Abstract
The identification of hereditary familial Alzheimer disease (FAD) mutations in the amyloid precursor protein (APP) and presenilin-1 (PS1) corroborated the causative role of amyloid-β peptides with 42 amino acid residues (Aβ42) in the pathogenesis of AD. Although most FAD mutations are known to increase Aβ42 levels, mutations within the APP GxxxG motif are known to lower Aβ42 levels by attenuating transmembrane sequence dimerization. Here, we show that aberrant Aβ42 levels of FAD mutations can be rescued by GxxxG mutations. The combination of the APP-GxxxG mutation G33A with APP-FAD mutations yielded a constant 60% decrease of Aβ42 levels and a concomitant 3-fold increase of Aβ38 levels compared with the Gly33 wild-type as determined by ELISA. In the presence of PS1-FAD mutations, the effects of G33A were attenuated, apparently attributable to a different mechanism of PS1-FAD mutants compared with APP-FAD mutants. Our results contribute to a general understanding of the mechanism how APP is processed by the γ-secretase module and strongly emphasize the potential of the GxxxG motif in the prevention of sporadic AD as well as FAD.
Keywords: Alzheimer Disease, Amyloid, Presenilin, Protein Motifs, Secretases, GxxxG, Dimer
Introduction
APP2 and APLPs were conventionally thought to exist and to act as monomers. However, biochemical and structural data have accumulated over the past few years, indicating that APP and APLPs exist as functional dimers or even are present in higher oligomeric units (1–6). Interactions of APP and APLPs were reported to promote cell adhesion in a homo- and heterotypic manner (7, 8). Among other mechanisms, the varying strength of APP dimerization mediated through N-terminal sites (5) or by the transmembrane sequence (TMS) (9) has been reported to influence APP processing.
APP is first cleaved by the β-site APP cleaving enzyme and is then sequentially processed by the γ-secretase complex to generate Aβ peptides of varying length (10, 11). γ-Secretase cleavage specificity is modulated by the GxxxG (“G-triple-x-G”) dimerization motif of the APP-TMS, and we showed previously that APP can be cleaved as a homodimer by β- and γ-secretases (9). APP, APLP1, and APLP2 share similar interaction motifs and can form APP-APLP1 and APP-APLP2 complexes (7). Co-transfections of APP with APLP1 or APLP2 influenced APP processing into Aβ leading to decreased Aβ40 and Aβ42 levels likely through an influence on γ-secretase cleavages (7).
According to the amyloid hypothesis, Aβ peptides represent the main culprit of Alzheimer disease (AD). Based on this assumption is the appealing prediction that reducing Aβ levels would ameliorate Alzheimer symptoms (12, 13). In the current model of Aβ generation, the initial cut at the ϵ-site is executed by the presenilins of the γ-secretase complex, leading to formation of the APP intracellular domain and Aβ49 or Aβ48 peptides (10, 14). The latter two likely remain bound to the active site and are successively cleaved every three to four residues at the ζ-site and at the γ-sites (11, 15). Most likely, two product lines exist. In the product line encompassing Aβ40, Aβ49 is trimmed to Aβ40, and in the other product line, Aβ48 is the precursor of Aβ42 and Aβ38 (10, 15). When we studied homodimerization of APP-TMS, a bacterial test system revealed that glycine residues 29 and 33 of the GxxxG amino acid motif represent the major helix-helix contact site (see Fig. 1A). Substitutions of either of the two glycine residues not only diminished APP-TMS homodimer stability but also drastically decreased Aβ42 and increased Aβ38 levels, among other changes (9). Our findings that the γ-secretase complex can cleave dimeric substrates complement the current model of successive cleavages producing Aβ peptides from two predominant product lines (15).
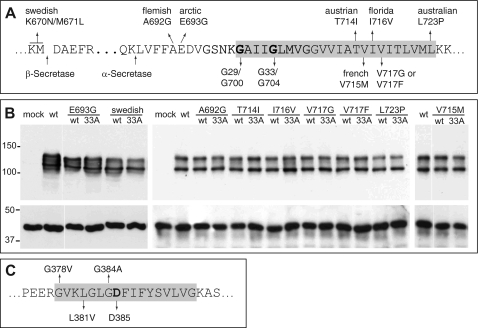
FIGURE 1.
APP- and PS1-FAD mutations. A, part of the APP sequence is shown. Indicated are the β- and α-secretase cleavage sites as well as all analyzed APP-FAD mutations. The glycine residues mediating the APP-TMS dimerization (9) are highlighted (Gly29/Gly700 and Gly33/Gly704 according to Aβ or APP770 numbering, respectively). B, APP-FAD and APP-FAD-G33A constructs are equally well and stably expressed in the SH-SY5Y cells. The double band represents mature, plasma membrane-residing (∼130 kDa) and immature (∼110 kDa) APP, and Western blot was stained with 22C11. As loading control, actin was stained as shown in the lower panel (∼40 kDa). C, an amino acid sequence of PS1 TMS-7 is shown. Analyzed FAD mutations and the catalytically active Asp385 are highlighted. The gray boxes in C mark residues embedded in the cell membrane.
Importantly, mutations in three different genes are associated with AD and were described to increase the ratio of Aβ42 to Aβ40 in mice and humans (16–18). Early onset dominant AD was first associated with the APP gene and found to alter the amount of Aβ produced and/or the ratio of Aβ42 to Aβ40 (16, 19). Subsequently, early onset familial mutations in the presenilins were also linked to AD (17, 20). Using some of these mutations, we further investigated the GxxxG-mediated effects on γ-secretase cleavage by combining GxxxG mutations with FAD mutations.
We found here a constant 60% decrease in Aβ42 and a 3-fold increase in Aβ38 levels for APP-FAD mutants when we impaired the APP-GxxxG-mediated interaction by introducing the mutation G33A. However, G33A only had mild effects in the presence of PS1-FAD mutations. Furthermore, we found that the analyzed FAD mutants can be divided into subgroups: (i) APP-FAD mutants that increase Aβ42 and Aβ38 levels and either show a concomitant increase or a decrease of Aβ40 levels, (ii) APP-FAD mutants that do not affect Aβ42 and Aβ38 levels but exclusively decrease Aβ40 levels, and (iii) PS1-FAD mutants that increase Aβ42, but decrease Aβ38 levels.
EXPERIMENTAL PROCEDURES
Plasmids and Transfections
The plasmid pCEP4 containing the coding sequence for APP695 with an N-terminal Myc tag and C-terminal FLAG tag was used as a template to introduce the APP-FAD mutations and G33A by site-directed mutagenesis. Either G33A or the FAD variants were used to generate the double mutant constructs APP-FAD-G33A in a second site-directed mutagenesis. The same method was applied to generate SPA4CT-G33A from pCEP4-SPA4CT with a C-terminal FLAG tag and the PS1-FAD mutants from pcDNA3.1/zeo (Invitrogen) containing the PS1-coding sequence with an N-terminal hemagglutinin tag. SPA4/1–52 and SPA4/1–51 were generated from pCEP4-SPA4CT by replacing codons 53 or 52 by stop codons, respectively. All sequences were confirmed by dideoxy sequencing and restriction digestion. For stable expression of APP or SPA4CT constructs, plasmids (3 μg) were transfected into SH-SY5Y cells (ATCC catalog no. CRL-2266) at 9 × 105 cells/6-well or 80% confluency using Transfectine (Bio-Rad) or Lipofectamine and Plus reagent (Invitrogen) following the manufacturer's instructions. Stably transfected cells were selected with hygromycin (250 μg/ml). SPA4CT-expressing cell lines were co-transfected with the PS1 constructs using Lipofectamine and Plus reagent and were additionally selected with zeocin (200 μg/ml). Two to three independent stable cell lines were generated.
Cell Culture, Sandwich ELISA, Immunoprecipitation, Western Blot, and MALDI-MS
SH-SY5Y cells routinely were cultured as described (9). Aβ40/Aβ42 ELISAs were performed according to the manufacturer's instructions (The Genetics Company). The following antibody combinations were used: for Aβ38, BA1–13 (Covance/Signet) and W0-2-biotin (TGC); for sAPPα, anti-Myc antibody (Cell Signaling Technology) and W0-2-biotin; and for total Aβ, W0-2 or 4G8 (Covance) and 4G8-horseradish peroxidase (Covance) or W0-2-biotin, respectively. W0–2-biotin was recognized by streptavidin-conjugated horseradish peroxidase, and ELISAs were developed with 1-Step Ultra-TMB (Pierce) and were measured at 450 nm in a microplate reader (Anthos). Western blot analyses and immunoprecipitations were performed as published (9). Briefly, 250 μl of conditioned media were diluted with phosphate-buffered saline and precipitated with polyclonal rabbit antibody 18-1 (generated to Aβ1-40) or W0-2 and protein A- or G-Sepharose (GE Healthcare), respectively. For Western blot analysis, cells were lysed in a buffer containing 50 mm Tris, pH 7.4, 150 mm NaCl, 2 mm EDTA, and 1% Nonidet P-40. Samples were separated by SDS-PAGE, transferred to nitrocellulose, and immunostained. For MALDI-MS analyses, Aβ was eluted twice from Sepharose with 500 μl 50% acetic acid, and the eluate was vacuum-dried. MALDI-MS analysis was carried out as described previously (9).
Structural Model
The APP-TMS model from previous analyses based on the tertiary structure of glycophorin A was used as a template (9). The FAD mutants were added to the structure using the Swiss-PdbViewer. The most likely rotamers were selected before the model was energetically minimized applying the GROMACS Software. All figures were produced with the PyMOL Molecular Graphics System.
RESULTS
Analysis of APP-FAD Mutants
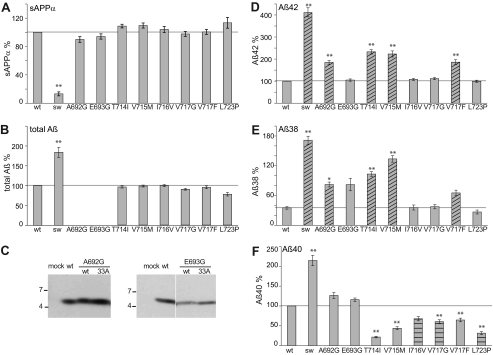
We analyzed APP processing of nine APP-FAD mutations scattered around the three secretase cleavage sites, i.e. K670N/M671L at the β-site (Swedish, hereafter referred to as “sw”), A692G (Flemish) and E693G (Arctic) near the α-site, and T714I (Austrian), V715M (French), I716V (Florida), V717G, V717F, and L723P (Australian) at the γ-secretase cleavage sites (Fig. 1A) (19, 21–28). The APP-FAD mutants were stably expressed in SH-SY5Y cells and yielded similar expression levels as determined by Western blot analysis (Fig. 1B). The levels of sAPPα, Aβ42, Aβ40, and Aβ38 were quantified by sandwich ELISA (Fig. 2). For the APP-sw mutation, sAPPα levels were found to be reduced by 80%, and total Aβ levels were increased, whereas all other APP-FAD mutants showed total Aβ levels comparable to APP-wt (Fig. 2, A–C). In five out of nine variants (sw, A692G, T714I, V715M, and V717F) increased Aβ42 levels were observed compared with APP-wt (Fig. 2D), whereas the other four FAD variants, i.e. E693G, I716V, V717G, and L723P only reached Aβ42 levels of APP-wt. Thus, increased Aβ42 levels are not a general feature of APP-FAD mutations. For six out of nine FAD variants, Aβ38 levels were increased compared with APP-wt (Fig. 2E). Interestingly, all five mutations that resulted in increased Aβ42 levels also showed increased Aβ38 levels (Fig. 2, D and E, shaded bars). Aβ40 levels were found elevated only for those FAD mutations that reside within the N-terminal half of the Aβ sequence. In contrast, a significant decrease of Aβ40 was observed for all six FAD mutations located within the C-terminal region of the TMS (Fig. 2F). FAD mutants that do not affect Aβ38 or Aβ42 levels, but solely decrease Aβ40 levels are I716V, V717G, and L723P (Fig. 2F, horizontal shaded bars). The Arctic APP mutation E693G is an exception insofar as it only increased Aβ38 levels. Interestingly, the APP-sw mutant yielded 4-fold enhanced Aβ42 and Aβ38 levels, but only 2-fold enhanced Aβ40 levels (9, 15). Thus, the sw mutation might not exclusively affect the β-secretase cut as predicted (19). We clearly observed that amino acid exchanges in the Aβ N-terminal region (sw, Flemish, and Arctic) can influence γ-secretase processing.
FIGURE 2.
Analysis of APP-FAD mutations. Quantification of secreted sAPPα, total Aβ, Aβ42, Aβ40, and Aβ38. A, sAPPα levels, APP-wt (without FAD) set as 100% (mean ± S.E., n = 5–16). B, total secreted Aβ levels, APP-wt set as 100% (mean ± S.E., n = 6–14). Note that A692G and E693G could not be analyzed by total Aβ ELISA as the FAD mutations alter the epitope of the antibody 4G8. C, total Aβ levels of A692G and E693G were analyzed by immunoprecipitation with antibody 18-1 and Western blot analysis with W0-2. D, Aβ42 levels, APP-wt set as 100% (mean ± S.E., n = 5–17). E, Aβ38 levels, APP-G33A set as 100%, see supplemental Fig. S1C and (mean ± S.E., n = 4–12). F, Aβ40 levels, APP-wt set as 100% (mean ± S.E., n = 7–16). A–E, asterisks indicate significant differences to APP-wt (*, p < 0.01, **, p < 0.001, one-way ANOVA type Dunnett). Shaded bars indicate mutations increasing Aβ42 and Aβ38. Horizontal shaded bars highlight mutations only decreasing Aβ40. Horizontal lines mark the fragment levels of APP-wt for better comparison with the mutants.
Effect of the GxxxG Motif on APP-FAD Processing
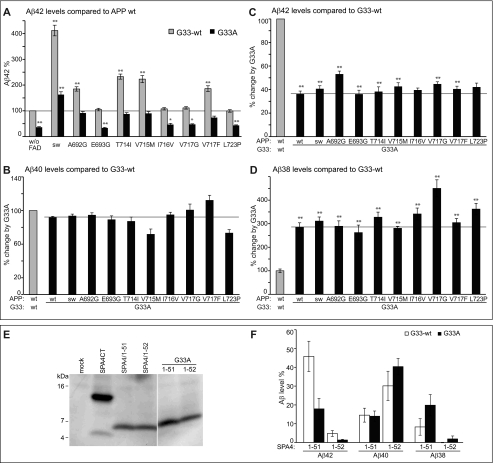
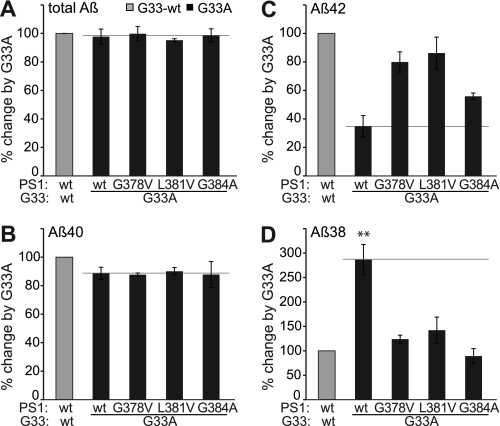
To investigate the role of the GxxxG motif known to mediate APP-TMS dimerization, we tested constructs encoding individual APP-FAD mutations in combination with the GxxxG mutation G33A (APP-FAD-G33A). All APP-FAD-G33A-derived constructs generated sAPPα and total Aβ levels similar to the respective APP-FAD variant alone, indicating that their processing is equally efficient as that of APP-wt or APP-FAD alone (supplemental Fig. S1). However, the striking difference is that the APP-FAD-G33A constructs drastically reduced Aβ42 levels compared with APP-FAD alone (Fig. 3A). Mutants A692G/G33A, T714I/G33A, and V715M/G33A yielded Aβ42 levels similar to APP-wt. The mutants E693G, I716V, V717G, V717F, and L723P in combination with G33A did not even reach Aβ42 levels of APP-wt. However, the APP-sw-G33A mutant still produced 1.6-fold higher Aβ42 levels than APP-wt (Fig. 3A). For better comparison and to calculate the percent impact of G33A on processing, data of Gly33-wt were each normalized to 100% (Fig. 3, B–D, gray bar) and are displayed for comparison to G33A (Fig. 3, B–D, black bars). For all nine APP-FAD mutations tested, the G33A mutation yielded a reduction of Aβ42 levels by ∼60% (Fig. 3C) and a 3-fold increase of Aβ38 levels (Fig. 3D), whereas Aβ40 levels remained unchanged (Fig. 3B). Thus, the G33A mutation within the GxxxG motif possesses the same strong impact on APP-FAD mutations as on APP-wt in terms of decreasing Aβ42 production and thus has a FAD-rescuing effect. By MALDI-MS analyses, we confirmed that no construct generated an aberrant pattern of Aβ species (supplemental Fig. S2). This data indicates that single amino acid changes within the GxxxG motif can rescue APP-FAD-related effects.
FIGURE 3.
Impact of the APP-GxxxG mutation G33A on APP-FAD processing. A, Aβ42 levels; APP-wt was set as 100% (mean ± S.E., n = 5–17). For better comparison, bars of APP-FAD mutations as in Fig. 2D are shown (G33-wt, gray bars). The mutation G33A leads to a relative decrease in Aβ42 levels (black bars). Asterisks indicate significant differences to APP-wt (*, p < 0.01, **, p < 0.001, one-way ANOVA type Dunnett). The horizontal line marks Aβ42 level of APP-wt for better comparison. Original data of sAPPα, total Aβ, Aβ40, and Aβ38 ELISA are shown in supplemental Fig. S1. B–D, Aβ levels of the G33A constructs (black bars) normalized to the respective Gly33-wt constructs, which were set as 100% (represented by gray bars). Asterisks indicate significant differences to the respective Gly33-wt construct (**, p < 0.001, one-way ANOVA type Bonferroni). B, relative Aβ40 levels (mean ± S.E., n = 7–16). C, relative Aβ42 levels (mean ± S.E., n = 5–17). D, relative Aβ38 levels (mean ± S.E., n = 4–12). B–D, horizontal lines mark the fragment levels of APP-G33A for better comparison with the FAD mutants. E, analysis of expression levels of SPA4CT-related constructs in SH-SY5Y cells. The fragments show comparable expression levels, although these are generally lower than for SPA4CT-wt. Western blot was stained with monoclonal W0-2. F, levels of secreted Aβ from SPA4-fragments are normalized to SPA4CT-wt, which was set as 100% (mean ± S.E., n = 3–9). The mutation G33A has no impact on Aβ40 levels but decreases Aβ42 and increases Aβ38 levels. w/o, without.
This observation raised the question of whether G33A affects the processing at early (ϵ-site) or subsequent (γ-sites) cleavage steps of γ-secretase. Therefore, two extended constructs were generated encoding elongated Aβ species that were shown to be either processed as precursors of the Aβ42 line, i.e. Aβ(1–51), or by the Aβ40 line, i.e. Aβ(1–52) (29). As these constructs were generated from the C-terminal 99 residues of APP (SPA4CT) (30), the fragments are named SPA4/1–51 and SPA4/1–52. The constructs are normally expressed in SH-SY5Y cells and processed like the C-terminal fragment of APP (Fig. 3E). The quantification of Aβ levels shows that SPA4/1–52 predominantly generates Aβ40 but neither Aβ42 nor Aβ38 (Fig. 3F). SPA4/1–51 generates mainly Aβ42 and Aβ38. SPA4/1–51 containing the mutation G33A shows 2.5-fold reduced Aβ42 levels and 2-fold increased Aβ38 levels compared with Gly33-wt. Aβ40 levels are not affected in both constructs. Thus, the mutation G33A similarly influences processing of SPA4/1–51, SPA4/1–52, APP-wt, and APP-FAD. This indicates that the mutation G33A has a universal effect on Aβ42 and Aβ38 generation. Importantly, the result also shows that the allocation to one or the other product line cannot be modulated by G33A mutation.
Analysis of PS1-FAD Mutants
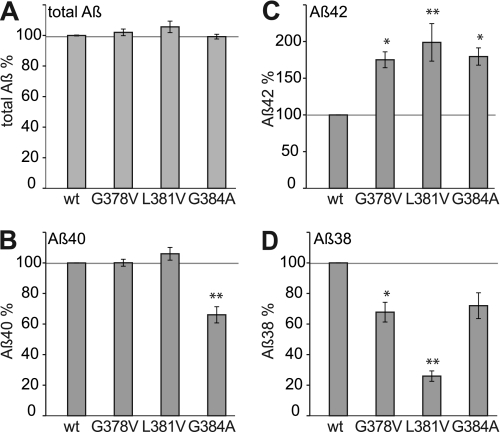
Three mutations, i.e. G378A, L381A, and G384A, located near or within the highly conserved GXGD motif of PS1 in close proximity to the catalytically critical aspartate residue 385 within TMS-7 (31–33) were analyzed (Fig. 1C). As substrate, we used SPA4CT instead of the full-length APP, as SPA4CT yields higher Aβ levels (9). Stably expressing SH-SY5Y cell lines were selected for expression of similar levels of holo-PS1, the N-terminal fragment of PS1, and SPA4CT by Western blot analysis (supplemental Fig. S3). Efficiency of SPA4CT processing by PS1-FAD mutants did not vary between cell lines as indicated by total Aβ levels measured (Fig. 4A). All three PS1-FAD mutants led to increased levels of Aβ42 and decreased levels of Aβ38 (Fig. 4, C and D). Aβ40 levels were found specifically reduced for PS1-G384A (Fig. 4B), a mutant that has already been described to selectively lower Aβ40 production (34). Thus, an obvious difference between the investigated PS1-FAD and APP-FAD mutations are the consistently increased Aβ42 levels of PS1-FAD at the expense of decreased Aβ38 levels, whereas APP-FAD mutants that increase Aβ42 also increase Aβ38 levels. This also indicates that PS1-FAD processing is divergent from APP-FAD.
FIGURE 4.
Analysis of PS1-FAD mutants in the Gly33-wt background. Quantification of secreted Aβ from SH-SY5Y cells coexpressing PS1-FAD mutants and SPA4CT-wt. PS1-wt was set as 100%. A, total Aβ levels (mean ± S.E., n = 3–10). B, Aβ40 levels (mean ± S.E., n = 3–10). C, Aβ42 levels (mean ± S.E., n = 3–8). D, Aβ38 levels (mean ± S.E., n = 3–9). Asterisks indicate significant differences to PS1-wt/SPA4CT-wt (*, p < 0.01, **, p < 0,001, one-way ANOVA type Dunnett).
Impact of the GxxxG Mutation G33A on PS1-FAD Processing
Effects of the GxxxG motif on APP and SPA4CT processing are very similar, as described previously (9). Now, to investigate possible GxxxG-mediated influences on PS1-FAD mutants, we stably coexpressed SPA4CT-G33A and PS1-FAD mutants in SH-SY5Y cells and determined levels of Aβ in the cell culture supernatants by ELISA (Fig. 5 and supplemental Fig. S4). Aβ levels generated from Gly33-wt substrates were set as 100%. Total Aβ levels were similar for all constructs tested indicating that the SPA4CT-G33A protein was efficiently processed by the respective PS1-FAD γ-secretase complex (Fig. 5A). As expected for the G33A mutant, Aβ40 levels were not affected (Fig. 5B). However, the G33A mutation showed the tendency to decrease Aβ42 levels (Fig. 5C) and to increase Aβ38 levels; the latter at least holds true for the G387V and L381V mutations compared with Gly33-wt (Fig. 5D). Attributable to the close proximity to the active site the G384A mutation might follow a different mode of action. There were no aberrant patterns of Aβ species (supplemental Fig. S5). Although the effect of G33A on Aβ42 and Aβ38 levels in the presence of PS1-FAD mutations was visible, it was not as strong as observed for the APP-FAD mutations.
FIGURE 5.
Impact of the GxxxG mutation G33A on PS1-FAD processing. ELISA data from PS1-FAD and SPA4CT-G33A coexpressing cells (black bars) in comparison to the respective SPA4CT-wt expressing lines, set as 100% (gray bars). A, total Aβ levels (mean ± S.E., n = 3–10). B, Aβ40 levels (mean ± S.E., n = 3–10). C, Aβ42 levels (mean ± S.E., n = 3–8). D, Aβ38 levels (mean ± S.E., n = 3–9). Asterisks indicate significant differences to PS1-wt/SPA4CT-wt (**, p < 0.001, one-way ANOVA type Bonferroni). Original data of total Aβ, Aβ42, Aβ40, and Aβ38 ELISA are shown in supplemental Fig. S4.
DISCUSSION
Processing of APP-FAD and PS1-FAD Mutants
Here, we describe a previously unrecognized relationship between mutations in juxtamembrane positions and γ-secretase cleavages, although the mutations are located ∼20–40 amino acid residues apart from the γ-cleavage sites. This is exemplified by the mixed effect found on Aβ species for the APP-sw mutant showing a 4-fold increase in Aβ42 and Aβ38 levels but an only 2-fold increase Aβ40 levels. So far, influences on Aβ42/Aβ40 ratios by APP-sw were only described to vary with the age of mice expressing APP-sw (35) or APP-sw and V717F (36).
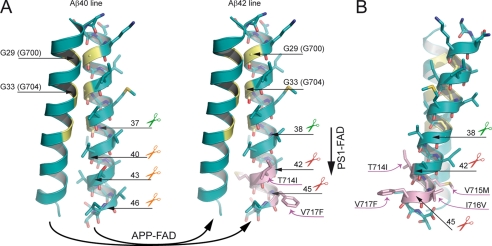
We also observed that most of the APP-FAD mutants either increase Aβ42 and Aβ38 levels or solely decrease Aβ40 levels. We explain this observation with the sequential cleavage model of γ-secretase proposing the existence of two product lines (9, 15), the Aβ40 line and the Aβ42 line (Fig. 6A). We suggest that APP-FAD mutations cause a general shift away from the Aβ40 line toward the Aβ42 line, explaining why Aβ40 levels were found decreased and both Aβ42 as well as Aβ38 levels were increased (Fig. 6A). Interestingly, molecular modeling reveals that the amino acid side chains of APP-FAD mutations stick out from the APP-TMS dimer interface and thus unlikely impair APP-TMS dimerization in a cellular environment (Fig. 6, A and B). However, Gorman et al. (37) observed an impact of FAD mutations on oligomerization of synthetic peptides in micelles. From our data, we speculate that APP-FAD mutations, especially those located in the C-terminal region, may shift the affiliation to the Aβ42 product line by affecting enzyme-substrate recognition rather than that they impair TMS dimerization itself. Thus, the initial recognition of APP-FAD mutations seems to decide on the pathway followed (Aβ42 or Aβ40 line). This view is supported by results describing that APP-FAD mutations in the C-terminal region affect ϵ-cleavage, leading to an increase of APP intracellular domain levels (APP intracellular domain) promoting the Aβ42 line (38, 39). Although the α-helices likely need to be unfolded prior to γ-secrease cleavages, it might be of interest that the peptide bonds cleaved in the Aβ42 line reside outside the dimer, whereas the peptide bonds of the Aβ40 line reside within the dimer interface.
FIGURE 6.
Model of the pathological mechanisms caused by APP-FAD and PS1-FAD mutations. Oxygen atoms are depicted in red, nitrogen atoms are depicted in blue, and sulfur atoms are colored dark yellow. The glycine residues of the interface are highlighted in yellow, and APP-FAD mutations are colored pink. Scissors indicate peptide bonds that are cleaved by the γ-secretase. A, dimeric wt substrates are degraded by the γ-secretase predominantly by the Aβ40 line. Left, corresponding peptide bonds are indicated and are located at the dimer interface. We propose that APP-FAD mutations cause a general shift between the two product lines so that the Aβ40 line is down-regulated, and the Aβ42 line becomes a major degradation pathway. Right, APP-FAD, peptide bonds of the Aβ42 line are indicated. Note, that amino acid side chains from FAD mutations do not reach into the dimer interface. PS1-FAD mutants seem to cause substrate flux inhibition leading to a retarded processing within the Aβ42 line causing increased Aβ42 and decreased Aβ38 levels (vertical arrow). The dimer crossing point mediated by Gly29 (Gly700) and Gly33 (Gly704) may cause a steric hindrance and inhibit the consecutive γ-secretase processing leading predominantly to Aβ40 (Aβ40 line) or to Aβ42 (Aβ42 line) (9, 10). Mechanistically, the effect of G33A occurs after the effects of APP-FAD mutations explaining why G33A causes a constant reduction by 60% for Aβ42 and a 3-fold increase of Aβ38 in the presence of all individual APP-FAD mutations analyzed. B, the APP-FAD-TMS dimer in side view for better illustration of the FAD-causing amino acid side chains. T714I, V715M, I716V, and V717F stick out of the dimer interface and thus likely affect processing by modulating the substrate-enzyme recognition.
For the PS1-FAD mutants analyzed, a reduction in Aβ38 but an increase in Aβ42 levels was the major change observed, indicating that PS1-FAD mutations generally act differently from APP-FAD mutations. In agreement with the sequential cleavage model, PS1-FAD mutations might lead to an inhibition of flux through the Aβ42 pathway, which could account for the decrease of Aβ38 and increase of Aβ42 levels (Fig. 6A). The inhibition of cleavage flux is in agreement with the loss-of-function hypothesis of PS1-FAD mutations (40). The apparently different APP-FAD and PS1-FAD mechanisms question cell culture or mouse models where these mutations are combined to accelerate the Aβ production and pathology.
Impact of the GxxxG Motif
We have reported previously that the sequence motif GxxxG within the APP-TMS has a regulatory impact on the Aβ species produced. The mutation G33A attenuated the TMS dimerization strength by 20%, specifically reduced the formation of Aβ42 by 60%, left Aβ40 levels unaffected, but increased the formation of Aβ38 (3-fold) and shorter Aβ species (9). When we analyzed the GxxxG motif in combination with APP-FAD mutations, we found that the G33A mutant yielded the same shift, i.e. a 60% decrease of Aβ42 levels and a concomitant 3-fold increase of Aβ38 levels for all APP-FAD mutants. Thus, G33A in combination with APP-FAD mutations affected γ-secretase processing in the same way as when combined with APP-wt. Furthermore, in protein constructs being degraded in a predetermined product line (SPA4/1–51, SPA4/1–52) G33A had the same strong effect on the consecutive processing. This implies that G33A particularly affects γ-cleavages rather than the primary ϵ-cleavage step. This also indicates that G33A exclusively influences processing within the Aβ42 line.
In the presence of PS1-FAD mutations, the impact of G33A on Aβ generation was diminished, which might be attributable to the possible inhibition of substrate flux by PS1-FAD mutations. Thus, the mutation G33A acts downstream of APP-FAD mutations but only partially downstream of PS1-FAD mutations.
In addition to this data, a product-precursor relationship of Aβ42 and Aβ38 was indicated by the effects on Aβ production by (i) nonsteroidal anti-inflammatory drugs or γ-secretase modulators (41), (ii) several γ-secretase inhibitors (42), (iii) N-terminal elongation of pen-2 (43), and (iv) GxxxG mutations (9) as well as direct detection (V) of the tetrapeptide V39VIA42 arising from the Aβ42 cleavage that generates Aβ38 (15). Page et al. (34) and Czirr et al. (44) concluded from their work that Aβ42 and Aβ38 are not related in their production as in the presence of PS1-FAD mutants nonsteroidal anti-inflammatory drugs sulindac sulfide and fenofibrate only had an attenuated effect on Aβ38 and Aβ42 levels. In agreement with this, we found only a tendency of G33A to change the Aβ38/Aβ42 levels in the presence of PS1-FAD mutants. We assumed that the attenuated effects are attributable to the inhibition of substrate flux by PS1-FAD mutations. Concordantly, we suggested previously that nonsteroidal anti-inflammatory drugs might act by modulating the substrate dimer stability (9), which recently has been supported by the finding that nonsteroidal anti-inflammatory drugs are substrate-targeted modulators, which possibly bind to the Aβ sequence (45).
CONCLUSION
APP-FAD and PS1-FAD mutations act differently on Aβ42/Aβ40 production. Mechanistically, the analyzed familial mutations can be divided into three subgroups: (i) APP-FAD mutants that increase Aβ42 and Aβ38, (ii) APP-FAD mutants that decrease Aβ40, and (iii) PS1-FAD mutants that increase Aβ42, but decrease Aβ38 levels. In the early steps of APP processing, APP-FAD mutations decide on the affiliation to the one or the other product line, and subsequently, the mutation G33A affects processing within the Aβ42 line.
An impairment of the GxxxG-mediated dimerization of APP was sufficient to rescue the pathological processing effects of APP-FAD mutants. On average, 60% less Aβ42 and 3-fold more Aβ38 were produced by the APP-FAD-G33A mutants. The effects of G33A were found attenuated with PS1-FAD mutants attributable to the different pathogenic mechanism of PS1-FAD. Thus, our data supports the idea that the APP GxxxG motif represents a new drug target site not only for sporadic AD but also for early onset FAD.
Supplementary Material
Acknowledgment
We thank Dr. Paul M. Mathews (Nathan Kline Institute for Psychiatric Research, Orangeburg, NY) for providing the NT1 antibody for detection of PS1-NTF.
This work was supported by the Alzheimer Forschung Initiative e.V. (to L.-M. M.), the Deutsche Forschungsgemeinschaft (MU901 (to G. M.), HI 1502/1-1 (to P. W. H.)), the SFB740, Kompetenznetz Degenerative Demenzen (Förderkennzeichen 01 GI 0723), and the Hans und Ilse Breuer Stiftung.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5 and additional references.
- APP
- amyloid precursor protein
- sw
- Swedish
- Aβ
- amyloid-β peptide
- AD
- Alzheimer disease
- ELISA
- enzyme-linked immunosorbent assay
- FAD
- familial Alzheimer disease
- GSM
- γ-secretase modulators
- sAPPα/-β
- secreted APP ectodomain α- or β-cleaved
- sw
- Swedish FAD mutation
- TMS
- transmembrane sequence
- PS
- presenilin
- wt
- wild-type
- MALDI-MS
- matrix-assisted laser desorption ionization mass spectrometry
- ANOVA
- analysis of variance
- APLP
- APP-like protein.
REFERENCES
- 1.Chen C. D., Oh S. Y., Hinman J. D., Abraham C. R. (2006) J. Neurochem. 97, 30–43 [DOI] [PubMed] [Google Scholar]
- 2.Gralle M., Botelho M. G., Wouters F. S. (2009) J. Biol. Chem. 284, 15016–15025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaden D., Munter L. M., Joshi M., Treiber C., Weise C., Bethge T., Voigt P., Schaefer M., Beyermann M., Reif B., Multhaup G. (2008) J. Biol. Chem. 283, 7271–7279 [DOI] [PubMed] [Google Scholar]
- 4.Rossjohn J., Cappai R., Feil S. C., Henry A., McKinstry W. J., Galatis D., Hesse L., Multhaup G., Beyreuther K., Masters C. L., Parker M. W. (1999) Nat. Struct. Biol. 6, 327–331 [DOI] [PubMed] [Google Scholar]
- 5.Scheuermann S., Hambsch B., Hesse L., Stumm J., Schmidt C., Beher D., Bayer T. A., Beyreuther K., Multhaup G. (2001) J. Biol. Chem. 276, 33923–33929 [DOI] [PubMed] [Google Scholar]
- 6.Wang Y., Ha Y. (2004) Mol. Cell 15, 343–353 [DOI] [PubMed] [Google Scholar]
- 7.Kaden D., Voigt P., Munter L. M., Bobowski K. D., Schaefer M., Multhaup G. (2009) J. Cell Sci. 122, 368–377 [DOI] [PubMed] [Google Scholar]
- 8.Soba P., Eggert S., Wagner K., Zentgraf H., Siehl K., Kreger S., Lower A., Langer A., Merdes G., Paro R., Masters C. L., Muller U., Kins S., Beyreuther K. (2005) EMBO J. 25, 653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Munter L. M., Voigt P., Harmeier A., Kaden D., Gottschalk K. E., Weise C., Pipkorn R., Schaefer M., Langosch D., Multhaup G. (2007) EMBO J. 26, 1702–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qi-Takahara Y., Morishima-Kawashima M., Tanimura Y., Dolios G., Hirotani N., Horikoshi Y., Kametani F., Maeda M., Saido T. C., Wang R., Ihara Y. (2005) J. Neurosci. 25, 436–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao G., Cui M. Z., Mao G., Dong Y., Tan J., Sun L., Xu X. (2005) J. Biol. Chem. 280, 37689–37697 [DOI] [PubMed] [Google Scholar]
- 12.Hardy J. (2009) J. Neurochem. 110, 1129–1134 [DOI] [PubMed] [Google Scholar]
- 13.Hardy J. A., Higgins G. A. (1992) Science 256, 184–185 [DOI] [PubMed] [Google Scholar]
- 14.Weidemann A., Eggert S., Reinhard F. B., Vogel M., Paliga K., Baier G., Masters C. L., Beyreuther K., Evin G. (2002) Biochemistry 41, 2825–2835 [DOI] [PubMed] [Google Scholar]
- 15.Takami M., Nagashima Y., Sano Y., Ishihara S., Morishima-Kawashima M., Funamoto S., Ihara Y. (2009) J. Neurosci. 29, 13042–13052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scheuner D., Eckman C., Jensen M., Song X., Citron M., Suzuki N., Bird T. D., Hardy J., Hutton M., Kukull W., Larson E., Levy-Lahad E., Viitanen M., Peskind E., Poorkaj P., Schellenberg G., Tanzi R., Wasco W., Lannfelt L., Selkoe D., Younkin S. (1996) Nat. Med. 2, 864–870 [DOI] [PubMed] [Google Scholar]
- 17.Borchelt D. R., Thinakaran G., Eckman C. B., Lee M. K., Davenport F., Ratovitsky T., Prada C. M., Kim G., Seekins S., Yager D., Slunt H. H., Wang R., Seeger M., Levey A. I., Gandy S. E., Copeland N. G., Jenkins N. A., Price D. L., Younkin S. G., Sisodia S. S. (1996) Neuron 17, 1005–1013 [DOI] [PubMed] [Google Scholar]
- 18.St George-Hyslop P. H. (2000) Biol. Psychiatry. 47, 183–199 [DOI] [PubMed] [Google Scholar]
- 19.Citron M., Oltersdorf T., Haass C., McConlogue L., Hung A. Y., Seubert P., Vigo-Pelfrey C., Lieberburg I., Selkoe D. J. (1992) Nature 360, 672–674 [DOI] [PubMed] [Google Scholar]
- 20.Sherrington R., Rogaev E. I., Liang Y., Rogaeva E. A., Levesque G., Ikeda M., Chi H., Lin C., Li G., Holman K., et al. (1995) Nature 375, 754–760 [DOI] [PubMed] [Google Scholar]
- 21.Ancolio K., Dumanchin C., Barelli H., Warter J. M., Brice A., Campion D., Frébourg T., Checler F. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 4119–4124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chartier-Harlin M. C., Crawford F., Houlden H., Warren A., Hughes D., Fidani L., Goate A., Rossor M., Roques P., Hardy J., et al. (1991) Nature 353, 844–846 [DOI] [PubMed] [Google Scholar]
- 23.Eckman C. B., Mehta N. D., Crook R., Perez-tur J., Prihar G., Pfeiffer E., Graff-Radford N., Hinder P., Yager D., Zenk B., Refolo L. M., Prada C. M., Younkin S. G., Hutton M., Hardy J. (1997) Hum. Mol. Genet. 6, 2087–2089 [DOI] [PubMed] [Google Scholar]
- 24.Hendriks L., van Duijn C. M., Cras P., Cruts M., Van Hul W., van Harskamp F., Warren A., McInnis M. G., Antonarakis S. E., Martin J. J., et al. (1992) Nat. Genet. 1, 218–221 [DOI] [PubMed] [Google Scholar]
- 25.Kamino K., Orr H. T., Payami H., Wijsman E. M., Alonso M. E., Pulst S. M., Anderson L., O'dahl S., Nemens E., White J. A. (1992) Am. J. Hum. Genet. 51, 998–1014 [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar-Singh S., De Jonghe C., Cruts M., Kleinert R., Wang R., Mercken M., De Strooper B., Vanderstichele H., Löfgren A., Vanderhoeven I., Backhovens H., Vanmechelen E., Kroisel P. M., Van Broeckhoven C. (2000) Hum. Mol. Genet. 9, 2589–2598 [DOI] [PubMed] [Google Scholar]
- 27.Kwok J. B., Li Q. X., Hallupp M., Whyte S., Ames D., Beyreuther K., Masters C. L., Schofield P. R. (2000) Ann. Neurol. 47, 249–253 [DOI] [PubMed] [Google Scholar]
- 28.Murrell J., Farlow M., Ghetti B., Benson M. D. (1991) Science 254, 97–99 [DOI] [PubMed] [Google Scholar]
- 29.Funamoto S., Morishima-Kawashima M., Tanimura Y., Hirotani N., Saido T. C., Ihara Y. (2004) Biochemistry 43, 13532–13540 [DOI] [PubMed] [Google Scholar]
- 30.Dyrks T., Dyrks E., Masters C., Beyreuther K. (1992) FEBS Lett. 309, 20–24 [DOI] [PubMed] [Google Scholar]
- 31.Cruts M., Backhovens H., Wang S. Y., Van Gassen G., Theuns J., De Jonghe C. D., Wehnert A., De Voecht J., De Winter G., Cras P., et al. (1995) Hum. Mol. Genet. 4, 2363–2371 [DOI] [PubMed] [Google Scholar]
- 32.Ikeuchi T., Kaneko H., Miyashita A., Nozaki H., Kasuga K., Tsukie T., Tsuchiya M., Imamura T., Ishizu H., Aoki K., Ishikawa A., Onodera O., Kuwano R., Nishizawa M. (2008) Dement Geriatr. Cogn. Disord. 26, 43–49 [DOI] [PubMed] [Google Scholar]
- 33.Janssen J. C., Beck J. A., Campbell T. A., Dickinson A., Fox N. C., Harvey R. J., Houlden H., Rossor M. N., Collinge J. (2003) Neurology 60, 235–239 [DOI] [PubMed] [Google Scholar]
- 34.Czirr E., Cottrell B. A., Leuchtenberger S., Kukar T., Ladd T. B., Esselmann H., Paul S., Schubenel R., Torpey J. W., Pietrzik C. U., Golde T. E., Wiltfang J., Baumann K., Koo E. H., Weggen S. (2008) J. Biol. Chem. 283, 17049–17054 [DOI] [PubMed] [Google Scholar]
- 35.McGowan E., Pickford F., Kim J., Onstead L., Eriksen J., Yu C., Skipper L., Murphy M. P., Beard J., Das P., Jansen K., Delucia M., Lin W. L., Dolios G., Wang R., Eckman C. B., Dickson D. W., Hutton M., Hardy J., Golde T. (2005) Neuron 47, 191–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chishti M. A., Yang D. S., Janus C., Phinney A. L., Horne P., Pearson J., Strome R., Zuker N., Loukides J., French J., Turner S., Lozza G., Grilli M., Kunicki S., Morissette C., Paquette J., Gervais F., Bergeron C., Fraser P. E., Carlson G. A., George-Hyslop P. S., Westaway D. (2001) J. Biol. Chem. 276, 21562–21570 [DOI] [PubMed] [Google Scholar]
- 37.Gorman P. M., Kim S., Guo M., Melnyk R. A., McLaurin J., Fraser P. E., Bowie J. U., Chakrabartty A. (2008) BMC Neurosci. 9, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kakuda N., Funamoto S., Yagishita S., Takami M., Osawa S., Dohmae N., Ihara Y. (2006) J. Biol. Chem. 281, 14776–14786 [DOI] [PubMed] [Google Scholar]
- 39.Sato T., Dohmae N., Qi Y., Kakuda N., Misonou H., Mitsumori R., Maruyama H., Koo E. H., Haass C., Takio K., Morishima-Kawashima M., Ishiura S., Ihara Y. (2003) J. Biol. Chem. 278, 24294–24301 [DOI] [PubMed] [Google Scholar]
- 40.De Strooper B. (2007) EMBO Rep. 8, 141–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weggen S., Eriksen J. L., Das P., Sagi S. A., Wang R., Pietrzik C. U., Findlay K. A., Smith T. E., Murphy M. P., Bulter T., Kang D. E., Marquez-Sterling N., Golde T. E., Koo E. H. (2001) Nature 414, 212–216 [DOI] [PubMed] [Google Scholar]
- 42.Zhao G., Tan J., Mao G., Cui M. Z., Xu X. (2007) J. Neurochem. 100, 1234–1246 [DOI] [PubMed] [Google Scholar]
- 43.Isoo N., Sato C., Miyashita H., Shinohara M., Takasugi N., Morohashi Y., Tsuji S., Tomita T., Iwatsubo T. (2007) J. Biol. Chem. 282, 12388–12396 [DOI] [PubMed] [Google Scholar]
- 44.Page R. M., Baumann K., Tomioka M., Pérez-Revuelta B. I., Fukumori A., Jacobsen H., Flohr A., Luebbers T., Ozmen L., Steiner H., Haass C. (2008) J. Biol. Chem. 283, 677–683 [DOI] [PubMed] [Google Scholar]
- 45.Kukar T. L., Ladd T. B., Bann M. A., Fraering P. C., Narlawar R., Maharvi G. M., Healy B., Chapman R., Welzel A. T., Price R. W., Moore B., Rangachari V., Cusack B., Eriksen J., Jansen-West K., Verbeeck C., Yager D., Eckman C., Ye W., Sagi S., Cottrell B. A., Torpey J., Rosenberry T. L., Fauq A., Wolfe M. S., Schmidt B., Walsh D. M., Koo E. H., Golde T. E. (2008) Nature 453, 925–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.