Abstract
Interferon regulatory factor (IRF) family members, especially interferon regulatory factor-1 (IRF-1) and interferon regulatory factor-8 (IRF-8 or ICSBP), play important roles in interferon signaling in a wide range of host responses to infection and tumor growth. Interleukin-27 (IL-27), as a member of the IL-12 cytokine family, not only acts as a proinflammatory cytokine that regulates the differentiation of naive T helper cells but also possesses anti-inflammatory properties. IL-27 consists of EBI3 (Epstein-Barr virus-induced gene 3) and p28 subunits. Our previous work has shown that IRF-1 regulates IL-27 p28 gene transcription by specifically binding to the IRF-1 response element in the p28 promoter. In this study, we found that IRF-8-deficient macrophages were highly defective in the production of IL-27 p28 at both mRNA and protein levels. Circulating IL-27 p28 in serum was also decreased in IRF-8−/− mice in a septic shock model. Lipopolysaccharide, as a potent inducer of IL-27 p28 expression, could activate IRF-8 expression in a MyD88-dependent pathway, which in turn induced p28 gene transcription through NF-κB and/or IRF-8. Transcriptional analyses revealed that IRF-8 activated p28 gene transcription through binding to a site located at −57 to −48 in the p28 promoter overlapping the IRF-1 binding site. Consistent with this observation, overexpression of both IRF-8 and IRF-1 additively activated IL-27 p28 promoter. This study provides further mechanistic information regarding how signals initiated during innate and adaptive immune responses synergize to yield greater IL-27 production and sustained cellular immunity.
Keywords: Cytokine, Gene Regulation, Innate Immunity, Interferon, Macrophage, IL-27, IRFs
Introduction
Interferons (IFNs),2 including type I and type II IFNs, are cytokines that effectively enhance host defenses against various infections and immunosurveillance against tumors (1). IFN-γ, the only member of the type II IFN family, is produced by a variety of cell types, including NK cells, NK T cells, and CD4+ and CD8+ T cells, and essential for mounting cellular immune responses against intracellular pathogens, such as Mycobacteria tuberculosis and Leishmania major. IFN-γ also plays an important role in the chronic nature of inflammatory responses associated with atopic dermatitis, rheumatoid arthritis, and systemic lupus erythematosus (1). IFN-γ acts on a remarkable range of distinct cell populations, including immune and nonimmune cells. Of these, macrophages are among the most important. IFN-γ can directly activate microbicidal functions and promote antigen processing and presentation in macrophages (2). The impact of IFN-γ on macrophage phenotype and function is achieved by a profound alteration of the macrophage transcriptional program in response to IFN-γ. It has been reported that exposure to IFN-γ results in changes in expression of 25% of the mouse genome in macrophages (3). Among those genes, interferon regulatory factors (IRFs) are major members activated by IFN-γ and essential for many IFN-γ-mediated responses.
The IRF family consists of nine transcription factors that commonly possess a unique helix-turn-helix DNA-binding motif at the N terminus and an activation domain in the C terminus (4). IRF-1, the first member of the IRF family to be identified, targets different sets of genes in various cell types in response to diverse cellular stimuli and evokes appropriate innate and adaptive immune responses (4). Our previous studies demonstrated that IFN-γ-induced IRF-1 differentially regulates IL-12 p35 and p40 gene expression in macrophages (5). IRF-1 has been well established as a critical effector molecule in IFN-γ-mediated signaling and in the development and function of NK cells, NK T cells, and cytotoxic T lymphocytes (6–11). IRF-1 also has direct antiproliferative effects, thus acting as a tumor suppressor and tumor susceptibility gene (12). IRF-8, also known as ICSBP (interferon consensus sequence-binding protein), is restricted in its expression to myeloid and lymphoid cell lineages (13) and seems to have broader effects on microbe-induced cytokine production than IRF-1. IRF-8 can function both as a transcriptional repressor and as an activator, depending on the partners that it interacts with, and plays crucial roles in myeloid differentiation, generation of plasmacytoid dendritic cells (14), macrophage activation, and tumor suppression (4, 14). Previous studies indicated that IRF-8 activates IL-12 p35 gene transcription in synergy with IRF-1 (15) and enhances CCL5 gene transcription in cooperation with PU.1, NF-κB, and IRF-1 (16), respectively. However, little is known about whether IRF-8 can regulate IL-27 expression.
IL-27 is a newly discovered IL-12 family cytokine. Like all members of the family, it is composed of two different subunits: EBI3 (Epstein-Barr virus-induced gene 3) and p28 (17). IL-27 is mainly produced by macrophages and dendritic cells in response to microbial infection and plays an important role in autoimmune disease and host defense against infection (18). In combination with IL-2 and/or IL-12, IL-27 enhances the production of IFN-γ by naive CD4+ T cells and NK cells (18). The molecular basis for IL-27-mediated proinflammatory events involves activation of STAT-1 and T-bet, which increase expression of the IL-12 receptor β2 chain on naive CD4+ T cells (17, 19, 20). These effects result in enhanced Th1 responses. Mice deficient in EBI3 or the IL-27 receptor WSX-1 displayed increased susceptibility to L. major (21–23) due to reduced IFN-γ-producing Th1 cells during the initial stages of parasite infection, demonstrating a direct role for IL-27 in promoting protective T cell responses. Furthermore, exacerbation of experimental allergic asthma was observed in WSX-1-deficient mice through facilitating the T helper cell differentiation into the Th2 lineage (24). The inhibitory effects of IL-27 on Th2 cells are mediated by inhibition of the expression of the Th2 master transcription factor, GATA3 (25). Recent studies also indicate that IL-27 has potent anti-tumor effects. The anti-tumor activities of IL-27 against colon carcinoma and neuroblastoma mainly depend on CD8+ T cells, IFN-γ, and T-bet (26–29), and the anti-B16 melanoma effect seemed to act through suppression of angiogenesis (30).
IL-27 has also been shown to have anti-inflammatory effects in several murine disease models (18, 31). IL-27 receptor WSX-1 knock-out mice infected with Trypanosoma cruzi and M. tuberculosis displayed lower pathogen tissue burdens postchallenge (32, 33). However, the WSX-1 knock-out mice in these studies and in another study with Toxoplasma gondii infection (34) developed increased pathologic lesions and mortality. Murine models of experimental autoimmune encephalomyelitis and Toxoplasma infection demonstrated that elevated Th17 cells exacerbated disease in IL-27 receptor WSX-1-deficient mice (35, 36). It was further demonstrated that IL-27 inhibited the Th17 cell differentiation driven by IL-6 and transforming growth factor-β in a STAT-1-dependent, IFN-γ-independent pathway (35, 36), indicating that suppression of Th17 cells by IL-27 can inhibit both autoimmunity and pathogenic responses to infection. A recent report indicates that during intestinal inflammation Th17 cells suppressed Th1 cell development by inhibiting T-bet, IL-12R-β2, and osteopontin expression (37), demonstrating a reciprocal inhibition between Th17 and Th1 cells. The anti-inflammatory effects of IL-27 can also be achieved through induction of IL-10 in T cells. Recent studies demonstrate that IL-27 can activate CD4+, CD8+, and Foxp3− T cells to produce IL-10 (36, 38–41), which may be important for the immunomodulatory function of IL-27 in certain autoimmune diseases, such as experimental autoimmune encephalomyelitis and collagen-induced arthritis.
IL-27 expression is regulated through both MyD88-dependent and -independent pathways. It has been reported that in human macrophages and dendritic cells, the adaptor protein Trif mediated TLR3-induced IL-27 production (42), whereas another adaptor protein, MyD88, mediated TLR4/8-induced IL-27 production (42, 43). TLRs, such as TLR3, TLR4, and TLR7/8, activated by pathogen-associated molecular patterns, could induce IL-27 production through induction of type I interferon, which further increased the binding of IRF-1 and IRF-3 but not IRF-8 to the p28 promoter and led to production of IL-27 (42, 44–45). Consistent with this notion, TLR2 agonist Pam3Cys, which does not induce IFN-α/β, failed to induce IL-27 gene expression. These studies suggest that type I and type II interferon regulate IL-27 expression through different pathways.
Given the importance of IL-27 in regulating host responses and lack of knowledge about how this cytokine is regulated by IRF-8, the goal of this study was to investigate the molecular mechanisms that regulate p28 subunit gene expression in macrophages activated by bacterial LPS and IFN-γ, with a focus on intracellular and nuclear effectors. Our study reveals that both MyD88-dependent and -independent mechanisms of IRF-8 expression activate IL-27 p28 gene expression in synergy with IRF-1 at the level of transcription.
EXPERIMENTAL PROCEDURES
Mice
IRF-8+/− mice breeders were originally obtained from Dr. Keiko Ozato (National Institutes of Health). c-Rel−/− mice were originally supplied by Dr. Hsiou-Chi Liou (Weill Medical College of Cornell University). Dr. Aihao Ding and Dr. Erik Falck-Pedersen (Weill Medical College of Cornell University) provided the MyD88−/− mice originated from Dr. Akira. All mice used in the experiments were female at 6–8 weeks old, housed in cages with filter tops in a laminar flow hood, and fed food and acid water ad libitum at Saint Louis University Animal Facilities in accordance with the principles of Animal Care (National Institutes of Health publication number 85-23, revised 1985). For inducing endotoxic shock, mice were injected intraperitoneally with Escherichia coli lipopolysaccharide (Sigma) (200 μg/mouse). 4 h later, mice were sacrificed for collection of serum and for isolation of spleens.
Reagents
Antibodies for IRFs used in this study were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Recombinant mouse IFN-γ was purchased from Genzyme (Boston, MA). LPS from Escherichia coli 0217:B8 was purchased from Sigma. M-CSF was purchased from R&D Systems.
Cells
The murine macrophage cell line RAW264.7 (RAW cells) was obtained from ATCC and maintained in complete RPMI 1640 supplemented with 2 mm glutamine, 100 units/ml penicillin and streptomycin, and 10% fetal bovine serum (Sigma; endotoxin NMT 10.0 enzyme units/ml). Mouse peritoneal macrophages were obtained by lavage 3 days after injection of sterile 3% thioglycolate broth (1 ml intraperitoneally per mouse). Cells were washed and resuspended in complete RPMI medium. Macrophages were plated in 24-well tissue culture plates (1 × 106 cells/well). After a 2-h incubation to allow for adherence of macrophages, monolayers were washed three times to remove nonadherent cells and incubated with complete RPMI medium. The next day, IFN-γ (10 ng/ml) and LPS (1 μg/ml) were added for different times. Mouse bone marrow-derived macrophages (BMDMs) were generated from bone marrow cells obtained from mouse femurs. After lysis of red blood cells, 3 × 106 bone marrow cells were inoculated in 60-mm Petri dishes with complete RPMI culture medium in the presence of 10 ng/ml mouse recombinant M-CSF. M-CSF-containing complete medium was replenished at day 3. After a 7-day culture, the fully differentiated and matured BMDMs were used for experiments.
Plasmids
Murine IL-27 p28 promoter constructs were generated as described previously (43). Expression vectors IRF-8 and control LK440 were originally provided by Dr. Keiko Ozato (National Institutes of Health). Expression vector pAct-1 (IRF-1) and control pAct-C were generously provided by Dr. T. Taniguchi (University of Tokyo). NF-κB plasmids were originally provided by Dr. Kenneth M. Murphy (Washington University, St. Louis, MO). All plasmid DNA were prepared with Qiagen Endo-free Maxi-Prep kits (Qiagen).
Retroviral Packaging and Transduction
GP2-293 packaging cells (Clontech) at 60–70% confluence in 100-mm culture plates were transfected with 5 μg of each plasmid (pVSV-G and IRF-8 or MyD88) using FuGENE 6 (Roche Applied Science). Two days after transfection, supernatants were collected and centrifuged at 1,000 × g for 10 min to remove cell debris, followed by ultraspeed centrifugation at 50,000 × g for 90 min to pellet the virus. The pelleted viruses were resuspended overnight at 4 °C in 100 μl of 50 mm Tris-HCl, pH 7.8, 130 mm NaCl, and 1 mm EDTA. Bone marrow cells were infected with retrovirus containing supernatant at day 2 during their maturation in the presence of M-CSF. On day 7, mature macrophages were stimulated by IFN-γ or LPS with fresh complete medium for 5 h, followed by RNA isolation to detect mRNA expression by RT-PCR.
Reverse Transcription-PCR
Reverse transcription (RT) reactions were carried out as follows. 1-μg aliquots of total RNA were mixed with 1 μl of oligo(dT) primers (0.5 mg/ml), 1 μl of 10 mm dNTPs, and double-distilled H2O to equalize volumes of all samples at 12 μl. The mixture was heated at 65 °C for 5 min, quenched on ice, and spun down briefly, and 8 μl of master mix was added. The RT master mix consisted of 4 μl of 5× first strand buffer (Invitrogen), 2 μl of 0.1 m DTT, 1 μl of RNase inhibitor (40 units/μl; Invitrogen), and 1 μl of Superscript II (200 μl/μl; Invitrogen). The reaction was incubated at 42 °C for 60 min and then at 70 °C for 15 min, followed by a 4 °C soak. To each sample (in a 20-μl total volume) 80 μl of double-distilled H2O was added. 5 μl of diluted cDNA was used for each PCR of 25-μl volume. The following primers were used: for PCR amplification of the mouse p28 cDNA, CTCTGCTTCCTCGCTACCAC (sense) and GGGGCAGCTTCTTTTCTTCT (antisense); for mouse IRF-8 cDNA, sense (TGACACCAACCAGTTCATCCGAGA) and antisense (CACCAGAATGAGTTTGGAGCGCAA); for mouse IRF-1 cDNA, sense (ACAGGCCGATACAAAGCAGGAGAA) and ACGGTGACAGTGCTGGAGTTATGT (antisense); and for mouse GAPDH cDNA, AACTTTGGCATTGTGGAAGG (sense) and ACACATTGGGGGTAGGAACA (antisense).
Quantitative Real-time PCR
To determine the levels of mRNA expression by quantitative real-time PCR (qRT-PCR), we used a modified protocol. Briefly, cDNA samples converted from 1 μg of total RNA were diluted and studied at several concentrations. Diluted cDNA was mixed with a pair of primers (10 μm) targeting mouse p28, IRF-8, IRF-1, or GAPDH cDNA sequences as described above and with SYBR Green PCR master mix (Applied Biosystem, CA) in a 15-μl volume. PCR cycling was as follows: 2 min at 50 °C, 10 min at 95 °C for 1 cycle, followed by 40 cycles at 15 s at 95 °C, 1 min at 60 °C.
Enzyme-linked Immunosorbent Assays (ELISAs)
Supernatants from murine peritoneal macrophage cultures were harvested at 24 h after IFN-γ and LPS stimulation and stored at −70 °C. Mouse IL-27 p28 was detected using Quantikine ELISA kits (R&D Systems, Minneapolis, MN) according to the manufacturer's instructions. Concentrations were calculated by regression analysis of a standard curve.
Transfection Assay
Transient transfections were performed by electroporation. Briefly, for each condition, 0.4 ml of a RAW cell suspension containing 1 × 107 cells was mixed with 16 μg of total DNA (including reporter, effector, internal control, and carrier DNA) and electroporated in 0.45-cm electroporation cuvettes (Gene Pulser II, Bio-Rad) at 975 microfarads and 300 V in RPMI 1640 medium without serum. The transfected cells from different cuvettes were resuspended in RPMI 1640 containing 10% fetal bovine serum, 2 mm glutamine, 10 μm chloroquine, and antibiotics and were added to 24-well plates and incubated for 48 h prior to harvesting. To measure luciferase activity, cells were pelleted by centrifugation and resuspended in 100 μl of lysis buffer containing 125 mm Tris-phosphate, pH 7.8, 10 mm DTT, 10 mm 1,2-diaminocyclohexane-tetraacetic acid, 50% glycerol, 5% Triton X-100. Luciferase activity was measured in cell lysates. Transfection efficiency was routinely monitored by a β-galactosidase assay by co-transfection with 3 μg of pCMV-β-galactosidase plasmid. Variability in β-galactosidase activity between samples was typically within 5%. Lysates were used for both luciferase and β-galactosidase assays.
Nuclear Extract Preparation
Nuclear extracts for EMSA were prepared according to the methods of Schreiber et al. (46). Briefly, 1 × 107 RAW cells were washed and resuspended in 400 μl of buffer containing 10 mm HEPES, pH 7.9, 10 mm KCl, 0.1 mm EDTA, 1 mm DTT, 0.5 mm phenylmethylsulfonyl fluoride for 15 min on ice. Cells were lysed in 0.6% Nonidet P-40 with inversion for 10 s. The homogenate was centrifuged for 5 min, and the nuclear pellet was resuspended in ice-cold buffer containing 20 mm HEPES, pH 7.9, 0.4 m NaCl, 1 mm EDTA, 1 mm DTT, 1 mm phenylmethylsulfonyl fluoride at 4 °C for 30 min with rotating. Following centrifugation for 10 min, the supernatant was either used immediately or frozen at −70 °C.
Light Shift Chemiluminescent EMSA
Light shift chemiluminescent EMSA was performed according to the manufacturer's protocol (Pierce). Briefly, single-stranded wild type and mutant oligonucleotides were labeled with biotin using a biotin 3′-end DNA labeling kit (Pierce). Equal amounts of labeled and complementary oligonucleotides were annealed to prepare double-stranded wild type and mutant probes by heating at 90 °C for 2 min, followed by slowly cooling to room temperature for 30 min. The labeled double-stranded probes were mixed with 10 μg of crude nuclear extracts and incubated at room temperature for 20–30 min in the presence of 1 μg of poly(dI-dC). The mixture was then fractionated through a 5% native polyacrylamide gel in 0.5× TBE buffer for about 1 h at 100 V. The gel was transferred to positively charged nylon membrane at 4 °C for 1 h at 100 V. When transfer was completed, the membrane was cross-linked at 120 mJ/cm2 using a commercial UV-light cross-linker instrument equipped with 254-nm bulbs. Then the membrane was blocked and applied with streptavidin-horseradish peroxidase conjugate (1:300 dilution) for 15 min, followed by thorough washing, and exposed to x-ray film after incubation with the chemiluminescence reagents. The x-ray film was developed according to the manufacturer's instruction.
DNA Affinity Binding Assay
Complementary biotinylated oligonucleotides encompassing the IRF-1 site in the mouse p28 promoter were synthesized and annealed. Two micrograms of biotinylated dsDNA were conjugated to 100 μl of streptavidin-bound magnetic beads (Dynabeads, M280, Dynal (Lake Success, NY)) in binding/washing buffer (10 mm Tris-HCl, pH 8.0, 1 mm EDTA, 0.1 m NaCl) for 30 min at room temperature. DNA-conjugated beads were then blocked by 0.5% bovine serum albumin in TGEDN buffer (120 mm Tris-HCl, pH 8.0, 1 mm EDTA, 0.1 m NaCl, 1 mm DTT, 0.1% Triton X-100, and 10% glycerol) at room temperature for 1 h. Beads were washed once in TGEDN buffer and resuspended in 50 μl of TGEDN. Ten microliters of beads conjugated to 2 μg of DNA were equilibrated with TGEDN buffer and incubated with 500 μg of RAW cell nuclear extracts and 20 μg of Herring sperm DNA (Sigma) at 4 °C for 2 h. Beads were washed in TGEDN buffer, and bound materials were eluted in 20 μl of the same buffer supplemented with 0.5% SDS and 1 m NaCl. Eluted materials were separated by 12% SDS-PAGE and detected by immunoblot analysis using rabbit anti-IRF-1 or anti-IRF-8 Ab with the enhanced chemiluminescence kit (PerkinElmer Life Sciences).
Western Blotting
SDS-PAGE was performed with 40–100 μg of nuclear extracts. Gels were transferred to PVDF membranes and blocked in 5% nonfat milk in Tris buffer, pH 8.0. Primary antibody was added at the concentration of 1 μg/ml in Tris buffer containing 5% milk powder and left overnight at 4 °C. After extensive washing, secondary antibody conjugated to horseradish peroxidase was added at a 1:5000 dilution in 5% milk. After extensive washing, blots were subjected to enhanced chemiluminescence detection (PerkinElmer Life Sciences).
Chromatin Immunoprecipitation (ChIP) Assay
The ChIP procedure was performed using an assay kit following the manufacturer's instructions (Upstate Biotechnology, Inc., Lake Placid, NY). Briefly, 1 × 107 peritoneal macrophages from wild type C57BL/6 mice were stimulated with IFN-γ (10 ng/ml) or LPS (1 μg/ml) for 2 h and then cross-linked by 1% formaldehyde for 10 min at 37 °C. Nuclei were prepared and subjected to sonication to obtain DNA fragments ranging from 200 to 1000 bp. Chromatin fractions were precleared with protein A-agarose beads, followed by immunoprecipitation overnight at 4 °C with 3 μg of anti-IRF-8, anti-IRF-1 or control antibody. Cross-linking was reversed for 4 h at 65 °C, followed by proteinase K digestion. DNA was purified and subjected to qRT-PCR. The input DNA was diluted 200 times prior to PCR amplification. The input and immunoprecipitated DNA were amplified by PCR using primers encompassing the IRF response element (RE) in the mouse p28 promoter (5′ primer, CCCTCTGGGAAGGGAAATTACGTT; 3′ primer, CCTGTCAAACTTTCCCAACC).
siRNA
IRF-8 siRNA expression plasmids were generated by cloning the siRNA corresponding to the coding sequence of the mouse IRF-8 gene (IRF-8 siRNA 1, gatccccACCACCACCTGCCTTGAAGttcaagagaCTTCAAGGCAGGTGGTGGTttttta; IRF-8 siRNA 2, gatccccACTCATTCTGGTGCAGGTAttcaagagaTACCTGCACCAGAATGAGTttttta) into an siRNA expression vector pSUPER.neo (OligoEngine, Seattle, WA) according to the manufacturer's instructions. A scramble sequence (gatccccTGTAGATGGGTACGCGCTCttcaagagaGAGCGCGTACCCATCTACAttttta) that targets no known mRNA sequences was cloned into the same vector and used as negative control. Briefly, equimolar amounts of complementary sense and antisense strands were separately mixed, annealed, and slowly cooled to 10 °C in a 50-μl reaction buffer (100 mm NaCl and 50 mm HEPES, pH 7.4). The annealed oligonucleotides were inserted into the BglII/HindIII sites of pSUPER.neo vector. IRF-8 siRNA plasmids and scramble control were transfected into RAW cells by Lipofectamine (Invitrogen). 48 h later, the transfected cells were stimulated with IFN-γ for additional 4 h, followed by collection of nuclear extract for Western blot and EMSA.
Statistical Analysis
Student's t test was performed wherever applicable. S.D. of the mean is shown unless otherwise indicated (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
RESULTS
IL-27 p28 Expression Is Defective in IRF-8-deficient Macrophages
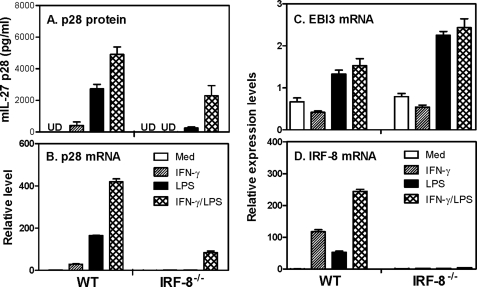
Our previous study demonstrated that IFN-γ-induced IL-27 p28 gene expression requires IRF-1 (43). Because IFN-γ also activates IRF-8 expression, we wanted to know if IRF-8 plays a role in IFN-γ-induced p28 gene expression. Mouse peritoneal macrophages elicited from wild type (WT) or IRF-8−/− mice were stimulated with IFN-γ, LPS, or IFN-γ plus LPS for 24 h, followed by collection of culture supernatants for measurement of IL-27 p28 protein secretion by ELISA. Meanwhile, cells treated with these stimuli for 4 h were used for RNA isolation and p28 mRNA detection by quantitative real-time PCR. The results showed that both p28 protein secretion (Fig. 1A) and mRNA expression (Fig. 1B) were almost completely abrogated in IRF-8−/− macrophages compared with the WT cells in response to IFN-γ or LPS. To determine if the other subunit of IL-27, EBI3 is also regulated by IRF-8, we measured EBI3 mRNA expression from the same cDNA used for the p28 detection. Consistent with our previous finding that the p28 subunit is the limiting factor for making biological IL-27, EBI3 mRNA was only slightly induced by LPS or IFN-γ plus LPS in WT cells. In contrast to p28, the EBI3 mRNA expression was even increased in IRF-8−/− cells (Fig. 1C), suggesting that IRF-8 specifically controls p28 gene expression. Our data further confirm that IRF-8 was induced by IFN-γ and LPS signals, and combination of these two signals further enhanced its expression in WT but not IRF-8−/− macrophages (Fig. 1D). Interestingly, combination of these two stimuli still induced a considerable amount of p28 protein production (Fig. 1A) and p28 mRNA expression (Fig. 1B) in IRF-8−/− cells, suggesting that other factors induced by IFN-γ and LPS may partially compensate for the impaired p28 expression in IRF-8-deficient macrophages.
FIGURE 1.
Role of IRF-8 in IL-27 p28 gene expression. A, 1 × 106 mouse peritoneal macrophages isolated from WT and IRF-8−/− mice were inoculated into each well of 24-well plates with 1 ml of RPMI 1640 complete medium. The cells were stimulated with IFN-γ (10 ng/ml), LPS (1 μg/ml), or IFN-γ plus LPS for 24 h, followed by collection of supernatant for measurement of IL-27 p28 protein secretion by ELISA. Results shown are mean plus S.D. (error bars) from three experiments. 3 × 106 peritoneal macrophages of WT and IRF-8−/− mice were stimulated with IFN-γ, LPS, and IFN-γ plus LPS for 4 h, followed by extraction of total RNA for detection of p28 mRNA expression (B), for measurement of EBI3 mRNA expression (C), and for evaluation of IRF-8 mRNA expression (D) by qRT-PCR. qRT-PCR data were normalized relative to GAPDH mRNA expression levels in each respective sample and further normalized to the results from the untreated group (medium; Med), which was set as 1.
Reconstitution of IRF-8 in IRF-8−/− Macrophages Rescues IL-27 p28 Expression
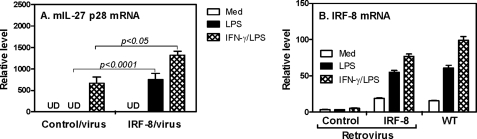
Because studies in knock-out mice can be complicated by developmental effects associated with the deleted gene, next we determined to exclude the possibility that other genes affected by IRF-8 deletion might contribute to the defect of p28 expression in IRF-8-deficient cells. We reconstituted IRF-8 in IRF-8-deficient BMDMs by retroviral transduction and then tested if the defect in p28 expression could be rescued. As shown in Fig. 2A, IRF-8−/− BMDM had greatly diminished responses to LPS, which, after reconstitution, were strongly enhanced compared with control retrovirus-transduced cells, indicating a direct effect of IRF-8 on p28 gene expression. The p28 mRNA expression was further enhanced in reconstituted cells upon IFN-γ and LPS co-stimulation (Fig. 2A). To assess the levels of IRF-8 expression reconstituted by IRF-8 retrovirus, we measured the IRF-8 mRNA expression in cells transduced with control and IRF-8 retrovirus as well as in WT cells. The data showed that IRF-8 mRNA expression in deficient BMDMs was reconstituted to a level similar to wild type cells in response to LPS (∼50-fold induction, Fig. 2B), further supporting the important role for IRF-8 in LPS-mediated p28 gene expression.
FIGURE 2.
Reconstitution of IRF-8 in IRF-8-deficient BMDMs. 1 × 106 bone marrow cells isolated from femurs of IRF-8−/− mice were inoculated into each well of 24-well plates with 1 ml of RPMI 1640 complete medium with 10 ng/ml M-CSF and 10% fetal bovine serum. At day 2 and day 4, the cells were infected with retroviruses encoding IRF-8 or green fluorescent protein for two rounds. The transduced BMDMs and BMDMs differentiated from WT mice were treated with LPS or IFN-γ plus LPS for 4 h. Then total RNA was extracted from the activated BMDMs, and 1 μg of RNA was used for reverse transcription, followed by measurement of mouse IL-27 p28 (A) and IRF-8 (B) mRNA expression by qRT-PCR. Data are representative of two experiments with similar results. UD, undetectable. Error bars, S.D.
IRF-8 Activates IL-27 p28 Gene Transcription
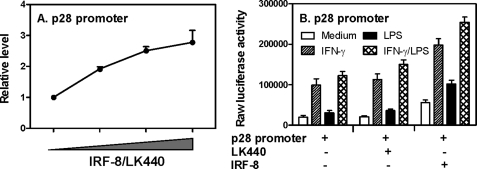
We next wanted to know if IRF-8 could induce p28 promoter activation. We transiently co-transfected a mouse p28 promoter-luciferase construct with different amounts of IRF-8 expression vector into RAW cells by electroporation, followed by measurement of luciferase activity in cell lysates. As shown in Fig. 3A, IRF-8 activated the p28 promoter in a dose-dependent manner, indicating that IRF-8-mediated p28 gene induction is indeed regulated at the level of transcription. Moreover, cells co-transfected with the p28 promoter and the IRF-8-expressing vector showed enhanced promoter activities in response to IFN-γ and LPS treatments compared with cells co-transfected with the empty vector, LK440 (Fig. 3B), further demonstrating that IRF-8 activates p28 gene transcription under both basal and inducible conditions.
FIGURE 3.
IRF-8 activates p28 promoter. A, the p28 promoter-luciferase reporter construct (−739) was transiently transfected into RAW cells together with increasing amounts of an IRF-8 expression vector or control vector (LK440), and the total amount of plasmids in the mixture was maintained at a constant 6.0 μg with varying portions of the effector and the control vector. Luciferase activities were measured in cell lysates 40 h after transfection and normalized to the activity obtained with cells only transfected with the empty vector (LK440), which was set as 1. Results shown are mean plus S.D. (error bars) of three independent experiments. B, the mouse p28 promoter-luciferase reporter construct was transiently co-transfected with IRF-8 and control vector into RAW cells by electroporation. The transfected cells were stimulated with IFN-γ (10 ng/ml), LPS (1 μg/ml), or IFN-γ plus LPS for 7 h, and the luciferase activity was measured in cell lysates. Results shown are mean plus S.D. of five independent experiments.
MyD88 and NF-κB Mediate LPS-induced IRF-8 and IL-27 p28 Gene Expression
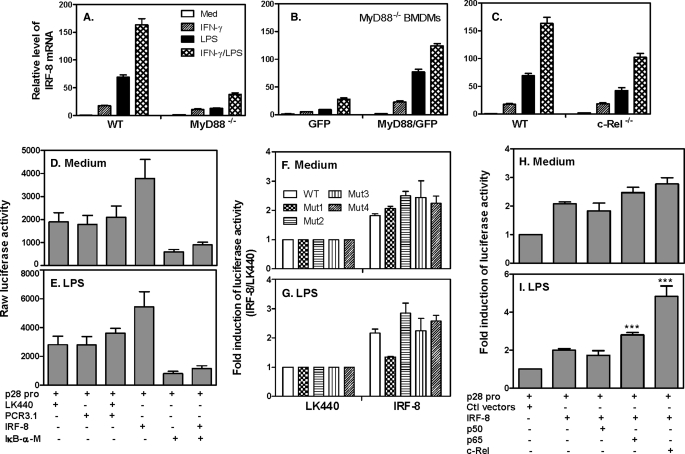
Because LPS-induced p28 gene expression depended on IRF-8 (Fig. 1A) and IL-27 p28 production is regulated through both MyD88-dependent and -independent pathways (42, 43), we decided to look at the role of MyD88 in LPS-induced IRF-8 expression. The results showed that IRF-8 expression induced by LPS was significantly reduced in MyD88−/− macrophages compared with WT control cells (Fig. 4A), and reconstitution of MyD88 by lentivirus completely rescued the IRF-8 gene expression in MyD88−/− cells to a level similar to WT control cells (Fig. 4B), indicating a critical role of MyD88 in signaling LPS-induced IRF-8 expression. Because NF-κB plays critical roles in LPS-induced production of many proinflammatory cytokines and c-Rel has been shown by us and others to be important for IL-12 and IL-27 production (5, 43), we next wanted to know if c-Rel was also involved in LPS-induced IRF-8 expression. The data indicated that NF-κB c-Rel only plays a partial role in LPS-induced IRF-8 expression because IRF-8 mRNA expression was reduced about 40% in c-Rel−/− cells treated with LPS or LPS plus IFN-γ (Fig. 4C).
FIGURE 4.
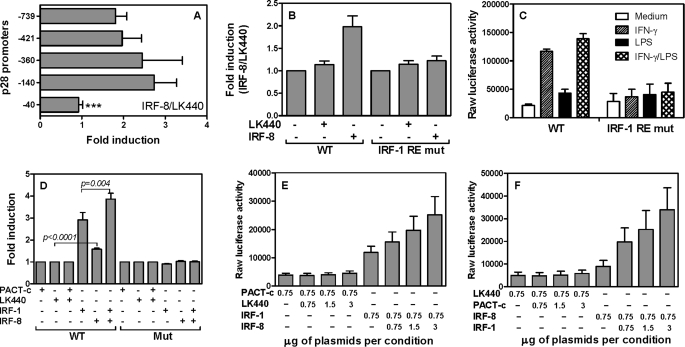
MyD88 and NF-κB mediate IRF-8-induced p28 gene transcription. Total RNA was extracted from WT and MyD88−/− peritoneal macrophages (A), MyD88−/− BMDMs infected with lentivirus encoding MyD88 or green fluorescent protein (B), and peritoneal macrophages of WT and c-Rel−/− mice (C). All macrophages were stimulated with IFN-γ, LPS, or IFN-γ plus LPS for 4 h, followed by measurement of IRF-8 mRNA expression by qRT-PCR. Data are from one of two experiments with similar results. RAW cells were transiently transfected with the p28 promoter (−3076/+129) and IkB-α mutant as well as IRF-8 expression vectors or their control vectors at a 1:1 molar ratio. Transfected cells were left either untreated (D) or treated with LPS (E) for 7 h, followed by measurement of luciferase activity in cell lysates. RAW cells were transiently tranfected with WT and four putative NF-κB mutant p28 promoter constructs. Transfected cells were left either untreated (F) or treated with LPS (G) for 7 h. Luciferase activity was normalized to the activity of cells transfected with the control vector (LK440) as -fold induction. RAW cells were transiently transfected with the p28 promoter together with an expression vector for NF-κB p50, p65, or c-Rel with IRF-8 or their control vectors at a 1:1 molar ratio (effector to reporter). Transfected cells were either untreated (H) or treated with LPS (I) for 7 h, followed by measurement of luciferase activity. Data are expressed as relative induction over the control vector-transfected cells of each cotransfection, which was set as 1. All results shown in D–I are mean plus S.D. (error bars) from three or four independent experiments.
To determine if NF-κB mediated IRF-8-activated p28 gene transcription, we co-transfected IRF-8 and IκB-α mutant with the p28 promoter into RAW cells, followed by measurement of luciferase activity. IκB-α mutant can effectively block nuclear translocation of NF-κB and suppress NF-κB activation (43). Suppression of NF-κB led to complete abrogation of p28 gene transcription induced by IRF-8 (Fig. 4, D and E). There are four putative NF-κB binding sites in the p28 promoter, which are located at −3051/−3042 (NF-κB 1), −1230/−1221 (NF-κB 2), −1189/−1178 (NF-κB 3), and −1146/−1137 (NF-κB 4) in the p28 promoter. We mutated each of these and designated them as mut1, mut2, mut3, and mut4, respectively (43). To further explore the molecular basis of NF-κB-mediated p28 gene transcription in response to IRF-8 stimulation, we cotransfected IRF-8 expression plasmid with these four NF-κB mutant p28 promoters into RAW cells and treated these cells with or without LPS (Fig. 4, F and G). Fig. 4, F and G, respectively, presents -fold inductions of p28 promoter activity in IRF-8-transfected versus LK440-transfected cells in unstimulated and LPS-stimulated conditions, with the LK440-transfected cell responses set arbitrarily as 1. In fact, the raw luciferase activities were increased about 2-fold by LPS compared with untreated cells transfected with the control vector LK440. In cells transfected with IRF-8, the raw luciferase activities were further enhanced about 2.5-fold compared with LK440-transfected cells (data not shown). As shown in Fig. 4G, the luciferase activity was almost completely abolished in NK-κB mut1-transfected cells treated with LPS in the presence of overexpressed IRF-8, suggesting that this NF-κB site located between −3051 and −3042 in the p28 promoter is critical for LPS/IRF-8-mediated p28 gene transcription. To further determine which component of the NF-κB family mediated IRF-8-activated p28 gene transcription, we co-expressed NF-κB p65, p50, and c-Rel expression vectors with the p28 promoter and IRF-8 and stimulated the transfected cells with or without LPS. The data indicated that NF-κB p50 did not increase IRF-8- and LPS-induced p28 promoter activity, whereas c-Rel and p65 significantly enhanced p28 promoter activity in IRF-8-overexpressing cells stimulated with LPS (Fig. 4I).
IRF-8 Activates the p28 Promoter in Synergy with IRF-1 through the IRF-1 Response Element
To elucidate the molecular basis of IRF-8-mediated transcriptional induction of p28, we decided to localize the functional IFN-γ/IRF-8 response element in the p28 promoter. Several 5′ deletion constructs of the p28 promoter were transiently co-transfected with the IRF-8 expression vector or empty control vector into RAW cells, followed by measurement of luciferase activity in cell lysates. As shown in Fig. 5A, the responses to IRF-8 were similar among all 5′ deletion constructs except the minimal construct −40, which completely lost responsiveness to IRF-8, indicating that a major IRF-8 RE in the p28 promoter was probably located between −140 and −40. Because our previous study identified an IRF-1 RE localized to this region at −57/−48, we reasoned that IRF-8 may activate the p28 promoter through this IRF-1 binding site. Then we transiently co-transfected the wild type and IRF RE mutant p28 promoter constructs with the IRF-8 expression or empty vectors into RAW cells, followed by measurement of luciferase activities. As shown in Fig. 5B, the response of the mutant promoter to overexpressed IRF-8 was completely abolished compared with the response detected with the wild type promoter. This was true in transfected cells stimulated with IFN-γ as well (Fig. 5C). These results indicate that this IRF-1 RE is critical for responsiveness of the p28 promoter to both IRF-8 and IRF-1.
FIGURE 5.
IRF-8 synergistically interacting with IRF-1 activates the p28 promoter through the IRF-1 binding site. A, 5 μg of WT p28 promoter-luciferase reporter construct (−739) or a series of 5′-end deletion constructs, as indicated, was transiently transfected into RAW cells by electroporation, together with 0.75 μg of IRF-8 or control vector LK440. 40 h later, luciferase activity was measured in cell lysates and expressed as relative activity (the activity of IRF-8-cotransfected reporter over that of control empty vector-cotransfected reporter, which was set as 1). B, RAW cells were transiently transfected with 5 μg of p28 promoter-luciferase WT (−739) or IRF-1 RE mutant promoter constructs with or without 0.75 μg of IRF-8 expression vector or its control vector (LK440). C, 5 μg of WT and IRF-1 RE mutant p28 promoter-luciferase reporter constructs were transiently transfected into RAW cells by electroporation. The transfected cells were stimulated with IFN-γ, LPS, or IFNγ plus LPS for 7 h, followed by measurement of luciferase activity in cell lysates. D, 5 μg of WT or IRF-1 mutant p28 promoter constructs was co-transfected into RAW cells with 1.5 μg of IRF-1 and/or 0.75 μg of IRF-8 expression vectors or their respective control vectors, PACT-C (empty vector for IRF-1) and LK440 (empty vector for IRF-8). 40 h later, luciferase activities were measured in cell lysates. E, 5 μg of WT p28 promoter (−739) was co-transfected with a fixed amount of IRF-8 (0.75 μg) and various amounts of IRF-1 (from 0.75 to 3 μg), and their respective control vectors into RAW cells as indicated, or vice versa (F). 40 h later, luciferase activity was measured in cell lysates. All results shown are mean plus S.D. (error bars) of 3–6 independent experiments.
Considering the above findings that IRF-1 and IRF-8 both activated the p28 promoter through the same site, we were interested to know if IRF-1 and IRF-8 could activate the p28 promoter synergistically. The IRF-1 and IRF-8 expression vectors or their empty control vectors PACT-c and LK440 were transiently co-transfected with the WT and IRF RE mutant p28 promoter constructs into RAW cells for luciferase measurement. As shown in Fig. 5D, IRF-1 and IRF-8 each individually activated the WT p28 promoter, with the former causing ∼3-fold induction and the latter causing ∼2-fold induction, indicating a stronger effect of IRF-1 than IRF-8 on p28 promoter activation. Moreover, co-transfection of both plasmids additively activated the p28 gene transcription associated with the WT promoter but not the IRF RE mutant promoter construct, demonstrating that IRF-8 and IRF-1 indeed work together to activate the p28 promoter through the same IRF RE (Fig. 5D). To further understand the additive effects between IRF-1 and IRF-8 on p28 gene transcription, we transfected the RAW cells with either a fixed amount of IRF-8 plus various amounts of IRF-1 or vice versa. The results confirm that IRF-1 and IRF-8 indeed additively activated the p28 promoter (Fig. 5, E and F).
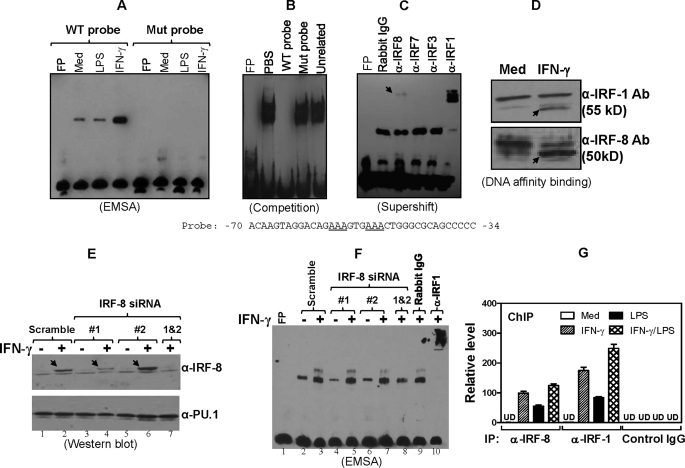
IRF-8 Binds to the p28 Promoter Both in Vitro and in Vivo
To further explore the molecular mechanisms of whether IRF-8-mediated p28 gene transcription is a direct or indirect event, we performed EMSA using nuclear extracts isolated from RAW cells stimulated with IFN-γ or LPS using a sequence of the p28 promoter harboring either WT or mutant IRF RE as probes. As shown in Fig. 6A, there was one major nuclear DNA-binding complex formed with the wild type probe. More importantly, the intensity of this complex was significantly increased upon IFN-γ but not LPS treatment. The latter is because this region in the p28 promoter only contains an IFN-γ but not LPS RE. This binding activity was completely abolished with the IRF RE mutant probe, indicating the binding specificity. The competition EMSA further confirmed the sequence specificity of the binding to the p28 promoter, because the binding was completely competed by WT cold probe but not the mutant cold probe (Fig. 6B). To further determine if IRF-8 was a part of the DNA-nuclear protein binding complex, we performed a “supershift” experiment with several IRF antibodies. As shown in Fig. 6C, this DNA-nuclear protein complex indeed contained IRF-8 because an anti-IRF-8 antibody (as indicated by the arrow) but not IRF-3- and IRF-7-specific antibodies was able to retard its mobility. Consistent with our previous study (43), IRF-1 was apparently a major component in the complex because anti-IRF-1 antibody caused a markedly supershifted band (Fig. 6C). Because IRF-8 was shown to bind weakly to the p28 promoter in the supershift assay (Fig. 6C), we performed an alternative DNA affinity binding assay to further confirm the IRF-8 binding. The binding nuclear proteins were pulled down by immunoprecipitation with biotin-labeled probe containing the IRF RE, followed by detection of the binding proteins by Western blot. As shown in Fig. 6D, antibodies against both IRF-1 and IRF-8 detected an IFN-γ-responsive band, with stronger binding intensity displayed with anti-IRF-8 than anti-IRF-1 antibody. We do not yet know the reason for these different binding patterns between conventional EMSA and DNA affinity binding assay. Nevertheless, both experiments (Fig. 6, C and D) demonstrate that both IRF-8 and IRF-1 bind to the p28 promoter in vitro.
FIGURE 6.
IRF-8 binds to the p28 promoter in vitro and in vivo. A, nuclear extracts were isolated from RAW cells stimulated with IFN-γ or LPS for 4 h. Light shift chemiluminescent EMSA was performed with 10 μg of nuclear extract for each sample and a double-stranded biotin-labeled oligonucleotide probe containing the IRF RE from the p28 promoter (sequence given with the critical IRF RE underlined) or the IRF RE mutated sequence. B, competitive EMSA was performed with biotin-labeled WT −70/−34 “hot” probe and various unlabeled “cold” competitors as indicated (molar ratio of cold to hot probes was 50:1) from IFN-γ-stimulated macrophages. C, supershift EMSA was performed with the WT probe and nuclear extracts from IFN-γ-stimulated macrophages. 2 μg of a series of IRF antibodies and isotype-matched control rabbit IgG were used. The IRF-8-related complex is indicated by an arrow. D, 10 × 106 RAW cells were stimulated with IFN-γ for 4 h, followed by isolation of nuclear extract for the DNA affinity binding assay. 500 μg of nuclear extract per condition was used for the assay. The bound proteins were analyzed by Western blot using both anti-IRF-1 and anti-IRF-8 antibodies. Data shown are one of two experiments with similar results. E, 8 μg of scramble siRNA and IRF-8 siRNA 1 and 2 plasmids were transfected into RAW cells by Lipofectamine. 48 h later, the transfected cells were stimulated with or without IFN-γ for 4 h, followed by nuclear extraction. 100 μg of nuclear extract per condition was used for Western blot with an anti-IRF-8 antibody (top). The IRF-8 is indicated by an arrow. The blot was stripped and reanalyzed with an anti-PU.1 antibody (bottom). F, 10 μg of nuclear extract per condition was used for EMSA as described previously, and 2 μg of anti-IRF-1 antibody and its control IgG were used to perform the supershift assay. G, 10 × 106 WT mouse peritoneal macrophages were treated with IFN-γ (10 ng/ml), LPS (1 μg/ml), and IFN-γ plus LPS for 2 h, followed by ChIP analysis performed according to the manufacturer's protocol. 3 μg of anti-IRF-8 antibody, anti-IRF-1 antibody, and control IgG were used in the intraperitoneal step. The control antibody was an isotype-matched IgG. Data were normalized relative to input in each respective sample and further normalized to the sample from the untreated group, which was set as 1. FP, free probe; Mut probe, mutant probe; UD, undetectable. Error bars, S.D.
Our data indicate that the binding activity following IFN-γ stimulation was predominantly composed of IRF-1. Thus, it is important to know if IRF-1 binds to the same site in the p28 promoter in IRF-8-deficient cells. We designed two IRF-8 siRNAs, cloned them into a pSUPPER vector, and validated their silencing efficiency by testing IRF-8 protein expression. The Western blot data (Fig. 6E) indicated that one of the IRF-8 siRNAs (siRNA 1) significantly silenced IRF-8 protein expression (lane 4), but the other IRF-8 siRNA (siRNA 2) showed little effect (lane 6). Co-transfection of cells with both IRF-8 siRNA 1 and 2 resulted in the most silencing effect on IRF-8 production (lane 7). Therefore, the nuclear extract isolated from these IRF-8 siRNA 1/2 co-transfected cells was used to test the IRF-1 binding. As shown in Fig. 6F, the binding activity was increased by IFN-γ treatment, and the binding intensity was similar among cells transfected with scramble (lane 3), IRF-8 siRNA 1 (lane 5), and IRF-8 siRNA 2 (lane 7). However, the binding intensity was reduced in cells transfected with both siRNA 1 and 2 (lane 8), which also showed the least IRF-8 expression, suggesting that IRF-8 partially regulates the IRF-1 binding to the p28 promoter. A complete supershift of the band by an anti-IRF-1 antibody in the same assay further confirmed the IRF-1 binding (lane 10).
For more biological relevance, we next tested if this binding activity between IRF-8 and the p28 promoter also existed in vivo. A ChIP assay was performed in mouse peritoneal macrophages. The region of the p28 promoter containing the IRF RE that was examined in the ChIP assay is illustrated in supplemental Fig. 3. Fig. 6G demonstrated that both IFN-γ and LPS could induce strong IRF-8 binding to this region of the p28 promoter, with the strongest binding in cells stimulated with LPS plus IFN-γ. The binding was detected specifically with the anti-IRF-8 antibody but not with the control IgG. For comparison, we also used an anti-IRF-1 antibody to pull-down the binding DNA. Consistent with our previous report (43), IRF-1 bound to the p28 promoter in cells treated by IFN-γ and LPS with enhanced binding upon LPS and IFN-γ co-stimulation (Fig. 6G).
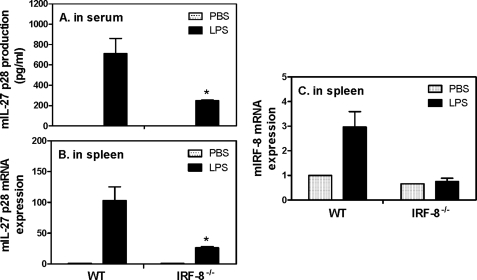
IL-27 p28 Production Is Decreased in IRF-8−/− Mice Challenged with LPS
To further confirm the inductive role of IRF-8 in IL-27 p28 production in vivo, we employed an endotoxic shock model by injecting LPS intraperitoneally as previously described (47). As shown in Fig. 7A, LPS induced significant amounts of IL-27 p28 in the serum of WT mice, whereas the levels of p28 were decreased about 65% in IRF-8−/− mice. Consistent with the reduced protein expression, the p28 mRNA expression was also decreased ∼70% in the spleens of IRF-8−/− mice compared with WT mice (Fig. 7B), suggesting a critical role for IRF-8 in IL-27 p28 production during host defense against microbial infection. In addition, our data showed that IRF-8 mRNA expression was induced by LPS in WT but not in IRF-8−/− mice in this endotoxic shock model (Fig. 7C), further supporting the inductive role of IRF-8 in IL-27 p28 gene expression in vivo.
FIGURE 7.
IL-27 p28 production is reduced in IRF-8−/− mice during endotoxic shock. Four female IRF-8−/− mice and their littermates (aged 6–8 weeks) were injected with LPS (200 μg/mouse) intraperitoneally. 4 h after LPS injection, mice were sacrificed and serums were collected for measurement of IL-27 p28 protein production by ELISA (A). Meanwhile, spleens were used to extract total RNA for measurement of IL-27 p28 (B) and IRF-8 (C) mRNA expression by qRT-PCR. Data were normalized relative to GAPDH mRNA expression levels in each respective sample and further normalized to the sample from the untreated group, which was set as 1. Mice injected with PBS served as controls. *, p < 0.05 between groups of WT and IRF-8−/− mice after LPS treatment. Error bars, S.D.
DISCUSSION
Since IL-12 was first discovered by Dr. George Trinchieri in 1989 (48), several other IL-12 related cytokines have been identified in recent years, including IL-23 (49), IL-27 (17), and IL-35 (50). Like IL-12, all IL-12 family members are heterodimeric cytokines consisting of two different subunits. Among those, IL-27 has been shown to have unique roles in controlling adaptive immune responses. It induces Th1 cell differentiation, inhibits Th2 and Th17 cell development, and possesses both pro- and anti-inflammatory properties (51), which make it an important immunomodulator for induction of host responses against invasive pathogens and for prevention of host tissue damage from excessive immune action (18).
In this study, we found that IRF-8 plays an essential role in IL-27 p28 production in that p28 mRNA expression and protein production were completely abolished in IRF-8-deficient macrophages in response to IFN-γ stimulation (Fig. 1, A and B). In contrast with IRF-1 deficiency that resulted in only moderate reduction of p28 expression in response to LPS (43), p28 mRNA expression and protein production were significantly impaired in IRF-8-deficient cells (Fig. 1, A and B), suggesting broader effects of IRF-8 than IRF-1 on IL-27 regulation. Consistent with our previous report that the p28 subunit controls biological IL-27 production (43), EBI3 was only slightly induced upon IFN-γ and LPS stimulation, and its expression did not require IRF-8 (Fig. 1C). It is worth noting that the levels of p28 expression did not correlate with the levels of IRF-8 expression induced by IFN-γ and LPS in that higher induction of IRF-8 induced by IFN-γ did not lead to higher p28 expression (Fig. 1D). This phenomenon suggests that LPS-triggered pathways, such as NF-κB, in cooperation with LPS-induced IRF signals, such as IRF-8, induce much more robust IL-27 p28 production than only IFN-γ/IRF signals, which further indicates the importance of NF-κB in p28 gene expression. Interestingly, combination of IFN-γ and LPS treatments could partially rescue the defect in p28 production in IRF-8-deficient cells (Fig. 1A), suggesting that other molecules induced by co-stimulation with these two stimuli can partially compensate for the deficiency of IRF-8. It is possible that other IRF members, such as IRF-1, may be involved in compensating for the IRF-8 deficiency. To address this possibility, we measured the IRF-1 mRNA expression in IRF-8−/− cells. The data indicated that IRF-1 could still be induced by LPS or IFN-γ plus LPS in IRF-8−/− cells but to reduced levels compared with WT cells (supplemental Fig. 1A). Silence of IRF-8 by siRNA in macrophages only slightly affected the IRF-1 binding to the p28 promoter (Fig. 6F). In addition, the ChIP assay demonstrated that both IRF-1 and IRF-8 bound to the p28 promoter (Fig. 6G). Collectively, these data suggest that the IRF-1 induced by LPS or LPS plus IFN-γ in IRF-8−/− cells might be responsible for the partially rescued p28 gene expression in IRF-8-deficient cells.
Previous studies reported that reconstitution of IRF-1 in IRF-1−/− BMDMs could not rescue the impaired p28 expression (43), suggesting that other developmental factors affected by IRF-1 deletion might be required for p28 expression. To elucidate if IRF-8 is the factor responsible for p28 expression, we reconstituted IRF-8 by retroviral transduction in IRF-8-deficient BMDMs. Our data demonstrate that reconstitution of IRF-8 rescued p28 mRNA expression in IRF-8-deficient cells (Fig. 2A). We notice that without IFN-γ and LPS stimulation, the IRF-8 expression was relatively low in cells reconstituted with the IRF-8/retrovirus, similar to the levels of IRF-8 expression in untreated WT cells (Fig. 2B). This low level of IRF-8 expression could not induce measurable amounts of p28 expression (Figs. Fig. 1A and 2A). However, after treatment with LPS or IFN-γ plus LPS, the levels of IRF-8 mRNA expression were enhanced in IRF-8-reconstituted cells to levels similar to those of wild type cells (Fig. 2B). The reason why IRF-8 in the retroviral vector needs to be induced to reach its maximal expression is currently unknown, but the same phenomenon with this vector was also reported by other laboratories (52). It is possible that the IRF-8 delivered by retrovirus randomly integrated into a genome harboring a transactivator that responded to LPS stimulation. This is not suppressing, because in the reconstitution experiment, we used macrophages that are highly responsive to LPS. Another possibility might be that the 5′-long terminal repeat of murine stem cell virus in the vector responded to factors induced by LPS in macrophages that indirectly activate the IRF-8 expression. It will be interesting to determine the exact reasons of the inducibility of inserted genes in this vector in future study.
Consistent with the impression that IL-27 p28 gene expression is regulated at the level of transcription, forced expression of IRF-8 dose-dependently activated the p28 promoter (Fig. 3A). Furthermore, the responses of the p28 promoter to IFN-γ and IFNγ plus LPS stimulation were significantly increased in IRF-8-transfected RAW cells (Fig. 3B). Taken together, it seems that both IRF-1 and IRF-8 regulate the p28 gene expression at the level of transcription.
So far, most studies have focused on IRF-8 targeting genes, and relatively little has been reported on how IRF-8 is regulated. The broader effects of IRF-8 than IRF-1 on IL-27 p28 gene expression made us wonder if the LPS signaling pathway also mediated IRF-8 expression. Our data showed that LPS activated IRF-8 gene expression through adaptor protein MyD88, which has never been reported before (Fig. 4, A and B). MyD88-mediated IRF-8 expression may act through interaction with the IFN-receptor 1 chain that triggers IFN-γ signaling leading to IRF-8 expression (53). NF-κB c-Rel partially mediated LPS-induced IRF-8 expression (Fig. 4C), suggesting that other members of the NF-κB family, such as p65, may also contribute to LPS-induced IRF-8 expression. It has been reported that IRF-8 could induce NF-κB activation in that NF-κB activation induced by TLR9 agonist CPG was completely abolished in IRF-8−/− dendritic cells as a result of the inability to phosphorylate IκB-α/β in the absence of IRF-8 (54). Macrophages treated with IFN-γ plus LPS displayed increased NF-κB activity compared with cells stimulated with solely IFN-γ or LPS (55). Our data also indicate that IRF-8 could induce NF-κB activation (supplemental Fig. 2), suggesting that IRF-8 induced by IFN-γ and LPS participates in activation of NF-κB. Indeed, blockade of NF-κB signaling by IκB-α mutant completely abolished both IRF-8 and LPS-induced p28 gene transcription (Fig. 4, D and E), further confirming the role of NF-κB in IRF-8-induced p28 gene expression. Our data also indicate that a previously identified NF-κB binding site in the p28 promoter (43) mediated LPS- and IRF-8-induced p28 promoter activity (Fig. 4G) but not to IRF-8 alone (Fig. 4F). This is not surprising because IRF-8 is a much weaker inducer for NF-κB activation compared with LPS, and its effect is mainly mediated through the IRF RE in the p28 promoter. Co-transfection of IRF-8 with different components of the NF-κB family, which mimic LPS signaling, indicates that NF-κB p65 and c-Rel mediated p28 gene expression (Fig. 4, H and I). Taken together, our data demonstrate that MyD88 mediates LPS-induced IRF-8 expression, which in turn activates NF-κB and induces IL-27 p28 gene expression.
By mapping the IRF-8 response element in the p28 promoter, we found that the IRF-8 RE seemed to be localized between −140 and −40 of the p28 promoter, exactly where the IRF-1 RE was identified previously (43). This made us think that IRF-8 might act through the IRF-1 site to regulate p28 gene activation. Indeed, the response of the p28 promoter to IRF-8 was completely abrogated in the IRF-1 RE-mutant p28 promoter. Because IRF-1 and IRF-8 could physically interact with each other to form a heterodimer (15), these data prompted us to further investigate if these two IRF members could synergistically activate the p28 promoter through the same site. Consistent with the earlier data and the previous study, IRF-8 and IRF-1 both activated the p28 promoter (Fig. 5D). Co-transfection of IRF-8 with IRF-1 or vice versa resulted in an additive effect on p28 promoter activation (Fig. 5, E and F). More importantly, the additive effects were completely abolished in IRF RE-mutant p28 promoter (Fig. 5D), demonstrating that IRF-8 and IRF-1 work together to activate p28 gene transcription.
Our previous studies showed that IRF-1 bound to the p28 promoter both in vitro and in vivo (43). Next, we wanted to know if IRF-8 could also physically bind to the p28 promoter. The EMSA data clearly showed an IFN-γ-inducible binding with the WT p28 promoter probe (Fig. 6A). This binding was completely abolished when used the IRF RE-mutated probe (Fig. 6A) and competed by the cold WT probe (Fig. 6B), indicating a sequence specificity of the binding. A supershift assay further demonstrated that IRF-8 is a member of the binding complex (Fig. 6C). Interestingly, IRF-1 appeared to be the major component of the complex because an anti-IRF-1 antibody supershifted the majority of the binding complex (Fig. 6C). However, when we used an alternative approach, a DNA affinity binding assay, to confirm the binding, it appeared that the binding intensity of IRF-8 to the p28 promoter was stronger than that of IRF-1 (Fig. 6D). The difference of binding between these two assays might be due to a conformation change of the complex or alteration of the epitopes. By using siRNA to silence IRF-8 expression, we were able to test whether IRF-8 deficiency could affect the IRF-1 binding. Our results indicate that IRF-1 binding to the p28 promoter is partially dependent on IRF-8 because IRF-1 binding was reduced in IRF-8-silenced cells (Fig. 6F). This is not surprising because IRF-1 expression was reduced in IRF-8−/− cells (supplemental Fig. 1A), and previous studies showed that IRF-8 and IRF-1/2 interact with each other to enhance DNA binding activity (15, 56). Interestingly, the IRF-8 mRNA expression appeared less affected by IRF-1 deficiency (supplemental Fig. 1B), supporting the idea that IRF-8 may have broader effects than IRF-1 on immune responses. In addition, the ChIP assay clearly demonstrated a strong IRF-8 and IRF-1 in vivo binding to the p28 promoter at the IRF RE region (Fig. 6G).
Cytokine production in vitro may not reflect overall responses in vivo that may determine the final outcome of immune responses to pathogens during infection. Therefore, we decided to test the levels of IL-27 p28 in serum between WT and IRF-8−/− mice challenged with LPS, a model of endotoxic shock. The levels of p28 protein in serum were significantly lower in challenged IRF-8−/− mice compared with WT mice. The impaired p28 protein secretion correlated with decreased mRNA expression in the spleens of IRF-8−/− mice. Because IRF-8 is an inducible gene and has low basal transcription, there was not much difference in IRF-8 expression between knock-out mice and WT mice under unstimulated conditions. However, after treatment with LPS, IRF-8 expression was induced only in WT and not IRF-8 knock-out mice. Given the fact that macrophages are the major producer of IL-27, these data suggest that during endotoxic shock, the reduced p28 production in IRF-8−/− mice may be due to a defect of p28 production in macrophages.
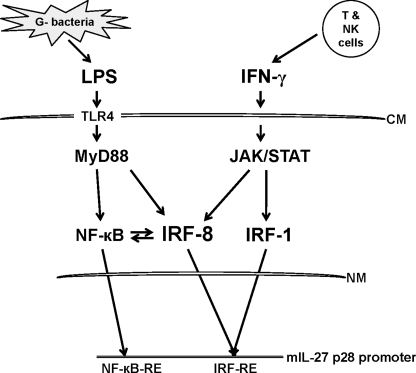
Based on our studies and others, we propose a model that represents the molecular events that lead to IL-27 production by antigen-presenting cells (Fig. 8). During the innate phase of an immune response, microbial antigens activate the NF-κB pathway via Toll-like receptors (TLR4 for LPS) in a MyD88-dependent manner. Activated NF-κB alone stimulates moderate transcription of the IL-27 p28 gene, primarily through c-Rel and p65, and low amounts of IRF-8 as well. The initial small amount of IL-27 is able to stimulate NK and Th1 cells to produce IFN-γ. IFN-γ then induces robust IRF-1 and IRF-8 expression, which bind to the p28 promoter, resulting in much greater levels of p28 transcription and IL-27 production. Combinations of signals derived from innate and adaptive immune events stimulate synergistically the production of IL-27 and sustain the inflammatory response as well as cell-mediated immunity against pathogens.
FIGURE 8.
Schematics of the role of IRF-8 in IL-27 production. LPS and IFN-γ induce IL-27 p28 gene expression and IL-27 production in macrophages through MyD88/NF-κB and JAK/STAT/IRF-1/IRF-8 signaling pathways either alone or together. See “Discussion” for a more detailed explanation. CM, cytoplasmic membrane; NM, nuclear membrane.
Supplementary Material
Acknowledgment
We thank Dr. Daniel F. Hoft for critical reading of the manuscript.
This work was supported in part by the Vasculitis Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
- IFN
- interferon
- IRF
- interferon regulatory factor
- IL
- interleukin
- LPS
- lipopolysaccharide
- BMDM
- bone marrow-derived macrophage
- M-CSF
- macrophage colony-stimulating factor
- RT
- reverse transcription
- qRT-PCR
- quantitative real-time PCR
- DTT
- dithiothreitol
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- ChIP
- chromatin immunoprecipitation
- WT
- wild type
- RE
- response element
- STAT
- signal transducers and activators of transcription
- ELISA
- enzyme-linked immunosorbent assay.
REFERENCES
- 1.Schroder K., Hertzog P. J., Ravasi T., Hume D. A. (2004) J. Leukoc. Biol. 75, 163–189 [DOI] [PubMed] [Google Scholar]
- 2.Boehm U., Klamp T., Groot M., Howard J. C. (1997) Annu. Rev. Immunol. 15, 749–795 [DOI] [PubMed] [Google Scholar]
- 3.Ehrt S., Schnappinger D., Bekiranov S., Drenkow J., Shi S., Gingeras T. R., Gaasterland T., Schoolnik G., Nathan C. (2001) J. Exp. Med. 194, 1123–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taniguchi T., Ogasawara K., Takaoka A., Tanaka N. (2001) Annu. Rev. Immunol. 19, 623–655 [DOI] [PubMed] [Google Scholar]
- 5.Liu J., Cao S., Herman L. M., Ma X. (2003) J. Exp. Med. 198, 1265–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsuyama T., Kimura T., Kitagawa M., Pfeffer K., Kawakami T., Watanabe N., Kündig T. M., Amakawa R., Kishihara K., Wakeham A. (1993) Cell 75, 83–97 [PubMed] [Google Scholar]
- 7.Duncan G. S., Mittrücker H. W., Kägi D., Matsuyama T., Mak T. W. (1996) J. Exp. Med. 184, 2043–2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taki S., Sato T., Ogasawara K., Fukuda T., Sato M., Hida S., Suzuki G., Mitsuyama M., Shin E. H., Kojima S., Taniguchi T., Asano Y. (1997) Immunity. 6, 673–679 [DOI] [PubMed] [Google Scholar]
- 9.Ogasawara K., Hida S., Azimi N., Tagaya Y., Sato T., Yokochi-Fukuda T., Waldmann T. A., Taniguchi T., Taki S. (1998) Nature 391, 700–703 [DOI] [PubMed] [Google Scholar]
- 10.Ohteki T., Yoshida H., Matsuyama T., Duncan G. S., Mak T. W., Ohashi P. S. (1998) J. Exp. Med. 187, 967–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lohoff M., Duncan G. S., Ferrick D., Mittrücker H. W., Bischof S., Prechtl S., Röllinghoff M., Schmitt E., Pahl A., Mak T. W. (2000) J. Exp. Med. 192, 325–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nozawa H., Oda E., Nakao K., Ishihara M., Ueda S., Yokochi T., Ogasawara K., Nakatsuru Y., Shimizu S., Ohira Y., Hioki K., Aizawa S., Ishikawa T., Katsuki M., Muto T., Taniguchi T., Tanaka N. (1999) Genes Dev. 13, 1240–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tamura T., Ozato K. (2002) J. Interferon Cytokine Res. 22, 145–152 [DOI] [PubMed] [Google Scholar]
- 14.Tsujimura H., Tamura T., Ozato K. (2003) J. Immunol. 170, 1131–1135 [DOI] [PubMed] [Google Scholar]
- 15.Liu J., Guan X., Tamura T., Ozato K., Ma X. (2004) J. Biol. Chem. 279, 55609–55617 [DOI] [PubMed] [Google Scholar]
- 16.Liu J., Ma X. (2006) J. Biol. Chem. 281, 19188–19195 [DOI] [PubMed] [Google Scholar]
- 17.Pflanz S., Timans J. C., Cheung J., Rosales R., Kanzler H., Gilbert J., Hibbert L., Churakova T., Travis M., Vaisberg E., Blumenschein W. M., Mattson J. D., Wagner J. L., To W., Zurawski S., McClanahan T. K., Gorman D. M., Bazan J. F., de Waal Malefyt R., Rennick D., Kastelein R. A. (2002) Immunity 16, 779–790 [DOI] [PubMed] [Google Scholar]
- 18.Kastelein R. A., Hunter C. A., Cua D. J. (2007) Annu. Rev. Immunol. 25, 221–242 [DOI] [PubMed] [Google Scholar]
- 19.Nieuwenhuis E. E., Neurath M. F., Corazza N., Iijima H., Trgovcich J., Wirtz S., Glickman J., Bailey D., Yoshida M., Galle P. R., Kronenberg M., Birkenbach M., Blumberg R. S. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 16951–16956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hibbert L., Pflanz S., De Waal Malefyt R., Kastelein R. A. (2003) J. Interferon Cytokine Res. 23, 513–522 [DOI] [PubMed] [Google Scholar]
- 21.Zahn S., Wirtz S., Birkenbach M., Blumberg R. S., Neurath M. F., von Stebut E. (2005) Eur. J. Immunol. 35, 1106–1112 [DOI] [PubMed] [Google Scholar]
- 22.Artis D., Johnson L. M., Joyce K., Saris C., Villarino A., Hunter C. A., Scott P. (2004) J. Immunol. 172, 4672–4675 [DOI] [PubMed] [Google Scholar]
- 23.Yoshida H., Hamano S., Senaldi G., Covey T., Faggioni R., Mu S., Xia M., Wakeham A. C., Nishina H., Potter J., Saris C. J., Mak T. W. (2001) Immunity 15, 569–578 [DOI] [PubMed] [Google Scholar]
- 24.Miyazaki Y., Inoue H., Matsumura M., Matsumoto K., Nakano T., Tsuda M., Hamano S., Yoshimura A., Yoshida H. (2005) J. Immunol. 175, 2401–2407 [DOI] [PubMed] [Google Scholar]
- 25.Lucas S., Ghilardi N., Li J., de Sauvage F. J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 15047–15052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hisada M., Kamiya S., Fujita K., Belladonna M. L., Aoki T., Koyanagi Y., Mizuguchi J., Yoshimoto T. (2004) Cancer Res. 64, 1152–1156 [DOI] [PubMed] [Google Scholar]
- 27.Chiyo M., Shimozato O., Iizasa T., Fujisawa T., Tagawa M. (2004) Anticancer Res. 24, 3763–3767 [PubMed] [Google Scholar]
- 28.Chiyo M., Shimozato O., Yu L., Kawamura K., Iizasa T., Fujisawa T., Tagawa M. (2005) Int. J. Cancer 115, 437–442 [DOI] [PubMed] [Google Scholar]
- 29.Salcedo R., Stauffer J. K., Lincoln E., Back T. C., Hixon J. A., Hahn C., Shafer-Weaver K., Malyguine A., Kastelein R., Wigginton J. M. (2004) J. Immunol. 173, 7170–7182 [DOI] [PubMed] [Google Scholar]
- 30.Shimizu M., Shimamura M., Owaki T., Asakawa M., Fujita K., Kudo M., Iwakura Y., Takeda Y., Luster A. D., Mizuguchi J., Yoshimoto T. (2006) J. Immunol. 176, 7317–7324 [DOI] [PubMed] [Google Scholar]
- 31.Hunter C. A. (2005) Nat. Rev. Immunol. 5, 521–531 [DOI] [PubMed] [Google Scholar]
- 32.Hamano S., Himeno K., Miyazaki Y., Ishii K., Yamanaka A., Takeda A., Zhang M., Hisaeda H., Mak T. W., Yoshimura A., Yoshida H. (2003) Immunity 19, 657–667 [DOI] [PubMed] [Google Scholar]
- 33.Hölscher C., Hölscher A., Rückerl D., Yoshimoto T., Yoshida H., Mak T., Saris C., Ehlers S. (2005) J. Immunol. 174, 3534–3544 [DOI] [PubMed] [Google Scholar]
- 34.Villarino A., Hibbert L., Lieberman L., Wilson E., Mak T., Yoshida H., Kastelein R. A., Saris C., Hunter C. A. (2003) Immunity. 19, 645–655 [DOI] [PubMed] [Google Scholar]
- 35.Batten M., Li J., Yi S., Kljavin N. M., Danilenko D. M., Lucas S., Lee J., de Sauvage F. J., Ghilardi N. (2006) Nat. Immunol. 7, 929–936 [DOI] [PubMed] [Google Scholar]
- 36.Stumhofer J. S., Laurence A., Wilson E. H., Huang E., Tato C. M., Johnson L. M., Villarino A. V., Huang Q., Yoshimura A., Sehy D., Saris C. J., O'Shea J. J., Hennighausen L., Ernst M., Hunter C. A. (2006) Nat. Immunol. 7, 937–945 [DOI] [PubMed] [Google Scholar]
- 37.O'Connor W., Jr., Kamanaka M., Booth C. J., Town T., Nakae S., Iwakura Y., Kolls J. K., Flavell R. A. (2009) Nat. Immunol. 10, 603–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stumhofer J. S., Silver J. S., Laurence A., Porrett P. M., Harris T. H., Turka L. A., Ernst M., Saris C. J., O'Shea J. J., Hunter C. A. (2007) Nat. Immunol. 8, 1363–1371 [DOI] [PubMed] [Google Scholar]
- 39.Fitzgerald D. C., Zhang G. X., El-Behi M., Fonseca-Kelly Z., Li H., Yu S., Saris C. J., Gran B., Ciric B., Rostami A. (2007) Nat. Immunol. 8, 1372–1379 [DOI] [PubMed] [Google Scholar]
- 40.Awasthi A., Carrier Y., Peron J. P., Bettelli E., Kamanaka M., Flavell R. A., Kuchroo V. K., Oukka M., Weiner H. L. (2007) Nat. Immunol. 8, 1380–1389 [DOI] [PubMed] [Google Scholar]
- 41.Diveu C., McGeachy M. J., Boniface K., Stumhofer J. S., Sathe M., Joyce-Shaikh B., Chen Y., Tato C. M., McClanahan T. K., de Waal Malefyt R., Hunter C. A., Cua D. J., Kastelein R. A. (2009) J. Immunol. 182, 5748–5756 [DOI] [PubMed] [Google Scholar]
- 42.Molle C., Nguyen M., Flamand V., Renneson J., Trottein F., De Wit D., Willems F., Goldman M., Goriely S. (2007) J. Immunol. 178, 7607–7615 [DOI] [PubMed] [Google Scholar]
- 43.Liu J., Guan X., Ma X. (2007) J. Exp. Med. 204, 141–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pirhonen J., Sirén J., Julkunen I., Matikainen S. (2007) J. Leukoc. Biol. 82, 1185–1192 [DOI] [PubMed] [Google Scholar]
- 45.Remoli M. E., Gafa V., Giacomini E., Severa M., Lande R., Coccia E. M. (2007) Eur. J. Immunol. 37, 3499–3508 [DOI] [PubMed] [Google Scholar]
- 46.Schreiber E., Matthias P., Müller M. M., Schaffner W. (1989) Nucleic Acids Res. 17, 6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu J., Mankani G., Shi X., Meyer M., Cunningham-Runddles S., Ma X., Sun Z. S. (2006) Infect. Immun. 74, 4750–4756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kobayashi M., Fitz L., Ryan M., Hewick R. M., Clark S. C., Chan S., Loudon R., Sherman F., Perussia B., Trinchieri G. (1989) J. Exp. Med. 170, 827–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oppmann B., Lesley R., Blom B., Timans J. C., Xu Y., Hunte B., Vega F., Yu N., Wang J., Singh K., Zonin F., Vaisberg E., Churakova T., Liu M., Gorman D., Wagner J., Zurawski S., Liu Y., Abrams J. S., Moore K. W., Rennick D., de Waal-Malefyt R., Hannum C., Bazan J. F., Kastelein R. A. (2000) Immunity 13, 715–725 [DOI] [PubMed] [Google Scholar]
- 50.Collison L. W., Workman C. J., Kuo T. T., Boyd K., Wang Y., Vignali K. M., Cross R., Sehy D., Blumberg R. S., Vignali D. A. (2007) Nature 450, 566–569 [DOI] [PubMed] [Google Scholar]
- 51.Stumhofer J. S., Hunter C. A. (2008) Immunol. Lett. 117, 123–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsujimura H., Tamura T., Gongora C., Aliberti J., Reis e Sousa C., Sher A., Ozato K. (2003) Blood 101, 961–969 [DOI] [PubMed] [Google Scholar]
- 53.Sun D., Ding A. (2006) Nat. Immunol. 7, 375–381 [DOI] [PubMed] [Google Scholar]
- 54.Tsujimura H., Tamura T., Kong H. J., Nishiyama A., Ishii K. J., Klinman D. M., Ozato K. (2004) J. Immunol. 172, 6820–6827 [DOI] [PubMed] [Google Scholar]
- 55.Zhao J., Kong H. J., Li H., Huang B., Yang M., Zhu C., Bogunovic M., Zheng F., Mayer L., Ozato K., Unkeless J., Xiong H. (2006) J. Biol. Chem. 281, 10073–10080 [DOI] [PubMed] [Google Scholar]
- 56.Levi B. Z., Hashmueli S., Gleit-Kielmanowicz M., Azriel A., Meraro D. (2002) J. Interferon Cytokine Res. 22, 153–160 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.