Abstract
R-spondin-1 (Rspo1) is an intestinal growth factor known to exert its effects through activation of the canonical Wnt (cWnt) signaling pathway and subsequent expression of cWnt target genes. We have detected Rspo1 mRNA in murine islets and the murine MIN6 and βTC β-cell lines, and Rspo1 protein in MIN6 β-cells. Rspo1 activated cWnt signaling in MIN6 β-cells by increasing nuclear β-catenin and c-myc, a cWnt target gene. Rspo1 also induced insulin mRNA expression in MIN6 cells. Analysis of MIN6 and mouse β-cell proliferation by [3H]thymidine and BrdU incorporation, respectively, revealed that Rspo1 stimulated cell growth. Incubation of MIN6 and mouse β-cells with cytokines (IL1β/TNFα/interferon-γ) significantly increased cellular apoptosis; this increase was abolished by pretreatment with Rspo1. Rspo1 also stimulated insulin secretion in a glucose-independent fashion. We further demonstrated that the glucagon-like peptide-1 receptor agonist, exendin4 (EX4), stimulated Rspo1 mRNA transcript levels in MIN6 cells in a glucose-, time-, dose-, and PI3-kinase-dependent fashion. This effect was not limited to this β-cell line, as similar time-dependent increases in Rspo1 were also observed in the βTC β-cell line and mouse islets in response to EX4 treatment. Together, these studies demonstrate that Rspo1 is a novel β-cell growth factor and insulin secretagogue that is regulated by EX4. These findings suggest that Rspo1 and the cWnt signaling pathway may serve as a novel target to enhance β-cell growth and function in patients with type 2 diabetes.
Keywords: Apoptosis, β-Catenin, Glucose, Growth Factors, Insulin, Insulin Secretion, Wnt Pathway, Glucagon-like Peptide-1, Rspo1
Introduction
Type 2 diabetes mellitus (T2DM)4 represents a serious and growing epidemic that poses a major public health threat in the 21st century. The development of T2DM usually requires the presence of both insulin resistance and impaired β-cell function, but also involves the loss of β-cells (1). Moreover, type 1 diabetes mellitus (T1DM) is characterized by the autoimmune-mediated destruction of β-cells. Therefore, novel therapeutic approaches that enhance β-cell mass expansion, as well as β-cell function, represent an exciting arsenal against diabetes.
Wnt signaling has been demonstrate to play important roles in development as well as in the pathogenesis of a variety of diseases, including diabetes (2). Activation of this pathway requires interaction between a secreted glycoprotein, Wnt, and a seven transmembrane receptor protein, Frizzled (Frz). There are at least three distinct intracellular Wnt pathways, including, most notably, the canonical Wnt (cWnt) cascade that leads to changes in intracellular β-catenin levels and is thought to be involved in cell fate specification and proliferation. β-catenin is normally phosphorylated and targeted for proteolysis by a complex of proteins, including adenomatosis polyposis coli (APC), axin and the serine/threonine kinase glycogen synthase kinase-3 β (GSK3β) (Fig. 1A). cWnt activation of the Frz and low density lipoprotein receptor-related protein (LRP) co-receptors results in dissociation of this degradation complex, permitting entry of β-catenin into the nucleus to activate cWnt target genes in conjunction with TCF/LEF family transcription factors and, possibly, other DNA-binding partners (3). cWnt target genes have been identified in different models and these include, but are not limited to, the cell-cycling genes, c-myc, and cyclin D1.
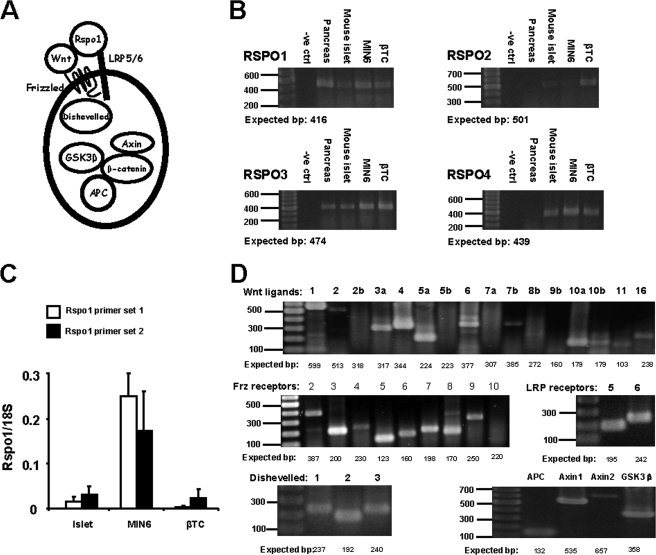
FIGURE 1.
Rspo1 and cWnt signaling molecules are expressed in murine β-cells. A, simplified schematic of cWnt and Rspo1 signaling showing cWnt ligand and Rspo1 binding to the Frz receptor and LRP5/6 co-receptor, as well as the intracellular protein Dishevelled and the β-catenin degradation complex consisting of APC, Axin, and GSK3β. B, RT-PCR analysis of Rspo1–4 mRNA in murine pancreas, islets, and MIN6 and βTC β-cell lines. A 100–1000-bp ladder was used. No RNA was used in the negative (-ve) control. C, relative qRT-PCR quantification of Rspo1 mRNA transcripts in murine islets, and MIN6 and βTC β-cells. Primer set 1, exons 2–3 and primer set 2, exons 3–4. Relative expression levels of Rspo1 were normalized to 18 S rRNA expression. (n = 5–30). D, RT-PCR for mRNA transcripts of various cWnt signaling molecules in MIN6 β-cells. A 100–1000-bp ladder was used.
Interestingly, mice that lack the gene encoding LRP5 show impaired glucose tolerance because of perturbed glucose-stimulated insulin secretion (GSIS) (4). Furthermore, adipocyte-secreted Wnts have been shown to stimulate insulin secretion and glucokinase gene transcription in INS1 cells in vitro through the activation of cWnt signaling (5). In contrast, transgenic mice that overexpress a dominant-negative form of mouse Frz8 under the Ipf-1/Pdx-1 promoter are normoglycemic and display normal GSIS (6). However, the β-cells of these mice produce four times more and secrete twice as much insulin as those of wild-type littermates, suggesting the presence of compensatory mechanisms to achieve and maintain normoglycemia (6). Finally, Rulifson et al. (7) demonstrated that conditional pancreatic β-cell specific expression of degradation-resistant β-catenin leads to β-cell expansion, increased insulin production and serum levels, and enhanced glucose handling. This observation is further strengthened by a recent study from Liu and Habener showing that exendin4 (EX4), a glucagon-like peptide-1 (GLP-1) receptor agonist, stimulates β-cell proliferation via activation of the cWnt signaling pathway (8).
The roof plate-specific spondin (R-spondin, Rspo) protein family consists of four structurally related members (Rspo1–4), with conserved cysteine-rich furin-like and thrombospondin domains. Several lines of evidence indicate that Rspo family members function as Frz and/or LRP receptor ligands in vitro: 1) Rspo is a secreted protein (9); 2) unlike Wnt ligands that form a ternary complex with Frz and LRP receptors, Rspo proteins failed to form a ternary complex but can nonetheless bind to both receptors (10); 3) there is a positive modulation of Wnt ligand activity by Rspo via direct interaction between the two ligands (11); and 4) Rspo prevents LRP6 internalization (12). Furthermore, transgenic mice expressing human Rspo1 exhibit a profound increase in proliferation of intestinal crypt epithelial cells, which correlates with the activation of β-catenin (9). Adenoviral-mediated transfection of each isoform of Rspo into mice also induces gastrointestinal proliferation in association with β-catenin activation (13). Unexpectedly, although expressed at high levels in the gut, Rspo1 has also been detected in human pancreatic islets by immunohistochemisty (9). However, no studies to-date have examined the role of Rspo1 in β-cell physiology. Therefore, in the present study, we have determined the role of Rspo1 in the mature pancreatic β-cell, through analysis of the effects of Rspo1 on β-cell proliferation, apoptosis and insulin secretion, as well as through analysis of the effects of known β-cell regulatory factors (i.e. glucose and GLP-1) on Rspo1 expression.
EXPERIMENTAL PROCEDURES
Cell Culture
MIN6 β-cells (mouse insulinoma cell line, a kind gift from Drs. J. Miyazaki, University of Tokyo and D. F. Steiner, University of Chicago) were maintained in Dulbecco's modified Eagle's medium (DMEM, Invitrogen) containing 25 mm glucose and supplemented with 2 mm l-glutamine, 10% heat-inactivated fetal bovine serum (FBS), penicillin (100 units/ml), streptomycin (100 μg/ml), and 71 μm 2-mercaptoethanol in humidified 5% CO2, 95% air at 37 °C. The βTC β-cell line was maintained in DMEM containing 25 mm glucose, 2 mm l-glutamine, 10% heat-inactivated FBS, penicillin (100 units/ml), and streptomycin (100 μg/ml).
Isolation and Culture of Intact and Dispersed Mouse Islets
Islets were isolated from 20–30 g of CD1 mice (Charles River, St. Constant, Quebec, Canada) by collagenase digestion, as previously described (14) and were cultured in RPMI 1640 containing 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin (Invitrogen) for 2 days after isolation. Mouse islet cells were dispersed by incubation with Dispase II (Roche Laboratories, Mississauga, Ontario, Canada) as previously described (15) and were plated on 35-mm Petri dishes (for Live-Cell Analyses, ibidi, Ingersoll, Ontario, Canada). Cells were then cultured overnight.
RNA Isolation
Animal tissues or cells grown to ∼80–90% confluence were lysed for preparation of RNA using either the RNeasy or RNeasy Micro Kit according to the manufacturer's instructions (Qiagen). RNA was quantified by spectrophotometry (absorbance at 260 nm) and stored at −80 °C until use.
RT-PCR
Equal amounts of RNA isolated from animal tissues, cells, or islets were analyzed by RT-PCR using a One-Step kit (Qiagen). RT-PCR primers and conditions have been reported previously (16–25) and are listed in Table 1. All primers were further verified using positive control samples selected based on previous reports listed in the expression data base (Roel Nusse, the Wnt home page (Stanford, Palo Alto, CA) and data not shown). Negative control reactions were performed using RNase-free water without template.
TABLE 1.
RT-PCR primers
Primers and conditions used to detect multiple cWnt signaling molecules as depicted in Fig. 1A. The RT-PCR primers were designed to recognize mouse sequences.
| Target | Primer sequence |
Size | Annealing temperature | Cycle # | Ref. | |
|---|---|---|---|---|---|---|
| Sense | Antisense | |||||
| Wnt1 | AAATCGCCCAACTTCTGCA | AATACCCAAAGAGGTCACAGC | 599 | 53.2 | 40 | 16 |
| Wnt2 | CGGCCTTTGTTTACGCCATC | TGAATACAGTAGTCTGGAGAA | 513 | 55.3 | 40 | 16 |
| Wnt2b | TGTACTCTGCGCACCTGCT | TGCACTCACACTGGGTGAC | 318 | 60.0 | 40 | 16 |
| Wnt3a | ATTGAATTTGGAGGAATGGT | CTTGAAGTACGTGTAACGTG | 317 | 57.5 | 40 | 16 |
| Wnt4 | TGTACCTGGCCAAGCTGTCAT | TCCGGTCACAGCCACACTT | 344 | 60.0 | 40 | 16 |
| Wnt5a | TCCTATGAGAGCGCACGCAT | CAGCTTGCCCCGGCTGTTGA | 224 | 60.0 | 40 | 16 |
| Wnt5b | TCGGAGGAGCAGGGCCGAGC | CAGCTTGCCCTGGCGGGTGA | 223 | 65.0 | 40 | 16 |
| Wnt6 | GCACCGAGTGTAAGTGCCAT | GAAGCGGCACAGACAGTTCT | 377 | 56.2 | 40 | 16 |
| Wnt7a | CAAGGCCAGTACCACTGGGA | GGCTCCACGTGGACGGCCTC | 307 | 56.5 | 40 | 16 |
| Wnt7b | ACCAAAACTTGCTGGACCAC | ACGTGTTGCACTTGACGAAG | 385 | 56.2 | 40 | 16 |
| Wnt8b | AACGTGGGCTTCGGAGAGGC | GCCCGCGCCTTGCAGCAGGT | 272 | 62.9 | 40 | 16 |
| Wnt9b | GTAAGTGCCATGGTGTGTC | GTGTCATAGCGTAGCTTCAG | 160 | 55 | 28 | 17 |
| Wnt10a | AAAGTCCCCTACGAGAGCCC | CAGCTTCCGACGGAAAGCTT | 179 | 52.3 | 40 | 16 |
| Wnt10b | CGGCTGCCGCACCACAGCGC | CAGCTTGGCTCTAAGCCGGT | 179 | 59.3 | 40 | 16 |
| Wnt11 | GTGGCTGCTGACCTCAAGACC | TTCTTCATGCAGAAGTCAGGAG | 103 | 55 | 28 | 17 |
| Wnt16 | ACAGCATCCAGATCTCAGAC | ACTACATGGGTGTTGTAGCC | 238 | 55 | 28 | 17 |
| Frz2 | CTAGCGCCGCTCTTCGTGTACCTG | CAGCGTCTTGCCCGACCAGATCCA | 387 | 60 | 25 | 18 |
| Frz3 | TAGCAATGGAGCCCTTCCACC | CTCCATATCTTCAGGCCACGG | 200 | 55 | 28 | 17 |
| Frz4 | GCTTGTGCTATGTTGGGAACCCAC | ACAGGTTGCAGGAACCGT | 230 | 55 | 28 | 17 |
| Frz5 | GTCTGTGCTGTGCTTCATC | AGTGACACACACAGGTAGCA | 123 | 55 | 28 | 17 |
| Frz6 | TGAAGGAGAGAAGCAATGGATC | TGAACAGGCAGAGATGTGGAG | 160 | 55 | 28 | 17 |
| Frz7 | GCCAGACCCACCTTTCACT | CGAACCGTCTCTCCTCTTCTT | 198 | 58 | 35 | 19 |
| Frz8 | TTTGTGCTGGCGCCACTGGTT | TAGAGCACGGTGAAGAGGCCC | 170 | 55 | 28 | 17 |
| Frz9 | CGGAGTCTTTTCCATCCTTTACAC | GCCATTTTTCGGTAGCACAGG | 250 | 58 | 35 | 20 |
| Frz10 | TGTCCGGTTGCTACACCATGGGCT | GCCAGGAACCAGGTGAGG | 220 | 55 | 28 | 17 |
| LRP5 | GATGTGCGGCTAGTGGATG | GCCCGAGATGACAATGTTCT | 195 | 58 | 35 | 21 |
| LRP6 | GGGCCGATGCAAAACTTAAT | CCTCTGTTGGCTGAAAGCAT | 242 | 58 | 35 | 21 |
| Dsh1 | GGGGGTAGTGGCAGTGAA | ACCTGTAAGTTCTGGAGGGACA | 237 | 58 | 35 | 21 |
| Dsh2 | GCAGTGGCAGTGAGTCAGAA | TCATGGGGTTATAGGGGAGAG | 192 | 58 | 35 | 21 |
| Dsh3 | CAAGGAGAAGGACCCAAAAG | ATCGGGGGACCATAGAGAG | 240 | 58 | 35 | 21 |
| APC | TCACCTAGCGGGACTGTTG | CTTCTTCGTGTTGGTGCTCA | 132 | 55 | 40 | Roel Nusse, The Wnt Homepage (Stanford, Palo Alto, CA) |
| Axin1 | CAGGGTTTCCCCTTGGACC | GGTCAAACATGGCAGGATC | 535 | 58 | 30 | 22 |
| Axin2 | CTCCTTGGAGGCAAGAGC | GGCCACGCAGCACCGCTG | 657 | 58 | 40 | 23 |
| GSK3β | CAGCAGCCTTCAGCTTTTGG | CCGGAACATAGTCCAGCACCAG | 358 | 60 | 35 | 24 |
| Rspo1 | TGTGAAATGAGCGAGTGGTCC | TCTCCCAGATGCTCCAGTTCT | 416 | 58 | 32 | 25 |
| Rspo2 | TTGCATAGAGGCCGCTGCTTT | CTGGTCAGAGGATCAGGAATG | 501 | 58 | 32 | 25 |
| Rspo3 | GTACACTGTGAGGCCAGTGAA | ATGGCTAGAACACCTGTCCTG | 474 | 58 | 32 | 25 |
| Rspo4 | CTGGAGTCCCTGCATACACAA | CACGGGGAGAAGGAAAGTTTC | 439 | 58 | 32 | 25 |
Real-time PCR
MIN6, βTC and islets were serum-starved overnight and then incubated with medium alone (containing the appropriate vehicle, PBS or DMSO), recombinant Wnt3a (641 pm, R&D Systems, Minneapolis, MN), recombinant mouse Rspo1 (34.5 pm to 34.5 nm, R&D Systems), or EX4 (1–100 nm, Bachem, Torrance, CA) with or without high glucose (25 mm) or inhibitors (LY294002 (50 μm, Sigma-Aldrich), wortmannin (100 nm, Sigma-Aldrich), H89 (10 μm, Sigma-Aldrich), SB239063 (10 μm, Calbiochem, Mississauga), PD98059 (20 μm, Sigma-Aldrich), or U0126 (1 μm, New England Biolabs, Mississauga)) for the indicated amount of time, ranging from 30 min to 24 h. 5 μg of total RNA from samples were reverse-transcribed with Superscript II Reverse Transcriptase (Invitrogen). Semi-quantitative RT-PCR (qRT-PCR) was performed in a Chromo4 Continuous Fluorescence Detection unit with Opticon Monitor 3 software (Bio-Rad) using Taqman Gene Expression Assays for specific primers (Applied Biosystems, Foster City, CA). All reactions were performed in duplicate, and control reactions were performed without RT enzyme and/or without template. The linearity of amplification of the Taqman primer-probe sets was verified over nine orders of magnitude (data not shown). Ribosomal protein 18 S RNA (no. Hs99999901_sl) was used as the endogenous control for all quantitative analyses of mRNA expression and was not found to change in response to any of the experimental treatments tested (data not shown). Relative quantification of Rspo1 (2 sets of primers used, set 1 no. Mm00507076_m1 and set 2 no. Mm00507077_m1), c-myc (no. Mm00487803_m1), cyclin D1 (no. Mm00432359_m1), and insulin2 (no. Mm00731595_gH) mRNA expression was calculated using the ▵▵ cycle threshold [▵▵C(t)] method (26).
Protein Extraction, Cell Fractionation, and Immunoblotting
Cells and islets were lysed with radioimmune precipitation assay buffer (50 mm β-glycerol phosphate, 10 mm HEPES (pH 7.4), 1% Triton X-100, 70 mm NaCl, 2 mm EGTA, 1 mm Na3VO4, 1 mm NaF, and EDTA-free protease inhibitors (Roche Laboratories)). Proteins of interest were detected with primary antibodies targeted against mouse Rspo1 (goat IgG, 1:1000, R&D Systems), cleaved caspase3 (rabbit IgG, 1:1000, New England Biolabs), or pan-actin (rabbit IgG, 1:1000, Sigma-Aldrich). Immunoblotted membranes were then probed with the appropriate secondary antibodies (horseradish peroxidase (HRP)-linked anti-rabbit (1:2000, New England Biolabs), and HRP-linked anti-goat (1:2000, Jackson ImmunoResearch Laboratories, West Grove, PA)) and visualized by electrochemical luminescence detection system (Amersham Biosciences, Baie d'Urfe, Quebec, Canada). Membranes were subsequently treated at 50 °C for 30 min with stripping buffer (62.5 mm Tris-HCl, pH 6.8, 2% SDS, and 100 mm β-mercaptoethanol) for a second round of immunoblotting.
To determine protein levels of nuclear β-catenin, MIN6 cells were grown to 80–90% confluency in 10-cm dishes and serum-starved overnight, followed by treatment with either medium alone (control), EX4 (10 nm), LiCl (20 mm), Wnt3a (641 pm), or Rspo1 (34.5 pm to 34.5 nm) for 30 min or 8 h. Lysates were then centrifuged at 2,000 × g for 5 min. The pellets were resuspended in 350 μl of Buffer A (20 mm HEPES (pH 7.5), 10 mm KCl, 0.1 mm EDTA, 0.1 mm EGTA, 1 mm dithiothreitol, and protease and phosphatase inhibitors (Roche Laboratories) and incubated on ice for 15 min. Cells were further lysed by addition of 10% Nonidet P-40 (to a final concentration of 1%, Sigma-Aldrich) and vortexed for 1 min. Nuclei were pelleted by centrifugation for 10 min at 1,600 × g at 4 °C. The pellets were washed once with 400 μl of Buffer A and the nuclear fraction was further pelleted for 10 min at 1600 × g at 4 °C. Nuclei were solubilized by addition of one pellet volume of NE buffer (20 mm Tris, pH 8.0), 420 mm NaCl, 1.5 mm MgCl2, 0.2 mm EDTA, 25% glycerol, and protease and phosphatase inhibitors). One-fourth pellet volume of 5 m NaCl and one pellet volume of NE buffer were further added to the resulting pellet. Nuclei were then homogenized by sonication for 5 s and were stored at −80 °C until used (27). 50 μg of nuclear protein, as measured by Bradford protein assay (Bio-Rad), was used for immunoblotting using antibodies against β-catenin (mouse IgG, 1:1000, BD Transduction Laboratories, Franklin Lakes, NJ) and poly-ADP-ribose polymerase (PARP, mouse IgG, 1:1000, nuclear fraction protein loading control (28), BD Transduction Laboratories) as described.
Cell Proliferation Assays
MIN6 cells were grown to 80–90% confluence in 24-well plates, serum-starved overnight, and then treated with medium alone (control), EX4 (10 nm) or various doses of recombinant Rspo1 (34.5 pm to 34.5 nm) overnight in the presence of serum-free medium with 25 mm glucose. Cell proliferation was measured as described (29). Briefly, cells were incubated with 37 kBq/ml [3H]methylthymidine (specific activity: 3000 GBq/mmol, Amersham Biosciences) for 4 h. Cells were then washed twice in cold PBS and incubated for 30 min in 1 ml of 5% trichloroacetic acid at 4 °C to precipitate the DNA. The liquid layer was removed by aspiration and 500 μl of 0.1 m sodium hydroxide was added to the cells for an additional 30 min at room temperature with gentle shaking. The solubilized material was then transferred to 4 ml of scintillant, and radioactive counts were determined by liquid scintillation counting.
For measurement of murine β-cell proliferation, dispersed islet cells were treated for 48 h with medium alone (control), EX4 (10 nm) or recombinant Rspo1 (34.5 nm) in the presence of serum-free medium with 20 mm glucose and, for the last 24 h, 5′-bromo-2′-deoxyuridine (BrdU, 10 μm). Cells were washed with PBS and fixed in 10% formalin and incubated with mouse anti-BrdU (1:200, Sigma-Aldrich) and guinea pig anti-insulin (1:200, Dako Diagnostics, Mississauga, ON, Canada) antibodies. Cells were then gently washed with PBS and incubated for 30 min with appropriate secondary antibodies (Texas-Red-conjugated anti-mouse (1:200) and Cy2-conjugated anti-guinea pig (1:200), Jackson ImmunoResearch Laboratories) and then mounted with mounting medium for fluorescence containing 4′,6-diamidino-2-phenylindole (VectaShield, Vector Laboratories, Inc., Burlingame, CA). Proliferative index is expressed as a percentage of BrdU- and insulin-positive cells over total insulin-positive cells analyzed under Zeiss Axioplan microscope with Axiovision software (Carl Zeiss Canada, Don Mills, Ontario, Canada). A minimum of 100 β-cells was counted per treatment.
Apoptosis Assays
MIN6 cells were grown to 80–90% confluency and apoptosis assay was performed as previously described (14). Briefly, cells were seeded in 12-well cell culture dishes for 24 h and subsequently preincubated with either medium alone (control), EX4 (10 nm), Wnt3a (641 pm), or Rspo1 (34.5 pm - 34.5 nm) for 18 h. The cells were then incubated with a mixture of cytokines (10 ng/ml IL-1β, 50 ng/ml TNFα, 50 ng/ml IFNγ, Sigma-Aldrich) in the absence or presence of treatment, as described above, for another 18 h. This incubation time was based on the results of (14). Apoptosis was measured by immunoblotting for cleaved caspase3 (14).
For measurement of murine β-cell apoptosis, after overnight serum starvation, cells were preincubated with either medium alone (control), EX4 (10 nm), or Rspo1 (34.5 nm) for 18 h. The cells were then incubated with a mixture of cytokines, as above, in the absence or presence of treatment, as described, for an additional overnight incubation. Dispersed cells were then washed and fixed in 10% formalin, and stained for insulin as above, with apoptosis detection performed using a TUNEL detection kit (Roche Laboratories). β-Cell apoptosis is expressed as a percentage of TUNEL- and insulin-positive cells over total number of insulin-positive cells, analyzed as above.
Insulin Secretion Assay
MIN6 cells were grown to 90% confluence in 24-well plates, and serum-starved overnight. Cells were then treated for 2 h with low (2 mm) or high glucose (25 mm) serum-free medium containing medium alone (control), EX4 (10 nm), Wnt3a (641 pm), or Rspo1 (34.5 pm to 34.5 nm) for an additional 2 h with serum-free medium. The medium was then transferred and spun at 2,000 × g at 4 °C for 1 min, and the supernatant was collected and placed on ice. Insulin secretion studies on isolated mouse islets were performed as previously described (30). Briefly, islets were cultured overnight in 2 mm glucose RPMI 1640 (Invitrogen) with 10% FBS and penicillin/streptomycin. Islets were then washed and incubated with experimental medium that consisted of either low (2 mm) or high glucose (20 mm) RPMI 1640 with or without Rspo1 (34.5 nm) for 2 h. Media samples were taken and centrifuged at 700 × g at 4 °C for 1 min, and the supernatant was collected. Samples were diluted into the assay buffer and assayed for insulin using an insulin RIA kit according to the manufacturer's instructions (Linco Research, St. Louis, MO). Cell protein content was determined by Bradford assay.
Statistical Analysis
All data are expressed as mean ± S.E. In some experiments, data were log10 transformed to normalize variance for statistical analysis. Data were analyzed by Student's t test or by one- or two-way ANOVA, followed by appropriate post-hoc testing using Statistical Analysis System software (SAS v 9.1.3, Cary, NC). Statistical significance was assumed at p < 0.05.
RESULTS
Expression of Rspo1 and cWnt Signaling Molecules in Murine β-Cells
Conventional RT-PCR demonstrated that Rspo1 mRNA is expressed in whole mouse pancreas as well as in isolated murine islets (Fig. 1B). Rspo1 mRNA was also detected in the murine MIN6 and βTC β-cell lines. Moreover, the Rspo3 and Rspo4, but not Rspo2, isoforms were detected in both mouse islets and MIN6 β-cells (Fig. 1B). Examination of the relative expression levels of Rspo1 by qRT-PCR using two different Rspo1 primer sets revealed that, although expression was lower in mouse islets and βTC β-cells, Rspo1 was highly expressed in the MIN6 β-cell line (Fig. 1C). Therefore, the MIN6 β-cell line was used as our main in vitro model to study Rspo1.
RT-PCR of total RNA from MIN6 β-cells was conducted to determine the expression of essential components of the cWnt pathway, including specific isoforms of Wnt ligands, Frizzled, and LRP receptors, and intracellular cWnt signaling molecules such as Axin, dishevelled, APC, and GSK3β (Fig. 1A). As shown in Fig. 1D, MIN6 β-cells expressed mRNA transcripts for the majority of Wnt ligands (except Wnt2b, Wnt5b, and Wnt9b) and Frz receptors (except Frz9 and Frz10). Both isoforms of the LRP co-receptors and all tested intracellular cWnt signaling molecules were also detected in MIN6 β-cells. Collectively, these observations imply that MIN6 β-cells are capable of a functional cWnt signaling response.
Rspo1 Stimulates cWnt Signaling and Insulin mRNA Expression in MIN6 β-Cells
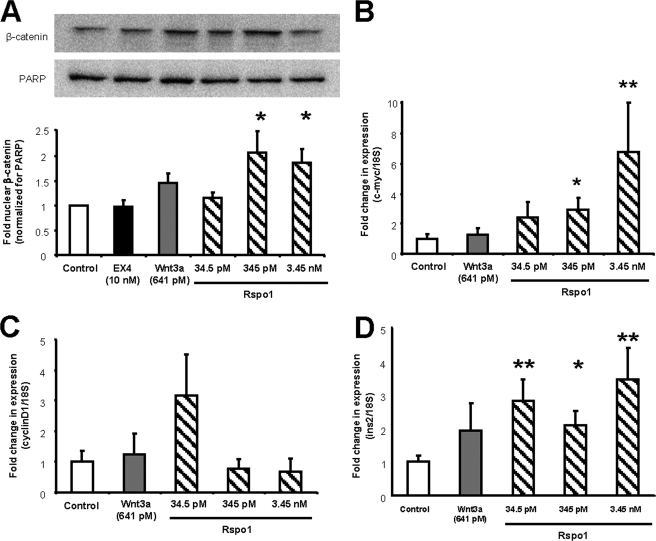
Activation of cWnt signaling involves the stabilization of β-catenin and its subsequent translocation to the nucleus where it interacts with the TCF/LEF family of transcription factors to initiate transcription of cWnt target genes. To determine whether Rspo1 induces cWnt signaling in MIN6 β-cells, cells were incubated for 30 min with medium alone or Rspo1 at 34.5 pm, 345 pm, and 3.45 nm, as well as with EX4 (10 nm) and Wnt3a (641 pm), positive controls, and nuclear lysates were used to immunoblot for β-catenin. Rspo1 at the 345 pm and 3.45 nm doses, but not Wnt3a, was found to significantly enhance nuclear β-catenin levels (p < 0.05). Although EX4 did not increase nuclear β-catenin within this time frame, we found that EX4 (and LiCl, positive control) significantly enhanced nuclear β-catenin after 8 h of incubation, by 1.5-fold (p < 0.05, data not shown).
To determine if Rspo1-induced increases in nuclear β-catenin translate to transcriptional output, two cWnt target genes, c-myc and cyclin D1, were analyzed by qRT-PCR. Rspo1, at concentrations of 345 pm and 3.45 nm, significantly, increased c-myc, but not cyclin D1 mRNA, after 12 h of incubation (Fig. 2B; p < 0.05–0.01), at which time, there was no effect of Wnt3a (641 pm). However, incubation with Wnt3a for 4 h stimulated a 3.5-fold increase in c-myc and a 3-fold increase in cyclin D1 mRNA levels in the MIN6 cells (p < 0.05 for both, data not shown). Together, these findings establish that Rspo1 induces cWnt signaling in MIN6 β-cells by increasing nuclear β-catenin levels, resulting in a subsequent elevation of c-myc mRNA levels, and that the timing and effects of Rspo1 on MIN6 β-cells differ from those of both EX4 and Wnt3a. Moreover, qRT-PCR revealed that, at all concentrations tested, Rspo1 also enhanced insulin mRNA levels after 12 h (Fig. 2D, p < 0.05–0.01). Interestingly, treatment with EX4 stimulated insulin mRNA levels only after 24 h of incubation by 2-fold (p < 0.01, data not shown), indicating that Rspo1 and EX4 may regulate insulin mRNA expression via different pathways.
FIGURE 2.
Rspo1 activates cWnt signaling and increases insulin mRNA levels in MIN6 β-cells. A, ratio of nuclear β-catenin to nuclear PARP in MIN6 cells treated with EX4, Wnt3a, and increasing doses of Rspo1 for 30 min. A representative blot is shown. All values are expressed as fold-relative to the control (medium alone, n = 4–6). B–D, relative expression analysis of c-myc (B), cyclin D1 (C), and insulin (D) mRNA levels by qRT-PCR in MIN6 β-cells treated with medium alone (control), Wnt3a, or increasing doses of Rspo1 for 12 h. Data were normalized to the housekeeping gene 18 S rRNA (n = 9–11) and are displayed relative to vehicle-treated controls. *, p < 0.05 and **, p < 0.01.
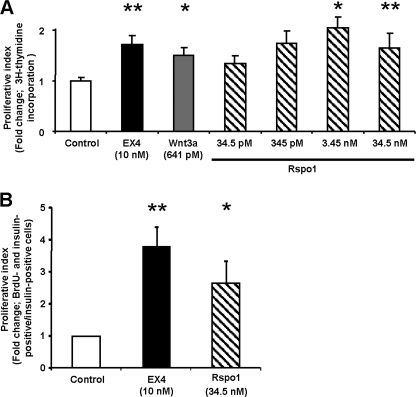
Rspo1 Stimulates β-Cell Proliferation
Cell proliferation assay using [3H]methylthymidine incorporation revealed that EX4 and Wnt3a (positive controls) stimulated MIN6 β-cell proliferation by nearly 2-fold compared with the control group (Fig. 3A, p < 0.05–0.01), consistent with prior reports (7, 8). Treatment with recombinant mouse Rspo1 at doses of 345 pm and 3.45 nm also stimulated MIN6 β-cell proliferation, reaching a maximum of 2.2-fold (p < 0.01) of controls. The highest dose of Rspo1 tested (34.5 nm) did not stimulate further proliferation. To assess whether Rspo1 can stimulate β-cell proliferation, dispersed mouse islet cells were incubated with EX4 (10 nm, positive control) and Rspo1 (34.5 nm) for 48 h, and BrdU was added for the last 24 h. Fig. 3B shows that Rspo1 at 34.5 nm induced a 2.5-fold increase in BrdU incorporation in insulin-positive cells (p < 0.01), whereas EX4 enhanced β-cell proliferation by 2.8-fold (p < 0.01).
FIGURE 3.
Rspo1 stimulates β-cell proliferation. A, MIN6 β-cells were treated with either medium alone (control), EX4, Wnt3a or increasing doses of Rspo1 overnight, and their proliferation index was determined by [3H]thymidine incorporation assay (n = 14–33). B, dispersed murine islet cells were treated with medium alone (control), EX4 or Rspo1for 48 h, and BrdU was added for the last 24 h. Cells were then fixed and co-stained for insulin and BrdU. Proliferative index was determined as the number of BrdU- and insulin-positive cells over total insulin-positive cells, and data are presented as fold of control (n = 4). *, p < 0.05; **, p < 0.01.
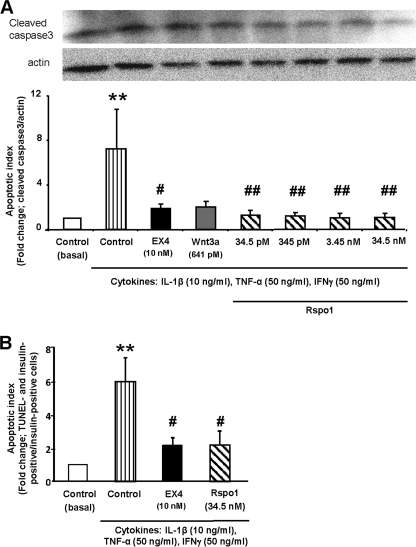
Rspo1 Prevents Cytokine-induced Apoptosis in β-Cells
In addition to the enhancement of cell growth, inhibition of apoptosis is another important variable in the β-cell growth equation. As shown in Fig. 4A, the level of activated, cleaved caspase3 was significantly increased by 7-fold (p < 0.05) following treatment of the MIN6 cells with a mixture of cytokines for 18 h, and this increase was completely prevented by pretreatment with EX4 (p < 0.05) or Wnt3a (641 pm), as well as by all doses of Rspo1 (p < 0.01). The level of activated caspase3 in the presence of cytokines was not further reduced when MIN6 cells were co-treated with both Wnt3a and Rspo1 (data not shown). A similar observation was observed in dispersed murine β-cells, such that treatment with cytokines for 18 h significantly increased the number of TUNEL-positive β-cells by 6-fold (p < 0.01); however pretreatment with either EX4 (10 nm) or Rspo1 (34.5 nm) significantly reduced cytokine-induced apoptosis (p < 0.05, Fig. 4B).
FIGURE 4.
Rspo1 inhibits cytokine-induced β-cell apoptosis. A and B, effects of Rspo1 on activated, cleaved caspase-3 in MIN6 β-cells (A) or TUNEL in dispersed murine β-cells (B). Cells were incubated in serum-free medium overnight, pretreated with medium alone (control), EX4, Wnt3a, or the specified doses of Rspo1 for 18 h, and then incubated without (basal) or with a combined cytokine mixture for a further 18 h. MIN6 β-cells were analyzed by immunoblotting for cleaved caspase-3 and pan-actin (n = 4–8). A representative blot is shown. Dispersed islet cells were fixed, and then co-stained for insulin and TUNEL (n = 6). Apoptotic index was expressed as fold change relative to the basal control group. *, p < 0.05 and **, p < 0.01 when compared with control (basal); #, p < 0.05 and ##, p < 0.01 when compared with control + cytokines.
Rspo1 Stimulates β-Cell Insulin Secretion
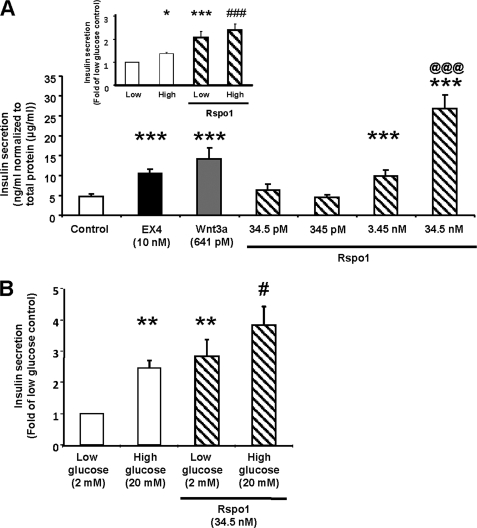
It is well established via knock-out of LRP5 that the manipulation of cWnt signaling induces changes in β-cell function (4). MIN6 β-cells were therefore treated for 2 h with either medium alone (control), EX4 (10 nm, positive control), Wnt3a (641 pm) or Rspo1 (34.5 pm to 34.5 nm, Fig. 5A) in the presence of high (25 mm) glucose. Not only EX4, but also Wnt3a stimulated insulin secretion under these conditions (p < 0.001). Furthermore, while no changes were seen with Rspo1 at the low doses tested (34.5 and 345 pm), insulin secretion from MIN6 β-cells treated with Rspo1 at higher doses (3.45 and 34.5 nm) was increased to 2- and 5-fold of control, respectively, in a dose-dependent fashion (Fig. 5A, p < 0.001 versus control, p < 0.001 for 34.5 nm versus 3.45 nm). Rspo1-stimulated insulin secretion in MIN6 β-cells was not glucose dependent as no difference in secretion was seen between low and high glucose in the presence of Rspo1 (Fig. 5A, inset). We next evaluated whether Rspo1 can regulate insulin secretion in mouse islets. Static incubation of islets with Rspo1 at 34.5 nm for 2 h induced a significant increase in insulin secretion, and this effect was glucose-independent (Fig. 5B).
FIGURE 5.
Rspo1 stimulates insulin secretion in MIN6 β-cells and isolated mouse islets. A, insulin secretory response to Rspo1 in MIN6 β-cells (n = 5 - 12) was tested by static incubation of medium containing medium alone (control), EX4, Wnt3a, or indicated doses of Rspo1 for 2 h with high glucose. Insulin in the medium was measured by radioimmunoassay, and the results were normalized to total protein content. (inset: MIN6 β-cells were treated with or without Rspo1 (34.5 nm) under low (2 mm) or high glucose (25 mm) conditions (n = 6–12). Data were normalized to total protein content and expressed as fold of low glucose alone). B, insulin secretion in isolated mouse islets was determined after 2 h incubation with low or high glucose and with or without Rspo1. The results were normalized to total protein content and expressed as fold of low glucose alone. *, p < 0.05; **, p < 0.01; ***, p < 0.001 compared with control values, @@@, p < 0.001 for 34.5 nm compared with 3.45 nm Rspo1, and #, p < 0.05 and ###, p < 0.001 for high glucose compared with high glucose in presence of 34.5 nm Rspo1.
EX4 Stimulates Rspo1 Expression in a Glucose-, Dose-, Time-, and PI3-kinase-dependent Manner
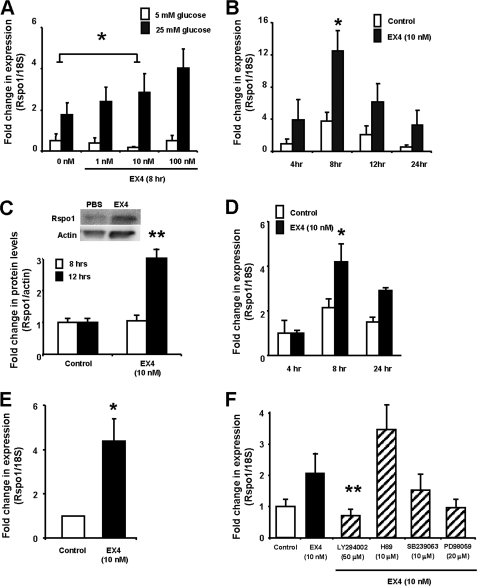
Finally, because β-cell behavior is regulated by both glucose and GLP-1, we determined whether Rspo1 is affected by these factors. Rspo1 mRNA levels were therefore examined in MIN6 cells treated for various times with either medium alone (control) or incremental doses of EX4 (1–100 nm) at either low (5 mm) or high (25 mm) glucose. Treatment with high glucose increased Rspo1 mRNA levels by 2-fold, while EX4 at 10 nm for 8 h induced a further increase in Rspo1 mRNA levels, an effect that was seen only under high glucose conditions (Fig. 6A, p < 0.05). A time course study demonstrated that Rspo1 mRNA levels peaked at 3-fold of control levels following 8 h of EX4 (10 nm) treatment with high glucose (Fig. 6B, p < 0.05) and returned back to basal levels at 12–24 h (Fig. 6B). Consistent with the mRNA findings, changes in Rspo1 protein levels were observed in response to EX4 treatment at 12 h but not at 8 h (Fig. 6C, p < 0.01). To determine if the observed effect of EX4 on Rspo1 mRNA is restricted to the MIN6 β-cells, the murine βTC β-cell line as well as isolated mouse islets were also tested. Treatment with EX4 at 10 nm stimulated Rspo1 mRNA by 4-fold at 8 h in the βTC cells (Fig. 6D, p < 0.05). A similar induction of Rspo1 mRNA was also observed in the isolated mouse islets (Fig. 6E, p < 0.05), albeit only at an earlier (i.e. 4 h) time point (preliminary 8 h and 12 h data not shown).
FIGURE 6.
Rspo1 is regulated by EX4 in the β-cell. A, MIN6 β-cells were treated with EX4 at indicated concentrations under low or high glucose conditions for 8 h. mRNA levels of Rspo1 were examined by relative qRT-PCR using 18 S as the internal control and then normalized to control (5 mm glucose without EX4 at the 0 h time point). B, MIN6 β-cells were incubated in high glucose conditions with medium alone (control) or EX4 for the indicated times. C, protein levels of Rspo1 and actin were determined by immunoblot of MIN6 β-cells treated with medium alone (control) or EX4 for 8 or 12 h. Optical densities of Rspo1 were normalized to that of pan-actin and were further normalized to their appropriate controls. A representative blot is shown for the 12-h time point. D, qRT-PCR for Rspo1 mRNA expression in βTC β-cells after incubation with medium alone (control) or EX4 for the indicated times. Relative expression values were normalized 18 S rRNA and then to the 4 h control group. E, qRT-PCR for Rspo1 mRNA levels in mouse islets after incubation with medium alone (control) or EX4 for 4 h. Relative expression values were normalized to the control group. F, MIN6 β-cells were treated with or without EX4 and with various inhibitors, as indicated, for 8 h. Relative expression values for Rspo1 were normalized to 18 S rRNA and then to the control treatment. *, p < 0.05; **, p < 0.01.
To delineate the mechanism of action whereby EX4 regulates Rspo1 mRNA expression, MIN6 cells were co-treated with EX4 and various inhibitors of the known GLP-1 receptor signaling pathway. Consistent with previous observations, EX4 treatment increased Rspo1 mRNA levels by 2-fold (Fig. 6F). Co-treatment with LY294002 (a PI3-kinase inhibitor) significantly attenuated Rspo1 mRNA expression (p < 0.01 versus EX4 alone), and a similar reduction was seen in cells cotreated with wortmannin (data not shown). In contrast, H89 (a PKA inhibitor), SB203580 (p38 MAPK inhibitor), and PD98059 and U0126 (MEK inhibitors) had no effect on EX4-induced Rspo1 mRNA levels. Treatment with each of these inhibitors alone did not alter Rspo1 mRNA levels (data not shown). These findings indicate that EX4 regulates Rspo1 mRNA expression in a PI3-kinase-dependent fashion.
DISCUSSION
Previous studies have demonstrated that the cWnt signaling pathway plays a crucial role in the maintenance of β-cell behavior. Recently, Rspo has been established as a novel family of secreted activators of cWnt signaling (13). Although Rspo1 has been detected in human pancreas (9), the effects of Rspo1 on the β-cell have not been explored. We now provide evidence that murine islets, MIN6 and βTC β-cells express Rspo1. We further found that Rspo1 activates cWnt signaling in the MIN6 β-cells, and that Rspo1 not only enhances β-cell growth and survival but is also an insulin secretagogue.
In the present study, we have found expression of Rspo1 in multiple β-cell models. It is interesting to note that the MIN6 and βTC β-cells as well as murine islets also expressed two other isoforms of Rspo (i.e. Rspo3 and Rspo4). However, βTC β-cells also expressed Rspo2, whereby this isoform was undetectable in the other models, raising the possibility that the βTC β-cell line may not be directly comparable to murine islets in vivo. Moreover, MIN6 β-cells were demonstrated to express high levels of Rspo1 mRNA, as well as Rspo1 protein, therefore serving as a useful model for the present investigation. The MIN6 β-cells were further found to express functional cWnt signaling, as indicated by expression of essential cWnt signaling molecules, as well as nuclear β-catenin translocation and cWnt target gene expression (i.e. c-myc and cyclin D1) in response to LiCl and Wnt3a respectively. In line with previous reports that Rspo1 can activate cWnt signaling (9, 11–13, 31), we also found that Rspo1 increased nuclear β-catenin as well as the cWnt target gene, c-myc in MIN6 β-cells. In contrast, we did not see any changes in expression levels of cyclin D1 after 12 h of treatment of Rspo1. This observation gives rise to the possibility of a differential responsiveness of these genes to Wnt3a and Rspo1 in the MIN6 β-cells, such as reported for their responses to other growth factors/hormones (e.g. estradiol versus insulin) (32).
β-Cell growth in vivo is determined by the rates of replication and apoptosis, as well as neogenesis (33). Several lines of evidence have established cWnt signaling as a pathway that regulates β-cell growth: 1) conditional pancreatic β-cell specific expression of degradation-resistant β-catenin leads to β-cell expansion, increased insulin production and serum levels, and enhanced glucose handling (7), and 2) endogenous Wnt3a is required for basal proliferation of INS-1 cells (8). Consistent with these findings, we found an enhancement of MIN6 β-cell proliferation in response to treatment with Wnt3a. Furthermore, Rspo1 was also found to induce significant growth of both the MIN6 cells and dispersed murine β-cells in vitro. Interestingly, the highest dose of Rspo1 tested did not stimulate any further proliferation in the MIN6 β-cells, and it remains possible that this cell line became desensitized by this recombinant protein. Further studies to examine the potential regulatory mechanisms induced by Rspo1 are crucial in understanding its role in cWnt signaling. Nevertheless, these findings are consistent with studies demonstrating that Rspo1 enhances intestinal growth through a cWnt-dependent pathway (9, 13).
Our finding that Rspo1 exerts proliferative effects on the β-cell prompted the question as to whether Rspo1 also protects the MIN6 β-cells from apoptosis. The cytotoxic effects of cytokines on β-cells have been demonstrated to include apoptosis, with caspase3 as the enzyme responsible for the features of cell death in this model (34). Consistent with previous results in INS-1E β-cells (14), we found that cytokine treatment increased cleaved caspase3 activity in the MIN6 β-cells, whereas treatment with EX4 decreased cytokine-induced caspase3 levels. However, in addition to proliferation, several downstream mediators of cWnt signaling has been found to regulate apoptosis in a variety of cell types, including β-cells (35–42). The present study shows, for the first time, that Rspo1 inhibits cytokine-induced apoptosis in the MIN6 β-cells. Consistent with this observation, we also report a parallel anti-apoptotic effect of Rspo1 in dispersed mouse β-cells treated with cytokines. Moreover, we found that the anti-apoptotic effect of Rspo1 in MIN6 β-cells is not further enhanced by the addition of the cWnt ligand, Wnt3a. However, given the possibility that one mechanism of action of Rspo1 involves enhanced Wnt ligand activity through stabilization of the Frz and LRP5/6 receptor complex (31), this observation does not preclude a requirement for endogenously secreted Wnt ligands for the actions of Rspo1. It remains possible that the MIN6 β-cells, like INS-1E β-cells (8), secrete endogenous Wnt ligands, in which case, further addition of the Wnt ligand my not be required for the anti-apoptotic effect of Rspo1.
To further establish the functional role of Rspo1 in regulating β-cell behavior, the effect of Rspo1 on insulin secretion was investigated. Although cWnt signaling molecules have been found to enhance insulin secretion from INS-1 β-cells (4, 5), the mechanism of action is unclear. Nonetheless, Fujino et al. (4) found impaired glucose-stimulated insulin secretion in LRP5 knock-out mice, in association with decreased expression of glucokinase. In this study, we demonstrated that Rspo1 enhances insulin secretion in MIN6 and dispersed mouse β-cells under acute conditions. Interestingly, Rspo1-induced insulin secretion in both MIN6 and dispersed β-cells was independent of glucose levels. Moreover, we also found that Rspo1 up-regulates insulin mRNA expression in vitro. In line with this observation, Loder et al. reported that silencing of TCF7L2, a crucial transcription factor in the cWnt signaling pathway, results in reduced levels of insulin mRNA (43). A recent study by da Silva et al. (44) has further demonstrated that the TCF7L2 gene is required for maintenance of β-cell genes regulating secretory granule fusion. It therefore remains possible that Rspo1 and the cWnt signaling pathway can also regulate insulin secretion chronically, at the level of insulin secretory granules. Given the importance of the TCF7L2 gene as a strong predictor for the development of T2DM (45), the regulation of insulin secretion and gene expression by Rspo1 warrants more detailed investigation.
β-Cell behavior is determined by many factors including glucose as a major regulator of insulin synthesis and release, as well as of β-cell mass (46–48). Furthermore, GLP-1 and its long-acting receptor agonist, EX4, have been well-characterized as both glucose-dependent insulin secretagogues and β-cell growth factors. We have now demonstrated that EX4 increases Rspo1 expression in the MIN6 β-cells in a dose- and time-dependent fashion and that this occurs only under high glucose conditions. This novel finding was not restricted to the MIN6 β-cell line, as similar effects of EX4 on Rspo1 mRNA were observed in mouse βTC cells, as well as in murine islets. Although interesting to note that glucose levels altered EX4-induced Rspo1 mRNA levels, this observation was not entirely surprising. Glucose regulation of β-cell behavior has been well-established and numerous studies have shown this nutrient can operate as a facilitator to enhance the actions of β-cell growth factors. Most notably, glucose confers β-cell responsivity by co-regulation with cAMP-increasing incretins such as GLP-1 that is especially vital for their mitogenic/anti-apoptotic actions (49–51).
Finally, the binding of GLP-1 or EX4 to the GLP-1R is known to stimulate adenylyl cyclase, leading to an increase in intracellular cAMP levels and activation of PKA (52–55). However, treatment with the PKA inhibitor H89 did not change basal or EX4-stimulated levels of Rspo1 mRNA, indicating that PKA is not required for this effect. GLP-1 has also been reported to stimulate a number of MAPK signaling pathways including ERK1/2 (56–60) and p38 MAPK (61, 62) to regulate β-cell behavior. Nonetheless, we found that inhibition of ERK1/2 with either PD98059 or U0126, or of p38 MAPK with SB203580, did not affect basal or EX4-induced changes in Rspo1 mRNA levels. In contrast, co-incubation of MIN6 β-cells with EX4 and the PI3-kinase inhibitors, LY294002 and wortmannin, abolished the EX4-induced increase in Rspo1 transcript levels. It is interesting to note that the PI3-kinase/Akt pathway appears to be involved in many pathways regulating β-cell behavior. Most notably, GLP-1 has been shown to exert its proliferative and anti-apoptotic effects in INS-1E cells via PI3-kinase/Akt, as these beneficial effects were abolished in the presence of wortmannin and by overexpression of a kinase-dead Akt construct (14, 29). Moreover, PI3-kinase γ knock-out mice demonstrate abnormal β-cell secretory responses that may involve downstream glucose-sensing pathways (63, 64). Our findings that EX4 regulates Rspo1 mRNA levels via a PI3-kinase-dependent pathway, therefore adds further evidence for a role of PI3-kinase in GLP-1 signaling in the β-cell.
Numerous investigations of transgenic mice expressing cWnt signaling molecules have provided clear evidence for the impact of cWnt signaling pathway in regulating β-cell biology. Our present data provides further support for this view by demonstrating, for the first time, the growth, survival and functional effects of Rspo1 on the β-cell. Studies of cWnt signaling in β-cells from insulin resistant or diabetic models have only been recently reported. Krützfeldt et al. (65) observed a relative increase in Wnt4, a specific inhibitor of cWnt signaling, in islets of insulin-resistant mice. Alternatively, Lee et al. (66) reported up-regulation of several cWnt signaling molecules, including β-catenin, TCF7L2, and cWnt ligand Wnt2b in islets from subjects with T2DM. The results of these studies suggest that cWnt signaling can be altered in insulin resistant and/or diabetic states. There are currently no reports to-date to examining the β-cell responses to and/or secretion of Rspo1 under such pathophysiological conditions. Future research in this area will therefore be important if Rspo1 is to be considered as a novel target for the therapeutic treatment of patients with T2DM.
Acknowledgments
We thank Drs. J. Miyazaki (University of Tokyo) and D. F. Steiner (University of Chicago) for the gift of MIN6 β-cells, and Angelo Izzo (University of Toronto) for technical expertise in mouse islet preparation.
This work was supported by an operating grant from the Canadian Diabetes Association (2374) and by an equipment grant from the Banting and Best Diabetes Centre (BBDC), University of Toronto.
- T2DM
- type 2 diabetes mellitus
- PI
- phosphatidylinositol
- FBS
- fetal bovine serum
- PBS
- phosphate-buffered saline
- TUNEL
- terminal deoxynucleotidyltransferase-mediated dUTP nick end-labeling
- Rspo
- R-spondin
- Frz
- Frizzled
- cWnt
- canonical Wnt
- LRP
- low density lipoprotein receptor-related protein
- qRT-PCR
- quantitative reverse transcription-PCR
- APC
- adenomatosis polyposis coli.
REFERENCES
- 1.Butler A. E., Janson J., Bonner-Weir S., Ritzel R., Rizza R. A., Butler P. C. (2003) Diabetes 52, 102–110 [DOI] [PubMed] [Google Scholar]
- 2.Welters H. J., Kulkarni R. N. (2008) Trends Endocrinol. Metab. 19, 349–355 [DOI] [PubMed] [Google Scholar]
- 3.Nusse R. (2005) Cell Res. 15, 28–32 [DOI] [PubMed] [Google Scholar]
- 4.Fujino T., Asaba H., Kang M. J., Ikeda Y., Sone H., Takada S., Kim D. H., Ioka R. X., Ono M., Tomoyori H., Okubo M., Murase T., Kamataki A., Yamamoto J., Magoori K., Takahashi S., Miyamoto Y., Oishi H., Nose M., Okazaki M., Usui S., Imaizumi K., Yanagisawa M., Sakai J., Yamamoto T. T. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 229–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schinner S., Ulgen F., Papewalis C., Schott M., Woelk A., Vidal-Puig A., Scherbaum W. A. (2008) Diabetologia 51, 147–154 [DOI] [PubMed] [Google Scholar]
- 6.Papadopoulou S., Edlund H. (2005) Diabetes 54, 2844–2851 [DOI] [PubMed] [Google Scholar]
- 7.Rulifson I. C., Karnik S. K., Heiser P. W., ten Berge D., Chen H., Gu X., Taketo M. M., Nusse R., Hebrok M., Kim S. K. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 6247–6252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Z., Habener J. F. (2008) J. Biol. Chem. 283, 8723–8735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim K. A., Kakitani M., Zhao J., Oshima T., Tang T., Binnerts M., Liu Y., Boyle B., Park E., Emtage P., Funk W. D., Tomizuka K. (2005) Science 309, 1256–1259 [DOI] [PubMed] [Google Scholar]
- 10.Nam J. S., Turcotte T. J., Smith P. F., Choi S., Yoon J. K. (2006) J. Biol. Chem. 281, 13247–13257 [DOI] [PubMed] [Google Scholar]
- 11.Wei Q., Yokota C., Semenov M. V., Doble B., Woodgett J., He X. (2007) J. Biol. Chem. 282, 15903–15911 [DOI] [PubMed] [Google Scholar]
- 12.Kim K. A., Wagle M., Tran K., Zhan X., Dixon M. A., Liu S., Gros D., Korver W., Yonkovich S., Tomasevic N., Binnerts M., Abo A. (2008) Mol. Biol. Cell 19, 2588–2596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim K. A., Zhao J., Andarmani S., Kakitani M., Oshima T., Binnerts M. E., Abo A., Tomizuka K., Funk W. D. (2006) Cell Cycle 5, 23–26 [DOI] [PubMed] [Google Scholar]
- 14.Li L., El-Kholy W., Rhodes C. J., Brubaker P. L. (2005) Diabetologia 48, 1339–1349 [DOI] [PubMed] [Google Scholar]
- 15.Gyulkhandanyan A. V., Lee S. C., Bikopoulos G., Dai F., Wheeler M. B. (2006) J. Biol. Chem. 281, 9361–9372 [DOI] [PubMed] [Google Scholar]
- 16.Mohamed O. A., Dufort D., Clarke H. J. (2004) Biol. Reprod. 71, 417–424 [DOI] [PubMed] [Google Scholar]
- 17.Heller R. S., Dichmann D. S., Jensen J., Miller C., Wong G., Madsen O. D., Serup P. (2002) Dev. Dyn. 225, 260–270 [DOI] [PubMed] [Google Scholar]
- 18.Rhee C. S., Sen M., Lu D., Wu C., Leoni L., Rubin J., Corr M., Carson D. A. (2002) Oncogene 21, 6598–6605 [DOI] [PubMed] [Google Scholar]
- 19.Chen Y., Stump R. J., Lovicu F. J., McAvoy J. W. (2004) Int. J. Dev. Biol. 48, 867–877 [DOI] [PubMed] [Google Scholar]
- 20.Ranheim E. A., Kwan H. C., Reya T., Wang Y. K., Weissman I. L., Francke U. (2005) Blood 105, 2487–2494 [DOI] [PubMed] [Google Scholar]
- 21.Stump R. J., Ang S., Chen Y., von Bahr T., Lovicu F. J., Pinson K., de Iongh R. U., Yamaguchi T. P., Sassoon D. A., McAvoy J. W. (2003) Dev. Biol. 259, 48–61 [DOI] [PubMed] [Google Scholar]
- 22.Lyu J., Costantini F., Jho E. H., Joo C. K. (2003) J. Biol. Chem. 278, 13487–13495 [DOI] [PubMed] [Google Scholar]
- 23.Jho E. H., Zhang T., Domon C., Joo C. K., Freund J. N., Costantini F. (2002) Mol. Cell. Biol. 22, 1172–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Etheridge S. L., Spencer G. J., Heath D. J., Genever P. G. (2004) Stem Cells 22, 849–860 [DOI] [PubMed] [Google Scholar]
- 25.Nam J. S., Turcotte T. J., Yoon J. K. (2007) Gene Expr. Patterns 7, 306–312 [DOI] [PubMed] [Google Scholar]
- 26.Pfaffl M. W. (2001) Nucleic Acids Res. 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Everly D. N., Jr., Kusano S., Raab-Traub N. (2004) J. Virol. 78, 11648–11655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meares G. P., Jope R. S. (2007) J. Biol. Chem. 282, 16989–17001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Q., Li L., Xu E., Wong V., Rhodes C., Brubaker P. L. (2004) Diabetologia 47, 478–487 [DOI] [PubMed] [Google Scholar]
- 30.MacDonald P. E., Ha X. F., Wang J., Smukler S. R., Sun A. M., Gaisano H. Y., Salapatek A. M., Backx P. H., Wheeler M. B. (2001) Mol. Endocrinol. 15, 1423–1435 [DOI] [PubMed] [Google Scholar]
- 31.Binnerts M. E., Kim K. A., Bright J. M., Patel S. M., Tran K., Zhou M., Leung J. M., Liu Y., Lomas W. E., 3rd, Dixon M., Hazell S. A., Wagle M., Nie W. S., Tomasevic N., Williams J., Zhan X., Levy M. D., Funk W. D., Abo A. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 14700–14705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mawson A., Lai A., Carroll J. S., Sergio C. M., Mitchell C. J., Sarcevic B. (2005) Mol. Cell. Endocrinol. 229, 161–173 [DOI] [PubMed] [Google Scholar]
- 33.Bonner-Weir S. (2000) Endocrinology 141, 1926–1929 [DOI] [PubMed] [Google Scholar]
- 34.Hengartner M. O. (2000) Nature 407, 770–776 [DOI] [PubMed] [Google Scholar]
- 35.Morin P. J., Vogelstein B., Kinzler K. W. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 7950–7954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He T. C., Chan T. A., Vogelstein B., Kinzler K. W. (1999) Cell 99, 335–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Orford K., Orford C. C., Byers S. W. (1999) J. Cell Biol. 146, 855–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Strovel E. T., Sussman D. J. (1999) Exp. Cell Res. 253, 637–648 [DOI] [PubMed] [Google Scholar]
- 39.Yun S. I., Yoon H. Y., Chung Y. S. (2009) Apoptosis. 14, 771–777 [DOI] [PubMed] [Google Scholar]
- 40.Torii K., Nishizawa K., Kawasaki A., Yamashita Y., Katada M., Ito M., Nishimoto I., Terashita K., Aiso S., Matsuoka M. (2008) Cell Signal. 20, 1256–1266 [DOI] [PubMed] [Google Scholar]
- 41.Vuga L. J., Ben-Yehudah A., Kovkarova-Naumovski E., Oriss T., Gibson K. F., Feghali-Bostwick C., Kaminski N. (2009) Am. J. Respir. Cell Mol. Biol. 41, 583–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Z., Habener J. F. (2009) Diabetologia 52, 1589–1598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Loder M. K., da Silva X. G., McDonald A., Rutter G. A. (2008) Biochem. Soc. Trans. 36, 357–359 [DOI] [PubMed] [Google Scholar]
- 44.da Silva X. G., Loder M. K., McDonald A., Tarasov A. I., Carzaniga R., Kronenberger K., Barg S., Rutter G. A. (2009) Diabetes 58, 894–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grant S. F., Thorleifsson G., Reynisdottir I., Benediktsson R., Manolescu A., Sainz J., Helgason A., Stefansson H., Emilsson V., Helgadottir A., Styrkarsdottir U., Magnusson K. P., Walters G. B., Palsdottir E., Jonsdottir T., Gudmundsdottir T., Gylfason A., Saemundsdottir J., Wilensky R. L., Reilly M. P., Rader D. J., Bagger Y., Christiansen C., Gudnason V., Sigurdsson G., Thorsteinsdottir U., Gulcher J. R., Kong A., Stefansson K. (2006) Nat. Genet. 38, 320–323 [DOI] [PubMed] [Google Scholar]
- 46.Bouwens L., Rooman I. (2005) Physiol. Rev. 85, 1255–1270 [DOI] [PubMed] [Google Scholar]
- 47.Vasavada R. C., Gonzalez-Pertusa J. A., Fujinaka Y., Fiaschi-Taesch N., Cozar-Castellano I., Garcia-Ocaña A. (2006) Int. J. Biochem. Cell Biol. 38, 931–950 [DOI] [PubMed] [Google Scholar]
- 48.Martens G. A., Pipeleers D. (2009) Vitam. Horm. 80, 507–539 [DOI] [PubMed] [Google Scholar]
- 49.Costes S., Broca C., Bertrand G., Lajoix A. D., Bataille D., Bockaert J., Dalle S. (2006) Diabetes 55, 2220–2230 [DOI] [PubMed] [Google Scholar]
- 50.Hussain M. A., Porras D. L., Rowe M. H., West J. R., Song W. J., Schreiber W. E., Wondisford F. E. (2006) Mol. Cell. Biol. 26, 7747–7759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jhala U. S., Canettieri G., Screaton R. A., Kulkarni R. N., Krajewski S., Reed J., Walker J., Lin X., White M., Montminy M. (2003) Genes Dev. 17, 1575–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ding W. G., Renström E., Rorsman P., Buschard K., Gromada J. (1997) Diabetes 46, 792–800 [DOI] [PubMed] [Google Scholar]
- 53.Lester L. B., Langeberg L. K., Scott J. D. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 14942–14947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dyachok O., Isakov Y., Sågetorp J., Tengholm A. (2006) Nature 439, 349–352 [DOI] [PubMed] [Google Scholar]
- 55.Wang X., Zhou J., Doyle M. E., Egan J. M. (2001) Endocrinology 142, 1820–1827 [DOI] [PubMed] [Google Scholar]
- 56.Briaud I., Lingohr M. K., Dickson L. M., Wrede C. E., Rhodes C. J. (2003) Diabetes 52, 974–983 [DOI] [PubMed] [Google Scholar]
- 57.Arnette D., Gibson T. B., Lawrence M. C., January B., Khoo S., McGlynn K., Vanderbilt C. A., Cobb M. H. (2003) J. Biol. Chem. 278, 32517–32525 [DOI] [PubMed] [Google Scholar]
- 58.Gomez E., Pritchard C., Herbert T. P. (2002) J. Biol. Chem. 277, 48146–48151 [DOI] [PubMed] [Google Scholar]
- 59.Trümper J., Ross D., Jahr H., Brendel M. D., Göke R., Hörsch D. (2005) Diabetologia 48, 1534–1540 [DOI] [PubMed] [Google Scholar]
- 60.Park S., Dong X., Fisher T. L., Dunn S., Omer A. K., Weir G., White M. F. (2006) J. Biol. Chem. 281, 1159–1168 [DOI] [PubMed] [Google Scholar]
- 61.Friedrichsen B. N., Neubauer N., Lee Y. C., Gram V. K., Blume N., Petersen J. S., Nielsen J. H., Møldrup A. (2006) J. Endocrinol. 188, 481–492 [DOI] [PubMed] [Google Scholar]
- 62.Kemp D. M., Habener J. F. (2001) Endocrinology 142, 1179–1187 [DOI] [PubMed] [Google Scholar]
- 63.Li L. X., MacDonald P. E., Ahn D. S., Oudit G. Y., Backx P. H., Brubaker P. L. (2006) Endocrinology 147, 3318–3325 [DOI] [PubMed] [Google Scholar]
- 64.Pigeau G. M., Kolic J., Ball B. J., Hoppa M. B., Wang Y. W., Ruckle T., Woo M., Manning Fox J. E., MacDonald P. E. (2009) Diabetes 58, 2084–2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Krützfeldt J., Stoffel M. (2010) Diabetologia 53, 123–127 [DOI] [PubMed] [Google Scholar]
- 66.Lee S. H., Demeterco C., Geron I., Abrahamsson A., Levine F., Itkin-Ansari P. (2008) Exp. Diabetes Res. 2008, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]