Abstract
Infection with the obligate bacterial intracellular pathogen Chlamydia trachomatis leads to the sustained activation of the small GTPase RAS and many of its downstream signaling components. In particular, the mitogen-activated protein kinase ERK and the calcium-dependent phospholipase cPLA2 are activated and are important for the onset of inflammatory responses. In this study we tested if activation of ERK and cPLA2 occurred as a result of RAS signaling during infection and determined the relative contribution of these signaling components to chlamydial replication and survival. We provide genetic and pharmacological evidence that during infection RAS, ERK, and, to a lesser extent, cPLA2 activation are uncoupled, suggesting that Chlamydia activates individual components of this signaling pathway in a non-canonical manner. In human cell lines, inhibition of ERK or cPLA2 signaling did not adversely impact C. trachomatis replication. In contrast, in murine cells, inhibition of ERK and cPLA2 played a significant protective role against C. trachomatis. We determined that cPLA2-deficient murine cells are permissive for C. trachomatis replication because of their impaired expression of β interferon and the induction of immunity-related GTPases (IRG) important for the containment of intracellular pathogens. Furthermore, the MAPK p38 was primarily responsible for cPLA2 activation in Chlamydia-infected cells and IRG expression. Overall, these findings define a previously unrecognized role for cPLA2 in the induction of cell autonomous cellular immunity to Chlamydia and highlight the many non-canonical signaling pathways engaged during infection.
Keywords: Cellular Immune Response, ERK, Glycerophospholipid, Interferon, MAP Kinases (MAPKs), Phospholipase A, Ras, Chlamydia, IRG Proteins, MAPK
Introduction
Mammalian cells detect microbial invaders and activate signaling pathways to restrict pathogen growth and to initiate the release of soluble mediators that recruit immune cells with antimicrobial functions (1). For example, infection with Chlamydia trachomatis, an obligate intracellular Gram-negative bacterial pathogen, leads to sustained activation of the MEK/ERK2 mitogen-activated protein kinases (MAPK) signaling pathway (2–5). ERK has emerged as a focal point of signaling during Chlamydia infection, with roles as a regulator of bacterial nutrient acquisition (5), IL-8 synthesis (2, 6) and the expression anti-apoptotic factors early in infection (4). Indeed, ERK-mediated signaling is required for the robust induction of inflammatory responses responsible for the bulk of the pathology (e.g. pelvic inflammatory disease and infertility) associated with recurrent and chronic genital chlamydial infections (2, 7).
Chlamydia infections begin with the attachment of an elementary body (EB), the invasive but non-replicative form of the pathogen, to epithelial cells of the genital tract. Shortly after entry, EBs transition into metabolically active reticulate bodies (RBs) that replicate within a membrane-bound bound pathogenic vacuole termed an “inclusion” (8). Midway through the infectious cycle, replication becomes asynchronous, with some RBs transitioning back to the infectious EB stage (8). Eventually, EBs are released into the extracellular space to initiate subsequent rounds of infection in adjacent cells. The entire infectious lifecycle is complete within 48–72h, depending on the C. trachomatis serovar. Throughout infection, Chlamydia translocates a diverse cohort of proteins into host membranes and cytoplasm to re-program membrane dynamics, protein transport, and cell signaling (9).
In canonical RAS signaling, growth factors bind to receptor-tyrosine kinases, leading to their dimerization and activation (10). Phosphorylated tyrosine residues on these receptors recruit adaptor proteins and RAS guanine nucleotide exchange factors (GEFs). Upon binding GTP, RAS undergoes a conformational switch that allows the binding and activation of downstream effectors via RAS-binding domains (RBD) (11). There are three main classes of RAS effectors: RAF kinases, PI(3)P-dependent kinases and a diverse family of proteins that regulate cytoskeletal organization and membrane dynamics (12). Activated RAF phosphorylates dual specificity MAPK/ERK (MEK) kinases, which in turn phosphorylate and activate the MAPKs ERK1 and ERK2. ERK phoshorylates transcription factors and other proteins (e.g. ribosomal protein kinases (RSK) and MAPK-interacting kinases (MNK)) that control cell growth and proliferation (10).
Epithelial cells infected with Chlamydia display sustained activation of RAS and many of its downstream effectors including the PI3P-dependent kinase AKT and components of the MEK/ERK MAPK signaling module (5, 13). The activation of both AKT and ERK enhance Chlamydia survival by protecting infected cells from apoptosis (4, 13). For example, ERK-dependent expression of Mcl-1 protects infected cells from apoptotic signal delivered by innate immune mediators early in infection (4).
One prominent target of ERK1/2 is the calcium-dependent cytoplasmic phospholipase A2 (cPLA2) which removes the acyl chain at the sn2 position of glycerophospholipids to generate a lysophospholipid and a free fatty acid (14). Lysophospholipids are readily transported to the inclusion, re-acylated with Chlamydia-derived branched fatty acids and incorporated into bacterial membranes (15). Because calcium chelators and specific inhibitors of cPLA2 significantly impair chlamydial replication and inhibit the transport of host-derived glycerophospholipids, it has been postulated that a host PLA2 activity is co-opted by Chlamydia to generate lipid precursors important for bacterial replication (5). In addition, cPLA2-mediated changes in the lysophospholipid content of the lipid bilayer can modulate membrane transport by influencing the curvature of phospholipid bilayers and the formation of membrane tubules (16) from endosomes and Golgi apparatus (17), both membrane transport events important for chlamydial replication (18, 19). Finally, activation of cPLA2 leads to pro-inflammatory responses, as its free fatty acid byproduct arachidonate is a substrate for lipoxygenases and prostaglandin generating enzymes (20). These observations combined suggest a prominent and central role for cPLA2 in chlamydial pathogenesis.
Chlamydia, like other bacterial pathogens, has evolved mechanisms to modulate signaling events to enhance their own replication and dissemination (21, 22). The observed activation of RAS signaling components in Chlamydia-infected cells and their apparent requirement for optimal bacterial replication has led to a model wherein activation of canonical RAS signaling may be important for bacterial survival and initiation of inflammatory responses (2, 4, 5). However, very recent evidence suggests that RAS may not be responsible for ERK activation (23). In this study we provide genetic and pharmacological evidence that confirms that components of the RAS signaling pathway are independently activated indicating that RAS and ERK signaling in response to chlamydial infection are uncoupled. Furthermore, we determined that in murine cells cPLA2, a downstream effector of ERK signaling, regulates cell autonomous innate immune responses by modulating the expression of type I interferons specifically in response to C. trachomatis infections.
EXPERIMENTAL PROCEDURES
Strains, Infections, and Cell Culture Reagents
HeLa and 293T cells were obtained from ATCC and maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Invitrogen). HEK-tTH cells were derived as described in Ref. 24. MEFs from cPLA2−/− knock-out mice were obtained from J. Bonaventre (Harvard Medical School) and have been previously described (25). All cells were maintained at 37 °C in a 5% CO2 incubator. Primary embryonic kidney cell lines were immortalized as previously described (24). C. trachomatis serovar LGV-L2 was propagated and stored as described in Ref. 26. Chlamydia muridarum was obtained from H. Caldwell (Rocky Mountain Laboratories, NIH). For infections, EBs were diluted in complete cell culture media and added to appropriate cells at an MOI of 1 and centrifuged at 1600 × g for 30 min at 4 °C. For experiments using chemical inhibitors, the medium was replaced 30 min after infection with media containing the appropriate inhibitor after which the infection was allowed to proceed for prescribed times. Other reagents: EGF (Sigma) was used at a concentration of 1–10 ng/ml for 5 min. IFN-γ 100 units/ml (Calbiochem 407303) and IFN-β (Calbiochem 407298) 100 units/ml were used for 5 h to overnight.
Inhibitors
The MEK 1/2 inhibitor U0126 (Cell Signaling Technologies 9903) was used at a concentration of 10 μm. The cPLA2 inhibitor AACOCF3 (Calbiochem 100109) was used at a concentration of 50 μm unless otherwise stated. The farnesyl transferase inhibitor L-744,832 (Calbiochem 422720) was used at a concentration of 10 μm. The p38 inhibitor SB203580 (Calbiochem 559389) was used at a concentration of 20 μm. All inhibitors were diluted in DMSO.
Virus Production
293T cells seeded in 10 cm plates for both lenti- and retrovirus production. For retroviral production, the pBabe retroviral system was used (27) and used as outlined in O'Hayer and Counter (24). Briefly, packaging plasmid (pCL-10A1) and transfer vector (pBabe) were transfected in a 1:1 ratio using Fugene6 (Roche) into 293T cells. The media was collected 40 h post-transfection, filtered with 0.44 μm filters (Whatman), supplemented with Polybrene (hexadimethrine bromide) (Sigma) at a concentration of 4 mg/ml and placed directly onto target cells. For lentivirus production (28), cPLA2 specific shRNA plasmid (Open Biosystems) was transfected with packaging plasmid (pCMV-dR8.74) and envelope protein pMD2.G (Micah Luftig, Duke University) into 293T cells. Virus-containing medium was collected 48 h post-transfection. Stable transductants were selected with puromycin 1–2 μg/ml 24 h post-infection.
Plasmids and Constructs
RBD-EGFP-cDNA5 and H-RAS(wt)- cDNA5 expression plasmids were obtained from Mark Phillips, laboratory (NYU). H-RAS17N was generated by site-directed mutagenesis with QuikChange (Invitrogen) as described by the manufacturer. cDNA5-EGFP was generated in our laboratory. For transient transfections, cells were seeded at 50–70% confluence and treated with FugeneHD (Roche)- DNA complexes as indicated by the manufacturer. Cells were analyzed 24 h post-transfection unless otherwise noted.
RAS-GTP Pulldown Assays
Cell lysates were collected in radioimmune precipitation assay buffer (1% Nonidet P-40, 20 mm Tris, pH 8.0, 137 mm NaCl, 10% glycerol, 2 mm EDTA) supplemented with phenylmethylsulfonyl fluoride (1 mm) and aprotinin (1%). Levels of GTP-bound RAS were assessed by incubating cell lysates with glutathione-agarose beads (29) bound to GST-tagged RAS binding domain of RAF (GST-RBD) for 45 min at 4 °C. Lysates were washed in lysis buffer and bound proteins were solublized in 2× Laemmli sample buffer. Bound RAS-GTP was detected by Western blot with a pan-RAS or isoform-specific anti-RAS antibodies.
Immunofluorescence Microscopy
Cells were fixed using either methanol or formaldehyde for 20 min and permeabilized using 0.1% Triton in phosphate-buffered saline for 5 min and then blocked with 5% bovine serum albumin (Sigma) in phosphate-buffered saline. Primary antibody staining at appropriate concentration was conducted at 4 °C for 1 h and followed by staining using fluorophore-conjugated secondary antibodies (Molecular Probes) for 30 min. DNA was labeled using either Hoechst or Topro (Molecular Probes) at a concentration of 1:10,000 or 1:1,000, respectively. Cells were mounted to slides using mounting media (Invitrogen) and were stored at 4 °C, prior to analysis by Laser Scanning Confocal Microscopy on a LEICA.
Protein Analysis/Western Blot and Antibodies
Protein lysates were generated from tissue cultures cells in freshly made cold lysis buffer (25 mm Tris pH 7.4, 150 mm NaCl, 5 mm EDTA, 1% Triton X-100) supplemented with Complete EDTA-free protease inhibitor mixture tablets (Roche), 1 mm Na3VO4, and 1 mm NaF. Samples were resolved by SDS-PAGE and blotted onto nitrocellulose membranes (Bio-Rad) for immunoblotting.
The following antibodies were used: Pan-RAS (Oncogene Ab-4), H-RAS (Santa Cruz Biotechnology sc-520), K-RAS (Santa Cruz Biotechnology sc-30), N-RAS (Santa Cruz Biotechnology sc-31). ERK 1/2 (9102), phospho-Erk 1/2 (Thr-202/Tyr-204) (9154), C-RAF (9422), phospho-c-RAF (Ser-338) (9427), cPLA2 (2832), phospho-cPLA2 (Ser-505) (2831), COX2 (4892), phospho-p38 (Thr-180/Tyr-182) (9211), and phospho-MEK (Ser-217/221) (9121) were purchased from Cell Signaling Technologies (Boston, MA). Anti-tubulin (B-5-1-2) and GAPDH (9483) antibodies were purchased from Sigma and Abcam, respectively. Anti-Irgm1, Irgm3, and Irgb6 have been previously described (30–32) and were given by G. Taylor (VA Durham). Anti-RpoD (M. Tang, UC Irvine) and IncA antibodies (33) were used to monitor Chlamydia replication.
Inclusion Forming Unit Assay/Inclusion Quantification
Cells were seeded at 2 × 105 cells/well in a 24-well plate. Infections were conducted in quadruplicate as described above at MOI <1. At 40 h, postinfection cells were lysed in SPG (sucrose-phosphate-glutamate) buffer by sonication. Serial dilutions were used to infect HeLa monolayers for 24 h. Cells were fixed with methanol and immunostained with anti-LGV-L2 antisera (P. Bavoil, University of Maryland) or anti-chlamydial LPS (H. Caldwell, RML/NIH). The number of inclusions per infection condition was determined with a Cellomics High Content Screening System (Thermo Scientific).
RT-PCR
Total RNA was collected from MEFs using Qiagen RNAeasy kit (Qiagen 74106). RNA (100 μg) from each sample was treated with Ambion DNA-free DNase kit (Ambion 1906) and cDNA generated using iScript cDNA synthesis kit (BioRad) as per the manufacturer's instructions. Relative transcript abundances were assessed by PCR using ChoiceTaq Blue Mastermix (Denville) with the following oligonucleotide primers: GAPHDH: 5′-AGG TCG GTG TGA ACG GAT TTG-3′ and 5′-TGT AGA CCA TGT AGT TGA GGT CA-3′; IFN-β: 5′-CAG CTC CAA GAA AGG ACG AAC-3′ and 5′-GGC AGT GTA ACT CTT CTG CAT-3′.
Type I Interferon Bioassay and Luciferase Reporter Assay
Lentiviruses containing and ISRE-luciferase reporter construct (SABioscience) were used to create stable cell lines that express luciferase in response to IFN via STAT1/STAT2 transcription factor. Cells were seeded in 6-well plates and infected with LGV-L2 for 24 h or stimulated by transfection of 8 μg of poly(I):poly(C) (Sigma P9043) for 6 h or IFN-γ treatment. Cells were washed twice in phosphate-buffered saline and lysed in Passive Lysis Buffer (Promega) as per the manufacturer's instructions. Lysates were collected, and luciferase activity assessed with firefly luciferase (Promega) as recommended by the manufacturer.
RESULTS
Chlamydia Infection Leads to Broad Activation of RAS GTPases in Human Cells
C. trachomatis infection leads to the sustained activation of RAS and its downstream effectors (2, 4, 5, 13). As a result, it has been proposed that a canonical RAS signaling pathway is engaged during infection (5). We tested this model by following the activation of selected components of the RAS signaling pathway in Chlamydia-infected cells. We first assessed levels of active RAS-GTP in cell lysates by co-precipitation with glutathione-Sepharose beads after incubation with recombinant glutathione S-transferase (GST) fused to the RAS-GTP binding domain of RAF (RAFRBD). As previously reported (4, 5), we observed a significant accumulation of GTP-loaded RAS in HeLa cells during the exponential growth phase of C. trachomatis serovar LGV-L2 at 24 and 36 h postinfection (Fig. 1A and not shown). Because different isoforms of RAS can differ in both their ability to activate effectors (34, 35) and subcellular localization (36), we next determined which RAS isoforms were activated. GTP-bound RAS was isolated from infected and uninfected HeLa lysates and H-RAS, N-RAS, and K-RAS were identified by immunoblot analysis with specific antibodies. All major RAS isoforms were activated, suggesting a broad engagement of RAS signaling during the replicative phase of infection (Fig. 1A).
FIGURE 1.
Chlamydia infection leads to the broad activation of RAS GTPases. A, HeLa cells were left untreated or infected with C. trachomatis (Ct-L2) for 24 h, and RAS-GTP levels were assessed by co-precipitation with GST-tagged RAS binding domain (RBD) on glutathione-agarose beads. RAS isoforms were detected with specific antibodies. B, the localization of active forms of RAS was assessed by monitoring the localization of EGFP-RBD in infected HeLa cells. Bacterial inclusions (arrows) were identified with anti-IncA antibodies. C, RAS is activated in primary cell lines (HEK-T/th) upon infection with L2. RAS-GTP pulldown assays were performed as in Fig. 1A. Epidermal growth factor (EGF) treatment for 5 min is shown as a control for RAS activation. D, total protein lysates from infected cells were harvested at various times postinfection, and the presence of active forms of the RAF, MEK, ERK, and cPLA2 were assessed by immunoblot analysis with antibodies specific for the phosphorylated forms of these proteins. The levels of the chlamydial RNA polymerase σ subunit (RpoD) and the host glyceraldehyde-3-phosphate dehydrogenase (GAPDH) are shown as markers for chlamydial replication and loading controls, respectively. Note activation of all components of the RAS signaling pathway. Immunoblots are representative of at least three independent experiments.
Next, we addressed if RAS was activated within specific subcellular compartments by monitoring the localization of RAFRBD-EGFP in infected cells (37). In uninfected cells treated with EGF or serum, RAS-GTP pools were observed at the plasma membrane, the Golgi apparatus and the endoplasmic reticulum. This activation pattern was largely unchanged in infected cells (Fig. 1B). We did not observe a distinct recruitment of active RAS to the inclusion membrane as has been described for other small GTPases like RhoA and Rabs (38–40), although a pool of RAFRBD-EGFP localized to the vicinity of the inclusion. It is unclear if this pool represents ER and Golgi, which envelope the inclusion (41, 42), or preferential RAS activation at these sites.
We tested if RAS activation also occurred in a genetically defined, recently immortalized cell line. Human embryonic kidney (HEK) cells were immortalized by retroviral delivery of SV40 large and small T-antigen (T/t), and the catalytic domain of human telomerase (hTert), (24) and tested for the activation of RAS during infection. As with HeLa cells, infection of HEK-t/TH cells led to increases in RAS-GTP and phosphorylation of ERK (Fig. 1C). These results indicate that the activation of RAS and ERK in Chlamydia-infected cells infection is not an artifact of the highly transformed status of HeLa cells.
In canonical RAS signaling, the activation of downstream components is dependent on RAS. We first determined the kinetics of how this signaling circuitry is engaged in Chlamydia-infected cells by monitoring the accumulation of activated forms of RAF1, MEK, ERK1/2, and cPLA2 by immunoblot analysis with antibodies specific for the activated forms of these signaling proteins. As previously reported (5), infected cells displayed increasing levels of phosphorylated RAF, MEK, ERK1/2, and cPLA2, indicating sustained engagement of this signaling branch (Fig. 1D). However, while phosphorylation of MEK and ERK1/2 increased throughout infection, the accumulation of active forms of RAF peaked at 12 h and then dropped abruptly. Phosphorylated forms of cPLA2 peaked at 24 h but did not increase significantly after that (Fig. 1D). This potential discrepancy in the kinetics and magnitude with which activate forms of RAF, cPLA2, and MEK/ERK accumulated, led us to postulate that the activation of individual components of this signaling pathway may not be linked during infection.
ERK and RAS Activation Are Uncoupled in Chlamydia-infected Cells
To determine if RAS signaling was required for ERK activation during infection, we manipulated RAS function by two methods. First, we used a farnesyl transferase inhibitor (FTI) to broadly inhibit the farnesylation and membrane anchoring of H-RAS (43), the primary RAS isoform present in HEK cells. FTI effectively inhibited RAS activation in HEK-t/TH cells, as assessed by the decreased efficiency of ERK phosphorylation in response to serum growth factors (Fig. 2A). Next, we tested if FTI-treatment prevented ERK activation during Chlamydia infection. FTI-treated infected HEK-t/TH cells had higher levels of phospho-ERK than uninfected cells, suggesting that ERK was activated despite the absence of proper RAS signaling (Fig. 2A). Treatment with FTI did not affect bacterial entry or replication (data not shown), indicating that any effects of FTI on ERK activation were not the result of lower bacterial loads. FTI treatment was ineffective in HeLa cells (data not shown), possibly because H-RAS is not the dominant RAS isoform in these cells (44).
FIGURE 2.
RAS and ERK activation are uncoupled in Chlamydia-infected cells. A, HEK-Tth cells were treated 30 min postinfection with the farnesyl transferase inhibitor (FTI) l-744/832 or the MEK inhibitor U0126. At 24 h, cells were harvested, and the levels of active ERK were assessed by immunoblots. Note increase in active ERK levels in response to infection despite FTI treatment. Control experiments show FTI-mediated inhibition of ERK phosphorylation in response to serum growth factors. B, HeLa cells were transfected with EGFP empty vector (DNA expression control), RAS, or dominant-negative RAS17N expression constructs for 20 h prior to infection with L2 or treatment with EGF. Transfection of RAS17N did not prevent the accumulation of active ERK in response to Chlamydia infection. C, infected HeLa cells were treated with U0126 and levels of RAS-GTP were assessed as in Fig. 1A. Inhibition of ERK activation did not prevent the accumulation of RAS-GTP in response to infection. All blots are representative from at least three independent experiments.
Because FTI does not inhibit N- and K-RAS it was formally possible that these minor RAS isoforms in HEK cells were capable of mediating ERK activation. As a complementary approach, we expressed a dominant negative allele of RAS (RAS17N), which broadly inhibits RAS activation by sequestering RAS-GEFs (45). Transient transfection of H-RAS17N effectively inhibited the ability of HeLa cells to activate ERK in response to EGF, while transfection of wild-type H-RAS led to increased basal levels of activated ERK (Fig. 2B). Next, we transfected infected HeLa cells with an empty vector control (as a control for DNA transfection), or vectors expressing H-RAS or H-RAS17N, and monitored the accumulation of activated ERK at 24 h postinfection. Consistent with our observations with FTI in HEK cells, activation of ERK in Chlamydia-infected HeLa cells was unaffected by expression of RAS17N (Fig. 2B). Overexpression of wild-type H-RAS only led to a modest increase in ERK activation.
Given that RAS function was not essential for ERK activation, we tested if the converse was true, and if RAS activation required ERK function. We performed RAS-GTP pulldowns in infected HeLa cells treated with the MEK inhibitor U0126. Inhibition of ERK phosphorylation and activation had minimal effect on the levels of RAS-GTP that accumulate in response to chlamydial infection (Fig. 2C). Overall these findings indicate that RAS and ERK activation are largely uncoupled during Chlamydia infection.
ERK Activation and cPLA2 Are Dispensable for Chlamydia Replication in Human Cells
The activation of ERK and cPLA2 have been linked to the generation of pro-inflammatory factors and lipid precursors important for bacterial replication (5). To assess the role of this branch of the ERK signaling pathway, we tested the effect of the MEK inhibitor U0126 and the cPLA2 inhibitor AACOCF3 on chlamydial replication. Treatment with AACOCF3 reduced the yield of chlamydial infectious units, although the concentrations of inhibitor required for >90% growth inhibition (Fig. 3A) were significantly higher than previously reported (5). These concentrations of inhibitor were not toxic to HeLa cells (data not shown) indicating that reduction of growth was not due to the loss of host cells. In contrast, U0126 only had a marginal effect on the yield of infectious units at concentrations that completely inhibited ERK phosphorylation and activation. These results are consistent with reports indicating mild effects for this inhibitor on C. trachomatis replication (2).
FIGURE 3.
C. trachomatis replicates in HeLa cells silenced for the expression of cPLA2. A, relative yield of infectious units of L2 was assessed in cells treated with different concentration of the cPLA2 inhibitor AACOCF3 (left panel), the MEK inhibitor U0126 and HeLa lines stably expressing a cPLA2 specific shRNA (right panel). B, cPLA2 silenced HeLa cells were infected with L2 and tested for the activation of ERK and COX2. Note normal activation of ERK in the absence of cPLA2. Error bars indicate one standard deviation from the mean.
Because cPLA2 is phosphorylated and activated by ERK, we were surprised that MEK inhibitors did not have a similar effect on bacterial replication as cPLA2 inhibitors. To directly address the role of cPLA2 during infection, we generated stable knockdown HeLa cell lines by lentiviral transduction of a cPLA2-specific shRNA. Expression of cPLA2 in these cell lines was <10% that of HeLa cells expressing a control shRNA. Control and cPLA2 shRNA knock-down cell lines were infected with C. trachomatis and bacterial replication was assessed by the generation of infectious units. As we observed with moderate concentrations of AACOCF3, cPLA2 knockdown cell lines did not exhibit any defects in the generation of infectious EBs, making it unlikely that this lipase contributes significantly to chlamydial replication in HeLa cells (Fig. 3B). Nonetheless, cPLA2 knockdown cell lines displayed enhanced levels of ERK phosphorylation and COX2 expression, another ERK-dependent response (6), in response to chlamydial infection, suggesting a potential regulatory role for cPLA2 in ERK signaling in HeLa cells (Fig. 3C).
C. trachomatis Displays Enhanced Replication in cPLA2-deficient Mouse Embryo Fibroblasts
To unambiguously determine the role of cPLA2 in Chlamydia infections, we tested MEFs derived from cPLA2−/− knockout mice for their ability to support chlamydial replication. Surprisingly, C. trachomatis LGV-L2 replicated in cPLA2−/− MEFs at levels 5–6-fold higher than MEFs derived from the same C57B/L6 mouse background (Fig. 4A). These cells are not broadly defective in the control of intracellular bacterial replication as Listeria monocytogenes replicated to similar levels in wild type and cPLA2−/− MEFs (not shown). To test if this replication defect was specific to human chlamydial species, we tested the yield of infectious units of C. muridarum, a Chlamydia that infects mice (46), in these MEFs. Unlike, C. trachomatis, C. muridarum replicated to similar levels in wild type and cPLA2−/− MEFs (Fig. 4B), indicating that cPLA2 plays a protective role for mouse cells against human but not mouse-adapted Chlamydia strains.
FIGURE 4.
cPLA2−/− MEFs are highly permissive for C. trachomatis replication. A, MEFs derived from cPLA2+/+ and cPLA2−/− mice were infected with C. muridarum or C. trachomatis and the relative yield of infectious units was assessed. Note enhanced replication of C. trachomatis in cPLA2-defficient MEFs. B, expression of IRG in wild-type and cPLA2−/− knockout MEFs in response to C. trachomatis and C. muridarum infection for 24 h was assessed by immunoblot analysis. Note impaired expression of Irgm1, Irgm3, Irgb6, and the IFN-inducible transcription factor STAT1. Similar results were obtained by inhibiting cPLA2 activity in wild-type MEFs with AACOCF3 (C). D, inhibition of cPLA2 activity (100 μm AACOCF3) or ERK activation (10 μm U0126) in wild-type MEFs also led to increase yield of infectious units. Error bars in all experiments indicate one standard deviation from the mean. All results are representative from at least three independent experiments.
cPLA2 Controls the Expression of the p47 Family of Immune-regulated GTPases (IRG)
The differential ability of mouse cells to contain the replication of human-adapted Chlamydia species has been linked to the expression of a family of immunity-related GTPases (IRG) (47). The immunoprotective function of a subset of these IRG proteins involves their recruitment to pathogenic vacuoles (48). We tested if the susceptibility of cPLA2−/− MEFs to C. trachomatis infection was linked to the expression or localization of IRGs to inclusions. Wild-type and cPLA2−/− MEFs were infected with C. trachomatis and the expression of a panel of IRG proteins was assessed by immunoblot analysis. Infection of wild-type MEFs with C. trachomatis led to the robust expression of Irgm1, Irgm3, and Irgb6. In contrast, Irgm1, Irm3, and, to a lesser extent, Irgb6 were poorly expressed in infected cPLA2−/− MEFs (Fig. 4C). Irgb10, an IRG that is recruited to inclusions (49), was also poorly expressed (not shown), suggesting a broad defect in the expression of IRG proteins in cPLA2-deficient MEFs.
Next, we tested if the phenotype of cPLA2−/− MEFs could be recapitulated by inhibiting cPLA2 enzymatic activity. Wild-type MEFs and mouse lung fibroblasts (not shown) treated with a sublethal dose (100 μm) of the cPLA2 inhibitor AACOCF3, were incapable of mounting a robust IRG response (Fig. 4C). This is likely due to the inability of cells lacking cPLA2 activity to properly sense Chlamydia and induce the expression of STAT1 (Fig. 4C), a transcription factor that regulates the expression of IRGs and other genes containing interferon-stimulated response elements (50, 51). Consistent with these observations, treatment of cPLA2+/+ MEFs with AACOCF3 led to a 4–5-fold increase in the replication of C. trachomatis in mouse cells (Fig. 4D). Overall these findings indicate that in murine cells cPLA2 plays a protective role in the control of C. trachomatis infections. We also determined that treatment with the MEK inhibitor U0126 increased the yield of infectious units >2.5-fold in MEFs. We postulate that the enhanced replication of Chlamydia in these cells is partially due to impaired activation of cPLA2 in these MEFs.
The Expression of Type I Interferons in Chlamydia-infected Murine Cells Is Regulated by cPLA2
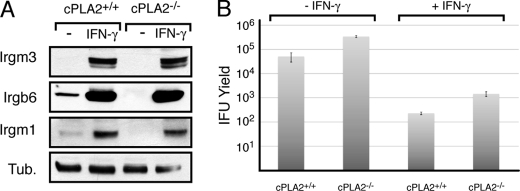
We next addressed whether decreased IRG expression in cPLA2-deficient cells was due to impaired autocrine production of interferons, which are known to regulate IRG expression (52), or whether signaling events distal to the activation of IFN receptors were impaired. To test if cPLA2-deficient cells were capable of responding to IFN-mediated signals, we treated wild-type and cPLA2−/− MEFs with IFN-γ and assessed IRG expression by immunoblot analysis. Both wild-type and cPLA2−/− MEFs expressed IRGs robustly in response to IFN-γ, indicating that cPLA2 is not required for the recognition of exogenous IFNs and IFN-mediated signaling (Fig. 5A). Not surprisingly, IFN-γ treatment effectively restricted C. trachomatis replication in cPLA2−/− MEFs (Fig. 5B). Irgb6 prominently localized to inclusions in IFN-γ treated wild type (53) and cPLA2−/− MEFs (not shown), suggesting that cPLA2 is unlikely to play a role in the localization of IRGs during infection.
FIGURE 5.
cPLA2−/− MEFs are responsive to exogenous interferon. cPLA2+/+ and cPLA2−/− MEFs were treated with 100 units/ml IFN-γ and tested for the expression of IRG proteins (A) and the ability contain chlamydial replication, as assessed by the yield of infectious units (B). Note that both wild-type and cPLA2−/− MEFs express IRGs and contain C. trachomatis replication. Error bars indicate one standard deviation from the mean.
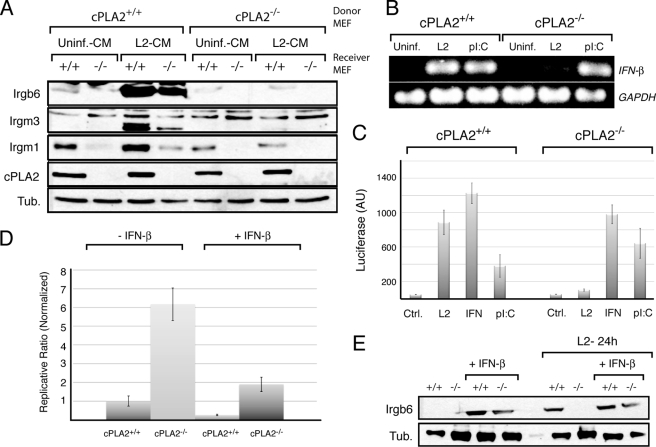
We next tested if C. trachomatis infected MEFs secrete factors that activate the expression of IRGs. Conditioned medium from infected and uninfected MEFS was filter sterilized and added to uninfected cells in which the expression of IRGs was monitored by immunoblot analysis. Conditioned medium from infected wild-type but not cPLA2−/− MEFs led to the expression of IRG proteins (Fig. 6A). These results strongly indicate that cPLA2-deficient cells cannot secrete a factor in response to Chlamydia infection that mediates the activation of anti-pathogen responses. Because recent reports indicate that murine epithelial cells secrete IFN-β in response to C. muridarum infection (54) we tested whether cPLA2 was required for the expression of IFN-β in MEFs. Total RNA was isolated from infected and uninfected wild-type and cPLA2−/− MEFs and the levels of IFN-β mRNAs assessed by RT-PCR. As previously reported, Chamydia infection led to the induction of IFN-β mRNAs (55, 56). However, this increase in expression was significantly lower in infected cPLA2−/− MEFs (Fig. 6B) even though the bacterial load in these cells was higher. This observation was independently confirmed by monitoring the expression of a luciferase reporter driven from an IFN-stimulated response element (ISRE) (Fig. 6C). As control for the ability of these cells to activate canonical type I interferon responses, we transfected double-stranded RNA (poly I:C) into wild-type and knock-out MEFs. Both wild-type and cPLA2−/− MEFs showed similar responses, indicating that cPLA2 is not required for the cytoplasmic-sensing mechanisms in classical antiviral responses or expression and assembly the core transcriptional machinery that controls type I IFN expression.
FIGURE 6.
cPLA2−/− MEFs do not express type I interferons in response to C. trachomatis. A, conditioned medium from infected and uninfected cPLA2+/+ and cPLA2−/− MEFs were placed onto uninfected cPLA2+/+ and cPLA2−/− cells. The ability of the medium to induce IRG expression in uninfected cells was assessed by immunoblots with specific antibodies. Note that conditioned medium from L2-infected cPLA2+/+, but not cPLA2−/− MEFs, activates IRG expression in both wild-type and knockout MEFs. Also note the lower basal levels of IRG expression in cPLA2-deficient cells. B, expression of IFN-β in response to C. trachomatis infection (L2) was assessed by RT-PCR. cPLA2-deficient cells were incapable of expression IFN-β during C. trachomatis infection, but retained the ability to respond to poly(I:C). C, cPLA2+/+ and cPLA2+/+ MEFs were transduced with an IFN-inducible luciferase reporter construct and tested for luciferase activity in response to infection, IFN-γ and transfection of poly(I:C). C. trachomatis-dependent induction of the luciferase reporter is dependent on cPLA2. D, cPLA2+/+ and cPLA2−/− MEFs were treated with 100 units/ml of IFN-β and assessed for expression of IRG proteins by immunoblot. Both wild-type and cPLA2 deficient MEFs are capable of responding to treatment with IFN-β. Irgm1 and Irgm3 were similarly induced (not shown). E, cPLA2+/+ and cPLA2−/− MEFs were infected C. trachomatis (L2), treated with 100 units/ml of IFN-β and bacterial replication was assessed by the generation of infectious units. Both wild-type and cPLA2-deficient cells are capable of containing bacterial growth after treatment with IFN-β. Results shown are representative of at least three independent experiments.
A prediction from our findings is that IRG expression in response to type I IFNs is responsible for the bulk of the cPLA2-mediated protection of murine cells from C. trachomatis infection. To test this premise, we assessed if the enhanced replication of C. trachomatis in cPLA2−/− MEFs could be suppressed by addition of exogenous IFN-β. Recombinant exogenous IFN-β (100 units) decreased C. trachomatis replication in both wild-type and cPLA2−/− MEFs (Fig. 6D). The addition of exogenous IFN-β to cPLA2-deficient MEFs led to Irgb6 levels similar to those of wild-type MEFs infected with C. trachomatis (Fig. 6E).
cPLA2 Is Activated by p38 MAPK during Chlamydia Infection
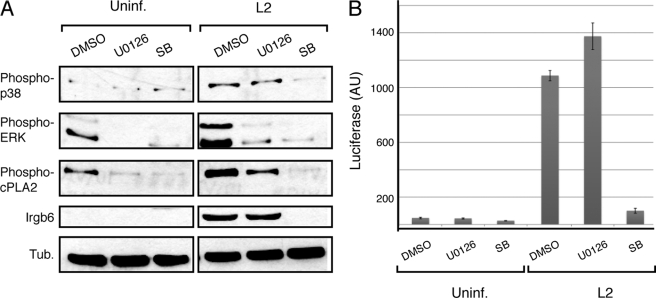
Pharmacological inhibition the ERK1/2 increased Chlamydia replication in MEFs to levels well below to those obtained by cPLA2 inhibition (Fig. 4D), suggesting that cPLA2 activity may be regulated independently of ERK1/2. Because cPLA2 is activated by phosphorylation at Ser-505 by both ERK1/2 and the MAPK p38 (14), we assessed the role of p38 in cPLA2 activation in Chlamydia-infected MEFs and in the expression of IRGs. Cells were infected with C. trachomatis for 24 h in the presence of U0126 or the p38 MEK inhibitor SB203580 and the levels of activated cPLA2, p38, and ERK1/2 were monitored by immunoblot analysis with antibodies specific for phospho-activated forms of these proteins. U0126 and SB203580 prevented the accumulation of activated forms of ERK1/2 and p38, respectively, in response to Chlamydia infection. Interestingly, U1026 partially decreased cPLA2 phosphorylation at Ser-505 while SB203580 completely abolished it (Fig. 7A). This finding suggests that cPLA2 activation in infected MEFs occurs primarily via a p38-dependent pathway. Consistent with this, SB203580-treated MEFs failed to express IRGs in response to infection (Fig. 7A) most likely due to the inability to these cells to activate Type I IFNs as assessed with IFN-inducible luciferase reporters (Fig. 7B). Our findings are consistent with very recent observations indicating a role for p38 in IFN-β expression during C. muridarum infection (57). Interestingly, while ERK1/2 contributes to cPLA2 phosphorylation, it is not required for activation of type IFN responses (Fig. 7B). Overall, these results establish that autonomous expression of type I IFNs in murine cells in response to Chlamydia infection requires cPLA2 activity and that multiple signaling events contribute independently to the activation of this lipase.
FIGURE 7.
cPLA2 activation in Chlamydia-infected MEFs is mediated by the MAPK p38. A, MEFs were infected with C. trachomatis and treated with inhibitors of the ERK (U0126) or p38 (SB203580) MAPK pathways and the levels of activated signaling and IRG proteins were determined by immunoblots. Note that inhibition of p38, but not ERK, completely abolished cPLA2 phosphorylation at Ser-505 and expression of the IRG protein Irgb6. B, MEFs transduced with an IFN-inducible luciferase reporter were treated with ERK or p38 inhibitors and the levels of luciferase expression in response to C. trachomatis infection was determined.
DISCUSSION
Upon encountering a pathogenic organism, mammalian cells activate signaling pathways that control cell autonomous microbicidal functions and alert the broader immune system to the presence of a foreign organism (1). If the invading pathogen is equipped with the proper arsenal of virulence factors, it will dampen these signaling events to avoid antimicrobial responses or harness them to further its own replication. During Chlamydia infections, activation of the RAS-ERK signaling pathway has been linked to the induction of pro-inflammatory cytokines and the generation of lipid precursors for the replicating bacteria (2, 5). In this study we confirmed the activation of various components of the RAS-RAF-MEK/ERK-cPLA2 signaling cascade and provide additional evidence through pharmacological and genetic approaches that individual components of this signaling circuitry are likely activated in a non-canonical manner.
We observed broad activation of all main RAS isoforms by 24 h postinfection (Fig. 1A); but this activation, as assessed by the localization of RAFRBD-EGFP expressed in infected cells, did not appear to be compartmentalized to any specific subcellular location (Fig. 1B). Although activated forms of ERK and cPLA2 have been reported to localize to the periphery of inclusions (5), the diffuse localization of RAFRBD-EGFP is more consistent with RAS signaling occurring at multiple areas within the infected cell. At this time we cannot exclude a role for RAS in the activation of ERK at the inclusion. However, a time course analysis of when activated forms of RAF, MEK, ERK, and cPLA2 accumulate during infection, suggested that these signaling components are not activated in a linear fashion as would be expected in response to serum growth factors (43). For example, RAF activation peaked at 12 h postinfection, while MEK, ERK and to a lesser extent cPLA2, activation increased throughout the infectious cycle (Fig. 1D). Consistent with this apparent temporal disconnect in the activation of RAS and its downstream signaling components, ERK activation was largely insensitive to pharmacological and genetic inhibitors of RAS (Fig. 2, A and B), indicating that ERK and RAS activation are uncoupled in Chlamydia infected cells. Our data are in agreement with recent work by Gurumurthy et al. (23) showing that activation of ERK can occur the absence of at least one isoform of RAS. We have also observed enhancement of replication of C. trachomatis during stable shRNA-mediated knock-down of all three isoforms of RAS (data not shown).
It is unclear what signals activate RAS in infected cells. Because cytokines and inflammatory factors can activate RAS (58) and Chlamydia-infected cells secrete IL-6, IL-8, and prostaglandins (2, 6, 59, 60), we postulate that RAS activation may occur in response to both endocrine and paracrine inflammatory signals. Consistent with this, we have observed that conditioned medium from infected cells can activate RAS in uninfected cells (not shown). Interestingly, the release of these extracellular RAS activating factors appear to be independent of ERK-dependent signaling as infected HeLa cells treated with levels of MEK inhibitor that block IL-8 expression still activated RAS-GTP (Fig. 2C).
ERK-dependent activation of cPLA2 has been postulated to generate essential lysophospholipid precursors for transport into the inclusion (5). Consistent with this, MEK and cPLA2 inhibitors inhibit the incorporation of host-derived glycerophospholipids into bacterial membranes and limit bacterial replication (5). To further define the role of the RAS-ERK-cPLA2 signaling pathway during infection, we tested the effect of specific inhibitors in Chlamydia replication. Treatment with the farnesyl transferase inhibitor L-744/832, did not have any negative effect on inclusion size in HEK cells (not shown), where FTI-sensitive H-RAS is the predominant RAS isoform. Similarly, the MEK inhibitor U0126 had a minimal effect on chlamydial replication at concentrations that abolish accumulation of phosphorylated forms of ERK and IL-8 expression (2). In contrast, treatment with the cPLA2 substrate analogue AACOCF3 resulted in a marked decrease in the yield of infectious units. However, we note that we found variability among different commercial batches of cPLA2 inhibitors3 and the concentrations required in our experiments to inhibit chlamydial replication were significantly higher than those previously reported as inhibitory to Chlamydia growth (5).
To independently address the role of cPLA2 in Chlamydia infection we generated stable knock-down cell lines by lentiviral transduction of cPLA2-specific shRNAs. Chlamydia replicated in cells expressing <10% of wild-type levels of cPLA2 to levels similar to control cell lines (Fig. 3A). Because it was formally possible that residual levels of cPLA2 in these knockdown cell lines was sufficient to support chlamydial replication, we tested if C. trachomatis replicated in fibroblasts derived from cPLA2 knock-out mice. Although mouse cells are less permissive for C. trachomatis replication than human cells, we reasoned that if cPLA2 was essential for chlamydial replication, these cells should be highly resistant to bacterial infection. Unexpectedly, mouse fibroblasts lacking cPLA2 were highly susceptible to C. trachomatis but not other intracellular pathogens (Fig. 4A). Similarly, treatment of cPLA2+/+ MEFs with low levels of AACOCF3 promoted C. trachomatis replication in these cells (Fig. 4D), indicating that cPLA2, at least in mouse cells, plays a protective role against C. trachomatis infections.
The differential susceptibility of mouse strains to C. trachomatis infections is linked to the expression of a family of IRGs important for the containment of intracellular parasites (47). How these GTPases exert their anti-parasitic functions is not well understood, although at least a subset of these proteins can be recruited to the surface of pathogenic vacuoles and cause a loss of membrane integrity and autophagic mediated clearance (49, 53, 61–63). Given that C. muridarum is resistant to this arm of the rodent's innate immune system, and that cPLA2-deficient MEFs were as permissive as wild-type MEFs for their replication, we explored the role that cPLA2 may play in the expression or function of IRGs. Indeed, we found that cPLA2−/− MEFs were impaired in IRG expression during C. trachomatis infection (Fig. 4B) and that this effect is mimicked by treating cPLA2+/+ MEFs with PLA2 inhibitors (Fig. 4C).
Because IRG expression is tightly controlled by IFNs, we tested if cPLA2 was necessary for the ability of MEFs to respond to exogenous IFNs. Both wild-type and cPLA2−/− MEFs treated with type I and type II interferons expressed IRGs (Figs. 5A and 6E) and contained C. trachomatis replication (Figs. 5B and 6D). Similarly, the intracellular signaling pathways required for induction of type I interferons in response to nucleic acids appear intact, as transfection of poly(I:C) induced a robust interferon response in both wild-type and cPLA2-deficient MEFs (Fig. 6, B and C). These findings suggest that cPLA2 is not required for the recognition of IFNs or the activation of IFN-regulated genes. Instead, it is likely that cPLA2 activity is required at some stage in the recognition of Chlamydia and the induction of type I interferon responses (Fig. 4, A–C).
It is increasingly clear that membrane bound intracellular pathogens can effectively activate type I interferon responses, an innate immune pathway that is normally associated with anti-viral responses (64). C. trachomatis and C. muridarum infection of both human and murine cells leads to the secretion of type I interferons (55, 56, 65–67). How chlamydial products activate this innate immune pathway is less clear. In HeLa cells it requires the JAK/STAT1 signaling pathway (55) and in murine cells the adaptor protein TRIF and the transcription factor IRF3 (54). TRIF can interact with TLR3 and TLR4 in endosomal compartments to activate IRFs in response to nucleic acids and lipopolysaccharide, respectively (68, 69) and Ly6C-positive monocytes, have been reported to induce the expression of type I interferons upon recognition of non-nucleic acid viral components by TLR2 (70).
During the review of our work, Pratner et al. (57) provided evidence that the initiation of IFN-β expression during C. muridarum infection of mouse macrophages is a TLR-independent process and that NOD1 and the endoplasmic reticulum/mitochondrial protein STING in conjunction with NF-κB, and p38 MAPK-mediated signaling are required for a full IFN-β response. We have confirmed the importance of p38 MAPK in IFN-β expression and shown that this kinase is required for the phosphorylation and presumably activation of cPLA2 during C. trachomatis infection of MEFs. It is unclear if cPLA2 plays a role in proper signaling by STING, NOD1, or the p38 MAPK cascade that is responsible for IFN-β expression in mice or in the proper localization or function of pattern recognition receptors within intracellular compartments (Fig. 8). Alternatively, like other intracellular pathogens, it is possible that many chlamydial compounds, ranging from metabolites to bacterial RNAs, leak into the cytoplasm and are sensed by pattern recognition receptors (71, 72) and that cPLA2 plays a role in their recognition by modulating bacterial or inclusion membrane stability.
FIGURE 8.
Model for the activation of RAS signaling and the cPLA2-dependent activation cell autonomous IFN responses in Chlamydia-infected cells. RAS and ERK activation are uncoupled during Chlamydia infection and RAS may be activated by exocrine factor released during infection independently of ERK. The MAPK p38 mediates the activation of cPLA2 to regulate the expression of type I interferons in response to C. trachomatis infection of mouse cells. This may be achieved by modulating pattern-recognition receptor signaling either directly or indirectly by altering membrane transport events or lipid signaling. Alternatively, cPLA2 activity may influence the permeability of the inclusion membrane and enhance leakage of microbial products that can be recognized by cytosolic PRR to initiate IFN responses. In murine cells, activation of IFN-β by Chlamydia leads to the expression of IRG proteins, which directly contribute to anti-chlamydial immune responses.
Acknowledgments
We thank Joern Coers for comments on this manuscript and numerous investigators for their kind gift of strains and reagents, especially Joseph Bonventre for the gift of cPLA2−/− MEFs.
This work was supported, in whole or in part, by National Institutes of Health Grant AI068032 and an award from the Burroughs Wellcome Program on the Pathogenesis of Infectious Diseases.
R. H. Valdivia, unpublished observations.
- ERK
- extracellular signal-regulated kinase
- IRG
- immunity-related GTPases
- MEF
- mouse embryonic fibroblast
- GST
- glutathione S-transferase
- MAPK
- mitogen-activated protein kinase
- IFN
- interferon
- IL
- interleukin
- cPLA
- calcium-dependent phospholipase
- EB
- elementary body.
REFERENCES
- 1.Mogensen T. H. (2009) Clin. Microbiol. Rev. 22, 240–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buchholz K. R., Stephens R. S. (2007) Infect. Immun. 75, 5924–5929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krüll M., Kramp J., Petrov T., Klucken A. C., Hocke A. C., Walter C., Schmeck B., Seybold J., Maass M., Ludwig S., Kuipers J. G., Suttorp N., Hippenstiel S. (2004) Infect. Immun. 72, 6615–6621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rajalingam K., Sharma M., Lohmann C., Oswald M., Thieck O., Froelich C. J., Rudel T. (2008) PLoS One 3, e3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Su H., McClarty G., Dong F., Hatch G. M., Pan Z. K., Zhong G. (2004) J. Biol. Chem. 279, 9409–9416 [DOI] [PubMed] [Google Scholar]
- 6.Fukuda E. Y., Lad S. P., Mikolon D. P., Iacobelli-Martinez M., Li E. (2005) Infect. Immun. 73, 4017–4024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stephens R. S. (2003) Trends Microbiol. 11, 44–51 [DOI] [PubMed] [Google Scholar]
- 8.Abdelrahman Y. M., Belland R. J. (2005) FEMS Microbiol. Rev. 29, 949–959 [DOI] [PubMed] [Google Scholar]
- 9.Valdivia R. H. (2008) Curr. Opin. Microbiol. 11, 53–59 [DOI] [PubMed] [Google Scholar]
- 10.Downward J. (2003) Nat. Rev. Cancer 3, 11–22 [DOI] [PubMed] [Google Scholar]
- 11.Donovan S., Shannon K. M., Bollag G. (2002) Biochim. Biophys. Acta 1602, 23–45 [DOI] [PubMed] [Google Scholar]
- 12.Schubbert S., Shannon K., Bollag G. (2007) Nat. Rev. Cancer 7, 295–308 [DOI] [PubMed] [Google Scholar]
- 13.Verbeke P., Welter-Stahl L., Ying S., Hansen J., Häcker G., Darville T., Ojcius D. M. (2006) PLoS Pathog. 2, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghosh M., Tucker D. E., Burchett S. A., Leslie C. C. (2006) Prog. Lipid Res. 45, 487–510 [DOI] [PubMed] [Google Scholar]
- 15.Wylie J. L., Hatch G. M., McClarty G. (1997) J. Bacteriol. 179, 7233–7242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown W. J., Chambers K., Doody A. (2003) Traffic 4, 214–221 [DOI] [PubMed] [Google Scholar]
- 17.San Pietro E., Capestrano M., Polishchuk E. V., DiPentima A., Trucco A., Zizza P., Mariggiò S., Pulvirenti T., Sallese M., Tete S., Mironov A. A., Leslie C. C., Corda D., Luini A., Polishchuk R. S. (2009) PLoS Biol. 7, e1000194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carabeo R. A., Mead D. J., Hackstadt T. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 6771–6776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robertson D. K., Gu L., Rowe R. K., Beatty W. L. (2009) PLoS Pathog. 5, e1000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wymann M. P., Schneiter R. (2008) Nat. Rev. Mol. Cell Biol. 9, 162–176 [DOI] [PubMed] [Google Scholar]
- 21.Betts H. J., Wolf K., Fields K. A. (2009) Curr. Opin. Microbiol. 12, 81–87 [DOI] [PubMed] [Google Scholar]
- 22.Saka H. A., Valdivia R. H. (2010) Curr. Opin. Microbiol. 13, 4–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gurumurthy R. K., Maurer A. P., Machuy N., Hess S., Pleissner K. P., Schuchhardt J., Rudel T., Meyer T. F. (2010) Sci. Signal 113, ra21. [DOI] [PubMed] [Google Scholar]
- 24.O'Hayer K. M., Counter C. M. (2006) Methods Enzymol. 407, 637–647 [DOI] [PubMed] [Google Scholar]
- 25.Bonventre J. V., Huang Z., Taheri M. R., O'Leary E., Li E., Moskowitz M. A., Sapirstein A. (1997) Nature 390, 622–625 [DOI] [PubMed] [Google Scholar]
- 26.Caldwell H. D., Kromhout J., Schachter J. (1981) Infect. Immun. 31, 1161–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morgenstern J. P., Land H. (1990) Nucleic Acids Res. 18, 3587–3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dull T., Zufferey R., Kelly M., Mandel R. J., Nguyen M., Trono D., Naldini L. (1998) J. Virol. 72, 8463–8471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Rooij J., Bos J. L. (1997) Oncogene 14, 623–625 [DOI] [PubMed] [Google Scholar]
- 30.Collazo C. M., Yap G. S., Sempowski G. D., Lusby K. C., Tessarollo L., Woude G. F., Sher A., Taylor G. A. (2001) J. Exp. Med. 194, 181–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uthaiah R. C., Praefcke G. J., Howard J. C., Herrmann C. (2003) J. Biol. Chem. 278, 29336–29343 [DOI] [PubMed] [Google Scholar]
- 32.Taylor G. A., Jeffers M., Largaespada D. A., Jenkins N. A., Copeland N. G., Woude G. F. (1996) J. Biol. Chem. 271, 20399–20405 [DOI] [PubMed] [Google Scholar]
- 33.Bannantine J. P., Stamm W. E., Suchland R. J., Rockey D. D. (1998) Infect. Immun. 66, 6017–6021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Voice J. K., Klemke R. L., Le A., Jackson J. H. (1999) J. Biol. Chem. 274, 17164–17170 [DOI] [PubMed] [Google Scholar]
- 35.Walsh A. B., Bar-Sagi D. (2001) J. Biol. Chem. 276, 15609–15615 [DOI] [PubMed] [Google Scholar]
- 36.Mor A., Philips M. R. (2006) Annu. Rev. Immunol. 24, 771–800 [DOI] [PubMed] [Google Scholar]
- 37.Bivona T. G., Quatela S., Philips M. R. (2006) Methods Enzymol. 407, 128–143 [DOI] [PubMed] [Google Scholar]
- 38.Kumar Y., Valdivia R. H. (2008) Cell Host Microbe 4, 159–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rzomp K. A., Moorhead A. R., Scidmore M. A. (2006) Infect. Immun. 74, 5362–5373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rzomp K. A., Scholtes L. D., Briggs B. J., Whittaker G. R., Scidmore M. A. (2003) Infect. Immun. 71, 5855–5870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heuer D., Rejman Lipinski A., Machuy N., Karlas A., Wehrens A., Siedler F., Brinkmann V., Meyer T. F. (2009) Nature 457, 731–735 [DOI] [PubMed] [Google Scholar]
- 42.Peterson E. M., de la Maza L. M. (1988) J. Bacteriol. 170, 1389–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roberts P. J., Der C. J. (2007) Oncogene 26, 3291–3310 [DOI] [PubMed] [Google Scholar]
- 44.Omerovic J., Hammond D. E., Clague M. J., Prior I. A. (2008) Oncogene 27, 2754–2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quilliam L. A., Kato K., Rabun K. M., Hisaka M. M., Huff S. Y., Campbell-Burk S., Der C. J. (1994) Mol. Cell. Biol. 14, 1113–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schachter J., Stephens R. S., Timms P., Kuo C., Bavoil P. M., Birkelund S., Boman J., Caldwell H., Campbell L. A., Chernesky M., Christiansen G., Clarke I. N., Gaydos C., Grayston J. T., Hackstadt T., Hsia R., Kaltenboeck B., Leinonnen M., Ojcius D., McClarty G., Orfila J., Peeling R., Puolakkainen M., Quinn T. C., Rank R. G., Raulston J., Ridgeway G. L., Saikku P., Stamm W. E., Taylor-Robinson D. T., Wang S. P., Wyrick P. B. (2001) Int. J. Syst. Evol. Microbiol. 51, 249. [DOI] [PubMed] [Google Scholar]
- 47.Bernstein-Hanley I., Coers J., Balsara Z. R., Taylor G. A., Starnbach M. N., Dietrich W. F. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 14092–14097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Howard J. (2008) Immunobiology 213, 367–375 [DOI] [PubMed] [Google Scholar]
- 49.Coers J., Bernstein-Hanley I., Grotsky D., Parvanova I., Howard J. C., Taylor G. A., Dietrich W. F., Starnbach M. N. (2008) J. Immunol. 180, 6237–6245 [DOI] [PubMed] [Google Scholar]
- 50.Collazo C. M., Yap G. S., Hieny S., Caspar P., Feng C. G., Taylor G. A., Sher A. (2002) Infect. Immun. 70, 6933–6939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shenoy A. R., Kim B. H., Choi H. P., Matsuzawa T., Tiwari S., MacMicking J. D. (2007) Immunobiology 212, 771–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taylor G. A., Feng C. G., Sher A. (2007) Microbes Infect. 9, 1644–1651 [DOI] [PubMed] [Google Scholar]
- 53.Al-Zeer M. A., Al-Younes H. M., Braun P. R., Zerrahn J., Meyer T. F. (2009) PLoS One 4, e4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Derbigny W. A., Hong S. C., Kerr M. S., Temkit M., Johnson R. M. (2007) Infect. Immun. 75, 1280–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lad S. P., Fukuda E. Y., Li J., de la Maza L. M., Li E. (2005) J. Immunol. 174, 7186–7193 [DOI] [PubMed] [Google Scholar]
- 56.Nagarajan U. M., Ojcius D. M., Stahl L., Rank R. G., Darville T. (2005) J. Immunol. 175, 450–460 [DOI] [PubMed] [Google Scholar]
- 57.Prantner D., Darville T., Nagarajan U. M. (2010) J. Immunol. 184, 2551–2560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nakafuku M., Satoh T., Kaziro Y. (1992) J. Biol. Chem. 267, 19448–19454 [PubMed] [Google Scholar]
- 59.Buchholz K. R., Stephens R. S. (2008) Infect. Immun. 76, 3150–3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cheng W., Shivshankar P., Zhong Y., Chen D., Li Z., Zhong G. (2008) Infect. Immun. 76, 942–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martens S., Parvanova I., Zerrahn J., Griffiths G., Schell G., Reichmann G., Howard J. C. (2005) PLoS Pathog. 1, e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhao Y. O., Khaminets A., Hunn J. P., Howard J. C. (2009) PLoS Pathog. 5, e1000288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tiwari S., Macmicking J. D. (2008) Methods Mol. Biol. 445, 407–415 [DOI] [PubMed] [Google Scholar]
- 64.Theofilopoulos A. N., Baccala R., Beutler B., Kono D. H. (2005) Annu. Rev. Immunol. 23, 307–336 [DOI] [PubMed] [Google Scholar]
- 65.Devitt A., Lund P. A., Morris A. G., Pearce J. H. (1996) Infect. Immun. 64, 3951–3956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Johnson R. M. (2004) Infect. Immun. 72, 3951–3960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Navarini A. A., Recher M., Lang K. S., Georgiev P., Meury S., Bergthaler A., Flatz L., Bille J., Landmann R., Odermatt B., Hengartner H., Zinkernagel R. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 15535–15539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fitzgerald K. A., Rowe D. C., Barnes B. J., Caffrey D. R., Visintin A., Latz E., Monks B., Pitha P. M., Golenbock D. T. (2003) J. Exp. Med. 198, 1043–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yamamoto M., Sato S., Hemmi H., Hoshino K., Kaisho T., Sanjo H., Takeuchi O., Sugiyama M., Okabe M., Takeda K., Akira S. (2003) Science 301, 640–643 [DOI] [PubMed] [Google Scholar]
- 70.Barbalat R., Lau L., Locksley R. M., Barton G. M. (2009) Nat. Immunol. 10, 1200–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McWhirter S. M., Barbalat R., Monroe K. M., Fontana M. F., Hyodo M., Joncker N. T., Ishii K. J., Akira S., Colonna M., Chen Z. J., Fitzgerald K. A., Hayakawa Y., Vance R. E. (2009) J. Exp. Med. 206, 1899–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Monroe K. M., McWhirter S. M., Vance R. E. (2009) PLoS Pathog. 5, e1000665. [DOI] [PMC free article] [PubMed] [Google Scholar]