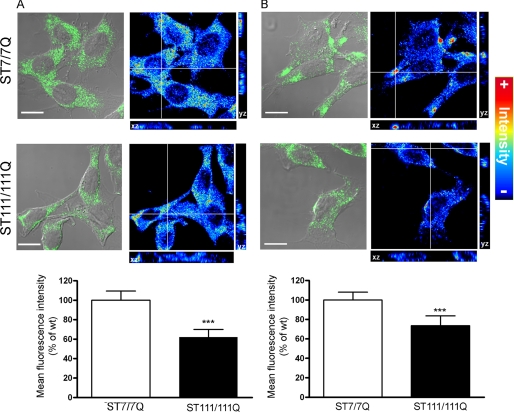
To further analyze the expression and distribution of cell surface TrkB receptors, immunocytochemistry analysis was performed in permeable (Fig. 2A) and non-permeable (Fig. 2B) wild type and mutant huntingtin striatal cells. According to the biochemical data, a significant reduction of total TrkB staining was revealed in mutant compared with wild type cells (40% ± 10%; p ≤ 0.001). Notably, when cell surface TrkB expression was examined using an antibody against the extracellular domain of TrkB in non-permeable wild type and mutant huntingtin striatal cells, a punctate TrkB staining that was significantly lower in mutant compare with wild type cells was revealed (29% ± 7%; p ≤ 0.001). Moreover, whereas a homogenous distribution of TrkB was observed in mutant cells, enriched TrkB membrane patches were detected in wild type cells (Fig. 2B). Altogether, these results indicate that mutant huntingtin leads to a specific reduction and altered distribution of cell surface TrkB receptors in striatal cells.
FIGURE 2.
Altered cell surface TrkB distribution in STHdhQ111 striatal cells. Overlay of confocal and corresponding differential interference contrast images of TrkB immunofluorescence in permeabilized (A) and non-permeabilized (B) wild type (ST7/7Q) and mutant (ST111/111Q) huntingtin striatal cells. TrkB distribution is showed in a pseudocolor scale. Warm colors represent maximum intensities, whereas cold colors are representative of low intensities (the intensity of each pixel was set to 255 levels of gray). Images represent the projection of the two slices containing the maximal cross-section of the cell nucleus. Confocal images show reduced total (A) and cell surface (B) TrkB immunoreactivity in mutant huntingtin striatal cells. Histograms represent the mean of the TrkB fluorescence intensity ± S.D. (error bars) of three independent experiments. ***, p ≤ 0.001. Bar scale, 10 μm.
Specific Reduction of TrkB-mediated ERK1/2 Activation in Mutant STHdhQ111 Striatal Cells
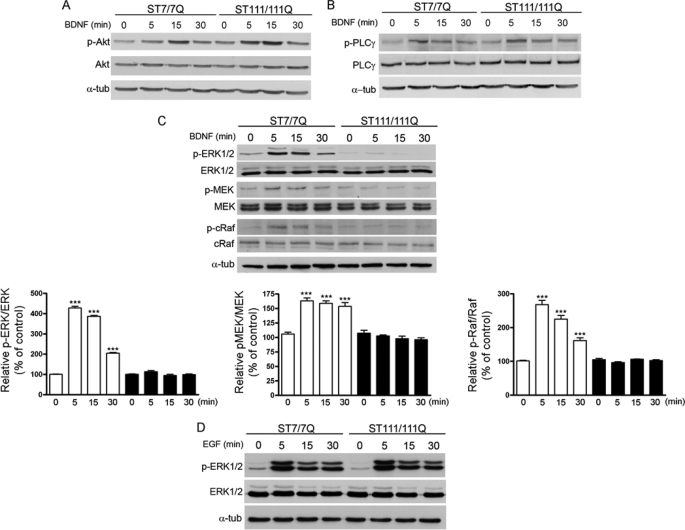
TrkB-mediated signaling involves the activation of the Ras/MAPK/ERK1/2, PI3K/Akt, and PLC-γ pathways (12–14, 19). To determine whether BDNF-induced TrkB signaling was also affected by mutant huntingtin expression, we evaluated BDNF-mediated ERK1/2, Akt, and PLC-γ activation in wild type STHdhQ7 and mutant STHdhQ111 huntingtin striatal cells. Cells were treated with BDNF for 0–30 min, and total cell extracts were collected and analyzed by Western blot using specific antibodies. BDNF treatment induced a similar increase of Akt (Ser473) and PLC-γ (Tyr783) phosphorylation in both wild type and mutant huntingtin striatal cells (Fig. 3, A and B, respectively), which suggests that both Akt and PLC-γ signaling are properly activated even in the presence of lower TrkB receptors in mutant huntingtin cells. In contrast to Akt and PLC-γ, BDNF-mediated activation of the MAPK/ERK1/2 pathway was significantly reduced in mutant compared with wild type cells. Thus, whereas in wild type cells BDNF treatment caused a robust and time-dependent activation of MAPK signaling detected as increased phosphorylation of ERK1/2, MEK, and c-Raf, in mutant huntingtin striatal cells, BDNF failed to increase phosphorylation of ERK1/2, MEK, and c-Raf (Fig. 3C).
FIGURE 3.
Specific inhibition of BDNF-induced MAPK signaling in STHdhQ111 striatal cells. Representative Western blots showing the time course of BDNF-induced phosphorylation of Akt (A) and PLC-γ (B) in wild type (ST7/7Q) and mutant (ST111/111Q) huntingtin striatal cells. Whole cell lysates were blotted with antibodies against phosphorylated Ser473 Akt (p-Akt), total Akt, phosphorylated Tyr783 PLC-γ (p-PLC-γ), total PLC-γ, and α-tubulin as a loading control, after stimulating with 100 ng/ml BDNF for the indicated time periods. The results are representative of three independent experiments. C, representative Western blot showing the time course of BDNF-induced phosphorylation of MAPK signaling proteins in wild type and mutant huntingtin striatal cells. Whole cell lysates were blotted with antibodies against phosphorylated ERK1/2 (p-ERK1/2), MEK (p-MEK), and cRaf (p-cRaf) as well as total ERK1/2, MEK, and c-Raf and α-tubulin as a loading control, after stimulating with 100 ng/ml BDNF for the indicated time periods. The histograms represent the relative phospho-ERK/ERK, phospho-MEK/MEK, and phospho-Raf/Raf ratios expressed as percentage versus control cells. Values are given as mean ± S.D. (error bars) of three independent experiments. ***, p ≤ 0.001 as determined by Student's t test. D, representative Western blot showing the time course of EGF-induced phosphorylation of ERK1/2 in wild type and mutant huntingtin striatal cells. Whole cell lysates were blotted with antibodies against phosphorylated ERK1/2 (p-ERK1/2) and total ERK1/2 after stimulating with 25 ng/ml EGF for the indicated time periods. The results are representative of three independent experiments.
We next examined whether impaired BDNF-mediated ERK1/2 activation was specific for TrkB or a general mechanism associated with activation of growth factor receptors. Wild type and mutant huntingtin striatal cells were treated with EGF, and the levels of p-ERK1/2 were evaluated by Western blot analysis (Fig. 3D). Importantly, mutant huntingtin striatal cells exhibited similar time-dependent EGF-induced ERK1/2 phosphorylation as wild type striatal cells. These findings demonstrate that mutant huntingtin interferes specifically with BDNF TrkB-mediated ERK1/2 signaling in striatal cells.
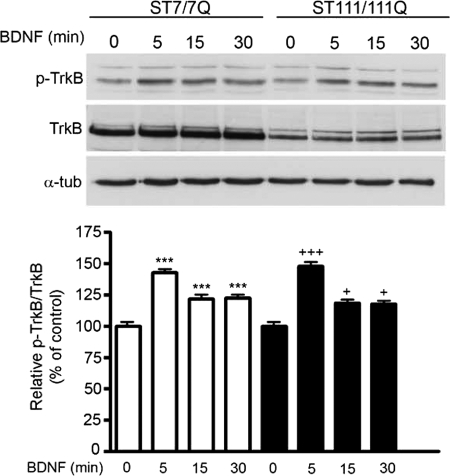
TrkB Phosphorylation Is Not Altered in Mutant STHdhQ111 Striatal Cells
The activation of BDNF-mediated TrkB signaling depends on TrkB phosphorylation (12–14, 19). To test whether reduced TrkB phosphorylation might explain the lack of BDNF-mediated ERK1/2 activation observed in mutant huntingtin striatal cells, we next analyzed the levels of phospho-Tyr490 TrkB, the Tyr residue critical for activation of Ras/MAPK/ERK1/2 and PI3K/Akt pathways. Western blot and quantitative analysis demonstrated similar time-dependent TrkB phosphorylation induced by BDNF in wild type and mutant huntingtin striatal cells (Fig. 4). These findings suggest that mutant huntingtin does not impair BDNF-induced TrkB signaling by altering TrkB phosphorylation and activation.
FIGURE 4.
Mutant huntingtin does not interfere with BDNF-induced TrkB phosphorylation in STHdhQ111 striatal cells. Representative Western blot showing levels of Tyr490-phosphorylated TrkB (p-TrkB), total TrkB (TrkB), and α-tubulin as a loading control from wild type (ST7/7Q) and mutant (ST111/111Q) cell extracts obtained after treatment with 100 ng/ml BDNF for the indicated time periods. The histogram represents the percentage of TrkB induction (p-TrkB/TrkB) relative to control wild type or mutant striatal cells. Values are given as mean ± S.D. (error bars) of three independent experiments. ***, p ≤ 0.001; +, p ≤ 0.05; +++, p ≤ 0.001 as determined by Student's t test.
Mutant Huntingtin Interferes with BDNF Signaling at the Level of Docking Proteins
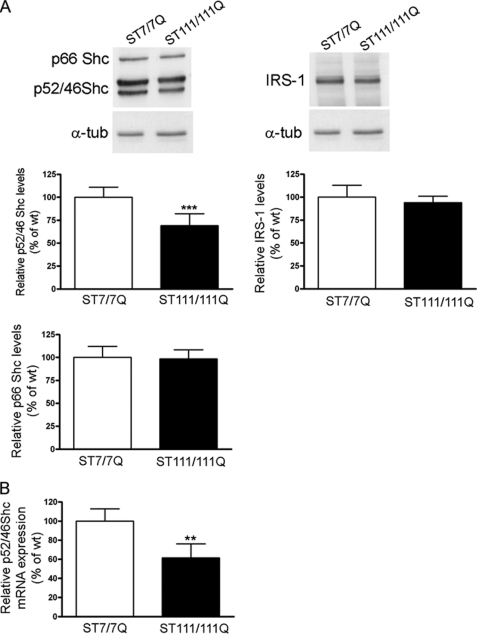
Phosphorylation of TrkB at Tyr490 provides critical binding sites to adapter proteins, such as Shc and IRS, involved on TrkB activation of Ras/MAPKs and PI3K/Akt, respectively (12, 19, 20). We then tested the hypothesis that mutant huntingtin could interfere with BDNF-ERK1/2-mediated activation by altering the levels of particular TrkB adapter proteins. Western blot analysis revealed that levels of the p52/p46 Shc isoforms were significantly reduced in mutant cells compared with those of wild type cells (30% ± 13%, p ≤ 0.001). By contrast, p66 Shc levels, which are not involved in Ras activation (21), and IRS-1, which participates in PI3K/Akt activation (12, 22), were similar in both genotypes (Fig. 5A).
FIGURE 5.
Specific reduction of protein adaptors p52/p46 Shc in STHdhQ111 striatal cells. A, representative Western blot showing levels of p52/p46 Shc, p66 Shc, and IRS-1 and α-tubulin as a loading control in total cell extracts obtained from wild type (ST7/7Q) and mutant (ST111/111Q) huntingtin striatal cells. The histograms represent the percentage of p52/p46 Shc, p66 Shc, and IRS-1 expression relative to wild type values. Values are given as mean ± S.D. of three independent experiments. ***, p ≤ 0.001 as determined by Student's t test. B, quantitative PCR analysis of p52/p46 Shc mRNA levels. The histogram represents the relative p52/p46 Shc mRNA levels expressed as a percentage versus wild type cells. Values are given as mean ± S.D. (error bars) of three independent experiments performed in duplicate. **, p ≤ 0.01 as determined by Student's t test.
To determine whether the reduction in p52/p46 Shc protein levels resulted from reduced mRNA expression, Q-PCR analysis of total mRNA derived from wild type and mutant huntingtin striatal cells was performed (Fig. 5B). Q-PCR analysis revealed a significant decrease (40 ± 6%, p ≤ 0.01) in p52/p46 Shc mRNA in mutant huntingtin compared with wild type striatal cells, which suggest that altered p52/p46 Shc transcription is involved in diminished p52/p46 Shc protein expression. All together, these results support the idea that impaired BDNF-mediated ERK1/2 activation caused by mutant huntingtin may involve the specific reduction of the adapter proteins p52/p46 Shc.
BDNF-mediated Ras Activation Is Impaired in Mutant STHdhQ111 Striatal Cells
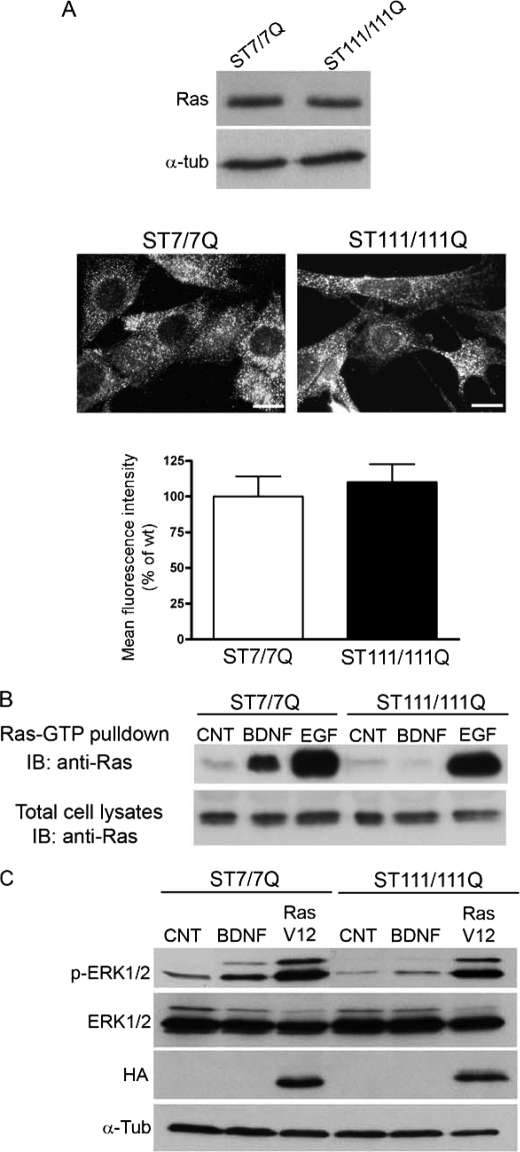
Activation of Ras by neurotrophic factors promotes neuronal cell survival and differentiation (12–14, 20). Following BDNF stimulation, the adaptor proteins p52/p46 Shc are recruited to phosphorylated Tyr490 on TrkB receptors, ensuring transient activation of Ras protein (14, 20). Therefore, we examined whether decreased expression of Shc proteins in mutant huntingtin striatal cells attenuates ERK1/2 signaling by altering Ras activation. We first determined total Ras expression in cell extracts obtained from wild type and mutant huntingtin striatal cells. As shown in Fig. 6A, the levels of Ras in wild type cells were similar to those in mutant cells. According to these biochemical data, immunocytochemistry analysis demonstrated similar levels and distribution of Ras in wild type and mutant huntingtin striatal cells (Fig. 6A).
FIGURE 6.
BDNF-induced Ras activation is impaired in STHdhQ111 striatal cells. A (top), representative Western blot showing levels of Ras and α-tubulin as a loading control in total cell extracts obtained from wild type (ST7/7Q) and mutant (ST111/111Q) huntingtin striatal cells. The results are representative of three independent experiments. Bottom, representative immunofluorescence images showing similar expression and distribution of Ras protein in wild type and mutant huntingtin striatal cells. Scale bar, 10 μm. The histogram represents the mean of the Ras fluorescence intensity expressed as a percentage versus wild type cells. Values are given as mean ± S.D. (error bars) of three independent experiments. B, representative Western blot showing levels of GTP-loaded Ras. Activated (GTP-bound) Ras pool was detected by immunoblotting (IB) of pull-down experiments from total cell lysates of untreated and BDNF- or EGF-treated wild type and mutant huntingtin striatal cells. Aliquots of the total cell extracts were analyzed in parallel to ascertain equal loading. The results are representative of three independent experiments. C, representative Western blot showing levels of phospho-ERK1/2, ERK1/2, and α-tubulin as a loading control in total cell extracts from wild type and mutant huntingtin striatal cells. Whole cell extracts were obtained from wild type and mutant huntingtin cells treated with BDNF (100 ng/ml 15 min) or transfected with a constitutively active form of Ras (HA-RasV12) and blotted with antibodies against phosphorylated ERK1/2 (p-ERK1/2), total ERK1/2 (ERK1/2), HA, and α-tubulin as a loading control. The results are representative of four independent experiments.
We then determined Ras activation induced by BDNF stimulation by measuring the levels of Ras-GTP (23). Active Ras-GTP was pulled down with a GST fusion protein containing the Ras binding domain of Raf-1 (GST-Raf1-RBS) and blotted with a Ras antibody. Ras activity measured in BDNF-treated wild type cells was increased by 7-fold compared with untreated wild type cells, whereas BDNF-induced Ras activation was undetectable in mutant huntingtin cells (Fig. 6B). By contrast, EGF-induced Ras activation was comparable between wild type and mutant huntingtin striatal cells (17- and 14-fold increase, respectively; Fig. 6B). Taken together, these results demonstrate that defective Ras activation is involved on altered BDNF-induced ERK1/2 activation in mutant huntingtin striatal cells.
Finally, to further evaluate the contribution of altered Ras activation to impaired BDNF-mediated ERK1/2 activation, wild type and mutant huntingtin striatal cells were transfected with a constitutively active HA-Ras (HA-RasV12), and levels of phosphorylated ERK1/2 were detected by Western blot analysis (Fig. 6C). Expression of Ras-V12 in mutant cells restored ERK1/2 phosphorylation to levels of wild type cells. These results indicate that defective Ras activation rather than downstream components of the TrkB pathway (Raf, MEK, or ERK1/2) underlies altered MAPK activation in mutant huntingtin striatal cells.
ERK1/2 Activation Is Required for Protection of Mutant STHdhQ111 Striatal Cells against Oxidative Stress
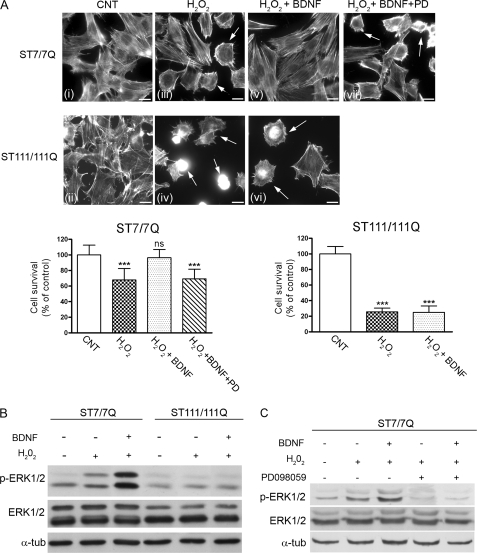
The Ras/ERK1/2 pathway is a critical signaling cascade mediating neuroprotection against oxidative stress-induced cell death (21, 24–28). To investigate the consequences of reduced ERK1/2 activation for the neuroprotective response to oxidative stress, wild type and mutant huntingtin striatal cells were exposed to H2O2 in the presence or absence of BDNF. Oxidative damage was determined by analysis of cell morphology changes on actin cytoskeleton as a measure of cell injury. Untreated wild type cells showed a well organized actin cytoskeleton (Fig. 7A, i), whereas treatment with H2O2 induced changes on the actin cytoskeleton detected as rearranged actin stress fibers and a significant reduction of cell survival (Fig. 7A, iii). Notably, BDNF treatment completely prevented H2O2-induced actin cytoskeleton alterations and cell death (Fig. 7A, v). Consistent with a neuroprotective role of ERK1/2 activation against oxidative damage, a significant increase of ERK1/2 phosphorylation was detected in BDNF-treated wild type cells compared with untreated or H2O2-treated wild type cells (Fig. 7B, 5- and 2-fold respectively, p ≤ 0.05). By contrast, mutant huntingtin striatal cells exhibited greater sensitivity to identical doses of H2O2 than wild type cells, demonstrated by reduced cell survival and a higher proportion of cells with altered actin cytoskeleton (Fig. 7A, ii and iv). Importantly, BDNF treatment did not protect mutant huntingtin cells from oxidative stress induced by H2O2 (Fig. 7A, vi). Accordingly, the phosphorylation of ERK1/2 following BDNF incubation was not significantly different from that detected in basal or H2O2-treated mutant huntingtin cells (Fig. 7B).
FIGURE 7.
ERK1/2 activation is required for BDNF-mediated neuroprotection against oxidative stress in STHdhQ111 striatal cells. A, representative immunofluorescence images (i–vii) showing rodamine-phalloidin staining to visualize the actin cytoskeleton in wild type (ST7/7Q) and mutant (ST111/111Q) huntingtin striatal cells. Wild type and mutant huntingtin striatal cells were exposed to H2O2 (200 μm for 15 min) in the absence (iii and iv) or presence of 100 ng/ml BDNF (v and vi), and cell viability was detected by actin cytoskeleton patterning. The inhibition of the neuroprotective effect of BDNF in the presence of the MEK inhibitor PD098059 is shown in vii. The preincubation period with BDNF provides protection against oxidative injury in wild type but not mutant huntingtin striatal cells. Notice the higher number of H2O2-treated mutant cells with altered cytoskeleton rearrangements compared with wild type cells (iv and vi, arrows). The results are representative of three independent experiments. Scale bar, 10 μm. The histograms represent the percentage of cell survival relative to control cells. ***, p ≤ 0.001 as determined by Student's t test. B, wild type and mutant huntingtin striatal cells were exposed to H2O2 (200 μm for 30 min) in the absence or presence of BDNF (100 ng/ml for 15 min), and total extracts were analyzed by Western blot, using antibodies against phosphorylated ERK1/2 (p-ERK1/2), total ERK1/2 (ERK1/2), or α-tubulin as a loading control. An example of Western blot is presented. The results are representative of three independent experiments. C, wild type (ST7/7Q) striatal cells were treated with BDNF (100 ng/m, 15 min) in the absence or presence of the MEK inhibitor PD098059 (10 μm, 15 min). The cells were then challenged with H2O2 (200 μm, 30 min), and total extracts were analyzed by Western blot, using antibodies against phosphorylated ERK1/2, total ERK1/2, or α- tubulin as a loading control. An example of Western blot is presented. The results are representative of three independent experiments.
To further confirm the possible role of ERK1/2 activation in the neuroprotection by BDNF, wild type striatal cells were preincubated with PD098059 to pharmacologically inhibit MEK, the upstream kinase of ERK1/2. In the presence of the MEK inhibitor, the neuroprotective effect of BDNF against H2O2-evoked cell death was completely abrogated (Fig. 7A, vii). Control experiments revealed that PD098059 completely inhibited BDNF-induced ERK1/2 phosphorylation in wild type cells (Fig. 7C). Altogether, these results suggest that the higher sensitivity of mutant huntingtin striatal cells to oxidative stress may be related to deficient activation of the ERK1/2 signaling pathway.
Phorbol Esters Rescue MAPK Signaling and Protect Mutant STHdhQ111 Striatal Cells from Oxidative Stress
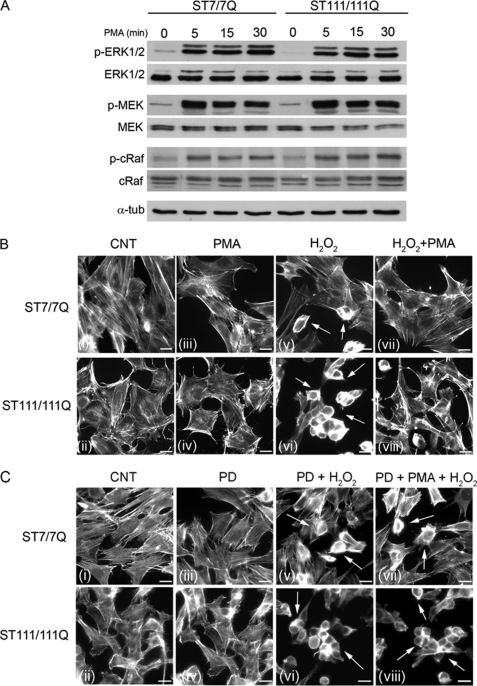
The diacylglycerol analogue PMA may activate Ras independently of the canonical Shc-Grb2-Sos pathway (29). We took advantage of this specific PMA action to investigate whether pharmacological manipulation of the MAPK pathway could rescue mutant huntingtin striatal cells from cell death induced by oxidative stress. As shown in Fig. 8A, PMA treatment induced similar activation of ERK1/2 monitored as levels of ERK1/2 phosphorylation in either wild type or mutant huntingtin striatal cells (Fig. 8A). In addition, PMA increased MEK and Raf phosphorylation with a similar kinetics of activation in wild type and mutant huntingtin striatal cells (Fig. 8A). These results indicate that activation of the MAPK cascade downstream of the Shc-Grb2-Sos pathway is not compromised in mutant huntingtin striatal cells.
FIGURE 8.
PMA treatment restores ERK1/2 activation and prevents cell injury induced by oxidative stress in STHdhQ111 striatal cells. A, representative Western blot showing the time course of PMA-induced phosphorylation of MAPK signaling proteins in wild type (ST7/7Q) and mutant (ST111/111Q) huntingtin striatal cells. Whole cell lysates were blotted with antibodies against phosphorylated ERK1/2 (p-ERK1/2), MEK (p-MEK), and cRaf (p-cRaf) as well as total ERK1/2, MEK, and c-Raf and α-tubulin as a loading control after stimulating with PMA for the indicated time periods. The results are representative of three independent experiments. B, representative immunofluorescence images (i–viii) showing rodamine-phalloidin staining to visualize the actin cytoskeleton in wild type and mutant huntingtin striatal cells. Wild type and mutant huntingtin striatal cells were exposed to H2O2 (200 μm, 30 min) in the absence (v and vi) or presence of PMA (vii and viii), and cell viability was detected by actin cytoskeleton patterning. The preincubation with PMA provides protection against oxidative injury in both wild type and mutant huntingtin striatal cells. C, representative immunofluorescence images (i–viii) showing rodamine-phalloidin staining to visualize the actin cytoskeleton in wild type and mutant huntingtin striatal cells. MAPK signaling was inhibited in wild type and mutant huntingtin striatal cells by incubation with the MEK inhibitor PD098059 prior to PMA treatment, and H2O2-induced cell death was determined by analysis of the actin cytoskeleton. The preincubation with PD98059 prevents PMA-mediated protection against oxidative injury in both wild type and mutant huntingtin striatal cells (vii and viii, arrows). The results are representative of three independent experiments. Scale bar, 10 μm.
We next investigated whether the recovery of MAPK signaling by PMA was able to protect mutant huntingtin striatal cells against oxidative stress induced by H2O2. Wild type and mutant cells were incubated with PMA prior to H2O2 treatment, and cell death was detected by analysis of the actin cytoskeleton pattern. As previously shown (Fig. 7A), mutant huntingtin striatal cells exhibited increased sensitivity to oxidative stress detected by a higher number of cells displaying an abnormal actin cytoskeleton pattern. Notably, PMA treatment prevented H2O2-induced toxicity either in wild type or mutant huntingtin striatal cells (Fig. 8B).
Given that PMA is a well known activator of protein kinase C, we then examined whether PMA-mediated neuroprotection from H2O2 involved protein kinase C activation. Wild type and mutant huntingtin striatal cells were exposed to PD098059 prior to PMA treatment, and H2O2-induced cell death was determined by analysis of actin cytoskeleton. Pretreatment of wild type cells with the MEK inhibitor PD098059 prior to H2O2 exposure induced higher cell death compared with those cells only treated with H2O2 (Fig. 8, B (v) and C (v)), which agrees with our biochemical data showing activation of the ERK1/2 pathway in wild type cells following H2O2 treatment (Fig. 7B). Notably, PMA-mediated neuroprotection from H2O2-induced toxicity was prevented by exposure of wild type and mutant huntingtin striatal cells to PD098059 (Fig. 8C, vii and viii), which is consistent with a specific role of ERK1/2 activation in the PMA neuroprotective effect. Taken together, these findings support the view that deficient BDNF-induced ERK1/2 activation underlies the lack of protection against oxidative damage in mutant huntingtin striatal cells.
DISCUSSION
Accumulating evidence suggests that altered neurotrophic support plays a critical role in the neurodegenerative process associated with HD. Reduced BDNF expression and transport have been proposed as the main mechanisms contributing to neurotrophic deficiency in HD (1, 4, 5, 8–10, 30). In addition, we have previously demonstrated a significant reduction of the specific BDNF receptor TrkB in HD cellular and mouse models and in HD human brain (11). Based on these previous observations, it is important to elucidate whether diminished TrkB expression may involve changes in BDNF-mediated signaling and neuroprotection. In this study, we have shown reduced levels of total and cell surface TrkB receptors as a result of mutant huntingtin expression. Notably, reduced TrkB expression was associated with decreased CRE-TrkB promoter activity, which suggests that diminished CREB-mediated transcription may account for altered TrkB expression in mutant huntingtin striatal cells.
Moreover, our results have shown that mutant huntingtin expression has differential effects on BDNF signaling. Thus, although BDNF-mediated Ras/MAPK/ERK1/2 activation was significantly attenuated in mutant compared with wild type cells, no differences were detected in the PI3K/Akt and PLC-γ pathways. This result, together with the unchanged phosphorylated levels of TrkB in mutant huntingtin striatal cells, suggests that reduced cell surface TrkB receptors in mutant cells cannot entirely account for the lack of ERK1/2 activation induced by BDNF treatment. Consistent with this idea, we found in mutant huntingtin striatal cells a specific reduction at the level of mRNA and protein of p52/p46Shc, the scaffolding proteins that couple the activated TrkB receptors to the Ras/MAPK/ERK1/2 pathway (12–14, 20). Importantly, p66Shc that is not involved in ERK1/2 activation was unaffected (31). Similar to p66Shc expression, the levels of IRS-1, a well known adaptor protein involved in BDNF-mediated PI3K/Akt activation (12, 14, 22), were unchanged. These findings, together with the fact that BDNF may also activate the PI3K/Akt pathway by direct association of PI3K with the phosphorylated form of TrkB receptors and that PLC-γ is also directly activated by binding to phospho-TrkB (12–14), may explain the selective disruption of the Ras/MAPK/ERK1/2 signaling in mutant huntingtin striatal cells. Notably, our studies revealed that in striatal cells, mutant huntingtin impairs TrkB-induced ERK1/2 activation without affecting EGFR-mediated MAPK signaling, although ERK1/2 by EGFR also involves activation of p52/p46 Shc proteins. Recruitment of Shc adapter proteins to tyrosine receptors is a key event to the subsequent interaction with Grb2-Sos complexes and the activation of the Ras-MAPK signaling pathway (20, 32). The specific mechanisms underlying recruitment of Shc proteins to tyrosine receptors remain poorly characterized. It has been suggested that the efficiency of tyrosine kinase receptors to recruit Shc proteins may be dependent on the Shc residues and/or domains phosphorylated by these receptors (20, 33–35). Thus, although insulin receptors can efficiently phosphorylate p52Shc but not p46Shc, EGFR leads to efficient phosphorylation and recruitment of both isoforms (20, 36). In addition, the phosphorylation of Shc at different residues/domains is also critical to recruit Grb2-Sos complexes to the membrane, which facilitates Ras/MAPK activation (20, 32–35, 37). Therefore, a possible scenario will be that in which mutant huntingtin specifically disrupts TrkB-mediated ERK1/2 signaling by 1) reducing levels of Shc adapter proteins and 2) preferentially interfering with the recruitment and phosphorylation of Shc-Grb2-Sos complexes to TrkB receptors. In agreement with this view, it has been reported that association of Grb2 with TrkA receptors following neural growth factor stimulation was disrupted by overexpressing full-length mutant huntingtin (38). Consistent with impaired coupling of TrkB to Shc complexes, we found reduced Ras activation following BDNF treatment in mutant huntingtin striatal cells. Moreover, overexpression of HA-RasV12, a constitutively active form of Ras, or PMA treatment, which directly activates c-Raf, restored MAPK signaling in mutant huntingtin striatal cells. These results support the idea that mutant huntingtin disrupts ERK1/2-mediated TrkB signaling upstream of Ras activation.
Our findings demonstrating diminished BDNF-mediated ERK1/2 activation in mutant huntingtin striatal cells raise the question whether lack of TrkB-dependent ERK1/2 signaling might contribute to HD pathology. Multiple lines of evidence have implicated oxidative stress linked to mitochondrial dysfunction in HD neuropathology (39, 40). Accumulation of oxidative markers ranging from DNA strand breaks to protein nitration or lipid oxidative damage has been demonstrated in several HD mouse models as in HD brain (39–42). Notably, ERK1/2 activation plays an important neuroprotective role mediating cell survival in response to oxidative stress (24–28). Therefore, we hypothesized that reduced BDNF-induced ERK1/2 activation could result in increased susceptibility of mutant huntingtin striatal cells to oxidative injury. We found that cell viability was significantly decreased in mutant huntingtin striatal cells compared with wild type cells when exposed to H2O2. Importantly, pretreatment with BDNF protected wild type but not mutant huntingtin striatal cells against oxidative stress. Indeed, the protective effect of BDNF was blocked by treatment with the MEK1/2 inhibitor PD098059. Together, these results demonstrate that the protective effect of BDNF against oxidative damage in striatal cells is mediated by activation of ERK1/2 and that activation of ERK1/2 in striatal cells is critical to mediate neuroprotection against oxidative stress. Accordingly, pharmacological activation of the MAPK pathway with PMA prevents cell death induced by oxidative stress in mutant huntingtin striatal cells. Consistent with a protective role of ERK1/2 in HD, it has been reported that activation of ERK1/2 protects against mutant huntingtin-associated toxicity in PC12 cells and in immortalized striatal neuronal cells that express exon-1 mutant huntingtin (43, 44). Moreover, orthovanadate treatment, a tyrosine phosphatase inhibitor that reduces the dephosphorylation of ERK1/2, decreases caspase-3 activation and cell death in PC12 cells expressing full-length mutant huntingtin (44).
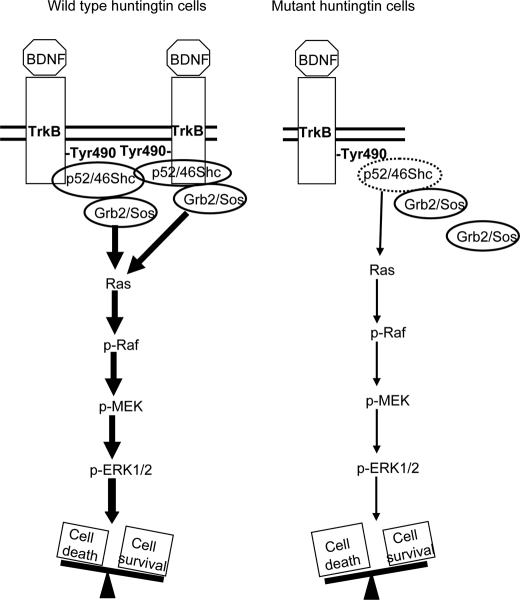
In summary, our studies have identified the reduction in p52/p46Shc adapter protein expression as the principal mechanism by which mutant huntingtin selectively impairs TrkB-mediated ERK1/2 activation. We propose that by reducing the expression of p52/p46Shc adapter proteins, mutant huntingtin might disturb the formation of TrkB-Shc signaling complexes, resulting in decreased Ras-mediated activation and decreased MAPK activation. We found that such inhibition increases mutant huntingtin striatal cell vulnerability to oxidative stress, suggesting that the lack of ERK1/2 activation might contribute to HD neurodegeneration (Fig. 9). Altogether our data strongly support the view that pharmacological strategies targeting BDNF but also targeting selectively the MAPK/ERK1/2 pathway might provide better benefits to HD treatment than those only aimed at raising BDNF levels.
FIGURE 9.
Proposed role of mutant huntingtin in reducing TrkB-mediated ERK1/2 activation in striatal cells. BDNF-TrkB activation involves ERK1/2 activation by recruitment of Shc-Grb2-Sos complexes to phosphorylated TrkB receptor (wild type cells). Mutant huntingtin disrupts BDNF-mediated induction of MAPK signaling by reducing expression of p52/p46 Shc adapter proteins and interfering with TrkB-Shc-mediated Ras activation. Reduced BDNF-mediated ERK1/2 activation in mutant huntingtin striatal cells leads to increased cell vulnerability to oxidative stress. This model is supported first by the ability of PD098059 to prevent BDNF-mediated neuroprotection in wild type cells and second by the capacity of PMA, which may activate Ras independently of Shc-Grb2-Sos complexes, to protect mutant huntingtin cells against oxidative damage.
Acknowledgments
We thank M. Macdonald for providing the huntingtin knock-in striatal cell lines and N. Agell for HA-RasV12. We are very grateful to Cristina Herranz, Ana Lopez, and M. Teresa Muñoz for technical assistance. We thank members of our laboratory for helpful discussion.
This work was supported by Ministerio de Ciencia e Innovación Grants SAF2008-00644 and SAF2009-7077 (to S. G.) and SAF2008-04360 (to J. A.), Centro de Investigaciones Biomédicas en Red sobre Enfermedades Neurodegenerativas (CIBERNED) Grant CB06/05/0054, Fondo de Investigaciones Sanitarias (Instituto de Salud Carlos III, RETICS) Grant RD06/0010/0006, and the Fundació La Marató de TV3.
- BDNF
- brain-derived neurotrophic factor
- HD
- Huntington disease
- MAPK
- mitogen-activated protein kinase
- PI3K
- phosphatidylinositol 3-kinase
- PLCγ
- phospholipase Cγ
- ERK
- extracellular signal-regulated kinase
- PMA
- phorbol 12-myristate 13-acetate
- TRITC
- tetramethylrhodamine isothiocyanate
- EGF
- epidermal growth factor
- MEK
- mitogen-activated protein kinase/extracellular signal-regulated kinase kinase
- PBS
- phosphate-buffered saline
- Q-PCR
- quantitative PCR
- CRE
- cAMP-response element
- CREB
- CRE-binding protein
- GST
- glutathione S-transferase
- HA
- hemagglutinin.
REFERENCES
- 1.Canals J. M., Pineda J. R., Torres-Peraza J. F., Bosch M., Martín-Ibañez R., Muñoz M. T., Mengod G., Ernfors P., Alberch J. (2004) J. Neurosci. 24, 7727–7739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fumagalli F., Molteni R., Calabrese F., Maj P. F., Racagni G., Riva M. A. (2008) CNS Drugs 22, 1005–1019 [DOI] [PubMed] [Google Scholar]
- 3.Pérez-Navarro E., Canudas A. M., Akerund P., Alberch J., Arenas E. (2000) J. Neurochem. 75, 2190–2199 [DOI] [PubMed] [Google Scholar]
- 4.Zuccato C., Cattaneo E. (2007) Prog. Neurobiol. 81, 294–330 [DOI] [PubMed] [Google Scholar]
- 5.Zuccato C., Ciammola A., Rigamonti D., Leavitt B. R., Goffredo D., Conti L., MacDonald M. E., Friedlander R. M., Silani V., Hayden M. R., Timmusk T., Sipione S., Cattaneo E. (2001) Science 293, 493–498 [DOI] [PubMed] [Google Scholar]
- 6.Zuccato C., Tartari M., Crotti A., Goffredo D., Valenza M., Conti L., Cataudella T., Leavitt B. R., Hayden M. R., Timmusk T., Rigamonti D., Cattaneo E. (2003) Nat. Genet. 35, 76–83 [DOI] [PubMed] [Google Scholar]
- 7.Duan W., Guo Z., Jiang H., Ware M., Li X. J., Mattson M. P. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 2911–2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferrer I., Goutan E., Marín C., Rey M. J., Ribalta T. (2000) Brain Res. 866, 257–261 [DOI] [PubMed] [Google Scholar]
- 9.Gines S., Seong I. S., Fossale E., Ivanova E., Trettel F., Gusella J. F., Wheeler V. C., Persichetti F., MacDonald M. E. (2003) Hum. Mol. Genet. 12, 497–508 [DOI] [PubMed] [Google Scholar]
- 10.Zuccato C., Marullo M., Conforti P., MacDonald M. E., Tartari M., Cattaneo E. (2008) Brain Pathol. 18, 225–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ginés S., Bosch M., Marco S., Gavaldà N., Díaz-Hernández M., Lucas J. J., Canals J. M., Alberch J. (2006) Eur. J. Neurosci. 23, 649–658 [DOI] [PubMed] [Google Scholar]
- 12.Kaplan D. R., Miller F. D. (2000) Curr. Opin. Neurobiol. 10, 381–391 [DOI] [PubMed] [Google Scholar]
- 13.Patapoutian A., Reichardt L. F. (2001) Curr. Opin. Neurobiol. 11, 272–280 [DOI] [PubMed] [Google Scholar]
- 14.Reichardt L. F. (2006) Philos. Trans. R. Soc. Lond. B Biol. Sci. 361, 1545–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trettel F., Rigamonti D., Hilditch-Maguire P., Wheeler V. C., Sharp A. H., Persichetti F., Cattaneo E., MacDonald M. E. (2000) Hum. Mol. Genet. 9, 2799–2809 [DOI] [PubMed] [Google Scholar]
- 16.Du J., Feng L., Yang F., Lu B. (2000) J. Cell Biol. 150, 1423–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barettino D., Pombo P. M., Espliguero G., Rodríguez-Peña A. (1999) Biochim. Biophys. Acta 1446, 24–34 [DOI] [PubMed] [Google Scholar]
- 18.Deogracias R., Espliguero G., Iglesias T., Rodríguez-Peña A. (2004) Mol. Cell Neurosci. 26, 470–480 [DOI] [PubMed] [Google Scholar]
- 19.Skaper S. D. (2008) CNS Neurol. Disord. Drug Targets 7, 46–62 [DOI] [PubMed] [Google Scholar]
- 20.Ravichandran K. S. (2001) Oncogene 20, 6322–6330 [DOI] [PubMed] [Google Scholar]
- 21.Arany I., Faisal A., Nagamine Y., Safirstein R. L. (2008) J. Biol. Chem. 283, 6110–6117 [DOI] [PubMed] [Google Scholar]
- 22.Yamada M., Ohnishi H., Sano S., Nakatani A., Ikeuchi T., Hatanaka H. (1997) J. Biol. Chem. 272, 30334–30339 [DOI] [PubMed] [Google Scholar]
- 23.Reynolds L. F., de Bettignies C., Norton T., Beeser A., Chernoff J., Tybulewicz V. L. (2004) J. Biol. Chem. 279, 18239–18246 [DOI] [PubMed] [Google Scholar]
- 24.Cavanaugh J. E., Jaumotte J. D., Lakoski J. M., Zigmond M. J. (2006) J. Neurosci. Res. 84, 1367–1375 [DOI] [PubMed] [Google Scholar]
- 25.Gu L., Cui T., Fan C., Zhao H., Zhao C., Lu L., Yang H. (2009) Biochem. Biophys. Res. Commun. 383, 469–474 [DOI] [PubMed] [Google Scholar]
- 26.Han B. H., Holtzman D. M. (2000) J. Neurosci. 20, 5775–5781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang H. Y., Lin S. Z., Kuo J. S., Chen W. F., Wang M. J. (2007) Neurobiol. Aging 28, 1258–1269 [DOI] [PubMed] [Google Scholar]
- 28.Jiang H., Zhang L., Koubi D., Kuo J., Groc L., Rodriguez A. I., Hunter T. J., Tang S., Lazarovici P., Gautam S. C., Levine R. A. (2005) J. Mol. Neurosci. 25, 133–140 [DOI] [PubMed] [Google Scholar]
- 29.Rubio I., Rennert K., Wittig U., Beer K., Dürst M., Stang S. L., Stone J., Wetzker R. (2006) Biochem. J. 398, 243–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gauthier L. R., Charrin B. C., Borrell-Pagès M., Dompierre J. P., Rangone H., Cordelières F. P., De Mey J., MacDonald M. E., Lessmann V., Humbert S., Saudou F. (2004) Cell 118, 127–138 [DOI] [PubMed] [Google Scholar]
- 31.Ventura A., Luzi L., Pacini S., Baldari C. T., Pelicci P. G. (2002) J. Biol. Chem. 277, 22370–22376 [DOI] [PubMed] [Google Scholar]
- 32.Luzi L., Confalonieri S., Di Fiore P. P., Pelicci P. G. (2000) Curr. Opin. Genet. Dev. 10, 668–674 [DOI] [PubMed] [Google Scholar]
- 33.Gotoh N., Muroya K., Hattori S., Nakamura S., Chida K., Shibuya M. (1995) Oncogene 11, 2525–2533 [PubMed] [Google Scholar]
- 34.Gotoh N., Tojo A., Shibuya M. (1996) EMBO J. 15, 6197–6204 [PMC free article] [PubMed] [Google Scholar]
- 35.Gotoh N., Toyoda M., Shibuya M. (1997) Mol. Cell. Biol. 17, 1824–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Okada S., Yamauchi K., Pessin J. E. (1995) J. Biol. Chem. 270, 20737–20741 [DOI] [PubMed] [Google Scholar]
- 37.Velazquez L., Gish G. D., van Der Geer P., Taylor L., Shulman J., Pawson T. (2000) Blood 96, 132–138 [PubMed] [Google Scholar]
- 38.Liu Y. F., Deth R. C., Devys D. (1997) J. Biol. Chem. 272, 8121–8124 [DOI] [PubMed] [Google Scholar]
- 39.Browne S. E., Beal M. F. (2006) Antioxid. Redox Signal. 8, 2061–2073 [DOI] [PubMed] [Google Scholar]
- 40.Butterworth N. J., Williams L., Bullock J. Y., Love D. R., Faull R. L., Dragunow M. (1998) Neuroscience 87, 49–53 [DOI] [PubMed] [Google Scholar]
- 41.Polidori M. C., Mecocci P., Browne S. E., Senin U., Beal M. F. (1999) Neurosci. Lett. 272, 53–56 [DOI] [PubMed] [Google Scholar]
- 42.Stack E. C., Matson W. R., Ferrante R. J. (2008) Ann. N.Y. Acad. Sci. 1147, 79–92 [DOI] [PubMed] [Google Scholar]
- 43.Apostol B. L., Illes K., Pallos J., Bodai L., Wu J., Strand A., Schweitzer E. S., Olson J. M., Kazantsev A., Marsh J. L., Thompson L. M. (2006) Hum. Mol. Genet. 15, 273–285 [DOI] [PubMed] [Google Scholar]
- 44.Wu Z. L., O'Kane T. M., Scott R. W., Savage M. J., Bozyczko-Coyne D. (2002) J. Biol. Chem. 277, 44208–44213 [DOI] [PubMed] [Google Scholar]