Abstract
The Bcr-Abl kinase inhibitor imatinib is remarkably effective in chronic myelogenous leukemia (CML), although drug resistance is an emerging problem. Myeloid Src family kinases such as Hck and Lyn are often overexpressed in imatinib-resistant CML cells that lack Bcr-Abl mutations. Here we tested whether Hck overexpression is sufficient to induce imatinib resistance using both wild-type Hck and a mutant (Hck-T338A) that is uniquely sensitive to the pyrazolo-pyrimidine inhibitor, NaPP1. Expression of either kinase in K562 CML cells caused resistance to imatinib-induced apoptosis and inhibition of soft-agar colony formation. Treatment with NaPP1 restored sensitivity to imatinib in cells expressing T338A but not wild-type Hck, demonstrating that resistance requires Hck kinase activity. NaPP1 also reduced Hck-mediated phosphorylation of Bcr-Abl at sites that may affect imatinib sensitivity exclusively in cells expressing Hck-T338A. These data show that elevated Src family kinase activity is sufficient to induce imatinib resistance through a mechanism that may involve phosphorylation of Bcr-Abl.
Keywords: Drug Resistance, Leukemia, Myeloid Cell, Src, Tyrosine-protein Kinase (Tyrosine Kinase), Hck, Chemical Genetics, Gatekeeper Mutation
Introduction
Chronic myelogenous leukemia (CML)2 is a hematopoietic stem cell disease with three clinical phases. The initial chronic phase involves expansion of myeloid cells that retain their ability to undergo terminal differentiation. As the disease progresses, patients enter an accelerated phase followed by a blast crisis characterized by differentiation arrest and accumulation of immature blast cells in the bone marrow and peripheral blood (1).
The cytogenetic hallmark of CML is the Philadelphia chromosome, which fuses the BCR (breakpoint cluster region) locus on chromosome 22 with the c-ABL proto-oncogene on chromosome 9. This translocation is present in greater than 90% of CML patients and leads to the expression of Bcr-Abl, a chimeric protein of 210 kDa (2, 3) with abnormal subcellular localization and constitutive protein-tyrosine kinase activity (4, 5). Bcr-Abl drives the pathogenesis of CML through the phosphorylation and activation of a broad range of downstream signaling proteins that increase cell survival and promote unregulated cell cycle progression (6). These pathways include but are not limited to the Ras/mitogen-activated protein kinase (MAPK), NF-κB, phosphatidylinositol 3-kinase/Akt, and Stat signaling cascades (7–9).
Bcr-Abl has been shown to activate other non-receptor protein-tyrosine kinases, particularly Src family kinases (SFKs) expressed in myeloid cells such as Hck and Lyn (10). A growing body of evidence supports the relevance of this interaction to CML pathogenesis. For example, expression of a kinase-defective mutant of Hck blocked Bcr-Abl-induced transformation of murine myeloid leukemia cells to cytokine independence (11). In addition, Hck was shown to couple Bcr-Abl to Stat5 signaling and to be required for Bcr-Abl-induced transformation of murine myeloid cells (12). Furthermore, SFK-selective inhibitors block proliferation and induce apoptosis in CML cells without affecting Philadelphia chromosome-negative myeloid leukemia cells (13). More recently, Hck has been shown to play a non-redundant role in Bcr-Abl survival signaling in CML cells (14). Taken together, these findings indicate that SFKs act as essential mediators of Bcr-Abl-induced leukemogenesis.
Because Bcr-Abl plays a critical role in the initiation and maintenance of the CML phenotype, targeting its tyrosine kinase activity is the therapeutic strategy of choice. Imatinib mesylate is a potent small molecule tyrosine kinase inhibitor relatively specific to Bcr-Abl that has become the frontline therapy for patients with CML (6, 15). Most CML patients in the chronic phase achieve and maintain significant hematologic and cytogenetic responses to imatinib treatment (16, 17). However, ∼4% of chronic phase patients treated with imatinib develop drug resistance per year. In addition, accelerated or blast crisis patients display higher rates of imatinib resistance (18, 19). Clinical resistance to imatinib can result from mutations in the Abl kinase domain at residues that directly contact imatinib or at positions that allosterically influence imatinib binding (20). Resistance can also arise in the absence of Bcr-Abl mutations. The present study is focused on the role of SFKs in Bcr-Abl mutation-independent imatinib resistance.
Overexpression of the myeloid SFKs Hck and Lyn has been associated with resistance to imatinib in the absence of Bcr-Abl kinase domain mutations. Selection of the CML cell line K562 for resistance to high levels of imatinib increased Lyn protein and activity levels (21). Exposure of these cells to imatinib resulted in an incomplete suppression of Bcr-Abl activity and was accompanied by persistent tyrosine phosphorylation of Bcr-Abl at Tyr-177, a binding site for Grb2 that links Bcr-Abl to the Ras signaling cascade (22, 23). Samples from patients after imatinib failure exhibited increased Hck and/or Lyn activity levels that were not affected by Bcr-Abl inhibition (21). In addition, no Bcr-Abl kinase domain mutations associated with imatinib resistance was present in these patients. However, small interfering RNA-mediated suppression of Lyn expression or treatment with dasatinib, a dual Abl/SFK inhibitor, induced apoptosis in these imatinib-resistant cells (24). Collectively, these findings point to a role for both Hck and Lyn in imatinib resistance in the absence of Bcr-Abl mutations. Although the role of Lyn has been pursued in some detail, the contribution of Hck to this type of imatinib resistance is less well understood.
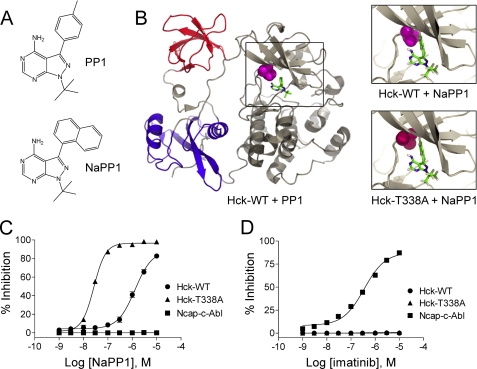
To address this question, we developed a system in which Hck activity can be inhibited in a highly specific manner. Typically, small molecule kinase inhibitors exhibit limited specificity, especially for individual members of a conserved kinase family. To circumvent this issue, we employed a chemical genetic approach originally described by Shokat and co-workers (for review, see Ref. 25). In this method, pyrazolopyrimidine 1 (PP1), a relatively potent yet non-selective SFK inhibitor, was modified to obtain a more bulky analog, naphthyl-pyrazolopyrimidine 1 (NaPP1), that is far less potent toward wild-type SFKs than the parent compound (Fig. 1A). Using site-directed mutagenesis, the ATP binding sites of SFKs were then modified to accommodate the bulkier naphthyl moiety of the NaPP1 molecule (25, 26). The resulting combination of inhibitor analog and mutant kinase allows for highly selective inhibition of the modified kinase without affecting the corresponding endogenous kinase or related kinases.
FIGURE 1.
Generation of NaPP1-sensitive Hck. A, structures of the non-selective SFK inhibitor PP1 and the bulky inactive analog, NaPP1 are shown. B, modeling of NaPP1 in the active site of wild-type Hck and the gatekeeper mutant, T338A is shown. The overall structure of Hck is shown on the left, with the SH3 domain in red, SH2 domain in blue, and the kinase domain in gray. The side chain of the gatekeeper residue (Thr-338) is highlighted in magenta, and its relationship to PP1 is shown in the boxed area. The model is based on the crystallographic coordinates of Schindler et al. (30) (PDB code 1QCF). Upper right, the spatial coordinates of the PP1 pyrazolo-pyrimidine were used to model the position of NaPP1 within the ATP binding site. This close-up view shows the clash of the naphthyl ring of NaPP1 with the side chain of the gatekeeper threonine. WT, wild type. Lower right, the T338A mutation was modeled in the Hck structure with the alanine side chain highlighted in red. This substitution creates a space that accommodates the naphthyl moiety of NaPP1 and sensitizes the kinase to this modified inhibitor. C and D, the Hck-T338A mutant is sensitive to NaPP1 but not to imatinib in an in vitro kinase assay. Recombinant wild-type Hck-YEEI (WT), Hck-T338A-YEEI, and Ncap-c-Abl were purified from Sf9 insect cells, and kinase activity was assessed in vitro using a fluorescence resonance energy transfer-based assay with a peptide substrate. Concentration-response curves are shown in the presence of NaPP1 (C) or imatinib (D). Percent inhibition is expressed as the mean ± S.D. from the results of four assay wells per condition. The entire experiment was repeated twice and produced comparable results; a representative example is shown.
In this study we used the chemical genetic principle described above to successfully engineer a Hck mutant (Hck-T338A) that is uniquely sensitive to NaPP1 inhibition. The validity of the mutant was confirmed in vitro using recombinant purified kinases as well as a Rat-2 fibroblast transformation assay. We then used this Hck mutant to address the role of Hck overexpression in imatinib resistance. Wild-type Hck and the T338A mutant were expressed in the CML cell line K562, which was derived from an erythroid blast crisis patient and does not express endogenous Hck (14). Remarkably, expression of either wild-type Hck or the T338A mutant rendered K562 cells resistant to imatinib-induced inhibition of colony formation and apoptosis. This effect correlated with sustained phosphorylation of Bcr-Abl at several regulatory tyrosine residues in the presence of imatinib. These findings suggest that Hck overexpression promotes Bcr-Abl activation, which correlates inversely with inhibition by imatinib (27). Furthermore, NaPP1 treatment restored imatinib sensitivity to cells expressing the Hck-T338A mutant but not the wild-type kinase. This effect correlated with inhibition of Hck-T338A activity and reduction of site-specific Bcr-Abl tyrosine phosphorylation. In contrast, NaPP1 did not affect cells expressing wild-type Hck or control cells. Taken together, these results establish the first causal relationship between Hck kinase activity and Bcr-Abl mutation-independent imatinib resistance.
EXPERIMENTAL PROCEDURES
Cell Culture
Sf9 insect cells were cultured in Grace's medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS) and 50 μg/ml gentamicin (Invitrogen). The K562 myeloid leukemia cell line, derived from a CML patient in erythroid blast crisis (28), was obtained from the ATCC and maintained in RPMI 1640 supplemented with 10% FBS (Atlanta Biologicals) and 0.25 μg/ml Antibiotic-antimycotic (Invitrogen). Rat-2 fibroblasts were purchased from the ATCC and maintained in Dulbecco's modified Eagle's medium containing 5% FBS, 100 units/ml penicillin G, 100 μg/ml streptomycin, and 0.25 μg/ml Antibiotic-antimycotic.
Protein Purification
Using site-directed mutagenesis, the Thr-338 to Ala (T338A) mutation was introduced into the coding sequence of human p59 Hck-YEEI (QuikChange XL site-directed mutagenesis kit, Stratagene) (29). The Hck-YEEI and Hck-T338A-YEEI constructs also contained N-terminal hexahistidine tags in place of the unique domain (30, 31). The Ncap-c-Abl construct encompasses residues 1–531 of human c-Abl-1b with residues 15–56 deleted and contains a C-terminal cleavage site for the tobacco etch virus (TEV) protease and a hexahistidine tag, both introduced by PCR (32, 33). Hck-YEEI, Hck-T338A-YEEI, and the Ncap-c-Abl constructs were cloned into pVL1392 (BD Biosciences), and each plasmid was used to create high titer recombinant baculovirus in Sf9 insect cells using BaculoGold DNA and the manufacturer's protocol (BD Biosciences). For protein production, Sf9 cells (1 liter) were cultured to a density of 2 × 106 cells/ml and then infected with either the Hck-YEEI or Hck-T338A-YEEI baculoviruses. For the Ncap-c-Abl construct, Sf9 cells were co-infected with Abl and YopH phosphatase baculoviruses at a multiplicity of infection of 10. YopH is a protein-tyrosine phosphatase that promotes a down-regulated conformation of Ncap-c-Abl, permitting high yield purification from Sf9 cells (32). Sf9 cells were grown for 48 h, centrifuged, washed in PBS, and then the pellets were resuspended in buffer A (20 mm Tris-HCl (pH 8.3), 10% glycerol, and 5 mm 2-mercaptoethanol), lysed by sonication, and centrifuged at 16,000 rpm for 30 min. The recombinant Hck or Ncap-c-Abl proteins were purified from the supernatant using a combination of ion exchange and affinity chromatography as originally described by Schindler et al. for Hck-YEEI (30). Upon purification, the proteins were dialyzed against 20 mm Tris-HCl (pH 8.3) containing 100 mm NaCl and 3 mm dithiothreitol.
In Vitro Kinase Assay
Kinase assays were performed using the fluorescence resonance energy transfer-based Z'-Lyte Src kinase assay kit and Tyr-2 peptide substrate according to the manufacturer's instructions (Invitrogen). All assays were performed in quadruplicate in low volume, non-binding 384-well plates (Corning). The assay was first optimized to determine the amount of each kinase and the incubation time necessary to phosphorylate 50–60% of the Tyr-2 peptide in the absence of inhibitor. Final ATP and Tyr-2 substrate concentrations were held constant at 50 and 2 μm, respectively. For inhibition experiments, the kinases were preincubated with NaPP1 or imatinib in kinase assay buffer for 30 min followed by incubation with ATP and Tyr-2 peptide for 1 h. Fluorescence was assessed on a Gemini XS microplate spectrofluorometer (Molecular Devices). IC50 values were calculated from the means of four wells using a sigmoidal curve fit and Prism software (GraphPad Software, Inc.).
Retroviral Transduction of Rat-2 Fibroblasts and K562 Cells
Using site-directed mutagenesis, an active form of Hck was constructed by replacing the negative regulatory tail tyrosine residue with phenylalanine (Hck-YF) (34). The wild-type Hck, Hck-T338A, Hck-YF, and Hck-T338A-YF constructs were subcloned into the retroviral vector pSRαMSVtkneo (35) and used to generate high titer retroviral stocks by co-transfection of 293T cells with an ecotropic packaging vector (29, 34, 36, 37). Control retroviruses were prepared using the empty parent vector. Rat-2 fibroblasts were infected as follows; 2.5 × 104 Rat-2 cells were plated per well in 6-well plates and incubated with viral stocks in a final volume of 5 ml in the presence of Polybrene (4 μg/ml final). To enhance infection efficiency, the plates were centrifuged at 3000 rpm for 4 h at 18 °C. After infection, the virus was replaced with fresh medium. G418 selection (800 μg/ml) was started 48 h after infection and continued for 14 days. At the end of the selection period, the G418 concentration was decreased to 400 μg/ml.
For K562 cells, wild-type Hck and the Hck-T338A mutant were subcloned into the retroviral expression vector pMSCV-IRES-neo (Clontech) between the MSCV promoter and internal ribosome entry site-neo sequence. Retroviral stocks were produced from the resulting constructs in 293T cells using an amphotropic packaging vector as described above for Rat-2 fibroblasts. K562 cells were plated in 6-well plates at 1 × 106 cells/well in 5 ml of undiluted viral supernatant in the presence of 4 μg/ml Polybrene and centrifuged at 3000 rpm for 3 h at room temperature. After infection, cells were washed, returned to regular medium for 48 h, and then placed under G418 selection (800 μg/ml) for 14 days. At the end of the selection period, cells were maintained in medium with 400 μg/ml G418.
Soft Agar Colony Assay
Rat-2 fibroblast or K562 cell colony-forming activity was assayed in triplicate 35-mm Petri dishes (Falcon) using Seaplaque-agarose (FMC Bioproducts). One ml of 0.5% bottom agarose layer in complete culture medium was poured in the presence of either vehicle (0.5% DMSO) or NaPP1/imatinib at twice the final desired concentration. After the hardening of the bottom layer, 1 × 104 Rat-2 cells or 2 × 103 K562 cells were mixed in culture medium containing 0.3% agarose, and 1 ml was added to the plates. 7–10 days later, the colonies were stained with 3-(4,5-dimethyl-thiazol-2-yl)-2,5-diphenyltetrazolium bromide and counted from scanned images using colony counting software (Bio-Rad QuantityOne).
Apoptosis Assay
Apoptosis was determined by measuring cell-surface phosphatidylserine using an Alexa Fluor 488-conjugated anti-phosphatidylserine antibody (Upstate Biotechnology) and flow cytometry. Cells (1 × 105/ml) were treated with vehicle alone (0.5% DMSO), imatinib, or combinations of NaPP1 and imatinib for 72 h at 37 °C. After incubation, cells were centrifuged at 1000 rpm for 5 min, washed 3 times with ice-cold PBS, and resuspended to 4 × 106 cells/ml in staining buffer (1% FBS in PBS). Aliquots (50 μl) were transferred to 96-well round-bottom tissue culture plates, mixed with the anti-phosphatidylserine antibody (0.21 μg/well), and incubated on ice for 1 h. Cells were washed three times in ice-cold PBS and analyzed using a FACSCalibur flow cytometer (BD Biosciences) and CellQuest software.
Antibodies
The following antibodies were used in this study: anti-actin (MAB1501; Chemicon), anti-Hck (N-30 polyclonal, Santa Cruz Biotechnology; monoclonal, Transduction Laboratories), anti-Lyn (Santa Cruz), anti-c-Abl (Calbiochem), anti-Src phosphospecific (Src phospho-Tyr-416; Upstate Biotechnology), anti-Bcr phospho-Tyr-177 (Cell Signaling), anti-c-Abl phospho-Tyr-89 (Cell Signaling), anti-c-Abl phospho-Tyr-245 (Cell Signaling), and anti-phosphotyrosine (PY99; Santa Cruz).
Immunoprecipitation and Immunoblotting
K562 cells (5 × 106) were incubated overnight in 0.5% FBS and then treated for 5 h with vehicle control (DMSO, 0.5%) or with imatinib and/or NaPP1. At the end of the incubation period, the cells were collected by centrifugation, washed twice with PBS, and lysed in ice-cold radioimmune precipitation assay buffer (34). For Hck or Lyn immunoprecipitation, total protein concentrations were first normalized with lysis buffer followed by the addition of 1 μg of anti-Hck or anti-Lyn antibody and 25 μl of protein G-Sepharose (50% slurry; GE Healthcare). After incubation for 2 h at 4 °C, immunoprecipitates were washed 3 times with 1.0 ml of radioimmune precipitation assay buffer and heated in SDS sample buffer. After SDS-PAGE, proteins were transferred to polyvinylidene difluoride or nitrocellulose membranes and incubated with the indicated primary antibodies. Immunoreactive proteins were visualized with appropriate secondary antibody-alkaline phosphatase conjugates and nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate colorimetric substrate (Sigma). For quantitative immunoblotting, the membranes were incubated with IRdye680- or IRdye800CW-conjugated secondary antibodies (LI-COR, Lincoln, NE) and scanned with the Odyssey infrared imaging system (LI-COR). Protein band intensities were quantitated using the Odyssey software.
Statistical Analysis
Data obtained from multiple independent experiments are reported as the mean ± S.D. values. Statistical tests were conducted using Student's t test or analysis of variance (ANOVA) where indicated. Statistical tests were performed using GraphPad Prism, Version 3.03.
RESULTS
Design of an Inhibitor Analog-sensitive Hck Allele
Crystal structures of various protein-tyrosine kinases have shown that a small hydrophobic pocket lies adjacent to the ATP binding site, access to which is controlled by a “gatekeeper residue.” The side chain of the gatekeeper (typically a bulky residue such as threonine, methionine, isoleucine, or phenylalanine) sterically hinders the access of many small molecule inhibitors to this pocket (38, 39). Based on these observations, several groups have successfully applied a kinase inhibitor sensitization approach to a number of protein kinase families by replacing the gatekeeper residue with a much smaller residue such as glycine or alanine (25, 40, 41). In particular, replacement of the v-Src gatekeeper (Ile-338; numbering is based on the crystal structure of human c-Src (42)) with either glycine or alanine rendered it susceptible to inhibition by NaPP1 (43), suggesting that this approach would work for Hck as well. Fig. 1B shows the crystal structure of Hck (PDB code 1QCF) in which NaPP1 was overlaid onto the crystal coordinates of PP1. Analysis of this structure reveals potential steric clash between the Hck gatekeeper (Thr-338) and the naphthyl moiety of NaPP1. Replacement of Thr-338 with Ala generates more space in the active site, allowing NaPP1 access to the hydrophobic pocket. We hypothesized that the T338A mutation would sensitize Hck to NaPP1 without disrupting normal kinase activity or signaling.
Replacement of Thr-338 with Ala Renders Hck Sensitive to NaPP1
To test whether the T338A mutation induces sensitivity to the bulky PP1 analog, NaPP1, the wild-type, and T338A forms of Hck were expressed as recombinant proteins in Sf9 insect cells and purified to homogeneity. To facilitate the purification of these proteins in the down-regulated conformation, we modified the natural C-terminal tail sequence, YQQQP, to YEEIP (referred to hereafter as “Hck-YEEI”). The YEEI modification promotes autophosphorylation of the tail tyrosine in the absence of Csk and increases affinity for the SH2 domain (30, 44, 45). Thus, the YEEI tail variant of Hck does not require Csk to adopt the down-regulated conformation, enabling high yield purification from Sf9 cells without the need for Csk co-expression (30, 31, 45). NaPP1 sensitivity of recombinant purified Hck-YEEI and Hck-T338A-YEEI protein kinases was then assayed using an in vitro kinase assay (Z'-Lyte) and a fluorescence resonance energy transfer-peptide substrate (a detailed explanation of the Z'-Lyte assay and the calculation of percent inhibition is provided in supplemental Fig. S1.) As shown in Fig. 1C, NaPP1 potently inhibited the Hck-T338A-YEEI mutant with an IC50 of 26.6 ± 1.26 nm. In contrast, the NaPP1 IC50 for Hck-YEEI was almost 50-fold higher (1.3 ± 0.39 μm). Importantly, NaPP1 did not inhibit a recombinant purified form of the c-Abl tyrosine kinase core consisting of the N-terminal cap (Ncap) region, the SH3 domain, the SH2 domain, and the kinase domain (referred to hereafter as “Ncap-c-Abl”) (33).
One possible issue concerning the introduction of the gatekeeper mutation in the ATP binding site of Hck is cross-sensitization to imatinib. To rule out this possibility, we also tested Hck-T338A sensitivity to imatinib in the in vitro kinase assay using the same set of purified recombinant kinases. As shown in Fig. 1D, imatinib did not inhibit wild-type Hck or the T338A mutant, further supporting the validity of this approach. In contrast, imatinib inhibited Ncap-c-Abl with an IC50 of 350 ± 53 nm, consistent with previous work (46).
NaPP1 Potently and Selectively Inhibits Hck-T338A in a Fibroblast Transformation Assay
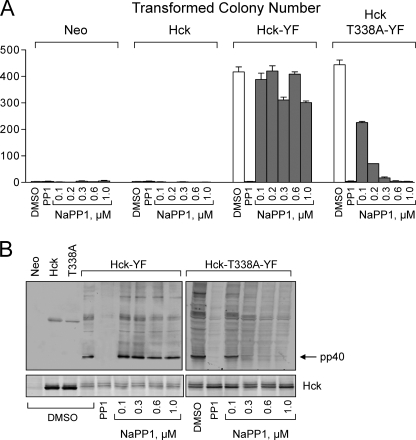
To determine whether the Hck-T338A mutant is sensitive to NaPP1 in a cell-based assay, we next tested the ability of NaPP1 to revert the transformed phenotype of fibroblasts expressing an active form of Hck-T338A. For these experiments, we replaced the C-terminal negative regulatory tail tyrosine residue (Tyr-527) with phenylalanine (“YF” mutation) in both wild-type Hck and the T338A mutant. This mutation was previously shown to prevent tail Tyr-527 phosphorylation by Csk and SH2 domain engagement, resulting in a marked increase in kinase activity and oncogenic transformation of fibroblasts (29, 34, 36, 37, 48). The resulting Hck-YF and Hck-T338A-YF mutants were expressed in Rat-2 fibroblasts, and their transforming potential and sensitivity to NaPP1 were compared in a soft-agar colony formation assay. Fibroblasts expressing wild-type Hck, Hck-T338A, or the G418 selection marker were used as negative controls. As shown in Fig. 2A, neither wild-type Hck nor the T338A mutant induced fibroblast transformation, demonstrating that alanine substitution at the gatekeeper position does not result in kinase activation. In contrast, both Hck-YF and Hck-T338A-YF had robust and comparable transforming activity, demonstrating that the T338A mutation does not interfere with the biological functions of the kinase in this system. Hck activity in each of the Rat-2 cell lines was then assessed by anti-phosphotyrosine immunoblotting of cell lysates, and the results are presented in Fig. 2B. Both Hck-YF and Hck-T338A-YF showed strong constitutive activity in this assay. In contrast, wild-type Hck and Hck-T338A showed very little kinase activity, consistent with their lack of transforming function.
FIGURE 2.
Hck-T338A is biologically active in fibroblasts and selectively sensitive to NaPP1. Rat-2 fibroblasts were infected with recombinant retroviruses carrying a neomycin selection marker (Neo control), wild-type Hck, Hck-T338A, Hck-YF, or Hck-T338A-YF and selected with G418. A, cell populations were plated in triplicate in soft agar in the presence of the indicated concentrations of NaPP1. The general SFK inhibitor and parent compound PP1 (3 μm) was used as a positive control. Transformed colonies were stained with 3-(4,5-dimethyl-thiazol-2-yl)-2,5-diphenyltetrazolium bromide after 10–14 days and enumerated from scanned images using QuantityOne colony-counting software (Bio-Rad). Results from a representative experiment are shown as the mean number of colonies of three plates ±S.D. The statistical significance of Hck expression on colony formation was determined using one-way ANOVA (p < 0.0001) followed by Bonferroni's post test for multiple comparisons (p < 0.001 for Neo versus Hck-YF, Neo versus Hck-T338A-YF, Hck versus Hck-YF, and Hck versus Hck-T338A-YF; p > 0.05 for Neo versus Hck and Hck-YF versus Hck-T338A-YF). The effect of NaPP1 on colony formation by Hck-YF and Hck-T338A-YF cells was assessed using a two-tailed unpaired Student's t test (normal distribution and unequal variance; p < 0.02). The complete experiment was repeated twice with comparable results. B, control or Rat-2 fibroblasts transformed by Hck-YF or Hck-T338A-YF were plated overnight with the indicated concentrations of NaPP1, PP1 (3 μm), or with the vehicle alone (DMSO). Lysates were probed with an anti-phosphotyrosine antibody to determine the phosphorylation status of the endogenous Hck substrate, pp40. Hck expression was confirmed in replicate immunoblots. The entire experiment was repeated twice with comparable results. A representative example is shown.
We next assessed the specificity of NaPP1 for Hck-T338A-YF in this system. As shown in Fig. 2A, NaPP1 significantly blocked colony formation by cells transformed with Hck-T338A-YF with an IC50 of ∼0.1 μm. This effect correlated with inhibition of Hck-T338A-YF activity as assessed by anti-phosphotyrosine immunoblotting, which reveals the endogenous Hck substrate protein, pp40 (Fig. 2B). Conversely, neither colony formation nor kinase activity was affected by NaPP1 in cells transformed by Hck-YF, even at concentrations as high as 1 μm. These results establish that the T338A mutation is functionally silent yet confers NaPP1 sensitivity on Hck in a cell-based assay.
Overexpression of Hck and Hck-T338A Protects K562 CML Cells from Imatinib-induced Apoptosis and Inhibition of Colony Formation
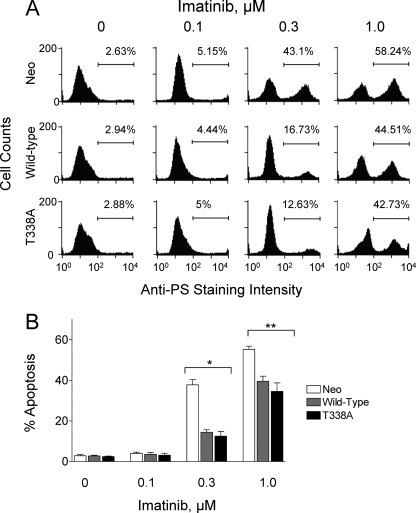
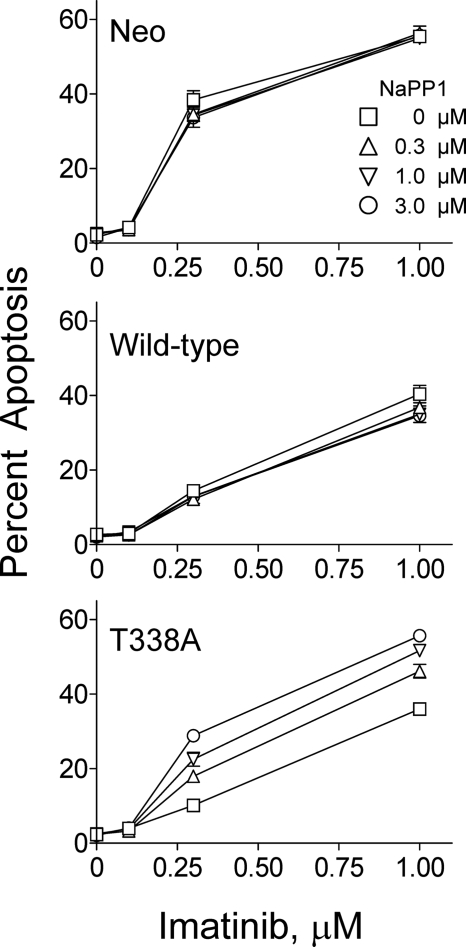
Previous studies have shown that a subset of imatinib-resistant blast crisis CML patients display increased Hck kinase protein expression and/or activity in the absence Bcr-Abl mutations (21, 22, 24). To determine whether Hck overexpression in CML cells is sufficient to induce imatinib resistance, wild-type Hck and Hck-T338A were expressed in K562 cells using recombinant retroviruses. Cells infected with a retrovirus carrying only the neomycin resistance marker served as a negative control. After selection with G418, the K562-Neo, K562-Hck, and K562-Hck-T338A cell populations were treated over a range of imatinib concentrations for 72 h. The fraction of apoptotic cells was then determined by anti-phosphatidylserine antibody staining and flow cytometry. As shown in Fig. 3, imatinib induced apoptosis in a dose-dependent manner in the K562-neo cell population, with apoptotic cells evident with as little as 0.3 μm imatinib. Strikingly, overexpression of wild-type Hck or Hck-T338A in K562 cells was sufficient to reduce the apoptotic effects of imatinib at both 0.3 and 1 μm (p < 0.05, one-way ANOVA).
FIGURE 3.
Expression of wild-type Hck or Hck-T338A protects K562 cells from imatinib-induced apoptosis. K562-Neo, K562-Hck, and K562-Hck-T338A cell populations were incubated for 72 h in the absence or presence of the indicated concentrations of imatinib. Apoptotic cells were stained with an anti-phosphatidylserine-Alexa Fluor 488-conjugated antibody and the percentage of apoptotic cells was determined by flow cytometry. A, shown are histograms from a representative flow cytometry experiment with the percentage of apoptotic cells shown above each plot. PS, phosphatidylserine. B, bar graph showing the average percentage of apoptotic cells from three independent experiments ±S.D. Two-way ANOVA showed significant effects of Hck or Hck-T338A expression (p < 0.0001) and of imatinib treatment (p < 0.0001). One-way ANOVA was performed separately for each concentration of imatinib across the three groups. The percent apoptosis among the three cell lines was statistically significant at 0.3 μm imatinib and 1 μm imatinib (*, p = 0.0003 and **, p = 0.006). Bonferroni's post test for multiple comparisons showed a p < 0.001 for Neo versus Hck and Neo versus Hck-T338A at 0.3 μm imatinib and a p < 0.05 for Neo versus Hck and Neo versus Hck-T338A at 1 μm imatinib.
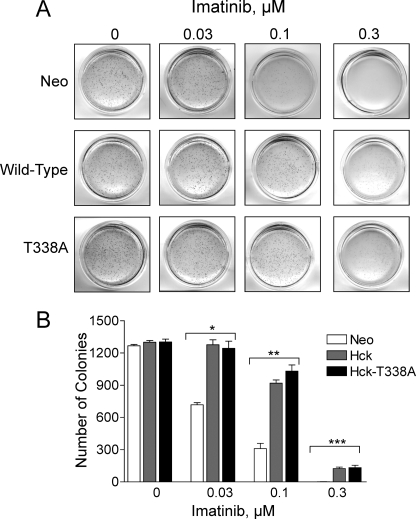
We next investigated whether expression of Hck in K562 cells reverses imatinib-induced inhibition of colony formation. To address this issue, all three K562 cell populations were plated in soft agar over a range of imatinib concentrations. As shown in Fig. 4, K562 cells expressing wild-type Hck or Hck-T338A yielded a comparable number of colonies as the vector control cells when plated in soft-agar in the absence of imatinib. This finding suggests that expression of wild-type Hck or Hck-T338A alone does not enhance the basal level of K562 cell colony-forming activity. Imatinib induced a dose-dependent inhibition of colony formation in all three cell lines. However, cells expressing wild-type Hck or Hck-T338A displayed substantial resistance to imatinib-induced inhibition of colony formation when compared with control cells. More specifically, although 0.03 μm imatinib reduced the number of K562-neo colonies by nearly 50%, it did not have a significant impact on cells expressing either form of Hck (p = 0.0003, one-way ANOVA). Furthermore, K562-Hck and K562-Hck-T338A cells were three times more resistant to 0.1 μm imatinib than K562-neo control cells in this assay (p < 0.0001, one-way ANOVA). Taken together, results from both the apoptosis and colony assays demonstrate that overexpression of Hck is sufficient to induce resistance to imatinib.
FIGURE 4.
Expression of wild-type Hck or Hck-T338A protects K562 cells from imatinib-induced inhibition of soft-agar colony formation. K562-Neo, K562-Hck, and K562-Hck-T338A cell populations were plated in soft agar colony assays in the presence of the concentrations of imatinib shown. A, colonies were stained 7–10 days later, and representative images of the plates are shown. B, the bar graph shows the average number of colonies from three independent experiments ±S.D. Two-way ANOVA showed significant effects of Hck or Hck-T338A expression (p < 0.0001) and of imatinib treatment (p < 0.0001). One-way ANOVA was performed separately for each concentration of imatinib, across the three groups. The effect of imatinib on colony formation by the three cell lines was statistically significant at 0.03, 0.1, and 0.3 μm (*, p = 0.0003; **, p < 0.0001; ***, p = 0.0015, respectively). Bonferroni's post test for multiple comparisons showed p < 0.001 for Neo versus Hck and Neo versus Hck-T338A at 0.03 and 0.1 μm imatinib and p < 0.01 for Neo versus Hck and Neo versus Hck-T338A at 0.3 μm imatinib.
Resistance to Imatinib-induced Apoptosis and Inhibition of Colony Growth Requires Hck Kinase Activity
To determine whether resistance to imatinib requires Hck kinase activity, we tested whether specific inhibition of Hck-T338A with NaPP1 restored sensitivity to imatinib treatment in both assays. For the apoptosis assay, K562-Neo, K562-Hck, and K562-Hck-T338A cells were incubated with imatinib over the same concentration range as in Fig. 3 in the presence of escalating concentrations of NaPP1. After 72 h, the percentage of apoptotic cells was determined by flow cytometry as before. The results presented in Fig. 5 show the apoptotic response to imatinib plotted for each concentration of NaPP1. Similar to the results presented in Fig. 3B, K562-Hck and K562-Hck-T338A cells displayed resistance to imatinib-induced apoptosis when compared with K562-Neo control cells. The addition of NaPP1 to K562-Neo cells as well as cells expressing wild-type Hck did not significantly affect the level of apoptosis induced by imatinib, demonstrating that NaPP1 alone is not cytotoxic and does not affect wild-type Hck at these concentrations. In contrast, the addition of NaPP1 to K562-Hck-T338A cells induced a statistically significant, concentration-dependent reversal of resistance to imatinib to a level similar to K562-neo control cells, demonstrating that resistance to imatinib-induced apoptosis requires the kinase activity of Hck.
FIGURE 5.
Inhibition of Hck-T338A kinase activity with NaPP1 selectively restores the apoptotic response to imatinib in K562-Hck-T338A cells. K562-Neo, K562-Hck, and K562-Hck-T338A cell populations were incubated with the indicated combinations of imatinib and NaPP1 for 72 h. Apoptotic cells were stained with an anti-phosphatidylserine antibody conjugated to Alexa Fluor 488, and the percentage of apoptotic cells was determined by flow cytometry. The apoptotic response to imatinib was plotted for each individual NaPP1 concentration. Each point represents the average percentage of apoptotic cells generated from three independent experiments ±S.D. Two-way ANOVA was performed to determine statistical significance of the effect of imatinib and NaPP1 on each individual cell line. For K562-Neo and K562-Hck cells, the apoptotic effect of imatinib was statistically significant with p < 0.0001, whereas NaPP1 had no significant effect (p > 0.05). For K562-Hck-T338A cells, both imatinib and NaPP1 had a significant effect (p < 0.0001).
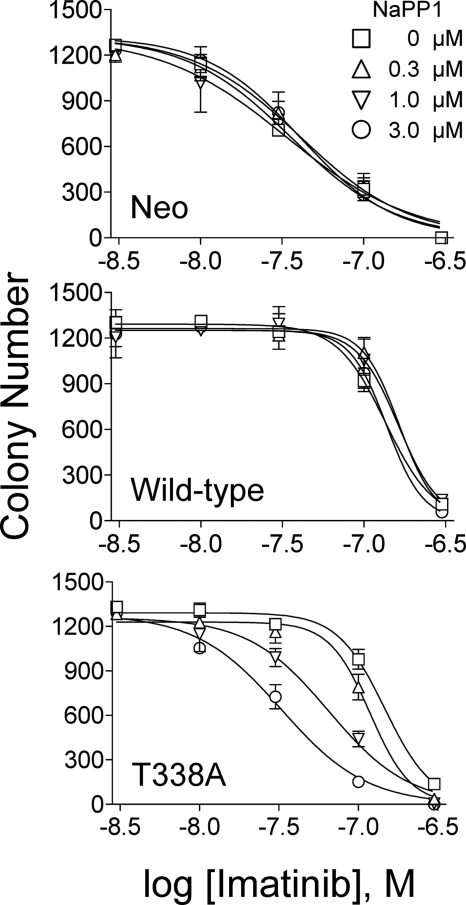
Next, we determined whether NaPP1 resensitizes K562-Hck-T338A cells to imatinib in the soft-agar colony assay. Each K562 cell population was plated in soft agar with various concentrations of imatinib and NaPP1 and incubated for 7 days to allow colony formation. Imatinib-induced inhibition of colony formation was determined for each NaPP1 concentration, and the results are plotted in Fig. 6. In addition, colony formation IC50 values for imatinib were determined by curve-fitting. Imatinib inhibited K562-Neo colony formation with an IC50 of 35.9 ± 6.8 nm in the absence of NaPP1, and this value was unaffected by NaPP1 treatment. Overexpression of wild-type Hck or Hck-T338A in K562 cells increased the IC50 value for the inhibition of colony formation by imatinib by about 4-fold, to 135 ± 11 and 146 ± 17.7 nm, respectively. Similar to the K562-Neo control cells, NaPP1 did not affect the inhibition of colony formation by imatinib in K562 cells expressing wild-type Hck, consistent with the lack of NaPP1 activity against the wild-type kinase. In contrast, the addition of NaPP1 to K562-Hck-T338A cells induced a dose-dependent reversal of the inhibition of colony formation by imatinib, with the IC50 value dropping from 146 ± 17.7 to 32 ± 6.6 nm in the presence of 3 μm NaPP1. These results demonstrate that selective inhibition of Hck-T338A kinase activity completely restores sensitivity to imatinib in terms of colony-forming activity.
FIGURE 6.
Inhibition of Hck-T338A kinase activity with NaPP1 selectively restores the sensitivity of K562-Hck-T338A cells to imatinib in the colony-forming assay. K562-Neo, K562-Hck, and K562-Hck-T338A cell populations were plated in colony-forming assays in the presence of the indicated combinations of imatinib and NaPP1. Colonies were stained 10–14 days later and were counted from scanned images of the plates. Each data point represents the average colony count from three replicate plates ±S.D. The entire experiment was repeated twice from independently derived cell populations and yielded comparable results. A representative experiment is shown. Two-way ANOVA was performed to determine statistical significance of the effect of imatinib and NaPP1 on each cell line. For K562-Neo and K562-Hck cells, the effect of imatinib on colony formation was statistically significant with p < 0.0001, whereas NaPP1 had no significant effect (p > 0.05). For K562-Hck-T338A cells, both imatinib and NaPP1 had a significant effect (p < 0.0001).
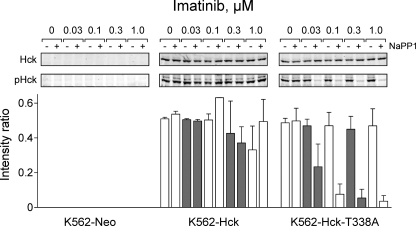
To determine whether the reversal of imatinib sensitivity was associated with inhibition of Hck-T338A tyrosine kinase activity, Hck was immunoprecipitated from cells after exposure to the same imatinib concentrations used in the apoptosis and colony assays in combination with 3 μm NaPP1 (Fig. 7). Quantitative immunoblot analysis using an antibody specific to phospho-Tyr-416 in the activation loop of the active form of Hck showed that NaPP1 completely inhibited the T338A mutant of the kinase, consistent with the complete reversal of imatinib sensitivity observed at this concentration in both the apoptosis and colony assays. In contrast, NaPP1 did not inhibit the activity of wild-type Hck. To confirm the specificity of NaPP1 further, we also immunoprecipitated endogenous Lyn and performed similar immunoblot analysis with the anti-phospho-Tyr-416 antibody. NaPP1 did not affect Lyn activation loop phosphorylation in the presence or absence of imatinib (data not shown).
FIGURE 7.
NaPP1 selectively inhibits Hck-T338A in K562 cells. K562-Neo, K562-Hck, and K562-Hck-T338A cell populations were grown in 0.5% FBS overnight and treated with the indicated concentrations of imatinib and NaPP1 (3 μm) for 5 h. Hck was immunoprecipitated from clarified cell lysates and immunoblotted with a phosphospecific antibody that recognizes the tyrosine-phosphorylated activation loop of Hck (pHck). Duplicate membranes were blotted for Hck as a loading control. Western blots were analyzed using the Odyssey Infrared Imaging System. The Hck phosphotyrosine signal intensities were normalized to the levels of Hck protein from the blots of two independent experiments, and the average intensity ratios ±S.D. are presented in the bar graph. The entire experiment was repeated twice from independently derived cell populations with comparable results; representative blots are shown at the top. Two-way ANOVA was performed to determine statistical significance of the effect of imatinib and NaPP1 on the phosphorylation of Hck within each cell line. For K562-Hck, neither imatinib nor NaPP1 had a significant effect (p > 0.3), whereas for K562-Hck-T338A, NaPP1 treatment produced a significant reduction in Hck phosphorylation (p < 0.0001).
Overexpression of Hck in K562 Cells Increases Tyrosine Phosphorylation of Multiple Proteins in the Presence of Imatinib
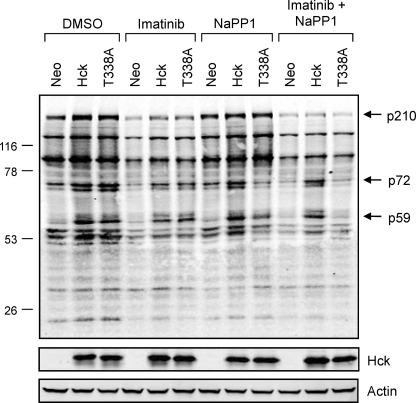
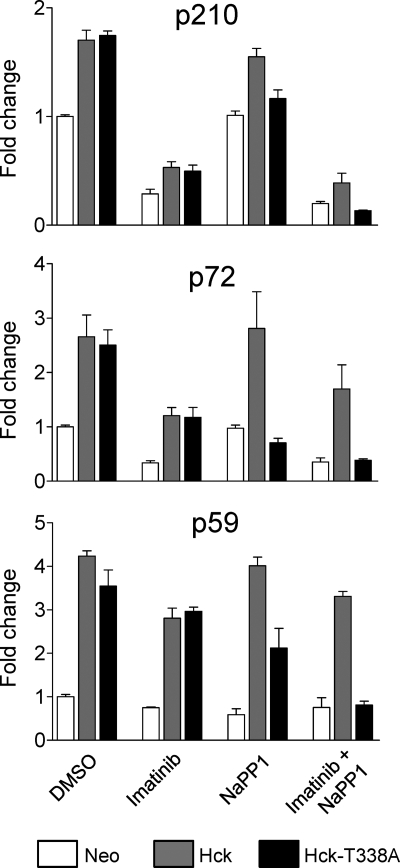
To explore the mechanism of Hck-induced imatinib resistance, we performed quantitative anti-phosphotyrosine immunoblotting on lysates from K562-Neo, K562-Hck, and K562-Hck-T338A cells after treatment with DMSO carrier solvent (control), 1 μm imatinib, 3 μm NaPP1, or a combination of both compounds. As shown in Fig. 8, expression of wild-type Hck and the Hck-T338A mutant enhanced the tyrosine phosphorylation of several proteins in K562 cells, the most prominent of which include a band of 210 kDa (most likely Bcr-Abl) as well as a second band of 72 kDa. A third band of 59 kDa is also readily apparent in the Hck-expressing cell lines (p59) and most likely represents tyrosine-phosphorylated Hck itself. The addition of NaPP1 decreased tyrosine phosphorylation of these proteins in cells expressing Hck-T338A but not wild-type Hck, demonstrating that Hck kinase activity is required for the enhanced phosphorylation observed (Fig. 9). Furthermore, the addition of NaPP1 in combination with imatinib decreased phosphorylation of these proteins in K562-Hck-T338A cells to levels observed in lysates from imatinib-treated K562-Neo cells. The observation that Hck overexpression in K562 cells does not induce tyrosine phosphorylation of new proteins supports the idea that Hck-induced resistance to imatinib occurs through enhanced phosphorylation of Bcr-Abl at sites that may influence imatinib binding (see the next paragraph). However, the possibility of Bcr-Abl-independent mechanisms of resistance cannot be completely excluded as Bcr-Abl and Src family kinases such as Hck are known to share common substrates (49–51) (for a more detailed description of the methods used for quantitative anti-phosphotyrosine immunoblotting, please see supplemental Fig. S2.)
FIGURE 8.
Wild-type Hck or Hck-T338A overexpression enhances tyrosine phosphorylation of several proteins in K562 cells without changing the overall tyrosine phosphoprotein banding pattern. K562 cells were infected with recombinant retroviruses carrying a neomycin selection marker (Neo), wild-type Hck or Hck-T338A and selected with G418. The resulting cell populations were plated in 0.5% FBS overnight and treated with the DMSO carrier solvent, imatinib (1 μm), NaPP1 (3 μm), or imatinib plus NaPP1 for 5 h. Cell lysates were resolved by SDS-PAGE, transferred to nitrocellulose membranes, and probed with anti-phosphotyrosine (top) and anti-Hck antibodies. Immunoreactive proteins were simultaneously detected using the Odyssey Infrared Imaging System (LI-COR). As a loading control, replicate blots were also probed with an anti-actin antibody. Using the Odyssey 3.0 software, the position of each molecular weight marker was used to estimate the molecular weights of each tyrosine phosphoprotein. The positions of the three bands showing the most prominent changes in phosphotyrosine content after Hck expression are indicated (p210, p72, and p59). This entire experiment was repeated three times from independently derived cell populations and produced comparable results in each case.
FIGURE 9.
Phosphorylation of p210 and p72 is Hck-dependent in K562 cells. Signal intensities for the three major NaPP1-sensitive tyrosine phosphoproteins shown in Fig. 8 (p210, p72, and p59) were quantitated from anti-phosphotyrosine immunoblots of three independent K562-Neo, K562-Hck, and K562-Hck-T338M cell populations after treatment with imatinib, NaPP1, or both as described in the legend to Fig. 8. The bar graphs show the -fold changes relative to DMSO-treated K562-Neo control cells of the average phosphotyrosine signal intensities ±S.D. for p210 (Bcr-Abl), p72, and p59 (Hck).
Overexpression of Hck Is Associated with Enhanced and Sustained Phosphorylation of Bcr-Abl at Multiple Regulatory Sites in the Presence of Imatinib
Previous studies have shown that Hck and other SFKs transphosphorylate c-Abl and Bcr-Abl (52–54). This may in turn stabilize the active conformation of the Abl kinase domain and, thus, decrease sensitivity to imatinib as well as increase Bcr-Abl-dependent signaling. For example, both c-Src and Hck phosphorylate c-Abl on its activation loop at Tyr-412 (53), which promotes kinase activation and induces imatinib resistance (Abl numbering is based on the crystal structure of the Ncap-c-Abl core; PDB code 2FO0) (32). Moreover, Hck and other SFKs phosphorylate Bcr-Abl at Tyr-89 in the SH3 domain and Tyr-245 in the SH2 kinase linker region both in vitro and in CML cells (54, 55). Phosphorylation at either of these residues may disrupt the negative regulatory interaction between the SH3 domain and the SH2 kinase linker (56). Furthermore, Hck phosphorylates Bcr-Abl in the Bcr-derived portion of the protein at Tyr-177 (Bcr numbering), a residue that links Bcr-Abl to Ras activation and is required for leukemogenesis (57–60).
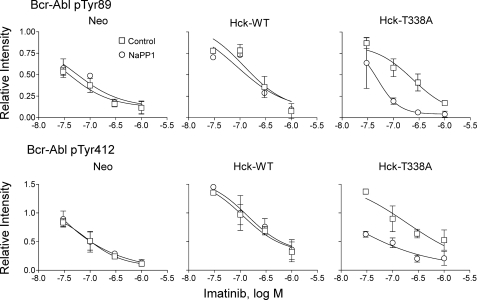
To test whether the imatinib resistance observed in response to Hck overexpression correlates with enhanced phosphorylation of Bcr-Abl at these key regulatory tyrosines, we performed quantitative immunoblotting with phosphospecific antibodies on lysates from K562-Neo, K562-Hck, and K562-Hck-T338A cells after treatment with imatinib and NaPP1. Fig. 10 shows that expression of wild-type or T338A Hck causes enhanced phosphorylation of the Bcr-Abl SH3 domain at Tyr-89 and that phosphorylation of this site is sustained in the presence of 0.03–0.3 μm imatinib. The addition of NaPP1 completely reversed this Hck-dependent phosphorylation of Tyr-89 in cells expressing Hck-T338A but not wild-type Hck. This effect of NaPP1 is particularly marked at 0.1 and 0.3 μm imatinib and suggests that Hck directly phosphorylates this key regulatory site in the Bcr-Abl SH3 domain. Tyr-89 phosphorylation is markedly reduced in all three cell lines at 1.0 μm imatinib in the presence or absence of NaPP1, suggesting that Bcr-Abl kinase activity may be required to stimulate Hck or endogenous SFK activation, which in turn leads to phosphorylation of this site. Bcr-Abl autophosphorylation may occur at Tyr-89 as well. Expression of wild-type or T338A Hck also increased phosphorylation of the Bcr-Abl activation loop at Tyr-412 (Fig. 10). Similar to Tyr-89, phosphorylation at this site was resistant to inhibition by lower concentrations of imatinib and dramatically reversed by NaPP1 exclusively in K562-Hck-T338A cells. Hck-mediated phosphorylation of Tyr-89 and Tyr-412 may induce an active conformation of Bcr-Abl due to the disruption of the intramolecular negative regulatory interaction between the SH3 domain and the linker or via direct phosphorylation of the activation loop (54, 56). In either case such changes favor an active Bcr-Abl kinase domain conformation less compatible with imatinib binding (27).
FIGURE 10.
Wild-type Hck or Hck-T338A overexpression increases Bcr-Abl phosphorylation at regulatory tyrosines 89 and 412 in a Hck kinase-dependent manner. K562-Neo, K562-Hck (wild-type (WT)), and K562-Hck-T338A cell populations were plated in 0.5% FBS overnight and treated with the concentrations of imatinib indicated and in the presence or absence of 3 μm NaPP1 for 5 h. Cell lysates were resolved by SDS-PAGE, transferred to nitrocellulose, and immunoblotted with phosphospecific antibodies for Abl pY89 or pY412. Immunoblots were analyzed using the Odyssey Infrared Imaging System. Site-specific phosphotyrosine signal intensities from two independent experiments were normalized to the corresponding levels of Bcr-Abl protein as determined by anti-Abl immunoblotting. The normalized phosphotyrosine band intensities are plotted relative to the ratios obtained from DMSO-treated K562-Neo control cell populations ±S.D. The data points were fitted by nonlinear regression analysis using GraphPad Prizm. Note that Hck expression shifts the imatinib concentration-response curves for both phosphorylation sites upward and to the right, indicative of enhanced phosphorylation and reduced drug sensitivity. These shifts are completely and exclusively reversed in Hck-T338A cells after treatment with NaPP1. Images of the immunoblots used to generate these curves, as well as additional data for Bcr-Abl Tyr-245 and Tyr-177 are shown in supplemental Fig. S3.
Phosphorylation of Bcr-Abl in the SH2 kinase linker region on Tyr-245 was also enhanced after expression of either wild-type Hck or Hck-T338A, and this effect was sustained in the presence of imatinib except at the highest concentration tested (1 μm; supplemental Fig. S3). In this case, however, the addition of NaPP1 did not significantly inhibit Tyr-245 phosphorylation in cells expressing HckT338A, suggesting that the enhancement of phosphorylation observed is independent of Hck kinase activity. One possible kinase-independent mechanism may involve binding of Hck to the regulatory SH2 and SH3 domains of Bcr-Abl (11).
Finally, we investigated the effect of Hck expression on Tyr-177 phosphorylation in the Bcr-derived portion of the oncoprotein (supplemental Fig. S3). Bcr-Abl from K562-Hck and K562-Hck-T338A cells demonstrated enhanced phosphorylation of this site that persisted even in the presence of 1 μm imatinib, whereas phosphorylation of Tyr-177 in control cells was inhibited by about 50% at this imatinib concentration. The addition of NaPP1 partially reduced Bcr-Abl Tyr-177 phosphorylation in K562-Hck-T338A but not K562-Hck cells. Because phosphorylation of Tyr-177 creates a binding site for Grb2 (22, 23) and other adaptors, it is possible that Hck may increase Ras-dependent signaling via Tyr-177 that contributes to imatinib resistance. The inability of imatinib to completely reverse Tyr-177 phosphorylation in control cells suggests that other SFKs coupled to Bcr-Abl or possibly other non-receptor tyrosine kinase families contribute to phosphorylation of this site.
DISCUSSION
Resistance to imatinib, the first-line therapy for CML, is typically associated with reactivation of Bcr-Abl signaling due to selection of drug-resistant Bcr-Abl mutants. However, emerging evidence shows that other mechanisms of clinical resistance can occur in the absence of Bcr-Abl mutations. Consistent with the role of SFKs in CML pathogenesis, recent evidence has linked Hck and Lyn overexpression or enhanced activity to imatinib resistance in the absence of Bcr-Abl mutations (21, 22, 24, 61). In this report, we establish a direct cause and effect relationship between Hck overexpression and resistance to imatinib in a CML cell line with wild-type Bcr-Abl and demonstrate that drug resistance requires Hck kinase activity.
To determine whether imatinib resistance requires Hck activity, we generated a Hck allele (Hck-T338A) that is uniquely targeted by NaPP1, a bulky analog of the non-selective SFK inhibitor PP1. The advantage of this chemical genetic approach is that it allows for specific inhibition of Hck activity while leaving the Hck protein intact. Thus, data derived using this approach may be more applicable to drug action than gene expression knockdown by small interfering RNA or other approaches. Since the original description of the method by Shokat and co-workers (43), this approach has been successfully applied to both serine/threonine and tyrosine kinases from diverse families (25).
As part of the Hck-T338A validation strategy, we first needed to establish that this mutant did not exhibit a loss or gain of function and that it was selectively sensitive to inhibition by NaPP1 but not affected by imatinib. Our results both in vitro and in cell-based assays show that the T338A mutation does not affect Hck catalytic or biological activities. Furthermore, we found that the T338A mutation made Hck almost 50 times more sensitive to NaPP1 than the wild-type kinase in vitro, without inducing sensitivity to imatinib. Importantly, NaPP1 did not inhibit a recombinant Abl kinase protein establishing the specificity of this compound for the Hck-T338A mutant.
Replacement of the corresponding gatekeeper residue in other protein-tyrosine kinases such as c-Abl, c-Src, or the platelet-derived growth factor receptor was recently reported to have kinase-activating effects as shown by the increased tyrosine phosphorylation of cellular proteins in transfected human 293T cells (62). However, these mutants were not able to transform murine BaF3 myeloid cells to interleukin-3 independence (62). In the case of Hck, evidence presented here shows that the replacement of the gatekeeper threonine with alanine does not induce a gain of function. Expression of Hck-T338A in Rat-2 fibroblasts did not induce cellular transformation or phosphorylation of endogenous substrates. Although these data indicate that the T338A mutation does not up-regulate Hck kinase activity, we cannot rule out the possibility that this gatekeeper mutation affects other biological functions.
Having validated the NaPP1-sensitive mutant, we next showed that both wild-type Hck and Hck-T338A overexpression are sufficient to restore K562 cell survival and colony-forming activity in the presence of imatinib. Colony formation in soft agar was strongly affected, with the IC50 value for inhibition of colony growth increased by 4-fold upon overexpression of either form of Hck. The resistance to imatinib induced by Hck-T338A overexpression was completely reversed by NaPP1 in a concentration-dependent manner in cells expressing Hck-T338A but not in cells expressing wild-type Hck. The reversal of imatinib sensitivity in terms of survival and colony growth correlated with a complete inhibition of Hck-T338A activity by NaPP1 treatment, whereas wild-type Hck activity was unaffected. These results show that imatinib resistance induced by Hck overexpression requires Hck kinase activity.
Hck-induced resistance to imatinib may occur through a feedback mechanism that sustains an active conformation of the Bcr-Abl tyrosine kinase domain and thereby promotes imatinib resistance. Using phosphospecific antibodies, we present evidence that both wild-type and T338A Hck expression enhance phosphorylation of Bcr-Abl Tyr-89 (SH3 domain) and Tyr-412 (activation loop) that is sustained in the presence of imatinib. Phosphorylation of both sites is markedly sensitive to NaPP1 solely in K562-Hck-T338A cells, strongly suggesting that Hck is responsible for phosphorylation of these sites in vivo. Recent work has shown that phosphorylation of Tyr-89, which localizes to the binding surface of the SH3 domain, disrupts its intramolecular interaction with the SH2 kinase linker and is predicted to promote an active conformation of the Abl core (56). Data presented here also show a Hck kinase-dependent increase in phosphorylation of the Abl activation loop tyrosine residue (Tyr-412), which may also increase Bcr-Abl activity and imatinib resistance. However, it is not clear whether this effect is due to direct Hck-mediated phosphorylation of Tyr-412 or disruption of intramolecular regulatory interactions within Bcr-Abl (e.g. via phosphorylation of Tyr-89 in the SH3 domain).
Our results also show an increase in Tyr-177 phosphorylation in the Bcr-derived part of Bcr-Abl that is partially sensitive to NaPP1 in K562-Hck-T338A cells. Because phosphorylation of Bcr-Abl Tyr-177 facilitates Ras signaling (22, 23), enhanced phosphorylation of this site may stimulate mitogenic and anti-apoptotic pathways downstream of Ras that contribute to the increased growth and survival of Hck-expressing K562 cells observed in the presence of imatinib. This finding may also help to explain the remarkable enhancement of K562 colony formation observed in the presence of imatinib after expression of either form of Hck in K562 cells.
Although our data suggest that Hck may directly influence Bcr-Abl activity or signaling to account for imatinib resistance, Bcr-Abl-independent mechanisms may play a role as well. For example, recent evidence shows that Bcr-Abl activates autocrine insulin-like growth factor-1 signaling through Hck and Stat5b (63). In other experimental systems, insulin-like growth factor-1 was shown to have anti-apoptotic effects due to β-catenin stabilization (64). Therefore, it is possible that Hck-induced imatinib resistance observed here may be mediated in part through insulin-like growth factor-1 release. In addition, several SFKs have been shown to phosphorylate the cytoplasmic domain of Muc-1, a transmembrane glycoprotein overexpressed in CML. Phosphorylation of this Muc-1 region promotes the binding, stabilization, and increased nuclear targeting of β-catenin (65, 66). Whether or not Hck-mediated stabilization of β-catenin through either of these pathways contributes to imatinib resistance will require further investigation.
Recently, several second-generation Bcr-Abl-tyrosine kinase inhibitors, such as nilotinib, have been developed to override the problem of imatinib resistance. This has led in turn to studies of nilotinib resistance mechanisms. One report describes the generation of a nilotinib-resistant K562 cell line by exposure to gradually increasing concentrations of the drug (47). In this cell line nilotinib resistance was linked to Lyn up-regulation. In addition, an increase in Lyn mRNA expression was found in two of seven patients that developed resistance to nilotinib (47). This report suggests that SFK up-regulation may represent a common mechanism of resistance to Bcr-Abl inhibitors in addition to selection of drug-resistant Bcr-Abl kinase domain mutants. This observation also suggests that patients developing imatinib resistance without Bcr-Abl mutations may also be cross-resistant to other Bcr-Abl inhibitors. In this context, it would be interesting to determine whether Hck overexpression also induces resistance to nilotinib.
In summary, imatinib is arguably the best known and most successful kinase inhibitor in clinical use, although resistance to this drug is a growing problem in CML. Mutations in the Bcr-Abl kinase domain account for many but not all cases of resistance. More vexing are a growing number of imatinib-resistant CML patients who do not exhibit mutations in the Abl kinase domain but instead show a correlation with overexpression of SFKs, which are not direct targets for imatinib action. Results presented in this report demonstrate for the first time that overexpression of the myeloid SFK Hck is sufficient to confer imatinib resistance to blast crisis CML cells that express wild-type Bcr-Abl and that resistance requires Hck kinase activity. Furthermore, our data suggest that the resistance mechanism may involve direct phosphorylation of Bcr-Abl by Hck, although enhanced phosphorylation of substrate proteins shared by these kinases is also a possibility. Inhibitors selective for Hck or other SFKs may be of particular clinical benefit in this type of imatinib-resistant CML.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants CA101828 and GM077629 (to T. E. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- CML
- chronic myelogenous leukemia
- SFK
- Src family kinases
- PP1
- pyrazolopyrimidine 1
- NaPP1
- naphthyl-pyrazolopyrimidine 1
- FBS
- fetal bovine serum
- ANOVA
- analysis of variance
- PBS
- phosphate-buffered saline.
REFERENCES
- 1.Deininger M. W., Goldman J. M., Melo J. V. (2000) Blood 96, 3343–3356 [PubMed] [Google Scholar]
- 2.Groffen J., Stephenson J. R., Heisterkamp N., de Klein A., Bartram C. R., Grosveld G. (1984) Cell 36, 93–99 [DOI] [PubMed] [Google Scholar]
- 3.Shtivelman E., Lifshitz B., Gale R. P., Canaani E. (1985) Nature 315, 550–554 [DOI] [PubMed] [Google Scholar]
- 4.Raitano A. B., Whang Y. E., Sawyers C. L. (1997) Biochim. Biophys. Acta 1333, F201–F216 [DOI] [PubMed] [Google Scholar]
- 5.Hantschel O., Superti-Furga G. (2004) Nat. Rev. Mol. Cell Biol. 5, 33–44 [DOI] [PubMed] [Google Scholar]
- 6.Faderl S., Talpaz M., Estrov Z., O'Brien S., Kurzrock R., Kantarjian H. M. (1999) N. Engl. J. Med. 341, 164–172 [DOI] [PubMed] [Google Scholar]
- 7.Kirchner D., Duyster J., Ottmann O., Schmid R. M., Bergmann L., Munzert G. (2003) Exp. Hematol. 31, 504–511 [DOI] [PubMed] [Google Scholar]
- 8.Danial N. N., Rothman P. (2000) Oncogene 19, 2523–2531 [DOI] [PubMed] [Google Scholar]
- 9.Voss J., Posern G., Hannemann J. R., Wiedemann L. M., Turhan A. G., Poirel H., Bernard O. A., Adermann K., Kardinal C., Feller S. M. (2000) Oncogene 19, 1684–1690 [DOI] [PubMed] [Google Scholar]
- 10.Danhauser-Riedl S., Warmuth M., Druker B. J., Emmerich B., Hallek M. (1996) Cancer Res. 56, 3589–3596 [PubMed] [Google Scholar]
- 11.Lionberger J. M., Wilson M. B., Smithgall T. E. (2000) J. Biol. Chem. 275, 18581–18585 [DOI] [PubMed] [Google Scholar]
- 12.Klejman A., Schreiner S. J., Nieborowska-Skorska M., Slupianek A., Wilson M., Smithgall T. E., Skorski T. (2002) EMBO J. 21, 5766–5774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson M. B., Schreiner S. J., Choi H. J., Kamens J., Smithgall T. E. (2002) Oncogene 21, 8075–8088 [DOI] [PubMed] [Google Scholar]
- 14.Pene-Dumitrescu T., Peterson L. F., Donato N. J., Smithgall T. E. (2008) Oncogene 27, 7055–7069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldman J. M., Melo J. V. (2003) N. Engl. J. Med. 349, 1451–1464 [DOI] [PubMed] [Google Scholar]
- 16.Hochhaus A., Druker B., Sawyers C., Guilhot F., Schiffer C. A., Cortes J., Niederwieser D. W., Gambacorti-Passerini C., Gambacorti C., Stone R. M., Goldman J., Fischer T., O'Brien S. G., Reiffers J. J., Mone M., Krahnke T., Talpaz M., Kantarjian H. M. (2008) Blood 111, 1039–1043 [DOI] [PubMed] [Google Scholar]
- 17.Druker B. J., Guilhot F., O'Brien S. G., Gathmann I., Kantarjian H., Gattermann N., Deininger M. W., Silver R. T., Goldman J. M., Stone R. M., Cervantes F., Hochhaus A., Powell B. L., Gabrilove J. L., Rousselot P., Reiffers J., Cornelissen J. J., Hughes T., Agis H., Fischer T., Verhoef G., Shepherd J., Saglio G., Gratwohl A., Nielsen J. L., Radich J. P., Simonsson B., Taylor K., Baccarani M., So C., Letvak L., Larson R. A. (2006) N. Engl. J. Med. 355, 2408–2417 [DOI] [PubMed] [Google Scholar]
- 18.Hochhaus A., Kreil S., Corbin A. S., La Rosée P., Müller M. C., Lahaye T., Hanfstein B., Schoch C., Cross N. C., Berger U., Gschaidmeier H., Druker B. J., Hehlmann R. (2002) Leukemia 16, 2190–2196 [DOI] [PubMed] [Google Scholar]
- 19.Gorre M. E., Mohammed M., Ellwood K., Hsu N., Paquette R., Rao P. N., Sawyers C. L. (2001) Science 293, 876–880 [DOI] [PubMed] [Google Scholar]
- 20.Azam M., Latek R. R., Daley G. Q. (2003) Cell 112, 831–843 [DOI] [PubMed] [Google Scholar]
- 21.Donato N. J., Wu J. Y., Stapley J., Gallick G., Lin H., Arlinghaus R., Talpaz M. (2003) Blood 101, 690–698 [DOI] [PubMed] [Google Scholar]
- 22.Wu J., Meng F., Lu H., Kong L., Bornmann W., Peng Z., Talpaz M., Donato N. J. (2008) Blood 111, 3821–3829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Warmuth M., Bergmann M., Priess A., Häuslmann K., Emmerich B., Hallek M. (1997) J. Biol. Chem. 272, 33260–33270 [DOI] [PubMed] [Google Scholar]
- 24.Wu J., Meng F., Kong L. Y., Peng Z., Ying Y., Bornmann W. G., Darnay B. G., Lamothe B., Sun H., Talpaz M., Donato N. J. (2008) J. Natl. Cancer Inst. 100, 926–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bishop A. C., Buzko O., Shokat K. M. (2001) Trends Cell Biol. 11, 167–172 [DOI] [PubMed] [Google Scholar]
- 26.Bishop A. C., Ubersax J. A., Petsch D. T., Matheos D. P., Gray N. S., Blethrow J., Shimizu E., Tsien J. Z., Schultz P. G., Rose M. D., Wood J. L., Morgan D. O., Shokat K. M. (2000) Nature 407, 395–401 [DOI] [PubMed] [Google Scholar]
- 27.Schindler T., Bornmann W., Pellicena P., Miller W. T., Clarkson B., Kuriyan J. (2000) Science 289, 1938–1942 [DOI] [PubMed] [Google Scholar]
- 28.Lozzio B. B., Lozzio C. B., Bamberger E. G., Feliu A. S. (1981) Proc. Soc. Exp. Biol. Med. 166, 546–550 [DOI] [PubMed] [Google Scholar]
- 29.Lerner E. C., Trible R. P., Schiavone A. P., Hochrein J. M., Engen J. R., Smithgall T. E. (2005) J. Biol. Chem. 280, 40832–40837 [DOI] [PubMed] [Google Scholar]
- 30.Schindler T., Sicheri F., Pico A., Gazit A., Levitzki A., Kuriyan J. (1999) Mol. Cell 3, 639–648 [DOI] [PubMed] [Google Scholar]
- 31.Trible R. P., Emert-Sedlak L., Smithgall T. E. (2006) J. Biol. Chem. 281, 27029–27038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagar B., Hantschel O., Seeliger M., Davies J. M., Weis W. I., Superti-Furga G., Kuriyan J. (2006) Mol. Cell 21, 787–798 [DOI] [PubMed] [Google Scholar]
- 33.Iacob R. E., Pene-Dumitrescu T., Zhang J., Gray N. S., Smithgall T. E., Engen J. R. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 1386–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Briggs S. D., Smithgall T. E. (1999) J. Biol. Chem. 274, 26579–26583 [DOI] [PubMed] [Google Scholar]
- 35.Muller A. J., Young J. C., Pendergast A. M., Pondel M., Landau N. R., Littman D. R., Witte O. N. (1991) Mol. Cell. Biol. 11, 1785–1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Briggs S. D., Sharkey M., Stevenson M., Smithgall T. E. (1997) J. Biol. Chem. 272, 17899–17902 [DOI] [PubMed] [Google Scholar]
- 37.Lerner E. C., Smithgall T. E. (2002) Nat. Struct. Biol. 9, 365–369 [DOI] [PubMed] [Google Scholar]
- 38.Liu Y., Bishop A., Witucki L., Kraybill B., Shimizu E., Tsien J., Ubersax J., Blethrow J., Morgan D. O., Shokat K. M. (1999) Chem. Biol. 6, 671–678 [DOI] [PubMed] [Google Scholar]
- 39.Blencke S., Zech B., Engkvist O., Greff Z., Orfi L., Horváth Z., Kéri G., Ullrich A., Daub H. (2004) Chem. Biol. 11, 691–701 [DOI] [PubMed] [Google Scholar]
- 40.Alaimo P. J., Knight Z. A., Shokat K. M. (2005) Bioorg. Med. Chem. 13, 2825–2836 [DOI] [PubMed] [Google Scholar]
- 41.Wong S., McLaughlin J., Cheng D., Zhang C., Shokat K. M., Witte O. N. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 17456–17461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu W., Doshi A., Lei M., Eck M. J., Harrison S. C. (1999) Mol. Cell 3, 629–638 [DOI] [PubMed] [Google Scholar]
- 43.Bishop A. C., Shah K., Liu Y., Witucki L., Kung C., Shokat K. M. (1998) Curr. Biol. 8, 257–266 [DOI] [PubMed] [Google Scholar]
- 44.MacAuley A., Okada M., Nada S., Nakagawa H., Cooper J. A. (1993) Oncogene 8, 117–124 [PubMed] [Google Scholar]
- 45.Porter M., Schindler T., Kuriyan J., Miller W. T. (2000) J. Biol. Chem. 275, 2721–2726 [DOI] [PubMed] [Google Scholar]
- 46.Rix U., Hantschel O., Dürnberger G., Remsing Rix L. L., Planyavsky M., Fernbach N. V., Kaupe I., Bennett K. L., Valent P., Colinge J., Köcher T., Superti-Furga G. (2007) Blood 110, 4055–4063 [DOI] [PubMed] [Google Scholar]
- 47.Mahon F. X., Hayette S., Lagarde V., Belloc F., Turcq B., Nicolini F., Belanger C., Manley P. W., Leroy C., Etienne G., Roche S., Pasquet J. M. (2008) Cancer Res. 68, 9809–9816 [DOI] [PubMed] [Google Scholar]
- 48.Choi H. J., Smithgall T. E. (2004) J. Mol. Biol. 343, 1255–1268 [DOI] [PubMed] [Google Scholar]
- 49.Howlett C. J., Bisson S. A., Resek M. E., Tigley A. W., Robbins S. M. (1999) Biochem. Biophys. Res. Commun. 257, 129–138 [DOI] [PubMed] [Google Scholar]
- 50.Sattler M., Salgia R., Okuda K., Uemura N., Durstin M. A., Pisick E., Xu G., Li J. L., Prasad K. V., Griffin J. D. (1996) Oncogene 12, 839–846 [PubMed] [Google Scholar]
- 51.Sattler M., Salgia R. (1997) Cytokine Growth Factor Rev. 8, 63–79 [DOI] [PubMed] [Google Scholar]
- 52.Plattner R., Kadlec L., DeMali K. A., Kazlauskas A., Pendergast A. M. (1999) Genes Dev. 13, 2400–2411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Furstoss O., Dorey K., Simon V., Barilà D., Superti-Furga G., Roche S. (2002) EMBO J. 21, 514–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meyn M. A., 3rd, Wilson M. B., Abdi F. A., Fahey N., Schiavone A. P., Wu J., Hochrein J. M., Engen J. R., Smithgall T. E. (2006) J. Biol. Chem. 281, 30907–30916 [DOI] [PubMed] [Google Scholar]
- 55.Brasher B. B., Van Etten R. A. (2000) J. Biol. Chem. 275, 35631–35637 [DOI] [PubMed] [Google Scholar]
- 56.Chen S., O'Reilly L. P., Smithgall T. E., Engen J. R. (2008) J. Mol. Biol. 383, 414–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pendergast A. M., Quilliam L. A., Cripe L. D., Bassing C. H., Dai Z., Li N., Batzer A., Rabun K. M., Der C. J., Schlessinger J., Gishizky M. L. (1993) Cell 75, 175–185 [PubMed] [Google Scholar]
- 58.Goga A., McLaughlin J., Afar D. E., Saffran D. C., Witte O. N. (1995) Cell 82, 981–988 [DOI] [PubMed] [Google Scholar]
- 59.Puil L., Liu J., Gish G., Mbamalu G., Bowtell D., Pelicci P. G., Arlinghaus R., Pawson T. (1994) EMBO J. 13, 764–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cortez D., Stoica G., Pierce J. H., Pendergast A. M. (1996) Oncogene 13, 2589–2594 [PubMed] [Google Scholar]
- 61.Donato N. J., Wu J. Y., Stapley J., Lin H., Arlinghaus R., Aggarwal B. B., Shishodia S., Albitar M., Hayes K., Kantarjian H., Talpaz M., Shishodin S. (2004) Cancer Res. 64, 672–677 [DOI] [PubMed] [Google Scholar]
- 62.Azam M., Seeliger M. A., Gray N. S., Kuriyan J., Daley G. Q. (2008) Nat. Struct. Mol. Biol. 15, 1109–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lakshmikuttyamma A., Pastural E., Takahashi N., Sawada K., Sheridan D. P., DeCoteau J. F., Geyer C. R. (2008) Oncogene 27, 3831–3844 [DOI] [PubMed] [Google Scholar]
- 64.Satyamoorthy K., Li G., Vaidya B., Patel D., Herlyn M. (2001) Cancer Res. 61, 7318–7324 [PubMed] [Google Scholar]
- 65.Kawano T., Ito M., Raina D., Wu Z., Rosenblatt J., Avigan D., Stone R., Kufe D. (2007) Cancer Res. 67, 11576–11584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li Y., Kuwahara H., Ren J., Wen G., Kufe D. (2001) J. Biol. Chem. 276, 6061–6064 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.