Abstract
β-Type phospholipase A2 inhibitory protein (PLIβ) from the serum of the venomous snake Gloydius brevicaudus neutralizes basic phospholipase A2 (PLA2) from its own venom, and it has 33% sequence homology with human leucine-rich α2-glycoprotein (LRG), which has been recently reported to bind cytochrome c (Cyt c) (Cummings, C., Walder, J., Treeful, A., and Jemmerson, R. (2006) Apoptosis 11, 1121–1129). In the present study, PLIβ was found to bind Cyt c. The interactions of LRG and PLIβ with Cyt c were compared by surface plasmon resonance analysis. Human LRG bound horse and snake Cyt c with dissociation constants of 1.58 × 10−13 m and 1.65 × 10−10 m, respectively, but did not bind yeast Cyt c, while G. brevicaudus PLIβ bound horse, snake, and yeast Cyt c with dissociation constants of 1.05 × 10−10 m, 2.37 × 10−12 m, and 1.67 × 10−6 m, respectively. On the other hand, LRG did not show any PLA2 inhibitory activity and did not bind G. brevicaudus basic PLA2, whereas PLIβ bound the basic PLA2 with a dissociation constant of 1.21 × 10−9 m, which is smaller than those with the Cyt c described above. The PLA2 inhibitory activity of PLIβ was also found to be suppressed by the binding of Cyt c to PLIβ. These results suggest that autologous Cyt c is an endogeneous ligand for LRG and PLIβ and that these serum proteins neutralize the autologous Cyt c released from the dead cells.
Keywords: Cytochrome c, Innate Immunity, Phospholipase A, Protein-Protein Interactions, Serum
Introduction
Venomous snakes have three distinct types of phospholipase A2 (PLA2)2 inhibitory proteins (PLIα, PLIβ, and PLIγ) in their blood to protect themselves from the leakage of their own venom PLA2s into the circulatory system (1, 2). PLIβ purified from the serum of Chinese mamushi Gloydius brevicaudus (renamed from Agkistrodon blomhoffii siniticus according to the present taxonomy) has nine tandem leucine-rich repeats (LRRs) and shows 33% sequence identity with human leucine-rich α2-glycoprotein (LRG) (3), one of the serum glycoproteins with unknown function. G. brevicaudus PLIβ inhibits specifically group II basic PLA2 from G. brevicaudus venom and does not inhibit group II acidic and neutral PLA2s from the same venom (1). PLIβs having similar inhibition spectra could also be purified from the sera of nonvenomous Colubridae snakes, Elaphe quadrivirgata (4) and Elaphe climacophora (5). Accordingly, we assumed that mammalian LRG might be a mammalian ortholog of PLIβ and function as a PLA2 inhibitory protein. We tried to produce the recombinant human LRG and mouse LRG, but the recombinant LRGs were expressed as inclusion body proteins in the Escherichia coli expression system and could not be refolded correctly.3 Therefore, this preparation of recombinant mouse LRG was used as an antigen to obtain antiserum in rabbit. By the immunological monitoring of LRG on Western blots, we could purify native LRG from mouse sera by sequential chromatography on Sephadex G-200, Q-Sepharose, phenyl-Sepharose, and Mono Q columns. However, the purified mouse LRG did not show any PLA2 inhibitory activities.3
Although LRG was first isolated from human serum in 1977 (6) and the first LRR protein discovered in 1985 (7), the physiological function of LRG is still unknown. In 2002, O'Donnell et al. (8) have reported that LRG expression was up-regulated during early neutrophilic granulocyte differentiation, suggesting its physiological role in neutrophilic granulocytes. In 2006, Cummings et al. (9) have found that the sera from different species contained the inhibitory factor that inhibited the cytochrome c (Cyt c) detection in the sandwich ELISA, which was finally identified as LRG. An ELISA for LRG using Cyt c as the capturing ligand was developed, and the serum LRG level was found to increase significantly in patients infected with bacteria (10). We have recently shown that LRG expression is up-regulated in HepG2 cells by IL-6 and suggested that LRG is one of the type 1 acute phase proteins (11). More recently, Codina et al. (12) have shown that the extracellular Cyt c produces a toxic effect on lymphocytes and that LRG protects against Cyt c-induced lymphotoxicity.
In the present study, we found that G. brevicaudus PLIβ could tightly bind Cyt c and compared its Cyt c binding activities with those of the purified human LRG. Tight binding of Cyt c with these serum proteins suggests that these proteins function as self-protective proteins against autologous Cyt c, which might function as a proinflammatory mediator and an endogenous danger signal.
EXPERIMENTAL PROCEDURES
Materials
The blood plasma and venom of Chinese mamushi (G. brevicaudus) and the heart of Erabu sea krait (Laticauda semifasciata) were obtained from the Japan Snake Institute (Gunma, Japan). G. brevicaudus basic PLA2 was purified from the venom as described previously (13). G. brevicaudus PLIβ was purified to homogeneity from blood plasma as described previously (1). Human sera were collected from healthy adult volunteers. Horse and yeast Cyt c were purchased from Sigma-Aldrich.
Purification of LRG from Human Sera
LRG was purified from human sera using the affinity resin coupled with horse heart Cyt c by a method similar to that of Cummings et al. (9). Horse Cyt c dissolved in 0.2 m NaHCO3 and 0.5 m NaCl (pH 8.3) was added to a 1-ml Hi-Trap NHS-activated HP column (GE Healthcare, Buckinghamshire, England) and incubated 1 h at room temperature. After deactivation of the remaining active groups by washing with 0.5 m ethanolamine containing 0.5 m NaCl (pH 8.3), the column was washed with equilibration buffer (20 mm sodium phosphate buffer containing 0.15 m NaCl, pH 7.4), then with the elution buffer (0.1 m acetate buffer containing 0.5 m NaCl, pH 4.0) and finally equilibrated with the equilibration buffer. About 5 ml of human sera were loaded into the column and washed with the equilibration buffer. Elution was performed with the elution buffer and fractions of 5 ml were collected in test tubes, each of which contained 1 ml of 1 m Tris-HCl buffer (pH 8.0). The fractions containing LRG were further purified by gel filtration chromatography on a Hi-Load 16/60 Superdex 200 pg column (GE Healthcare) that had been equilibrated with 50 mm Hepes buffer (pH 7.5, ionic strength 0.2). Because the LRG-containing fraction thus obtained gave a single homogeneous band on SDS-PAGE, this fraction was used as a purified LRG preparation. The N-terminal sequence of the purified LRG was determined by use of a protein sequencer (Procise 491HT; Applied Biosystems, Foster City, CA). The concentration of LRG was determined spectrophotometrically by using the extinction coefficients calculated from their amino acid sequence data (14).
SDS-PAGE and Immunodetection
SDS-PAGE was carried out on 10% gels. Samples were boiled in the SDS sample buffer containing 50 mm dithiothreitol for 5 min and then loaded onto the stacking gel. After electrophoresis, protein bands were stained with Coomassie Blue or periodic acid-Schiff (PAS). For immunodetection, proteins on the gel were transferred electrophoretically onto a polyvinylidene difluoride membrane (Bio-Rad). The blot was then probed with the rabbit antiserum against recombinant human LRG prepared as described previously (11). After the blot had been washed, the proteins on the blot were detected with anti-rabbit IgG donkey antibodies conjugated with horseradish peroxidase and ECL plus detection reagents (GE Healthcare).
Circular Dichroism (CD) Measurements
CD spectra in the far-ultraviolet region were recorded at 20 °C and ionic strength 0.1 with a Jasco model J-500A spectropolarimeter which had been calibrated with a standard solution of ammonium d-camphorsulfonate. A cell of 0.2 or 0.1-cm path length was used for the measurement. The mean residue ellipticity, [θ], was obtained from the equation: [θ] = (100 × θ)/(l × c), where θ is the observed ellipticity in degrees, c the residue molar concentration of the protein, and l the path length of the cell in centimeters.
Purification of Sea Krait Cyt c
Cyt c was purified by the method similar to that described previously (15). A frozen heart of a Erabu sea krait (L. semifasciata) was homogenized with 0.5% Al2(SO4)3 and was allowed to stand for 3 h on ice. After the centrifugation at 10,000 × g for 20 min, the supernatant was neutralized by addition of NH4OH and then centrifuged to discard aluminum hydroxide precipitated. The supernatant thus obtained was passed through a CM-Sephadex C50 column (1 × 5 cm) which had been equilibrated with 20 mm sodium phosphate buffer (pH 8.0). Cyt c adsorbed on the column was eluted with 20 mm sodium phosphate buffer (pH 8.0) containing 0.5 m NaCl. The eluate was applied to a Cosmosil Protein-R column (Nacalai Tesque, Kyoto, Japan) that had been equilibrated with 0.1% trifluoroacetic acid. Cyt c was eluted with a linear gradient of acetonitrile from 0% to 48% in 0.1% trifluoroacetic acid. The peak containing Cyt c was collected and lyophilized. The concentration of Cyt c was determined by measuring the absorbance at 550 nm (ϵmm at 550 nm = 27.7) after the Cyt c had been fully reduced with Na2S2O4 (16).
Detection of the LRG-Cyt c Complex by Gel Filtration Chromatography and Native PAGE
A 0.29-nmol amount of LRG was incubated for 1 h at room temperature with 0, 0.29, and 0.58 nmol of horse Cyt c in phosphate-buffered saline. The mixture was then applied to a Superdex 200 HR10/30 column (GE Healthcare), which had been equilibrated with 50 mm Hepes buffer (pH 7.5, ionic strength 0.2). Thyroglobulin (670 kDa), bovine γ-globulin (158 kDa), chicken ovalbumin (44 kDa), and equine myoglobin (17 kDa) were used as molecular mass standards (Bio-Rad). The mixtures of a 12-pmol amount of LRG with various amounts of horse Cyt c were subjected to native PAGE using the Ornstein-Davis discontinuous buffer system (17). Proteins in the gel were stained with a silver staining kit (Nag Research Laboratories, Fremont, CA).
Binding Analysis by Surface Plasmon Resonance (SPR)
The binding of Cyt c and PLA2 to PLIβ and LRG was analyzed with a BIAcore X system (GE Healthcare). G. brevicaudus PLIβ or human LRG was directly linked to the carboxymethylated dextran matrix of a CM5 sensor chip surface at a flow rate of 5 μl/min by using an amine coupling kit (GE Healthcare) according to the manufacturer's instruction. Various concentrations of Cyt c or PLA2 were then injected at a flow rate of 10 μl/min at 25 °C with running buffer (50 mm Hepes buffer containing 0.05% Tween 20, pH 7.5, prepared with NaCl added to give an ionic strength of 0.2). Sensor chips were regenerated at the end of each run by the injection of 10 mm sodium acetate buffer (pH 4.0) containing 0.5 m NaCl. Analysis of the association and dissociation curves was performed with the BIAevaluation 3.0 software (GE Healthcare) using the 1:1 Langmuir binding model with drifting baseline (global fitting), and the mean values of the apparent dissociation constants (Kd) were calculated from the association rate constant (kassoc) and the dissociation rate constant (kdiss) (Kd = kdiss/kassoc).
Inhibition of PLA2 Enzymatic Activity
Enzymatic activities of G. brevicaudus basic PLA2 were measured fluorometrically using 1-palmitoyl-2-pyrenedecanoylphosphatidylcholine (Cayman Chemical, Ann Arbor, MI) as a substrate, according to the method of Radvanyi et al. (18) with some modifications as described previously (19), in the presence of various concentrations of the PLIβ.
RESULTS
Purification and Fundamental Characterization of Human LRG
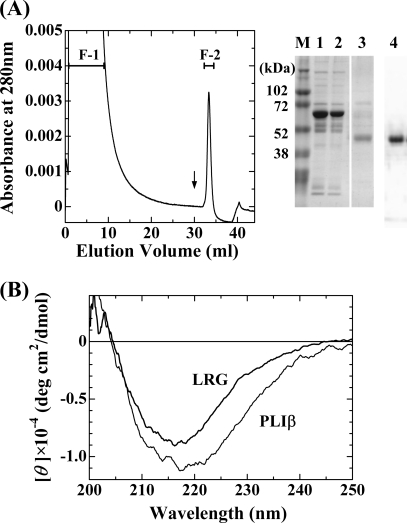
Cummings et al. (9) have found that serum LRG binds Cyt c from the observation that the serum inhibitory factor, which inhibited the Cyt c detection in the sandwich ELISA, was purified and identified as LRG. They purified the factor using the Cyt c-coupled Affi-Gel column. In the present study, we could purify LRG from human sera using a Cyt c-coupled Hi-Trap NHS-activated HP column as shown in Fig. 1A. LRG specifically adsorbed to the column, and the adsorbed fraction F-2 gave a nearly single band with an apparent molecular mass of 50 kDa on SDS-PAGE, which could be detected by Western blotting using a rabbit antiserum raised against recombinant human LRG. Its N-terminal sequence was determined to be VXLSPKDXQ, which agreed with that of human LRG. In contrast to the molecular mass of 34,336 based on the amino acid sequence of human LRG, the molecular mass of the purified LRG was determined to be 42,778 Da by mass spectrometry, indicating the presence of carbohydrate chains as already reported (7). The presence of carbohydrate chains was also confirmed by PAS staining of the LRG band on SDS-PAGE (data not shown).
FIGURE 1.
Purification of human LRG by Cyt c affinity chromatography and CD spectrum of the purified human LRG. A, Cyt c affinity column chromatography of human serum. Human serum was applied to a Cyt c-coupled Hi-Trap NHS-activated HP column, which had been equilibrated with 20 mm phosphate buffer (pH 7.4) containing 0.15 m NaCl. The column was washed with the same buffer, and the adsorbed proteins were eluted with 100 mm acetate buffer (pH 4.0) containing 0.5 m NaCl. SDS-PAGE of the whole serum (lane 1) and the fractions (lane 2, F-1; lane 3, F-II) obtained after Cyt c affinity column chromatography is shown. M, molecular weight markers. LRG was detected by Western blot analysis using a rabbit antiserum raised against human LRG (lane 4). B, CD spectra of human LRG and G. brevicaudus PLIβ. The spectra were measured at 20 °C, pH 7.5, and ionic strength 0.1.
Previously, we found that a β-type PLA2 inhibitory protein (PLIβ), which had been purified from the blood plasma of Chinese mamushi snake G. brevicaudus, exhibited 33% sequence identity with that of human LRG (3). Presence of the significant sequence homology between them suggested that they were orthologous serum proteins. Therefore, we investigated the PLA2 inhibitory activity of human LRG. But the purified human LRG did not show any inhibitory activities against G. brevicaudus type II basic PLA2, human type IIA PLA2, and human type V PLA2 (data not shown). On the other hand, when the purified PLIβ was applied to a Cyt c-coupled Hi-Trap NHS-activated HP column, it was found that PLIβ as well as human LRG tightly adsorbed to the column, indicating that PLIβ had Cyt c binding activity (data not shown). Therefore, the Cyt c binding activity of PLIβ was further investigated as described below.
Fig. 1B shows the far-ultraviolet CD spectrum of the LRG preparation that was finally purified by a Superdex 200 column. The CD spectrum of LRG is very similar to that of PLIβ. They are characteristic of proteins with a high content of β-sheet structure. LRG and PLIβ share similarities in the primary and secondary structures, which have common structural features characteristic of LRR proteins.
Direct Interaction of Human LRG with Horse Cyt c
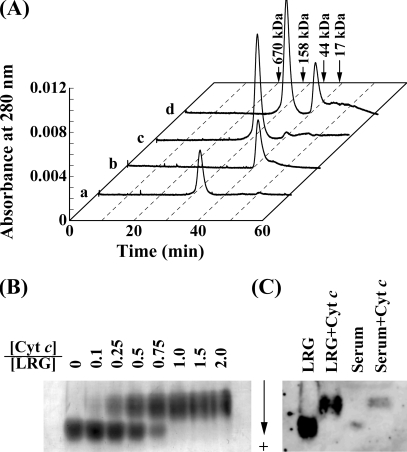
As shown in Fig. 2A, complex formation of human LRG with horse Cyt c was investigated by gel filtration on a Superdex 200 HR10/30 column. When 0.29 nmol of LRG and 0.29 nmol of Cyt c were separately applied to the column, they were eluted with a single peak at 31.6 min and 40.7 min, corresponding to their molecular masses of 72.5 and 12.2 kDa, respectively. However, when their mixture was applied to the column, the latter peak disappeared and the former peak increased in height, suggesting the formation of a stable complex between LRG and Cyt c. When the mixture of 0.29 nmol of LRG and 0.58 nmol of Cyt c was applied to the column, the former peak of the complex between LRG and Cyt c showed the same height as that in the previous case, indicating that the LRG was saturated with Cyt c; and the latter peak of the resultant excess amount of Cyt c showed the same height as that of 0.29 nmol of Cyt c. These results suggest that one LRG molecule bind one Cyt c molecule. The apparent molecular mass value of 72.5 kDa estimated from molecular mass standards seemed to be much higher than that of 42.8 kDa determined from mass spectrometry. This apparent discrepancy might be due to the structural difference between LRG and normal globular proteins. Like many LRR proteins, LRG could be expected to form a horseshoe-like curved solenoid structure where each repeat is a turn of the solenoid, and this horseshoe-like structure of LRG probably resulted in the increase in the apparent molecular size of LRG on size-exclusion chromatography. The higher apparent molecular masses on size-exclusion chromatography have also been observed for many LRR proteins (20). The apparent molecular mass of the LRG-Cyt c complex on a Superdex 200 column could be estimated to be 75.2 kDa, which is slightly higher than that of LRG. It is likely that Cyt c may be tightly bound to the concave side of horseshoe-like LRG and deeply buried within the horseshoe structure, which cause only a slight increase in the apparent molecular size of the complex on size-exclusion chromatography.
FIGURE 2.
Direct interaction of human LRG with horse Cyt c. A, gel filtration of the mixture of human LRG and horse Cyt c on Superdex 200 HR10/30 column. Samples were applied onto a Superdex 200 column that had been equilibrated with 50 mm Hepes buffer (pH 7.5 and ionic strength of 0.2). a, 0.29 nmol of LRG; b, 0.29 nmol of Cyt c; c, a mixture of 0.29 nmol of LRG and 0.29 nmol of Cyt c; d, a mixture of 0.29 nmol of LRG and 0.58 nmol of Cyt c. B, native PAGE of the mixture of human LRG and horse Cyt c. 5 μg each of the purified human LRG samples mixed with indicated molar ratios of Cyt c were applied to native PAGE on a 5% gel. C, Western blot analysis of native PAGE. The purified human LRG and whole human serum samples, and those mixed with excess amounts of horse Cyt c were applied to 5% native PAGE. After the proteins on the gel were transferred onto polyvinylidene difluoride membrane, LRG was detected immunochemically with the rabbit antiserum against human LRG.
As shown in Fig. 2B, the complex formation of human LRG with horse Cyt c was also investigated by native PAGE. The LRG samples mixed with various molar ratios of horse Cyt c were subjected to 5% native PAGE by using the Ornstein-Davis discontinuous buffer system (17). Because LRG is an acidic glycoprotein having negative charges and Cyt c is a basic protein having positive charges at neutral pH values, LRG bands on native PAGE are expected to be shifted toward the cathode by forming a complex with Cyt c. Two protein bands corresponding to LRG and LRG-Cyt c complex were observed for the mixtures containing 0.1, 0.25, 0.5, and 0.75 equivalents of Cyt c, and only one band of the complex was observed for the mixtures containing 1, 1.5, and 2.0 equivalents of Cyt c. These results further confirmed that one LRG molecule tightly bind one Cyt c molecule.
Because we found that the native PAGE could quantify the free and bound forms of LRG separately, LRG in the intact serum was examined by native PAGE, followed by immunodetection. As shown in Fig. 2C, the bound forms of LRG could not be detected in normal human serum. Weivoda et al. (10) have already reported that LRG was not bound to Cyt c to any significant extent even in the sera of patients with a bacterial infection.
SPR Analyses of the Interaction of LRG and PLIβ with Various Cyt c
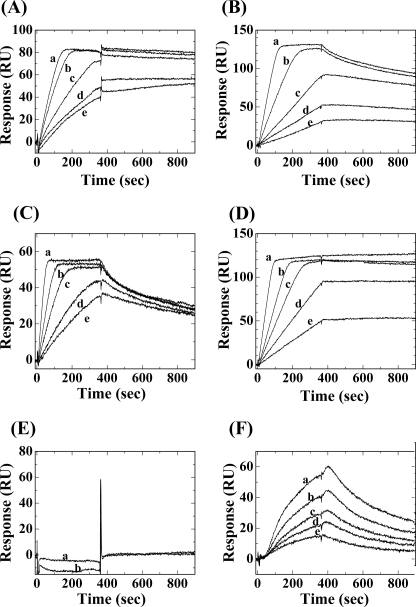
Because LRG and PLIβ shared common structural similarities and common functional properties in Cyt c binding, PLIβ could be regarded as a snake ortholog of human LRG. To compare their Cyt c binding activities in detail, we immobilized human LRG or G. brevicaudus PLIβ on biosensor chips and measured their interactions with horse, snake, and yeast Cyt c in real-time by monitoring the changes in SPR signals after the injection of various concentrations of Cyt c (Fig. 3). The dissociation constants (Kd) and the association and dissociation rate constants (kassoc and kdiss) were calculated from the curve fitting analysis of the sensorgrams (Table 1). As shown in Fig. 3A, the sensorgrams showed that LRG had very high affinity for horse Cyt c with a binding manner characterized by fast association and slow dissociation rates. The Kd value could be calculated to be 1.58 × 10−13 m, which reflected one of the highest affinity interactions among known protein-protein interactions. This is inconsistent with a recently reported result that human LRG bound human Cyt c with a Ka value of 4.5 × 107 m−1 (namely, a Kd value of 2.2 × 10−8 m) by SPR analysis (12). This large difference in Kd values could not be explained by the difference in amino acid sequence between human and horse Cyt c, but could probably be ascribed to the fact that different immobilized ligands were used for these two SPR analyses; we immobilized human LRG, whereas Codina et al. (12) immobilized human Cyt c to the biosensor chip. In their SPR analyses, some of the lysine residues in a human Cyt c molecule, which are responsible for the interaction with a LRG molecule, might be used up for the immobilization on the sensor chip, and its binding affinity to human LRG might therefore be reduced. As shown in Fig. 3B, G. brevicaudus PLIβ had reduced affinity for horse Cyt c with a Kd value of 1.05 × 10−10 m. The difference between human LRG and snake PLIβ in the affinities for horse Cyt c might be due to the difference in the species specificity. Therefore, we purified snake Cyt c from the heart of a sea krait Laticauda semifasciata. As shown in Fig. 3, C and D, G. brevicaudus PLIβ had a greater affinity for snake Cyt c with a Kd value of 2.37 × 10−12 m than human LRG, namely, human LRG had a higher affinity for horse Cyt c than for snake Cyt c and snake PLIβ had a higher affinity for snake Cyt c than for horse Cyt c. In view of the species specificity of LRG-Cyt c interaction, it is assumed that the endogenous ligands for LRG and PLIβ are autologous Cyt c. As shown in Fig. 3, E and F, yeast Cyt c did not bind to LRG, but bound with low affinity to PLIβ (a Kd value of 1.67 × 10−6 m). It has recently been reported that LRG did not bind to yeast iso-1 Cyt c nor iso-2 Cyt c by indirect ELISA (12). Lys-72 in the Cyt c molecule, which is trimethylated in yeast Cyt c, was suggested to be responsible for interaction with LRG. Because PLIβ could bind to yeast Cyt c, Lys-72 might not be critical to the interaction with PLIβ.
FIGURE 3.
Binding curves for the interactions of horse, sea krait, and yeast Cyt c with the immobilized human LRG and G. brevicaudus PLIβ, measured in real-time by surface plasmon resonance. A, sensorgrams for interaction of immobilized LRG with various concentrations of horse Cyt c. a, 10 nm; b, 5.0 nm; c, 2.5 nm; d, 1.0 nm; e, 0.5 nm. B, sensorgrams for interaction of immobilized PLIβ with various concentrations of horse Cyt c. a, 10 nm; b, 5.0 nm; c, 2.5 nm; d, 1.5 nm; e, 1.0 nm. C, sensorgrams for interaction of immobilized LRG with various concentrations of sea krait Cyt c. a, 10 nm; b, 5.0 nm; c, 3.0 nm; d, 1.5 nm; e, 1.0 nm. D, sensorgrams for interaction of immobilized PLIβ with various concentrations of sea krait Cyt c. a, 10 nm; b, 5.0 nm; c, 3.0 nm; d, 2.0 nm; e, 1.0 nm. E, sensorgrams for interaction of immobilized LRG with various concentrations of yeast Cyt c. a, 50 nm; b, 100 nm. F, sensorgrams for interaction of immobilized PLIβ with various concentrations of yeast Cyt c. a, 200 nm; b, 150 nm; c, 100 nm; d, 75 nm; e, 50 nm.
TABLE 1.
Dissociation constants (Kd) and association and dissociation rate constants (kassoc and kdiss) for the interaction of horse, snake, and yeast Cyt c, and G. brevicaudus basic PLA2 with immobilized human LRG or G. brevicaudus PLIβ
| Ligand | Analyte | Kd | kassoc | kdiss |
|---|---|---|---|---|
| m | m−1s−1 | s−1 | ||
| Human LRG | Horse Cyt c | 1.58 ± 9.92 × 10−13 | 2.00 ± 0.02 × 106 | 3.15 ± 19.8 × 10−7 |
| Snake PLIβ | Horse Cyt c | 1.05 ± 0.19 × 10−10 | 2.53 ± 0.05 × 107 | 4.18 ± 0.08 × 10−3 |
| Human LRG | Snake Cyt c | 1.65 ± 0.06 × 10−10 | 3.90 ± 0.36 × 107 | 4.08 ± 0.35 × 10−3 |
| Snake PLIβ | Snake Cyt c | 2.37 ± 0.48 × 10−12 | 7.86 ± 0.12 × 106 | 1.86 ± 0.35 × 10−5 |
| Human LRG | Yeast Cyt c | NBa | NB | NB |
| Snake PLIβ | Yeast Cyt c | 1.67 ± 0.05 × 10−6 | 1.26 ± 0.04 × 103 | 2.10 ± 0.01 × 10−3 |
| Human LRG | Basic PLA2 | NB | NB | NB |
| Snake PLIβ | Basic PLA2 | 1.21 ± 0.03 × 10−9 | 8.69 ± 0.13 × 105 | 1.05 ± 0.01 × 10−3 |
a NB, no binding.
SPR Analyses of the Interaction of LRG and PLIβ with Type II Basic PLA2
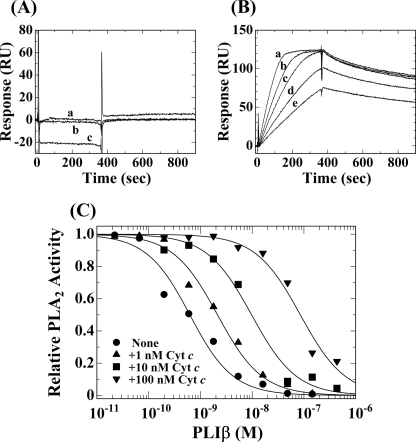
As mentioned above, human LRG did not inhibit any PLA2 activities examined, while G. brevicaudus PLIβ specifically inhibited the enzyme activity of G. brevicaudus basic PLA2 (1). Interactions of LRG and PLIβ with G. brevicaudus basic PLA2 were measured by SPR analysis. As shown in Fig. 4, A and B, the basic PLA2 did not bind to human LRG, but bound tightly to PLIβ with a Kd value of 1.21 × 10−9 m (Table 1), which was comparable with the apparent inhibition constant (Kiapp) value of 7.5 × 10−10 m, which had previously been estimated from the measurement of its inhibitory activity (1). Because the binding affinity of G. brevicaudus PLIβ for the G. brevicaudus basic PLA2 was lower than those for the horse and snake Cyt c, the endogenous ligand of PLIβ might be autologous Cyt c rather than the venom PLA2.
FIGURE 4.
Interaction of G. brevicaudus basic PLA2 with human LRG and G. brevicaudus PLIβ. A, sensorgrams for interaction of immobilized LRG with various concentrations of G. brevicaudus basic PLA2. a, 28 nm; b, 7 nm; c, 110 nm. B, sensorgrams for interaction of immobilized PLIβ with various concentrations of G. brevicaudus basic PLA2. a, 40 nm; b, 30 nm; c, 20 nm; d, 15 nm; e, 10 nm. C, effect of horse Cyt c on the inhibition by PLIβ against enzymatic activity of G. brevicaudus basic PLA2.
The Effect of Cyt c on the PLA2 Inhibitory Activity of PLIβ
Fig. 4C shows the PLA2 inhibitory activity of G. brevicaudus PLIβ in the presence of horse Cyt c. In the absence of Cyt c, PLIβ inhibited the enzymatic activity of G. brevicaudus basic PLA2 with a Kiapp value of 0.57 nm. But the inhibitory activity of PLIβ was suppressed by horse Cyt c; the Kiapp values were shifted to 2.1, 9.7, and 85 nm in the presence of the Cyt c at its concentrations of 1, 10, and 100 nm, respectively. These results suggest that the Cyt c binding site on the PLIβ molecule closely overlaps the PLA2 binding site, and hence the bound Cyt c molecule would suppress the PLA2 inhibitory activity of PLIβ.
DISCUSSION
The present study demonstrates, for the first time, that PLIβ tightly binds Cyt c. Because snake PLIβ and human LRG are homologous serum glycoproteins with the Cyt c binding activity, PLIβ could be deemed to be a snake ortholog of LRG. PLIβ is one of the three types of PLA2 inhibitory proteins purified from serum of Chinese mamushi snake G. brevicaudus (1). Because PLIβ binds specifically basic PLA2 included in its own venom and inhibits its enzymatic activity, it appears that PLIβ is involved in the clearance of venom PLA2 for self-protection against its own venom. PLIβs have also been purified from the sera of nonvenomous snakes E. quadrivirgata (4) and E. climacophora (5), although their endogenous PLA2s targeted have not been identified yet. The E. quadrivirgata PLIβ inhibited specifically G. brevicaudus basic PLA2 and its inhibition spectrum was quite similar to that of G. brevicaudus PLIβ (4). Because PLA2-rich tissues, such as the venom glands of venomous snakes, are not present in the nonvenomous snakes, the nonvenomous snake PLIβ might function as a Cyt c-binding protein rather than a PLA2 inhibitory protein. The fact that the affinity of G. brevicaudus PLIβ for horse and snake Cyt c is higher than that for the autologous venom PLA2 lends support to the hypothesis that PLIβ has been evolved to acquire a new function as a PLA2 inhibitory protein in addition to the ancestral function as a Cyt c-binding protein during evolution of the venomous snake.
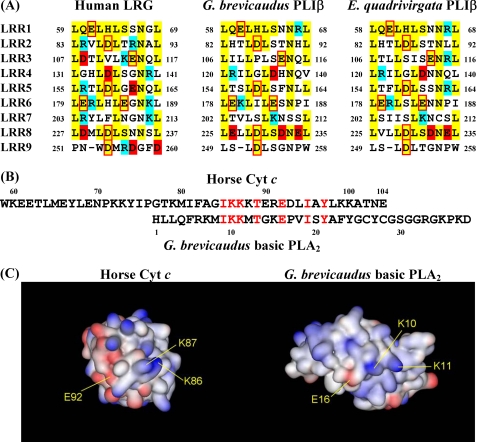
LRG is a serum protein in which LRRs were first discovered, and it contains eight typical 24-residue LRRs with the consensus sequence of LXXLXLXXNXLXXLPXXLLXXLXX (X being any amino acid) (7). LRG and PLIβ belong to the typical LRR subfamily based on their consensus sequences according to Kobe and Kajava (21). LRR domains are built of tandem repeats, and they form curved solenoid structures that are particularly suitable for protein-protein interactions (21). The concave space defined by the curvature of LRR structures is suited for the ligand binding. Three-dimensional structures of the complexes between LRR proteins and their ligands have shown that the ligands were surrounded at least in part by the concave surface of the parallel β-sheet in the LRR domain (22–25). It is most likely that the charged amino acid residues included in the concave surface are involved in the interaction of LRG and PLIβ with Cyt c and PLA2. Therefore, the amino acid sequences of LRG and PLIβs, which correspond to the parallel β-sheet on the concave surface, were compared (Fig. 5A). Some of the eight negatively charged residues conserved among all three sequences might be responsible for the binding of Cyt c, while some of the negatively charged residues only conserved between two PLIβ sequences, Asp-126, Asp-232, and Glu-234 in the PLIβ sequence, might be responsible for specific binding to the G. brevicaudus basic PLA2.
FIGURE 5.
Sequence comparison between LRG and PLIβ on the concave surface of LRR structures and sequence homology between Cyt c and PLA2. A, sequence comparison between LRG and PLIβ on the concave surface of LRR structures. The residues conserved among these proteins are shown in yellow. The residues with negative and positive charges are in red and blue, respectively. Conserved residues with negative charges are in red boxes. B, sequence homology found between the C-terminal region of horse Cyt c and the N-terminal region of G. brevicaudus basic PLA2. C, solid molecular surfaces of Cyt c (PDB ID: 1HRC) and basic PLA2 (PDB ID: 1B4W) colored by charge (blue, positive charge; red, negative charge).
PLIβ was found to bind exclusively to either horse Cyt c or G. brevicaudus basic PLA2. When the amino acid sequence of horse Cyt c is compared with that of G. brevicaudus basic PLA2, there is a significant sequence homology between the C-terminal region of horse Cyt c and the N-terminal region of the basic PLA2 (Fig. 5B). Previously, based on the results of the inhibition specificity and the sequence comparison of 14 PLA2s from the various sources (1), we have anticipated that PLIβ would recognize the specific amino acid residues, such as His-1, Arg-6, Glu-16, and Trp-61, located in or close to the interfacial binding surface of the basic PLA2. Because Glu-16 of the PLA2 is included in the homologous sequence between Cyt c and PLA2, the N-terminal region of the basic PLA2 might be responsible for the interaction with PLIβ. Snake venom basic PLA2 might have evolved to acquire the structure analogous to that of Cyt c to utilize a Cyt c-binding protein, LRG, as a PLA2-binding protein, PLIβ, in the evolutional process of venomous snakes. As shown in Fig. 5C, the homologous regions between Cyt c and PLA2 were compared on the three-dimensional structures of Cyt c and PLA2. However, we could not find any significant similarities in the spatial patterns of charge distributions between them. Because human LRG does not bind basic PLA2 but binds tightly to horse Cyt c, the homologous region from residues 85–97 of Cyt c would be excluded from the main LRG binding site on a Cyt c molecule if human LRG and G. brevicaudus PLIβ have the same binding site on Cyt c. But it is possible that this homologous region on the Cyt c molecule functions as a sub-binding site for LRG and PLIβ and functions as the main binding site for PLIβ. Because trimethylation of Lys-72 in Cyt c prevented Apaf-1 activation, Lys-72 was suggested to be responsible for the interaction between Apaf-1 and Cyt c (26). Recently, Codina et al. (12) have shown that LRG competed with Apaf-1 for binding horse Cyt c and that LRG did not bind the recombinant rat and pigeon Cyt c in which Lys-72 was trimethylated specifically by the post-translational modification in yeast cells, suggesting that Lys-72 is also responsible for the interaction between human LRG and Cyt c. PLIβ could bind yeast Cyt c, albeit with lower affinity, indicating that Lys-72 is not an essential residue for the interaction between G. brevicaudus PLIβ and Cyt c. Because the discussion about the binding sites described above remains speculative, further studies are required to identify the LRG and PLIβ binding site on the Cyt c molecule.
Snake venom PLA2s exhibit a wide variety of pharmacological activities including neurotoxicity and myotoxicity (27). Venomous snakes must neutralize and eliminate their own venom PLA2s, which are entered accidentally into their circulatory system. G. brevicaudus PLIβ is one of these antitoxic serum proteins, and it inhibits and neutralizes specifically the basic PLA2 from its own venom (1). The present study shows that PLIβ binds more tightly to autologous Cyt c than the basic PLA2, suggesting that the original function of PLIβ may be the clearance of autologous Cyt c from the circulatory system. Cyt c is known to be critical to the induction of apoptosis in eukaryotic cells where it is released from the intermembrane space of the mitochondria to the cytosol. The released Cyt c binds to Apaf-1, and this eventually leads to an apoptosome formation and activation of the caspase cascade that triggers apoptosis (28–30). Then, Cyt c is released at least in part from the apoptotic cells to the extracellular space in an intact monomeric form, because the Cyt c release is initiated very soon after its translocation into the cytosol (31, 32). Extracellular Cyt c has been reported to induce apoptosis in J774 cells (33), lymphocytes (12), and dendritic cells (34), although it has no apparent effects on the apoptosis of most other cells. Furthermore, a single intra-articular injection of Cyt c into a mouse knee joint has been reported to induce an acute inflammatory synovitis (35). Extracellular Cyt c has also been reported to activate NF-κB and induce cytokine and chemokine production in mouse splenocytes (35). Therefore, extracellular Cyt c acts as a proinflammatory mediator and an endogenous danger signal called DAMPs (damage-associated molecular patterns). Previously, we found that LRG was a secretory type 1 acute-phase protein whose expression was up-regulated by the mediators of acute-phase response (11). Extracellular Cyt c released from cell death by an acute inflammation could be trapped to form a tight complex with LRG and be excluded from circulation. Recently, mitochondrial DAMPs including formyl peptides and mitochondrial DNA have been shown to activate human polymorphonuclear neutrophils (PMNs) through the pattern recognition receptors and lead to PMN migration and degranulation in vitro and in vivo (36). Because LRG was shown to be up-regulated during neutrophilic differentiation (8) and be localized in the primary (azurophilic) granule compartment (37), it is possible that LRG functions as one of the pattern recognition receptors of PMN and modulates the neutrophilic function through an innate immune pathway.
R. Shirai, Y. Morimoto, F. Hirano, K. Ikeda, and S. Inoue, unpublished results.
- PLA2
- phospholipase A2
- PLI
- phospholipase A2 inhibitor
- PLIβ
- β-type phospholipase A2 inhibitor
- LRR
- leucine-rich repeat
- LRG
- leucine-rich α2-glycoprotein
- Cyt c
- cytochrome c
- SPR
- surface plasmon resonance
- PDB
- Protein Data Bank
- PAS
- periodic acid-Schiff.
REFERENCES
- 1.Ohkura N., Okuhara H., Inoue S., Ikeda K., Hayashi K. (1997) Biochem. J. 325, 527–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lambeau G., Lazdunski M. (1999) Trends. Pharmacol. Sci. 20, 162–170 [DOI] [PubMed] [Google Scholar]
- 3.Okumura K., Ohkura N., Inoue S., Ikeda K., Hayashi K. (1998) J. Biol. Chem. 273, 19469–19475 [DOI] [PubMed] [Google Scholar]
- 4.Okumura K., Inoue S., Ikeda K., Hayashi K. (2002) Arch. Biochem. Biophys. 408, 124–130 [DOI] [PubMed] [Google Scholar]
- 5.Shirai R., Toriba M., Hayashi K., Ikeda K., Inoue S. (2009) Toxicon 53, 685–692 [DOI] [PubMed] [Google Scholar]
- 6.Haupt H., Baudner S. (1977) Hoppe Seylers Z. Physiol. Chem. 358, 639–646 [PubMed] [Google Scholar]
- 7.Takahashi N., Takahashi Y., Putnam F. W. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 1906–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Donnell L. C., Druhan L. J., Avalos B. R. (2002) J. Leukoc. Biol. 72, 478–485 [PubMed] [Google Scholar]
- 9.Cummings C., Walder J., Treeful A., Jemmerson R. (2006) Apoptosis. 11, 1121–1129 [DOI] [PubMed] [Google Scholar]
- 10.Weivoda S., Andersen J. D., Skogen A., Schlievert P. M., Fontana D., Schacker T., Tuite P., Dubinsky J. M., Jemmerson R. (2008) J. Immunol. Methods 336, 22–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shirai R., Hirano F., Ohkura N., Ikeda K., Inoue S. (2009) Biochem. Biophys. Res. Commun. 382, 776–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Codina R., Vanasse A., Kelekar A., Vezys V., Jemmerson R. (2010) Apoptosis 15, 139–152 [DOI] [PubMed] [Google Scholar]
- 13.Inoue S., Shimada A., Ohkura N., Ikeda K., Samejima Y., Omori-Satoh T., Hayashi K. (1997) Biochem. Mol. Biol. Int. 41, 529–537 [DOI] [PubMed] [Google Scholar]
- 14.Gill S. C., von Hippel P. H. (1989) Anal. Biochem. 182, 319–326 [DOI] [PubMed] [Google Scholar]
- 15.Inoue S., Hiroyoshi T., Matsubara H., Yamanaka T. (1984) Biochim. Biophys. Acta 790, 188–195 [DOI] [PubMed] [Google Scholar]
- 16.Margoliash E., Frohwirt N. (1959) Biochem. J. 71, 570–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ornstein L. (1964) Ann. N. Y. Acad. Sci. 121, 321–349 [DOI] [PubMed] [Google Scholar]
- 18.Radvanyi F., Jordan L., Russo-Marie F., Bon C. (1989) Anal. Biochem. 177, 103–109 [DOI] [PubMed] [Google Scholar]
- 19.Okumura K., Inoue S., Ikeda K., Hayashi K. (1999) Biochim. Biophys. Acta 1441, 51–60 [DOI] [PubMed] [Google Scholar]
- 20.Stumpp M. T., Forrer P., Binz H. K., Plückthun A. (2003) J. Mol. Biol. 332, 471–487 [DOI] [PubMed] [Google Scholar]
- 21.Kobe B., Kajava A. V. (2001) Curr. Opin. Struct. Biol. 11, 725–732 [DOI] [PubMed] [Google Scholar]
- 22.Schubert W. D., Urbanke C., Ziehm T., Beier V., Machner M. P., Domann E., Wehland J., Chakraborty T., Heinz D. W. (2002) Cell 111, 825–836 [DOI] [PubMed] [Google Scholar]
- 23.Fan Q. R., Hendrickson W. A. (2005) Nature 433, 269–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson R. J., McCoy J. G., Bingman C. A., Phillips G. N., Jr., Raines R. T. (2007) J. Mol. Biol. 368, 434–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huizinga E. G., Tsuji S., Romijn R. A., Schiphorst M. E., de Groot P. G., Sixma J. J., Gros P. (2002) Science 297, 1176–1179 [DOI] [PubMed] [Google Scholar]
- 26.Kluck R. M., Ellerby L. M., Ellerby H. M., Naiem S., Yaffe M. P., Margoliash E., Bredesen D., Mauk A. G., Sherman F., Newmeyer D. D. (2000) J. Biol. Chem. 275, 16127–16133 [DOI] [PubMed] [Google Scholar]
- 27.Kini R. M. (2003) Toxicon 42, 827–840 [DOI] [PubMed] [Google Scholar]
- 28.Liu X., Kim C. N., Yang J., Jemmerson R., Wang X. (1996) Cell 86, 147–157 [DOI] [PubMed] [Google Scholar]
- 29.Zou H., Li Y., Liu X., Wang X. (1999) J. Biol. Chem. 274, 11549–11556 [DOI] [PubMed] [Google Scholar]
- 30.Jiang X., Wang X. (2004) Annu. Rev. Biochem. 73, 87–106 [DOI] [PubMed] [Google Scholar]
- 31.Renz A., Berdel W. E., Kreuter M., Belka C., Schulze-Osthoff K., Los M. (2001) Blood 98, 1542–1548 [DOI] [PubMed] [Google Scholar]
- 32.Jemmerson R., LaPlante B., Treeful A. (2002) Cell Death. Differ. 9, 538–548 [DOI] [PubMed] [Google Scholar]
- 33.Hiraoka Y., Yamada T., Goto M., Das Gupta T. K., Chakrabarty A. M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 6427–6432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin M. L., Zhan Y., Proietto A. I., Prato S., Wu L., Heath W. R., Villadangos J. A., Lew A. M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 3029–3034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pullerits R., Bokarewa M., Jonsson I. M., Verdrengh M., Tarkowski A. (2005) Rheumatology 44, 32–39 [DOI] [PubMed] [Google Scholar]
- 36.Zhang Q., Raoof M., Chen Y., Sumi Y., Sursal T., Junger W., Brohi K., Itagaki K., Hauser C. J. (2010) Nature 464, 104–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ai J., Druhan L. J., Hunter M. G., Loveland M. J., Avalos B. R. (2008) J. Leukoc. Biol. 83, 1277–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]