Abstract
Paclitaxel (Taxol) is an effective chemotherapeutic agent for treatment of cancer patients. Despite impressive initial clinical responses, the majority of patients eventually develop some degree of resistance to Taxol-based therapy. The mechanisms underlying cancer cells resistance to Taxol are not fully understood. MicroRNA (miRNA) has emerged to play important roles in tumorigenesis and drug resistance. However, the interaction between the development of Taxol resistance and miRNA has not been previously explored. In this study we utilized a miRNA array to compare the differentially expressed miRNAs in Taxol-resistant and their Taxol-sensitive parental cells. We verified that miR-125b, miR-221, miR-222, and miR-923 were up-regulated in Taxol-resistant cancer cells by real-time PCR. We further investigated the role and mechanisms of miR-125b in Taxol resistance. We found that miR-125b was up-regulated in Taxol-resistant cells, causing a marked inhibition of Taxol-induced cytotoxicity and apoptosis and a subsequent increase in the resistance to Taxol in cancer cells. Moreover, we demonstrated that the pro-apoptotic Bcl-2 antagonist killer 1 (Bak1) is a direct target of miR-125b. Down-regulation of Bak1 suppressed Taxol-induced apoptosis and led to an increased resistance to Taxol. Restoring Bak1 expression by either miR-125b inhibitor or re-expression of Bak1 in miR-125b-overexpressing cells recovered Taxol sensitivity, overcoming miR-125-mediated Taxol resistance. Taken together, our data strongly support a central role for miR-125b in conferring Taxol resistance through the suppression of Bak1 expression. This finding has important implications in the development of targeted therapeutics for overcoming Taxol resistance in a number of different tumor histologies.
Keywords: Apoptosis, Breast Cancer, Drug Resistance, MicroRNA, Oncogene, Bak1, Chemotherapy, Paclitaxel, Taxol, miR-125b
Introduction
Breast cancer is the most common cancer and the second leading cause of cancer-related deaths in American women. Paclitaxel (Taxol) has recently emerged as an important chemotherapeutic agent in the treatment of breast cancer as well as other tumor histologies, such as ovarian, prostate, and non-small cell lung cancers (1, 2). The resistance of cancer cells to Taxol and other chemotherapeutic agents frequently results in the subsequent recurrence and metastasis of cancer (3). One known mechanism involved with cancer cell resistance to Taxol is the high expression of the membrane P-glycoprotein that functions as a drug-efflux pump (4). Other mechanisms include the alterations of tubulin structure (5, 6), changes in the drug binding affinity of the microtubules (7), and cell cycle deregulation (8, 9). However, the detailed molecular mechanisms that may contribute to Taxol resistance of cancer cells are still not fully understood.
MicroRNA (miRNA)3 is the non-coding, single-stranded RNA of ∼22 nucleotides, which regulates gene expression and constitutes a novel class of gene regulators (10). Mature miRNA molecules are partially complementary to one or more messenger RNA (mRNA) molecules and translationally down-regulate gene expression (11, 12) or induce the degradation of mRNAs bearing fully complementary target sites (13). Among hundreds of miRNAs, some have been shown to play a role in a variety of biological processes including proliferation, differentiation, migration, cell cycle, and apoptosis (14, 15). In recent years, some miRNAs have been reported to be involved in drug-resistance, acting as potential oncogenes or tumor suppressors (16–18). The “knockdown” and inhibition of miR-221 or miR-222 expression has been shown to sensitize MDA-MB-468 cells to tamoxifen-induced cell growth arrest and apoptosis (17). Ectopic miR-34a expression resulted in cell cycle arrest and growth inhibition and attenuated chemoresistance to the anti-cancer drug, camptothecin, by inducing apoptosis (18). To date, however, the interaction between miRNAs and Taxol resistance in cancer cells has not been explored.
We aimed to examine the role of miRNAs in Taxol resistance in breast cancer cells through the screening of differentially expressed miRNAs between Taxol-resistant and Taxol-sensitive cancer cells. We found that miR-125b is up-regulated in Taxol-resistant cancer cells, with its overexpression inhibiting Taxol-induced cytotoxicity and apoptosis, subsequently increasing the resistance of cancer cells to Taxol. Moreover, we demonstrate that Bak1 is a direct target of miR-125b in breast cancer cells. Restoring the expression of Bak1 recovered Taxol sensitivity in counteracting miR-125b-mediated Taxol resistance. Taken together, miR-125b plays a critical role in Taxol resistance through suppression of Bak1 expression, and it may serve as a potential target for overcoming Taxol resistance in human breast cancer.
EXPERIMENTAL PROCEDURES
Cells and Cell Culture
Human breast cancer cell lines, MDA-MB-435 (MDA-435), MDA-MB-231 (MDA-231), MDA-MB-436 (MDA-436), MCF7, SKBr3, and BT474 were purchased from American Type Culture Collection (ATCC). Taxol-resistant 435TR1 (single clone) and 435TRP (pooled clones) cells were developed from MDA-435 cells by treatment with gradually increasing concentrations of Taxol in cell culture medium. Cell viability assay showed that 435TR1 and 435TRP cells could tolerate much higher concentrations of Taxol compared with MDA-435 cells, with their IC50 concentrations found to be more than 30-fold higher than those of parental MDA-435 cells (19). Two more Taxol-resistant cell variants, 436TRP and SKBr3-TRP (both are pooled clones), were developed from MDA-436 and SKBr3 cell lines, respectively, using the same methods as for 435TRP. The cells were cultured in Dulbecco's modified Eagle's medium/F-12 (Mediatech, Inc.) supplemented with 10% fetal bovine serum and penicillin/streptomycin.
miRNA Microarray Expression Analysis
Total RNA was extracted from MDA-435, 435TR1, and 435TRP cells using TRIzol reagent (Invitrogen), and the total RNA was labeled with Hy3 and Hy5 fluorescent dyes. Pairs of labeled samples were hybridized to miRCURY LNATM microRNA array slides with coverage of 428 unique human microRNAs (Exiqon, 208201-A) by using miRCURY LNATM Power labeling kit and miRCURY LNATM microRNA array kit (Exiqon) according to the manufacturer's protocol. A miRNA detection signal threshold was defined as twice the maximum background signal. Normalization was done using a cyclic LOWESS (locally weighted regression) method to remove the system-related variations. Data adjustments included data filtering, log2 transformation, and gene centering and normalization. Student's t test analysis was conducted between MDA-435 and 435TR1 and MDA-435 and 435TRP samples, and miRNAs with p values < 0.05 were selected for cluster analysis. The clustering analysis was done using a hierarchical method and average linkage and Euclidean distance metrics.
Pre-miRNA or Anti-miRNA Transfection
miRNA precursors (pre-miRNAs) and miRNAs antisense RNAs (anti-miRNAs) were purchased from Applied Biosystems. Pre-miR-negative and anti-miR-negative were used as negative controls. Oligofectamine transfection reagent or Lipofectamine 2000 (Invitrogen) was used for the transfection of pre-miRNAs or anti-miRNAs. Forty-eight hours after transfection, the expression of miR-125b was detected by real-time PCR, and the expression of Bak1, a target of miR-125b, was tested by real-time PCR and/or Western blotting.
Quantitative Real Time PCR (qRT-PCR) and RT-PCR
Total RNA was isolated from cultured cells using the TRIzol reagent (Invitrogen). For miRNA expression analysis, qRT-PCR was done by using the TaqMan® microRNA reverse transcription kit (Applied Biosystems) and TaqMan® microRNA assays kit (Applied Biosystems) following the manufacturer's protocols. RNU6B served as an internal control. End-point PCR was performed to analyze the expression of miR-125b by using a mirVana qRT-PCR miRNA detection kit and mirVana qRT-PCR Primer Sets (Applied Biosystems) according to the manufacturer's protocols. Human U6 served as an internal control. For the expression of Bak1 and glyceraldehyde-3-phosphate dehydrogenase, cDNAs were synthesized from 1 μg of total RNA using Cloned AMV First-Strand cDNA Synthesis kit (Invitrogen). For quantitative PCR, cDNA was mixed with 2× SYBR Green PCR Master Mix (Applied Biosystems) and various sets of gene-specific primers and then subjected to RT-PCR quantification by using the iQ5 real time PCR system (Bio-Rad). The sequences of the primers used were as follows: Bak1, 5′ primer (5′-GCTCCCAACCCATTCACTAC-3′) and 3′ primer (5′-TCCCTACTCCTTTTCCCTGA-3′); glyceraldehyde-3-phosphate dehydrogenase, 5′ primer (5′-ATCCCATCACCATCTTCCAG-3′) and 3′ primer (5′-ATGAGTCCTTCCACGATACC-3′). For semiquantitative PCR, the cDNA was mixed with PCR SuperMix (Invitrogen) and gene-specific primers. The PCR conditions were 25–30 cycles at 95 °C for 30s, 56 °C for 30s, and 72 °C for 1 min. The PCR products were separated on a 2% agarose gel. All reactions were performed in triplicate. The relative amounts of mRNA were calculated by using the comparative CT method. The results are presented as –fold change of each miRNA or mRNA in the 435TR1 and 435TRP cells relative to the parental MDA-435 cells.
siRNA Experiments
siRNA oligonucleotides for Bak1 were purchased from Sigma, with a scrambled siRNA (Sigma) used as a control. Transfection was performed using the Oligofectamine Transfection reagent (Invitrogen) according to the manufacturer's protocol. Forty-eight hours after transfection, the cell lysates were prepared for further analysis by Western blotting.
mRNA Stability
MDA-435 cells were transfected with 100 nm pre-miR-125b or pre-miR-negative. After 12 h, cells were treated with actinomycin (5 μg/ml). The stability of endogenous Bak1 mRNA was examined with real-time RT-PCR by measuring Bak1 mRNA levels at the indicated time.
Activity of Caspase 3
MDA-435 cells were transfected with 100 nm pre-miR-125b or pre-miR-negative and seeded into 96-well plates at 8 × 103 cells per well in triplicates. After 8 h, cells were treated with or without 20 nm Taxol for 24 h. Caspase 3 activity was measured by Caspase-Glo® 3/7 assay kit (Promega) according to the manufacturer's instructions.
Apoptosis Assay
Cells were transfected with 100 nm miR-125b precursor (pre-miR-125b), 100 nm pre-miR-125b in combination with 100 nm miR-125b inhibitor (anti-miR-125b), or 4 μg of Bak1-expressing vector, respectively, and 24 h after transfection the cells were followed with the indicated concentrations of Taxol treatment for 48 h and then were subjected to an apoptosis assay. Apoptosis was determined by annexin V/propidium iodide staining with the apoptosis detection kit (BD Pharmingen). Briefly, 1 × 105 treated cells were incubated with annexin V/propidium iodide for 15 min at room temperature. The cells were then analyzed by flow cytometry using two-color fluorescence-activated cell sorting analysis (BD LSR II).
Luciferase Reporter Assay
The pMIR-reporter luciferase vector containing the 3′-UTR of Bak1 and the empty vector were generous gifts from Dr. deVere White (20). For the luciferase assay, cells at the density of 1.2 × 105 per well in 24-well plates were cotransfected with pMIR-REPORT luciferase reporters with or without 3′-UTR of Bak1, pre-miR-125b, or pre-miR-negative using Lipofectamine 2000 reagent. Twenty-four hours later, cells were harvested and lysed with passive lysis buffer (Promega). Luciferase activity was measured by using a dual luciferase reporter assay (Promega). The pRL-TK vector (Promega) was used as an internal control. The results were expressed as relative luciferase activity (firefly Luc/Renilla Luc).
Western Blotting
Cells were harvested and lysed in buffer containing 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 2 mm EDTA, 1% Triton, 1 mm phenylmethylsulfonyl fluoride, and protease inhibitor mixture (Sigma) for 20 min on ice. Lysates were cleared by centrifugation at 14,000 rpm at 4 °C for 10 min. Supernatants were collected, and protein concentrations were determined by the Bradford assay (Bio-Rad). The proteins were then separated with a SDS/polyacrylamide gel and transferred to a nitrocellulose membrane (Bio-Rad). After blocking in phosphate-buffered saline with 5% nonfat dry milk for 1 h, the membranes were incubated overnight at 4–8 °C with the primary antibodies in phosphate-buffered saline with 5% nonfat dry milk. The following antibodies were utilized: anti-Bak1 monoclonal antibody (1:1000, Cell Signaling), anti-cleaved PARP monoclonal antibody (1:1000, Cell Signaling), and anti-β-actin monoclonal antibody (1:2000, Sigma). Membranes were extensively washed with phosphate-buffered saline and incubated with horseradish peroxidase-conjugated secondary anti-mouse antibody or anti-rabbit antibody (dilution 1:2000, Bio-Rad). After additional washes with phosphate-buffered saline, antigen-antibody complexes were visualized with the enhanced chemiluminescence kit (Pierce).
Cell Viability Assay
A total of 5 × 103∼1 × 104 cells/well were seeded in 96-well plates. Twenty-four hours later, the medium was replaced with fresh medium with or without Taxol and incubated for 48 h. Cell viability was determined using CellTiter 96 Aqueous One Solution Cell Proliferation Assay kit (Promega) as previously reported (21).
Statistical Analysis
Statistical evaluation for data analysis was determined by unpaired Student's t test. All data were shown as the means ± S.E.. A statistical difference of p < 0.05 was considered significant.
RESULTS
Profiles of miRNAs in Taxol-sensitive and Resistant Cancer Cells
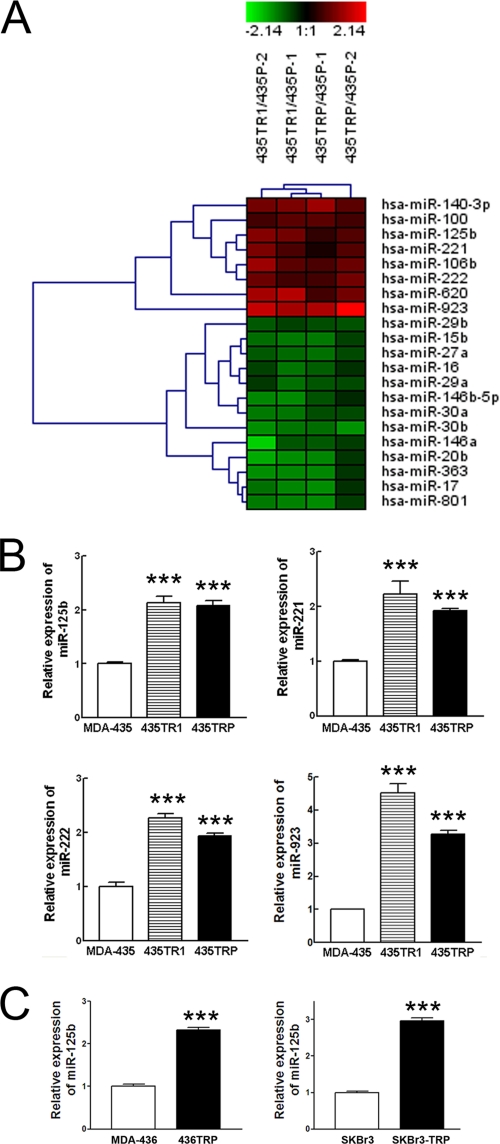
To search for the critical miRNAs involved in Taxol resistance, the total RNA extracted from Taxol-resistant 435TR1 and 435TRP cells and their Taxol-sensitive parental MDA-435 cells were hybridized with miRNA array chips. Eight miRNAs were up-regulated in Taxol-resistant cells by more than 1.5-fold, and 13 miRNAs were down-regulated in Taxol-resistant cells by more than 1.5-fold (Fig. 1A and supplemental Table S1). Notably, the profiling results are correlative with the reported functions of miRNAs. For example, miRNAs with oncogenic function, such as miR-100 (22), miR-125b (20, 23–26), miR-106b (27), miR-221, and miR-222 (28, 29), were up-regulated, whereas miRNAs with tumor suppressor functions, such as miR-16 (30, 31), miR-27a (32), miR-20a (33), miR-29a/b (34), and miR-146a/b (35), were down-regulated in Taxol-resistant cells. To further validate the miRNA array results, we randomly selected four miRNAs, miR-923, miR-125b, miR-221m, and miR-222, from the up-regulated miRNAs and measured their expression levels by real-time PCR. The results of real-time PCR for all of the four selected miRNAs were consistent with the miRNA array results (Fig. 1B). Furthermore, we examined the expression of miR-125b in two more Taxol-resistant cancer cell variants, 436TRP and SKBr3-TRP, that were derived from MD-MB-436 and SKBr3 breast cancer cell lines, respectively. More than 2-fold higher expression of miR-125b was found in Taxol-resistant 436TRP and SKBr3-TRP cells compared with their parental MDA-436 and SKBr3 cells, respectively (Fig. 1C), indicating that the correlation between miR-125b and Taxol resistance is not a single cell line-specific effect. These results suggested that the differential expression profiles screened by the miRNA array are reliable, and some of them may play a role in Taxol resistance.
FIGURE 1.
Profiles of miRNAs in Taxol-sensitive and -resistant cancer cells. A, shown is a Heatmap of a total of 21 miRNAs that were significantly up-regulated (n = 8) or down-regulated (n = 13) in Taxol-resistant 435TR1 and 435TRP cells compared with those in their parental MDA-435 cells (p < 0.05), which are listed in supplemental Table S1. Heatmap colors represent relative signal ratio of miRNA expression between Taxol-resistant 435TR1 or 435TRP to their parental MDA-435 cells as indicated in the color key for each panel. The expression ratio values for a given sample group of interest were summarized by their mean. Rows, miRNAs; columns, signal ratio of miRNA expression of Taxol-resistant cells to their parental cells. For each miRNA the red color means an expression of miRNA in Taxol-resistant cells higher than their parental cells, and the green color means a lower expression value. B, qRT-PCR was performed to validate the expression of miR-125b, miR-221, miR-222, and miR-923 in Taxol-resistant 435TR1 and 435TRP and their parental MDA-435 cells. RNU6B was used as an internal control and for normalization of the data. C, qRT-PCR was performed to validate the expression of miR-125b in Taxol-resistant 436TRP and SKBr3-TRP and their parental MDA-436 and SKBr3 cells. RNU6B was used as an internal control and for normalization of the data. Columns, mean of three independent experiments; bars, S.E. ***, p < 0.001.
Involvement of miR-125b in Taxol Resistance in Breast Cancer Cells
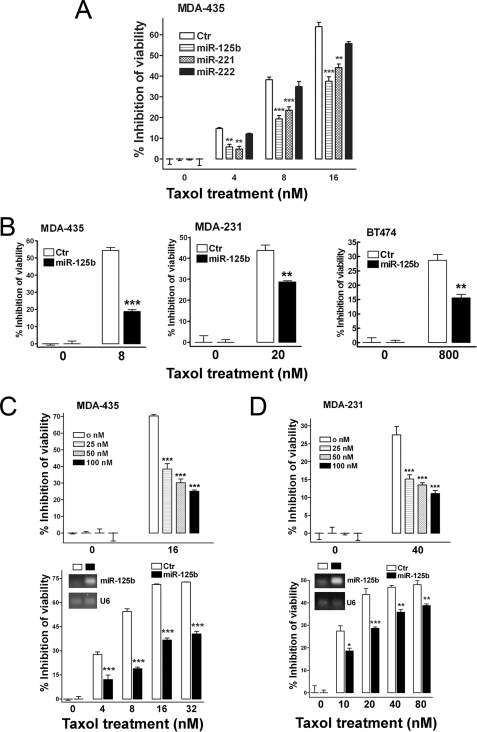
We investigated the role of miR-221, miR-222, and miR-125b in Taxol-induced cytotoxicity. Compared with negative control, overexpression of miR-221 and miR-125b markedly inhibited Taxol-induced cell cytotoxicity in MDA-435 cells (Fig. 2A). Because miR-125b showed a stronger inhibition to Taxol-induced cytotoxicity, we first focused on miR-125b and further investigated its role and mechanism in Taxol resistance.
FIGURE 2.
Involvement of miR-125b in Taxol resistance in breast cancer cells. A, MDA-435 cells were transfected with 100 nm pre-miR-negative (Ctr), pre-miR-125b, pre-miR-221, and pre-miR-222. 24 h after transfection, cells were seeded into 96-well plates at the density of 8 × 103 per well. Twelve hours later cells were treated with 0, 4, 8, and 16 nm Taxol for 48 h. Then the cell viability was detected using a MTS reagent. B, breast cancer cells, MDA-435, MDA-231, and BT474, were seeded into 96-well plates 24 h after the transfection of 100 nm pre-miR-125b and then followed with the indicated concentrations of Taxol for 48 h. Pre-miR-negative served as a negative control. The cell viability was detected using MTS reagent. C, MDA-435 cells were transfected with 0, 25, 50, and 100 nm pre-miR-125b (top). Twenty-four hours after transfection, the cells were seeded into 96-well plates at the density of 8 × 103 cells/well. Twelve hours later cells were treated with 16 nm Taxol for 48 h. Then, the cell viability was detected. MDA-435 cells were transfected with 100 nm pre-miR-negative or pre-miR-125b (bottom). Twenty-four hours after transfection, some of cells were collected for detection of miR-125b level by end-PCR (inset). Some of cells were seeded into 96-well plates at the density of 8 × 103 cells/well. Twelve hours later cells were treated with 0, 4, 8, 16, and 32 nm Taxol for 48 h. Then the cell viability was detected. D, similar experiments were performed in MDA-231 cells as described in panel C. Data are presented as the percentage of viability inhibition measured in cells treated without Taxol. Columns, mean of three independent experiments; bars, S.E. *, p < 0.05, **, p < 0.01, ***, p < 0.001.
To confirm and extend our findings, two more breast cancer cell lines, MDA-231 and BT474, were used to investigate the effect of overexpression of miR-125b on Taxol resistance. Overexpression of miR-125b markedly inhibited Taxol-induced cytotoxicity in all of the cell lines tested (Fig. 2B). Moreover, overexpression of miR-125b dramatically decreased Taxol-induced cytotoxicity and increased Taxol resistance in a miR-125b dose-dependent manner in MDA-435 cells (Fig. 2C, top). Furthermore, the expression of miR-125b (Fig. 2C, bottom inset) decreased Taxol-induced cytotoxicity and increased Taxol resistance under various concentrations of Taxol treatment in MDA435 cells (Fig. 2C, bottom). Similar experiments were performed in MDA-231 cells with similar results obtained (Fig. 2D).
Although overexpression of miR-125b confers Taxol resistance to cells, we asked whether down-regulation of miR-125b may increase Taxol sensitivity. miR-125b antisense (anti-miR-125b) was expressed in Taxol-resistant 435TR1 cells followed by treatment with increasing concentrations of Taxol for 48 h. We found that the knockdown of miR-125b by anti-miR-125b increased Taxol-induced cytotoxicity in these cells known to be highly resistant to Taxol (supplemental Fig. S1). These results clearly demonstrate that miR-125b plays an important role in Taxol resistance in breast cancer cells.
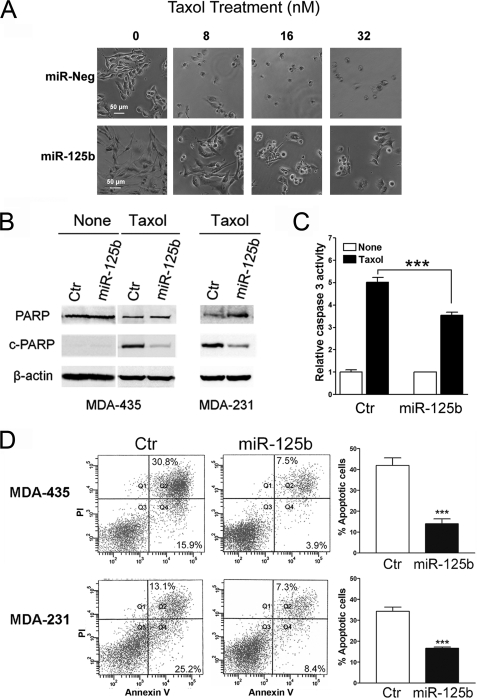
Involvement of miR-125b in Taxol-induced Apoptosis
Taxol is known to exert its cancer cell killing effect through induction of apoptosis (36). Next, we examined whether overexpression of miR-125b was capable of inhibiting Taxol-induced apoptosis. MDA-435 cells were transfected with pre-miR-negative or pre-miR-125b and then treated with increasing concentrations of Taxol. Cell morphological change was observed with a microscope, showing markedly less cellular debris and non-viable cells in miR-125b-transfected cells compared with negative controls (Fig. 3A). Poly (ADP-ribose) polymerase (PARP) is an important marker of apoptosis, and it is cleaved downstream of caspases when cellular apoptosis happens (37, 38). We next detected the protein levels of the cleaved PARP and total PARP by Western blotting in pre-miR-125b transfected MDA-435 and control cells after Taxol treatment. Compared with the negative control, the protein level of cleaved PARP was dramatically reduced in pre-miR-125b-transfected cells (Fig. 3B). The activity of caspase 3 was further detected using the Caspase-Glo® 3/7 assay system in pre-miR-125b-transfected MDA-435 cells after Taxol treatment for 24 h. Lower caspase 3 activity was found in miR-125b-transfected cells compared with the control cells (Fig. 3C). Moreover, apoptosis assays with annexin V staining revealed that significantly less apoptotic cells were detected in pre-miR-125b-transfected MDA-435 and MDA-231 cells compared with negative controls (Fig. 3D). Furthermore, knockdown of endogenous miR-125b enhanced Taxol-induced apoptosis in Taxol-resistant 435TRP cells (supplemental Fig. S2). These results indicated that miR-125b confers Taxol resistance in cancer cells by blocking Taxol-induced apoptosis.
FIGURE 3.
Overexpression of miR-125b inhibits Taxol-induced apoptosis. A, MDA-435 cells with transfection of 100 nm pre-miR-125b or pre-miR-negative were seeded into 96-well plates at 8 × 103 cells per well in triplicate. After 12 h of incubation, cells were treated with 0, 8, 16, and 32 nm Taxol for 48 h. The cells were visualized using phase-contrast microscope. B, MDA-435 and MDA-231 cells were transfected with 100 nm pre-miR-negative (Ctr) or pre-miR-125b and then treated with 20 or 40 nm Taxol for 48 h, respectively. Cell lysates were for Western blotting using antibodies against cleaved PARP or total PARP. β-Actin was used as a loading control. C, MDA-435 cells transfected with 100 nm pre-miR-125b or pre-miR-negative were seeded into 96-well plates at 8 × 103 cells per well in triplicates. After 8 h cells were treated with or without 20 nm Taxol for 24 h. Caspase 3 activity was measured by using Caspase-Glo® 3/7 assay kit (Promega). D, MDA-435 (top) and MDA-231 (bottom) cells were transfected with 100 nm pre-miR-negative (Ctr) or pre-miR-125b, respectively. Twenty-four hours later the cells were treated with 8 or 40 nm Taxol for 48 h. Then the cells were collected for apoptosis analysis by annexin V staining and flow cytometry. The percentage of apoptotic cells is represented in a bar diagram from three independent experiments (right). Columns, mean of three independent experiments; bars, S.E. *, p < 0.05, **, p < 0.01, ***, p < 0.001.
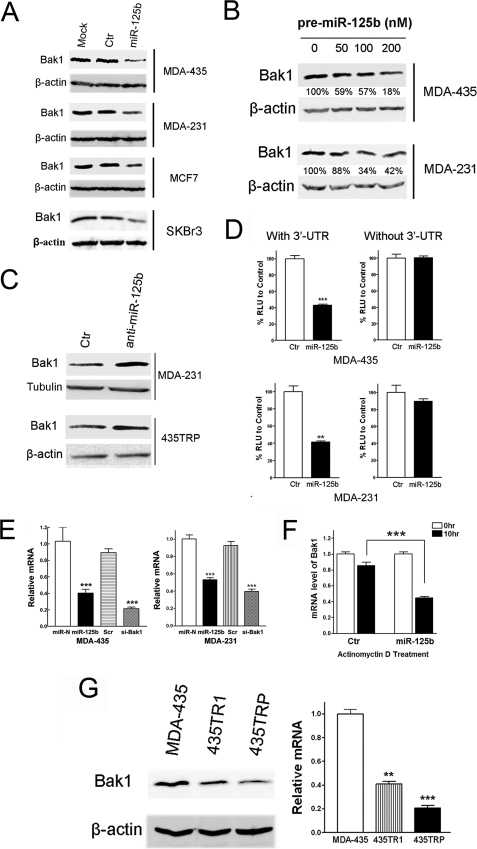
Bak1 Is a Direct Target of miR-125b in Breast Cancer Cells
Because miR-125b is capable of suppressing Taxol-induced apoptosis and increasing the resistance of cancer cells to Taxol, we searched miRNA databases for potential miR-125b targets that may possibly contribute to Taxol resistance. The three public miRNA databases (TargetScan, Pictar, and MicroRNA) all predicted that Bak1 might be a target for miR-125b, and the 3′-UTR of Bak1 contains a highly conserved binding site from position 629 to 635 for miR-125b (supplemental Fig. S3). To determine whether miR-125b could target Bak1 in breast cancer cells, we transfected the pre-miR-125b into MDA-435, MDA-231, MCF7, and SKBr3 cells. The expression of miR-125b significantly down-regulated Bak1 protein in a dose-dependent manner in all tested cells (Fig. 4, A and B). Moreover, knockdown of miR-125b up-regulated the expression of Bak1 in both MDA-231 and 435TRP cells with high expression of endogenous miR-125b (Fig. 4C). These results demonstrate that Bak1 is a target of miR-125b in breast cancer cells.
FIGURE 4.
Bak1 is a direct target of miR-125b. A, breast cancer cells, MDA-435, MDA-231, MCF7, and SKBr3 were transfected with 100 nm pre-miR-negative (Ctr) or pre-miR-125b. Twenty-four hours after transfection, cell lysates were prepared for Western blotting with an antibody against Bak1, and β-actin was used as a loading control. B, increasing concentrations (0, 50, 100, 200 nm) of pre-miR-125b were transfected into MDA-435 and MDA-231 cells, and then the cells were collected for Western blotting. The membranes were incubated with an antibody against Bak1. β-Actin was used as a loading control. The gray density was quantified using Scion image software and normalized to the β-actin. C, MDA-231 and 435TRP cells were transfected with 100 nm anti-miR-negative (Ctr) or anti-miR-125b, respectively. Twenty-four hours after transfection, cell lysates were extracted for Western blotting with an antibody against Bak1. β-Actin was used as a loading control. D, MDA-435 cells (top) and MDA-231 cells (bottom) were co-transfected with luciferase reporter plasmids with or without 3′-UTR of Bak1, pre-miR-125, or pre-miR-negative (Ctr) by using Lipofectamine 2000 reagent. Thirty-six hours post-transfection, cells were harvested and lysed with passive lysis buffer. Luciferase activity was measured by using a dual luciferase reporter assay. The pRL-TK vector was used as an internal control. The results were expressed as relative luciferase activity (firefly LUC/Renilla LUC). RLU, relative luciferase units. E, MDA-435 cells (left) and MDA-231 cells (right) were transfected with pre-miR-negative, pre-miR-125b, Scramble siRNA, or siRNA to Bak1. Twenty-four hours after transfection, the cells were collected, and Bak1 mRNA levels were measured by qRT-PCR. The relative Bak1 mRNA levels were normalized to glyceraldehyde-3-phosphate dehydrogenase siRNA to Bak1 served as a positive control. F, MDA-435 cells were transfected with 100 nm pre-miR-125b or pre-miR-negative. After 12 h, cells were treated with actinomycin D (5 μg/ml) 10 h. The stability of endogenous Bak1 mRNA was determined by real-time RT-PCR at the indicated time. G, cell lysate or cDNA was prepared from 435TR1, 435TRP, and their parental MDA-435 cells, and then the protein or cDNAs were subjected to Western blotting or qRT-PCR assays for detection of Bak1 protein (left) or mRNA expression (right). For qRT-PCR assay, relative mRNA expression of Bak1 was shown in the bar diagram from three independent experiments, and glyceraldehyde-3-phosphate dehydrogenase was used as a loading control. Columns, mean of three independent experiments; bars, S.E. *, p < 0.05, **, p < 0.01, ***, p < 0.001.
We, therefore, investigated whether miR-125b directly targets the 3′-UTR of Bak1 mRNA, and we performed luciferase reporter analysis by co-transfecting a vector containing pMIR reporter-luciferase fused with or without the 3′-UTR of Bak1 mRNA and pre-miR-125b or pre-miR-negative. Overexpression of miR-125b decreased the luciferase activity of the reporter with 3′-UTR of Bak1 by about 60% in both MDA-435 and MDA-231 cells (Fig. 4D, left). However, no inhibitory effect of miR-125b on the activity of the reporter without 3′-UTR of Bak1 was detected (Fig. 4D, right), indicating that Bak1 mRNA is a direct target of miR-125b.
miR-125b was examined for its capacity to suppress the expression of Bak1 at the mRNA level. Quantitative PCR was performed to detect Bak1 mRNA expression in MDA-435 and MDA-231 cells after transfection with pre-miR-125b or pre-miR-negative. We found that miR-125b decreased the expression of Bak1 mRNA with a similar inhibitory efficiency to the Bak1-specific siRNA in both cell lines, indicating that miR-125b inhibits the expression level of Bak1 mRNA (Fig. 4E). To explore the possibility that miR-125b decreases the stability of Bak1 mRNA, MDA-435 cells were transfected with or without pre-miR-125b and then treated with transcription inhibitor actinomycin D. The endogenous Bak1 mRNA level was determined by qRT-PCR. We found that after treatment of actinomycin D for 10 h, the Bak1 mRNA expression level was lower in miR-125b-overexpressing cells compared with that of the control cells (Fig. 4F), indicating that miR-125b decreases the stability of Bak1 mRNA. To test whether an increased expression of miR-125b would correlate with a decreased expression of Bak1 protein and mRNA levels in Taxol-resistant cancer cells, we performed Western blotting and quantitative real-time PCR to detect the expression of Bak1 in 435TR1, 435TRP, and their parental MDA-435 cells. Compared with the parental cells, decreased expression of Bak1 was found in Taxol-resistant 435TR1 and 435TRP cells at both protein and mRNA levels (Fig. 4G). These results further support that Bak1 is a target of miR-125b, and the loss of Bak1 expression in Taxol-resistant cells may mediate the role of miR-125b in Taxol resistance.
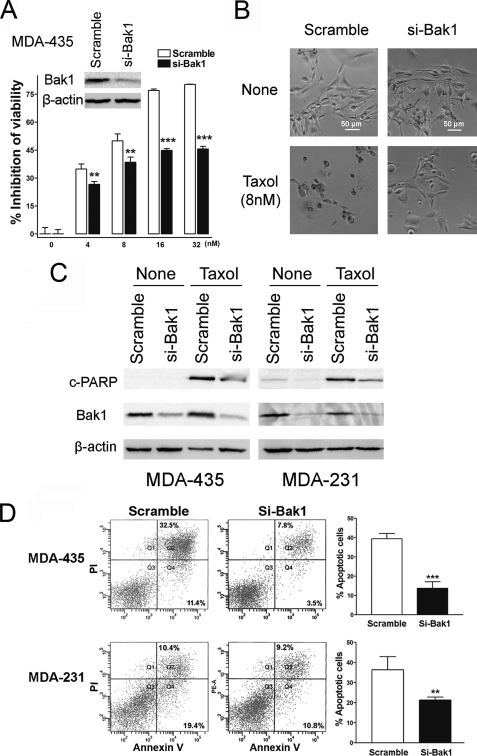
Bak1 Plays a Critical Role in Taxol-induced Apoptosis
Bak1, together with Bax, is an ultimate death effecter of apoptosis triggering cues (39). Although Bak1 has been proven to play a critical role in apoptosis, its role in Taxol-induced cell cytotoxicity and Taxol sensitivity has not been studied. To investigate the association of Bak1- and Taxol-induced cell cytotoxicity, we transfected Bak1-specific siRNA into MDA-435 cells and then treated these cells with increasing concentrations of Taxol. Compared with negative control, Bak1 siRNA efficiently knocked down Bak1 expression (Fig. 5A, inset) and decreased the inhibition of cell viability by Taxol (Fig. 5A). Similar experiments were performed in MDA231 cells, and the results were similar (supplemental Fig. S4). Moreover, compared with control cells, si-Bak1-transfected MDA-435 cells showed much less cellular debris and non-viable cells (Fig. 5B). PARP cleavage was also dramatically reduced in si-Bak1-transfected MDA-435 and MDA-231 cells after treatment with 20 or 40 nm Taxol for 48 h (Fig. 5C). Cellular apoptosis were further detected in MDA-435 and MDA-231 cells by annexin V staining, and the results revealed markedly less apoptotic cells in si-Bak1-transfected cells (Fig. 5D). These results indicate that the knockdown of Bak1 could efficiently suppress Taxol-induced cell cytotoxicity via the inhibition of Bak1-mediated apoptosis.
FIGURE 5.
Bak1 plays a critical role in Taxol-induced apoptosis. A, MDA-435 cells were transfected with scramble siRNA or Bak1 siRNA (si-Bak1). Forty-eight hours after transfection, lysates were prepared, and Western blotting was carried out with antibody against Bak1 (inset). β-Actin was used as a loading control. MDA-435 cells with knockdown of Bak1 were seeded into 96-well plate at the density of 8 × 103 per well and then followed with 0, 4, 8, 16, and 32 nm Taxol treatment for 48 h. Scramble siRNA served as a negative control. The inhibition of cell viability was detected. Columns, mean of three independent experiments; bars, S.E. *, p < 0.05, **, p < 0.01, ***, p < 0.001. B, MDA-435 cells with Bak1 knockdown by specific siRNA to Bak1 were seeded into 96-well plates at 8 × 103 cells per well. After 12 h incubation cells were treated with 8 nm Taxol for 48 h. The cells were visualized using a phase-contrast microscope. Scramble siRNA-transfected MDA-435 cells served as a negative control. C, MDA-435 and MDA-231 cells were transfected with 100 nm Scramble siRNA (Ctr) or si-Bak1, 24 h after transfection, and cells were treated with 20 lysates were prepared for Western blotting with the antibodies against cleaved PARP (top) and Bak1 (middle). β-Actin was used as a loading control (bottom). D, MDA-435 and MDA-231 cells were transfected with 100 nm Scramble siRNA (Ctr) or si-Bak1, and 24 h after transfection cells were treated with 20 or 40 nm Taxol for 48 h, respectively. Then the cells were collected for apoptosis analysis by annexin V staining and flow cytometry. The percentage of apoptotic cells are represented in bar diagram from three independent experiments.
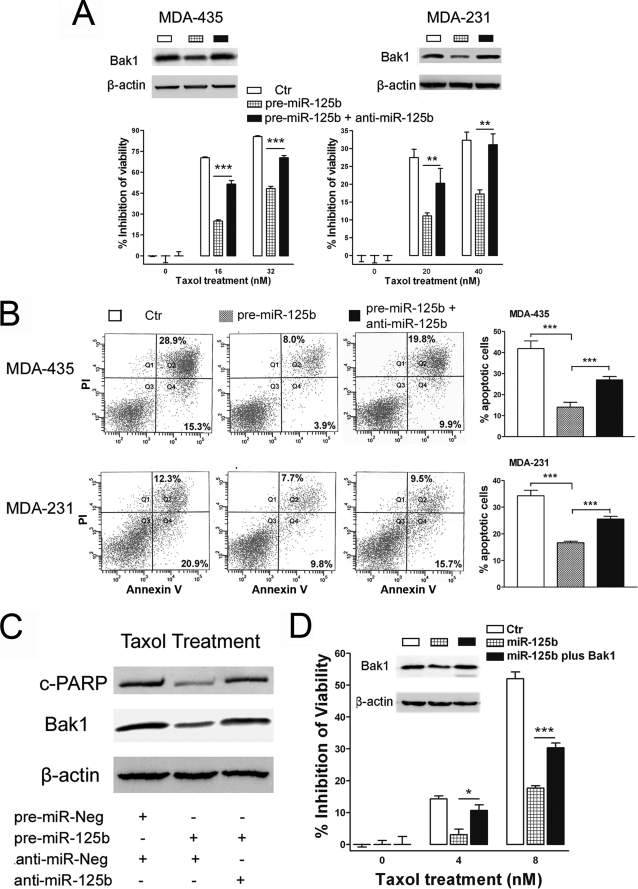
Restoring the Expression of Bak1 Recovers Taxol Sensitivity in Counteracting miR-125-mediated Taxol Resistance
Because Bak1, a direct target of miR-125b, plays a critical role in Taxol-induced apoptosis, we next investigated whether miR-125b affects Taxol resistance through suppression of Bak1 expression. We, therefore, transfected pre-miR-125b or pre-miR-125b in combination with anti-miR-125b into MDA-435 and MDA-231 cells, and then we treated the cells with indicated concentrations of Taxol for 48 h. Transfection with the pre-miR-125b alone decreased the expression of Bak1 (Fig. 6A, top) and increased the resistance of these cells to Taxol and suppressed apoptosis in both of cell lines (Fig. 6A, bottom). Interestingly, transfection with the miR-125b antisense, anti-miR-125b, restored the expression of Bak1 (Fig. 6A, top) and recovered Taxol-induced cytotoxicity (Fig. 6A, bottom) and apoptosis (Fig. 6B, and supplemental Fig. S5). Moreover, with recovery of Bak1 expression by miR-125b antisense, PARP cleavage was also restored in pre-miR-125b-expressing cells when treated with Taxol (Fig. 6C).
FIGURE 6.
Restoring the expression of Bak1 recovers Taxol sensitivity. A, MDA-435 (left) and MDA-231 (right) cells were co-transfected with pre-miR-125b alone or in combination with anti-miR-125b. Western blotting was performed to detect the expression of Bak1 (top). β-Actin was used as a loading control (Ctr). MDA-435 and MDA-231 cells with transfection of pre-miR-125b alone or in combination with anti-miR-125b were seeded into 96-well plate at the density of 8 × 103 per well and then treated with the indicated concentrations of Taxol for 48 h. The cell viability was detected. Data are presented as the percentage of viability inhibition measured in cells treated wi3thout Taxol. Columns, mean of three independent experiments; bars, S.E. *, p < 0.05, **, p < 0.01, ***, p < 0.001. B, MDA-435 or MDA-231 cells were transfected with 100 nm pre-miR-125b alone or in combination with 100 nm anti-miR-125b, and then the transfected cells were treated with 8 or 40 nm Taxol for 48 h. Then the cells were collected for apoptosis analysis by annexin V staining and flow cytometry. Percentages of apoptotic cells are represented in bar diagram from three independent experiments (right). C, MDA-435 cells were transfected with 100 nm pre-miR-Neg plus anti-miR-Neg (Ctr), pre-miR-125b plus anti-miR-Neg, or pre-miR-125b plus anti-miR-125b; 24 h after transfection, cells were treated with 20 nm Taxol for 48 h. Cell lysates were prepared for Western blotting using antibodies against cleaved PARP (top) or Bak1 (middle). β-Actin was used as a loading control (bottom). D, MDA-435 and MDA-231 cells were co-transfected with 100 nm pre-miR-125b alone or in combination with 4 μg of pIRES2-EGFP/Bak1 or pIRES2-EGFP. Western blotting was performed to detect the expression of Bak1 (inset). β-Actin was used as a loading control. The transfected MDA-435 were seeded into a 96-well plate at the density of 8 × 103 per well and then treated with 0, 4, and 8 nm Taxol for 48 h. The cell viability was detected. Data are presented as the percentage of viability inhibition measured in cells treated without Taxol. Columns, mean of three independent experiments; bars, S.E. *, p < 0.05, **, p < 0.01, ***, p < 0.001.
To further confirm these results, we co-transfected pre-miR-125b plus the control vector or a Bak1 expression vector into MDA-435 cells and then treated these cells with Taxol. Transfection of pre-miR-125b alone decreased the expression of Bak1 and increased the resistance of these cells to Taxol (Fig. 6D); however, restoring expression of Bak1 by co-transfecting pIRES2-EGFP/Bak1 recovered the Taxol sensitivity (Fig. 6D). Of note, the overexpression of miR-125b had no effect upon the expression of exogenously expressed Bak1. This can be explained by the lack of 3′-UTR in the exogenously expressed Bak1 mRNA. Taken together, these results clearly demonstrate that Bak1 plays an important role in breast cancer cell Taxol sensitivity, and overexpression of miR-125b confers Taxol resistance at least partially through the direct suppression of Bak1 expression.
DISCUSSION
Many miRNAs have been identified as having an oncogenic or tumor suppressor-like function shown to be involved in cell proliferation, differentiation, apoptosis, and drug resistance (14–18). However, little is known about their role in chemoresistance, such as to Taxol. In this study we compared the differential profiles of miRNAs between Taxol-resistant and their parental Taxol-sensitive MDA-435 cells. We found that most of the miRNAs that are highly expressed in Taxol-resistant cells are previously reported to function as oncogenes, such as miR-221 (27, 28), miR-222 (28, 29), miR-100 (22), and miR-106b (27). Of the lower expressed miRNAs, most of them are previously reported as tumor suppressors, such as miR-16 (30, 31), miR-20a (33), miR-29a/b (34), and miR-146a/b (35). Taxol resistance is a phenotype of malignant cancer cells; therefore, the findings of our microRNA assay are in line with the reported functions of miRNAs, suggesting our profiling results are reliable.
We verified the differential expression levels of miR-221, miR-222, and miR-125b and examined the effects on Taxol-induced cytotoxicity. We found that miR-125b and miR-221 markedly inhibited Taxol-induced cytotoxicity in breast cancer cell lines. The inhibitory effect was found to be dose-dependent over a wide range of Taxol concentrations. Moreover, antisense to miR-125b increased Taxol sensitivity in Taxol-resistant cells, clearly demonstrating that miR-125b plays a critical role in the development of Taxol resistance. It was reported that miR-125b could be transactivated by the NF-κB p65 subunit through binding to the promoter of miR-125 (40, 41). We also found that compared with that of the parental MDA-435 cells, Taxol-resistant cells showed increased transcriptional activity of NF-κB (supplemental Fig. S6), implying that activation of NF-κB by long term Taxol treatment might be a mechanism for Taxol-mediated up-regulation of miR-125b. Further study is needed to validate this potential mechanism.
We also identified a lower percentage of apoptotic cells in miR-125b-transfected cells by annexin V staining after treatment with Taxol, further confirmed by immunoblotting of the apoptotic marker, cleaved PARP, and through detection of caspase 3 activity. These results suggest that the anti-apoptotic effect is a key mechanism of miR-125b-mediated Taxol resistance in breast cancer cells.
Apoptosis is a predominant mechanism by which cancer chemotherapeutic agents such as Taxol kill cancer cells (42). Bak1 is an ultimate death effecter of apoptosis-triggering cues. It is localized to the cytoplasmic side of the mitochondrial membrane, tethered by a membrane-bound C-terminal helix, and is held inactive by forming heterodimeric complexes with the anti-apoptotic Bcl2 family member Mcl1 (39). Bak1 was recently found to be a target of miR-125b and implicated in androgen-independent growth in prostate cancer cells (20). Here, we demonstrate that Bak1 is a direct target of miR-125b in breast cancer cells. Moreover, we show that Bak1 enhanced Taxol-induced cytotoxicity and apoptosis. To our knowledge, this is the first report to demonstrate a direct link between Bak1 expression and cancer cell Taxol sensitivity. Importantly, we found that restoring the expression of Bak1 either by anti-miR-125b or exogenously Bak1 expression recovered Taxol sensitivity in counteracting miR-125-mediated down-regulation of Bak1 and Taxol resistance. Thus, miR-125b expression contributes to Taxol resistance through suppression of Bak1 expression. These results indicate that miR-125b-Bak1 axis is critical for Taxol resistance in breast cancer cells and may potentially serve as a therapeutic target to overcome Taxol resistance. This study represents one of the first attempts to explore the role of microRNA in cancer cell Taxol resistance. This novel finding provides a unique insight into the molecular mechanism of miR-125b-mediated Taxol resistance.
To date, several targets of miR-125b have been identified. One recent study showed that tumor suppressor p53 is a direct target of miR-125b, and suppression of p53 gene expression through overexpression of miR-125b inhibited p53-dependent apoptosis in human neuroblastoma cells and human lung fibroblast cells (43). Another report showed that ErbB2 and ErbB3 were both targets of miR-125b in breast cancer SKBr3 cells (44). Conversely, Shi et al. (20) showed that there was no miR-125b-mediated down-regulation of ErbB2 and ErbB3 in miR-125b-transfected LNCaP and cds1 prostate cancer cells. Consistent with the latter report, we also did not find the down-regulation of ErbB2 by miR-125b in breast cancer BT-474, BT-474M1, and SKBr3 cells (supplemental Fig. S7). In line with these findings, miR-125b was reported to have complicated functions as either oncogene-like or tumor suppressor-like in different cancer types or cell lines. miR-125b is not only elevated in pancreatic cancer, oligodendroglia tumors, prostate cancer, myelodysplastic syndromes, and acute myeloid leukemia but also promotes cell proliferation in prostate cancer cells, enhances invasive potential in urothelial carcinomas, and suppresses p53-dependent apoptosis in human neuroblastoma cells (20, 23–26, 45). However, miR-125b has been reported to be down-regulated in ovarian carcinoma, thyroid carcinoma, and oral squamous cell carcinoma and has been shown to inhibit cell proliferation and cell cycle progression in these cancers (46–48). Similar phenomenon of duality of function for miRNA in different types of cancers has also been found on miR-29a (49) and miR-17–92 clusters (50). One possible explanation is the known diverse nature of miRNA target genes. The net effect of changes in the expression of a miRNA appears to be the sum of all impacts on its targets in a cell type-specific and phenotype-specific manner (49). Thus, overexpression or knockdown of a miRNA might have diverse effects that depend on the types of cancer cells.
Recently, miR-125b was reported to exhibit under-expressed in breast cancer tissues (51); however, the exact association of miR-125b expression and breast tumor progression is still unclear. Two recent studies reported that miR-125b may act as an oncogene in human breast cancer cells. One study reported that miR-125b promoted cell proliferation and decreased the antitumor effects of 1,25(OH)(2)D(3) in breast cancer MCF7 cells (52). Another study showed that compared with nontumorigenic cancer cells, miR-125b is overexpressed in highly tumorigenic human breast cancer stem cells (53). In our study we show that miR-125b inhibited Bak1 and increased Taxol resistance, acting primarily as an oncogene in breast cancer MDA-435, MDA-231, SKBr3, and BT474 cells. However, further investigations with large patient samples and well designed laboratory studies should be performed in the future to clarify the role of miR-125b in human breast cancer development.
The origin of MDA-MB-435 cells has recently been called into question (54, 55). However, a latest literature indicated that MDA-MB-435 cells are in fact breast cancer cells and not of melanoma origin (56). Nevertheless, we confirmed all of the results from MDA-MB-435 cells with another breast cancer cell line MDA-MB-231, and we further confirmed the key findings with breast cancer cell lines MDA-MB-436, SKBr3, BT474, and MCF-7.
Taken together, we have shown for the first time that miR-125b is up-regulated in Taxol-resistant cells, with the overexpression of miR-125b causing a marked inhibition of Taxol-induced apoptosis and increased resistance to Taxol. We demonstrate that Bak1 is a direct target of miR-125b in breast cancer cells. Moreover, we first showed that Bak1 plays an important role in Taxol sensitivity of breast cancer cells. Thus, miR-125b confers Taxol resistance through suppression of Bak1 expression and may potentially serve as a therapeutic target for overcoming Taxol resistance in human breast cancer. These novel findings have important implications in the development of targeted therapeutics for overcoming Taxol resistance in human breast cancer.
Supplementary Material
Acknowledgments
We thank Dr. de Vere White for providing the pMIR-reporter luciferase vector containing the 3′-UTR of Bak1, Dr. Stanley N. Cohen for cell lines, Dr. William T. Gerthoffer and Dr. Chunxiang Zhang for invaluable discussion, and Amy Brown for the editorial assistance.
This work was supported by the Vincent F. Kilborn, Jr., Cancer Research Foundation (to M. T.) and The Norwegian Radiumhospitalet Legater (Project 334003, to M. T. and O. F.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S7.
- miRNA
- microRNA
- pre-miR
- miRNA precursor
- anti-miR
- miRNA antisense
- Scr
- scramble
- PARP
- poly(ADP-ribose) polymerase
- si-Bak1
- Bak1 siRNA
- qRT-PCR
- quantitative real time PCR
- RT
- reverse transcription
- siRNA
- small interfering RNA
- UTR
- untranslated region
- MTS
- 2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide.
REFERENCES
- 1.Henley D., Isbill M., Fernando R., Foster J. S., Wimalasena J. (2007) Cancer Chemother. Pharmacol. 59, 235–249 [DOI] [PubMed] [Google Scholar]
- 2.Tan M., Yu D. (2007) Adv. Exp. Med. Biol. 608, 119–129 [DOI] [PubMed] [Google Scholar]
- 3.Chen L. P., Cai S. M., Fan J. X., Li Z. T. (1995) Gynecol. Oncol. 56, 231–234 [DOI] [PubMed] [Google Scholar]
- 4.Gottesman M. M., Pastan I. (1993) Annu. Rev. Biochem. 62, 385–427 [DOI] [PubMed] [Google Scholar]
- 5.Panda D., Miller H. P., Banerjee A., Ludueña R. F., Wilson L. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 11358–11362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martello L. A., Verdier-Pinard P., Shen H. J., He L., Torres K., Orr G. A., Horwitz S. B. (2003) Cancer Res. 63, 1207–1213 [PubMed] [Google Scholar]
- 7.Gonçalves A., Braguer D., Kamath K., Martello L., Briand C., Horwitz S., Wilson L., Jordan M. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 11737–11742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan M., Jing T., Lan K. H., Neal C. L., Li P., Lee S., Fang D., Nagata Y., Liu J., Arlinghaus R., Hung M. C., Yu D. (2002) Mol. Cell 9, 993–1004 [DOI] [PubMed] [Google Scholar]
- 9.Lu J., Tan M., Huang W. C., Li P., Guo H., Tseng L. M., Su X. H., Yang W. T., Treekitkarnmongkol W., Andreeff M., Symmans F., Yu D. (2009) Clin. Cancer Res. 15, 1326–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartel D. P. (2004) Cell 116, 281–297 [DOI] [PubMed] [Google Scholar]
- 11.Reinhart B. J., Slack F. J., Basson M., Pasquinelli A. E., Bettinger J. C., Rougvie A. E., Horvitz H. R., Ruvkun G. (2000) Nature 403, 901–906 [DOI] [PubMed] [Google Scholar]
- 12.Olsen P. H., Ambros V. (1999) Dev. Biol. 216, 671–680 [DOI] [PubMed] [Google Scholar]
- 13.Zeng Y., Yi R., Cullen B. R. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 9779–9784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kobayashi T., Lu J., Cobb B. S., Rodda S. J., McMahon A. P., Schipani E., Merkenschlager M., Kronenberg H. M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 1949–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang Q., Gumireddy K., Schrier M., le Sage C., Nagel R., Nair S., Egan D. A., Li A., Huang G., Klein-Szanto A. J., Gimotty P. A., Katsaros D., Coukos G., Zhang L., Puré E., Agami R. (2008) Nat. Cell. Biol. 10, 202–210 [DOI] [PubMed] [Google Scholar]
- 16.Sorrentino A., Liu C. G., Addario A., Peschle C., Scambia G., Ferlini C. (2008) Gynecol. Oncol. 111, 478–486 [DOI] [PubMed] [Google Scholar]
- 17.Zhao J. J., Lin J., Yang H., Kong W., He L., Ma X., Coppola D., Cheng J. Q. (2008) J. Biol. Chem. 283, 31079–31086 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Fujita Y., Kojima K., Hamada N., Ohhashi R., Akao Y., Nozawa Y., Deguchi T., Ito M. (2008) Biochem. Biophys. Res. Commun. 377, 114–119 [DOI] [PubMed] [Google Scholar]
- 19.Zhou M., Zhao Y., Ding Y., Liu H., Liu Z., Fodstad O., Riker A. I., Kamarajugadda S., Lu J., Owen L. B., Ledoux S. P., Tan M. (2010) Mol. Cancer 9, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi X. B., Xue L., Yang J., Ma A. H., Zhao J., Xu M., Tepper C. G., Evans C. P., Kung H. J., deVere White R. W. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 19983–19988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao Y. H., Zhou M., Liu H., Ding Y., Khong H. T., Yu D., Fodstad O., Tan M. (2009) Oncogene 28, 3689–3701 [DOI] [PubMed] [Google Scholar]
- 22.Henson B. J., Bhattacharjee S., O'Dee D. M., Feingold E., Gollin S. M. (2009) Genes Chromosomes Cancer 48, 569–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bloomston M., Frankel W. L., Petrocca F., Volinia S., Alder H., Hagan J. P., Liu C. G., Bhatt D., Taccioli C., Croce C. M. (2007) JAMA 297, 1901–1908 [DOI] [PubMed] [Google Scholar]
- 24.Bousquet M., Quelen C., Rosati R., Mansat-De Mas V., La Starza R., Bastard C., Lippert E., Talmant P., Lafage-Pochitaloff M., Leroux D., Gervais C., Viguié F., Lai J. L., Terre C., Beverlo B., Sambani C., Hagemeijer A., Marynen P., Delsol G., Dastugue N., Mecucci C., Brousset P. (2008) J. Exp. Med. 205, 2499–2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xia H. F., He T. Z., Liu C. M., Cui Y., Song P. P., Jin X. H., Ma X. (2009) Cell. Physiol. Biochem. 23, 347–358 [DOI] [PubMed] [Google Scholar]
- 26.Ozen M., Creighton C. J., Ozdemir M., Ittmann M. (2008) Oncogene 27, 1788–1793 [DOI] [PubMed] [Google Scholar]
- 27.Kan T., Sato F., Ito T., Matsumura N., David S., Cheng Y., Agarwal R., Paun B. C., Jin Z., Olaru A. V., Selaru F. M., Hamilton J. P., Yang J., Abraham J. M., Mori Y., Meltzer S. J. (2009) Gastroenterology 136, 1689–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galardi S., Mercatelli N., Giorda E., Massalini S., Frajese G. V., Ciafrè S. A., Farace M. G. (2007) J. Biol. Chem. 282, 23716–23724 [DOI] [PubMed] [Google Scholar]
- 29.le Sage C., Nagel R., Egan D. A., Schrier M., Mesman E., Mangiola A., Anile C., Maira G., Mercatelli N., Ciafrè S. A., Farace M. G., Agami R. (2007) EMBO J. 26, 3699–3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bonci D., Coppola V., Musumeci M., Addario A., Giuffrida R., Memeo L., D'Urso L., Pagliuca A., Biffoni M., Labbaye C., Bartucci M., Muto G., Peschle C., De Maria R. (2008) Nat. Med. 14, 1271–1277 [DOI] [PubMed] [Google Scholar]
- 31.Calin G. A., Cimmino A., Fabbri M., Ferracin M., Wojcik S. E., Shimizu M., Taccioli C., Zanesi N., Garzon R., Aqeilan R. I., Alder H., Volinia S., Rassenti L., Liu X., Liu C. G., Kipps T. J., Negrini M., Croce C. M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 5166–5171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ichimi T., Enokida H., Okuno Y., Kunimoto R., Chiyomaru T., Kawamoto K., Kawahara K., Toki K., Kawakami K., Nishiyama K., Tsujimoto G., Nakagawa M., Seki N. (2009) Int. J. Cancer 125, 345–352 [DOI] [PubMed] [Google Scholar]
- 33.Pickering M. T., Stadler B. M., Kowalik T. F. (2009) Oncogene 28, 140–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park S. Y., Lee J. H., Ha M., Nam J. W., Kim V. N. (2009) Nat. Struct. Mol. Biol. 16, 23–29 [DOI] [PubMed] [Google Scholar]
- 35.Hurst D. R., Edmonds M. D., Scott G. K., Benz C. C., Vaidya K. S., Welch D. R. (2009) Cancer Res. 69, 1279–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ofir R., Seidman R., Rabinski T., Krup M., Yavelsky V., Weinstein Y., Wolfson M. (2002) Cell Death Differ. 9, 636–642 [DOI] [PubMed] [Google Scholar]
- 37.Wu W., Hodges E., Höög C. (2006) Nucleic Acids Res. 34, e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boulares A. H., Yakovlev A. G., Ivanova V., Stoica B. A., Wang G., Iyer S., Smulson M. (1999) J. Biol. Chem. 274, 22932–22940 [DOI] [PubMed] [Google Scholar]
- 39.Cuconati A., Mukherjee C., Perez D., White E. (2003) Genes Dev. 17, 2922–2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou R., Hu G., Gong A. Y., Chen X. M. (2010) Nucleic Acids Res. 38, 3222–3232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou R., Hu G., Liu J., Gong A. Y., Drescher K. M., Chen X. M. (2009) PLoS Pathog. 5, e1000681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fisher D. E. (1994) Cell 78, 539–542 [DOI] [PubMed] [Google Scholar]
- 43.Le M. T., Teh C., Shyh-Chang N., Xie H., Zhou B., Korzh V., Lodish H. F., Lim B. (2009) Genes Dev. 23, 862–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scott G. K., Goga A., Bhaumik D., Berger C. E., Sullivan C. S., Benz C. C. (2007) J. Biol. Chem. 282, 1479–1486 [DOI] [PubMed] [Google Scholar]
- 45.Veerla S., Lindgren D., Kvist A., Frigyesi A., Staaf J., Persson H., Liedberg F., Chebil G., Gudjonsson S., Borg A., Månsson W., Rovira C., Höglund M. (2009) Int. J. Cancer 124, 2236–2242 [DOI] [PubMed] [Google Scholar]
- 46.Iorio M. V., Visone R., Di Leva G., Donati V., Petrocca F., Casalini P., Taccioli C., Volinia S., Liu C. G., Alder H., Calin G. A., Ménard S., Croce C. M. (2007) Cancer Res. 67, 8699–8707 [DOI] [PubMed] [Google Scholar]
- 47.Nam E. J., Yoon H., Kim S. W., Kim H., Kim Y. T., Kim J. H., Kim J. W., Kim S. (2008) Clin. Cancer Res. 14, 2690–2695 [DOI] [PubMed] [Google Scholar]
- 48.Visone R., Pallante P., Vecchione A., Cirombella R., Ferracin M., Ferraro A., Volinia S., Coluzzi S., Leone V., Borbone E., Liu C. G., Petrocca F., Troncone G., Calin G. A., Scarpa A., Colato C., Tallini G., Santoro M., Croce C. M., Fusco A. (2007) Oncogene 26, 7590–7595 [DOI] [PubMed] [Google Scholar]
- 49.Gebeshuber C. A., Zatloukal K., Martinez J. (2009) EMBO Rep 10, 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Calin G. A., Croce C. M. (2006) Nat. Rev. Cancer 6, 857–866 [DOI] [PubMed] [Google Scholar]
- 51.Iorio M. V., Ferracin M., Liu C. G., Veronese A., Spizzo R., Sabbioni S., Magri E., Pedriali M., Fabbri M., Campiglio M., Ménard S., Palazzo J. P., Rosenberg A., Musiani P., Volinia S., Nenci I., Calin G. A., Querzoli P., Negrini M., Croce C. M. (2005) Cancer Res. 65, 7065–7070 [DOI] [PubMed] [Google Scholar]
- 52.Mohri T., Nakajima M., Takagi S., Komagata S., Yokoi T. (2009) Int. J. Cancer 125, 1328–1333 [DOI] [PubMed] [Google Scholar]
- 53.Shimono Y., Zabala M., Cho R. W., Lobo N., Dalerba P., Qian D., Diehn M., Liu H., Panula S. P., Chiao E., Dirbas F. M., Somlo G., Pera R. A., Lao K., Clarke M. F. (2009) Cell 138, 592–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sellappan S., Grijalva R., Zhou X., Yang W., Eli M. B., Mills G. B., Yu D. (2004) Cancer Res. 64, 3479–3485 [DOI] [PubMed] [Google Scholar]
- 55.Welch D. R. (1997) Clin. Exp. Metastasis 15, 272–306 [DOI] [PubMed] [Google Scholar]
- 56.Chambers A. F. (2009) Cancer Res. 69, 5292–5293 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.