Abstract
UvrD (DNA helicase II) has been implicated in DNA replication, DNA recombination, nucleotide excision repair, and methyl-directed mismatch repair. The enzymatic function of UvrD is to translocate along a DNA strand in a 3′ to 5′ direction and unwind duplex DNA utilizing a DNA-dependent ATPase activity. In addition, UvrD interacts with many other proteins involved in the above processes and is hypothesized to facilitate protein turnover, thus promoting further DNA processing. Although UvrD interactions with proteins bound to DNA have significant biological implications, the effects of covalent DNA-protein cross-links on UvrD helicase activity have not been characterized. Herein, we demonstrate that UvrD-catalyzed strand separation was inhibited on a DNA strand to which a 16-kDa protein was covalently bound. Our sequestration studies suggest that the inhibition of UvrD activity is most likely due to a translocation block and not helicase sequestration on the cross-link-containing DNA substrate. In contrast, no inhibition of UvrD-catalyzed strand separation was apparent when the protein was linked to the complementary strand. The latter result is surprising given the earlier observations that the DNA in this covalent complex is severely bent (∼70°), with both DNA strands making multiple contacts with the cross-linked protein. In addition, UvrD was shown to be required for replication of plasmid DNAs containing covalent DNA-protein complexes. Combined, these data suggest a critical role for UvrD in the processing of DNA-protein cross-links.
Keywords: DNA/Helicase, DNA/Repair, DNA/Replication, Bacterial Genetics, Protein-DNA Interaction, DNA Damage, DNA-Protein Cross-links, UvrD, Helicase
Introduction
Escherichia coli UvrD (DNA helicase II) is a member of the SF1 superfamily of DNA helicases, and its activity has been implicated in many genome maintenance functions including replication, methyl-directed mismatch repair, homologous recombination, and nucleotide excision repair (NER).3 UvrD functions as a 3′ to 5′ helicase and translocates via a single-stranded DNA-dependent ATPase activity (1). UvrD-deficient E. coli cells are hyper-recombinogenic (2), have increased levels of spontaneous mutagenesis (3), and show reduced survival after exposure to ultraviolet radiation (UV) and methyl methanesulfonate (4, 5). UvrD helicase activity can be significantly stimulated by interactions with the NER UvrAB complex (6) or methyl-directed mismatch repair protein, MutL (7, 8).
UvrD is involved in several replication-associated processes and has been shown to co-purify with the DNA polymerase III holoenzyme complex (9). UvrD is also implicated in the processing of Okazaki fragments, such that E. coli cells that are deficient in polymerase I are unable to grow on rich media but do survive when grown on defined minimal media (10). Although these cells are extremely defective in the joining of Okazaki fragments, an alternative UvrD-dependent replication pathway is hypothesized to remove RNA primers and fill in these gaps. This alternative pathway requires not only UvrD but also UvrA and UvrB (not UvrC) (6, 11) such that double mutants of polymerase I and UvrA, -B, or -D are lethal. It is hypothesized that the UvrABD complex can replace the strand-displacement function of polymerase I, whereas alternative exonucleases and polymerases complete lagging strand DNA synthesis (6).
UvrD is also essential for removal of Tus protein bound at Ter sites and has been proposed to promote recombination-dependent replication restart from forks arrested by Ter-Tus complexes (12). In temperature-sensitive dnaN and dnaE cells, UvrD has been implicated in the removal of RecA molecules that are bound to the arrested replication forks (13). Similarly, UvrD disrupts RecA bound to synthetic recombination intermediates, and it is hypothesized that in the absence of UvrD, the inability to disrupt RecA filaments leads to the hyper-recombinogenic phenotype (14). In agreement with these conclusions, cells expressing the hyper-helicase mutant of UvrD (UvrD303) have anti-recombinogenic and anti-mutator phenotypes (15, 16). Despite the hyper-helicase activity, UvrD303 cells remain UV-sensitive, and this phenotype is epistatic with RecA but additive with UvrA (16).
A possible function of UvrD in NER is to facilitate the release of the UvrABC complex, thus accelerating protein turnover (17–19). The specific role for the helicase activity has been assumed to involve strand displacement of the incised damage-containing strand. However, it was subsequently shown that the addition of UvrD to a bound UvrAB complex did not change the UvrAB footprint, with a slight hint of UvrD binding near the 5′ incision site (20). This same study also showed that UvrD did not affect the ability of DNA polymerase I to catalyze strand displacement synthesis at the dual incision sites and release of UvrABC. These data were in good agreement with the earlier studies that demonstrated that tracks of NER-dependent repair synthesis were normal in UvrD-deficient cells and that the incision steps were not affected in this strain (21, 22). Further, Matson et al. (1) concluded that UvrD may participate in the turnover of UvrABC through protein-protein interactions rather than its DNA unwinding activity. Thus, the exact role of UvrD in NER is not precisely defined.
In addition to the various functions described above, the UvrABC complex-independent role of UvrD was demonstrated in modulating cytotoxicity of norfloxin, an inhibitor of gyrase and topoisomerase IV (23). Because gyrase and topoisomerase IV function ahead of and behind the replication fork, respectively, it was interesting that the uvrD mutant showed increased sensitivity to norfloxin when topoisomerase IV, but not gyrase, was targeted. Thus, UvrD appears to be specifically involved in facilitating the repair of the topoisomerase IV but not gyrase-norfloxacin-DNA complexes.
Overall, these data suggest that UvrD may be important for displacing non-covalent protein-DNA interactions. However, at present no data are available concerning how UvrD interacts with DNA containing covalent DNA-protein complexes (DPCs). In this regard, we have recently reported that a yeast helicase, Sgs1, is important for cellular tolerance to formaldehyde-induced DPCs (24). Additionally, a recent report has investigated the role of DNA helicases as sensors for DNA damage (25). Although several helicases were differentially affected by the relative positioning of a thymine glycol lesion, the UvrD helicase activity was not inhibited by this lesion on either the translocating or complementary DNA strand. The present work addresses a biological role for UvrD in processing of DPCs and is the first study that characterizes the in vitro function of a helicase on DNA substrates containing a site-specific, structurally well defined DPC.
EXPERIMENTAL PROCEDURES
Design and Generation of Substrates
The oligodeoxynucleotides were synthesized by Invitrogen. One of the two DNA strands of the partial duplexes used in the helicase reactions was 32P-labeled by incubation with T4 polynucleotide kinase (New England BioLabs, Inc., 10 units) and 10 μCi of [32P]ATP in a total volume of 20 μl for 1 h at 37 °C. The annealing reactions were performed by incubating the 32P-labeled strand and the complementary strand (2–3-fold excess) in TM buffer (Tris-HCl (pH 8.0), 100 mm MgCl2) containing 50 mm NaCl. The resultant partial duplex DNAs contained deoxyuridine (dU) either on the translocating or non-translocating strand (Fig. 1). To form the site-specific DPC, an apurinic/apyrimidinic (AP) site was created by reaction with uracil DNA glycosylase followed by a reaction with T4 pyrimidine dimer glycosylase/AP site lyase (T4-pdg) protein in the presence of NaBH4 as previously described (26).
FIGURE 1.
DNA substrates. Sequence and structural representation of the double-stranded DNA substrates, 50C-30C and 50T-30T, containing a covalently linked DPC either on the complementary (non-translocating) or the translocating strand. As described under “Experimental Procedures,” the 50-mer oligodeoxynucleotide (50C or 50T) was annealed to the complementary 30-mer oligodeoxynucleotide (30C or 30T) to generate a partially complemented duplex substrate. U is a uracil base and marks the site where the DPC was formed (indicated by the filled circle). To facilitate the loading of UvrD on these substrates, the length of the 3′ single-stranded arm of the fork was kept longer than the 5′ single-stranded arm.
Helicase Assay
Purification of UvrD was done as described previously (27). For the helicase assay, UvrD was preincubated with DNA substrates (1 nm) in 10 μl of buffer T20 (10 mm Tris-HCl (pH 8.3), 20 mm NaCl, 20% (v/v) glycerol). The helicase reaction was initiated by adding 10 μl of buffer T20 supplemented with 2 mm ATP, 1 mm MgCl2 (final concentration) and an excess of unlabeled DNA strand (10-fold excess over the 32P-labeled strand). This DNA trap was added to minimize any re-annealing of the unwound DNA products from the helicase reaction. Reactions were carried out at room temperature for the indicated times. The reactions were terminated by adding 2.5 μl of quenching solution (0.4 m EDTA and 10% glycerol) and 2.5 μl of loading dye (40% (w/v) sucrose and 0.25% bromphenol blue (w/v)). DNAs were separated by electrophoresis through a 5% native gel in 0.5× Tris borate EDTA for 1.5 h at 190 V. The results were visualized and quantified by PhosphorImager analyses.
Sequestration Assay
For helicase sequestration studies, UvrD (10 nm) was preincubated in standard helicase reaction buffer in the presence of the indicated amounts (0–8 nm) of unlabeled partial duplex DNA that were either unadducted or contained a DPC. After incubation in the presence of 2 mm ATP for 5 min at room temperature, 1 nm 32P-labeled non-damaged partial duplex substrate (50T-30T) was subsequently added, and the reaction mixtures were incubated for 5 min at room temperature. Reactions were then quenched and resolved on native polyacrylamide gels as described above. Typically, 70–75% of the 32P-labeled substrate was unwound in reactions lacking the competitor DNA molecule. The extent of strand separation was calculated relative to the control reactions lacking the competitor DNA.
ATPase Assay
ATPase assays were carried out using the colorimetric ATPase assay kit purchased from Innova Biosciences and performed according to the manufacturer's protocol. Reaction mixtures (100 μl) were prepared in T20 buffer, 2 nm duplex DNA substrate and varying concentrations of UvrD helicase (5–20 nm). Reactions were initiated by the addition of a mixture (100 μl) containing 2.5 mm MgCl2 and 0.5 mm ATP and incubated for 5 min at room temperature. Reactions were terminated by the addition of Lock Gold and accelerator mixture (50 μl and 0.5 μl, respectively). After 2 min, 20 μl of stabilizer was added, and after 30 min of incubation at room temperature, the absorbance was measured at a wavelength of 650 nm using an Spectra Max 190 microplate reader (Molecular Devices). Absorbance (A650) values were plotted as a function of increasing UvrD concentrations.
Bacterial Strains and Culture Conditions
Bacterial strains used in this study are described in Table 1. All strains were maintained in Luria-Bertani (LB) broth that was supplemented with kanamycin (kan) (50 μg/ml) for the ΔuvrD strain and chloramphenicol (cam) (50 and 20 μg/ml, respectively) for the AK100 and AK101 strains. The deletion in the uvrD gene was confirmed by PCR (supplemental Fig. S1).
TABLE 1.
E. coli strains used in this study
| Strains | Genotype | Source |
|---|---|---|
| BW25113 | Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), LAM-, rph-1, Δ(rhaD-rhaB)568, hsdR514 | E. coli Genetic Stock Center, Yale University |
| JW3786-5 | Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), LAM-, rph-1, ΔuvrD769::kan, Δ(rhaD- rhaB)568, hsdR514 | E. coli Genetic Stock Center, Yale University |
| DY329 | W3110 ΔlacU169 nadA::Tn10 gal490 λcl857 Δ(cro-bioA) | J. Courcelle, Portland State University |
| AK100 | W3110 ΔlacU169 nadA::Tn10 ΔuvrA::cam, gal490 λcl857 Δ(cro-bioA) | This study |
| AK101 | Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), LAM-, rph-1, Δ(rhaD-rhaB)568, hsdR514, ΔuvrA::cam | This study |
Strains AK100 and AK101 were generated utilizing the previously developed strategy for chromosome engineering in E. coli (28). Strain AK100 was constructed by transformation of DY329 with a linear donor DNA that was designed to disrupt the uvrA gene by replacing it with a cam selective marker. The donor DNA was obtained by PCR amplification using the DNA template that was derived from a cam-resistant cassette (Stratagene). Primer sequences were 5′-TGTGACGGAAGATCACTTCGGGGCGCCCGCACCCATAATCTCAAAAACATGTGACGGAAGATCACTTCG-3′ (uvrA-cam forward primer) and 5′-CGGCTTAAGGAAGCGTGCCGTGTGTGATGCTTCGCACTCCACCAGCAATAGACACCAGCAATAGACATAAGCG-3′ (uvrA-cam reverse primer). These primers were designed such that the 5′ 20-nucleotide (nt)-long ends were homologous to the flanking regions of the target gene followed by the 40-nt-long stretches priming the cam cassette DNA for replication. The reactions were carried out using Fermentas PCR master mix. The conditions for PCR were as follows: initial denaturation at 94 °C for 10 min followed by 29 cycles at 94 °C for 30 s, 57 °C for 1 min, and 72 °C for 2 min and a final extension step of 72 °C for 10 min. The PCR product was purified from a 1% agarose gel using a Qiagen PCR purification kit.
Electroporation of the donor DNA into DY329-competent cells was performed as described (28). The electroporated cells were diluted with 1 ml of LB broth and incubated overnight at 30 °C, and the recombinant cells were selected on LB-cam plates. To generate strain AK101, P1 phage was passed through the strain AK100, and the phage lysate was used for transduction of the BW25113 strain. The extractions of genomic DNA and RNA were done using Qiagen genomic DNA and RNA extraction kits, respectively. The deletion in the uvrA gene was confirmed by PCR amplification in four independent colonies (supplemental Fig. S2).
Survival Assay
Fresh overnight cultures (grown in the presence of appropriate antibiotics) were diluted 1:100 in LB medium and grown to an A595 ∼ 0.3. The culture was diluted 1:500 in LB, and 100 μl aliquots were incubated with varying concentrations of formaldehyde for 30 min at 37 °C, followed by plating on LB agar plates.
Generation of Plasmids Containing DPCs
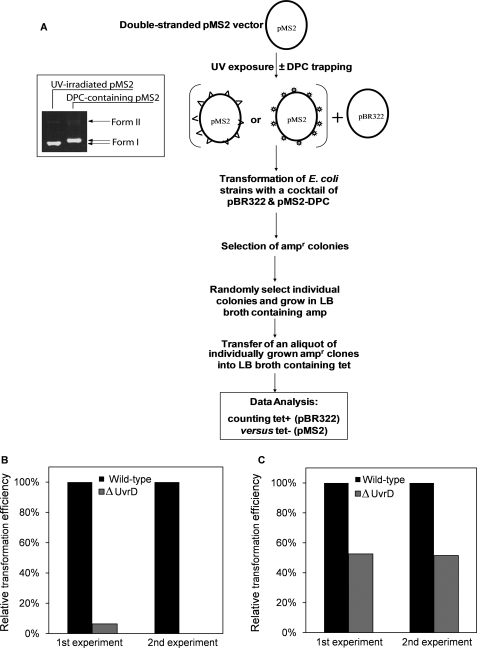
To introduce DPCs into plasmid DNA, the catalytic chemistry of T4-pdg was exploited in which T4-pdg proceeds via a covalent DNA-protein intermediate. Specifically, after scission of the glycosidic bond at the cyclobutane pyrimidine dimer (CPD) site, T4-pdg forms an imine intermediate with the C1′ of the deoxyribose at an AP site and generates a nick in the DNA strand (29, 30). Under reducing conditions, the imine intermediate can be trapped to produce a stable complex between DNA and protein (29, 30). Initially, the number of CPDs per plasmid molecule was determined empirically. The pMS2 vector (31) (100 ng/μl) in 10 mm Tris-HCl (pH 7.0), 1 mm EDTA was exposed to 254-nm UV light at 100 microwatts/cm2, and 2 μl aliquots of DNA were collected during the course of exposure to measure the rate of CPD formation. Subsequently, these DNAs were incubated with T4-pdg in 25 mm sodium phosphate buffer (pH 6.8) containing 100 μg/ml bovine serum albumin, 100 mm NaCl, and 1 mm EDTA to generate single-stranded breaks at the CPD sites, converting plasmids from the supercoiled form (Form I) to the nicked relaxed (Form II) and linear (Form III) forms. Incubations were carried out for 1 h at 37 °C, and the various topological forms of the DNA were separated by electrophoresis through a 1% agarose gel. The gel was stained with ethidium bromide (0.2 μg/ml), and the intensity of each band was determined using an Alpha Innotech Imaging system. Based on an assumption of a Poisson distribution of CPDs throughout the plasmid population, the number of CPDs per plasmid molecule was calculated using the equation, CPD number = −ln(1.42IForm I/(1.42IForm I + IForm II + IForm III)), where I is the band intensity, and 1.42 is a factor to correct for the reduced intercalation of ethidium bromide into the supercoiled DNA (32). The results of the time course experiment are shown in supplemental Fig. S3, panels A and B. After these initial analyses, the pMS2 vector was exposed under the same conditions as above for 4 min to introduce ∼8 CPDs per plasmid molecule.
To generate plasmids containing DPCs (pMS2-DPC), 300 ng of UV-irradiated pMS2 were reacted with T4-pdg (0.4 μm) and freshly prepared NaBH4 (100 mm) in a total volume of 40 μl. The T4-pdg and NaBH4 were mixed first and immediately added to the plasmid DNA. Formation of DPCs was confirmed by the decreased electrophoretic mobility of the vector DNA through a 1% agarose gel. As a non-DPC control, UV-irradiated pMS2 was treated with NaBH4 in the absence of T4-pdg; these plasmids (pMS2-UV) along with pMS2-DPC were used in the plasmid reactivation assays.
Plasmid Reactivation Assays
The pMS2-UV and pMS2-DPC plasmids were individually mixed with the non-damaged pBR322 plasmid at a 15 to 1 weight ratio, and the mixture was used to transform wild-type and ΔuvrD E. coli strains. The pBR322 plasmid served as an internal control for measuring transformation efficiency. Although pMS2 confers resistance to ampicillin (amp) only, pBR322 encodes for two selective markers, amp and tetracycline (tet); thus, the pMS2- and pBR322-containing bacterial clones can be easily distinguished. The transformation of bacteria was done by electroporation using the conditions described previously (33), and the transformants were selected on LB agar plates containing amp (100 μg/ml). For further screening, the transformants were individually grown first in LB broth containing amp (100 μg/ml) in 96-well plates at 37 °C for 4–6 h. A 20-μl aliquot from each 96-well was transferred to another 96-well plate containing LB broth with tet (5 μg/ml) and grown overnight at 37 °C.
Statistical Analyses
Separate generalized linear models were fit to the data depending on the particular outcome being modeled. Pixel density as a function of UvrD concentration (helicase reactions) was best described using a gamma distribution. The number of surviving colonies as a function of formaldehyde concentration (survival assays) followed a negative binomial distribution, while absorbance as a function of UvrD enzyme concentration (ATPase assays) followed a normal distribution. All models also included indicator variables identifying the particular treatment (e.g. damaged versus non-damaged) along with interactions between concentration (of UvrD or formaldehyde) and treatment. Models also employed a log link between the particular outcome and the explanatory variables, thereby allowing model parameters to be interpreted as log ratios of the mean response. In particular, fitted values measured relative to the model's intercept term reflect the (log) mean response relative to control conditions (where formaldehyde or UvrD concentration is 0). Exponentiation of these log ratios re-expresses the relative change in the mean response on the original scale of measurement, with 95% confidence intervals for the corresponding effect similarly back-transformed. All models were fit using R (Version 2.10.1; R Development Core Team, 2009) with the associated MASS package (34) to fit the negative binomial model.
RESULTS
Inhibition of the Helicase Activity of UvrD by a DPC
Several strategies have been used to create site-specific DNA-protein cross-links (35–37). In this study, synthetic oligodeoxynucleotides were prepared in which a dU had been introduced either at position 16 from the 5′ end of the non-translocating strand or at position 14 from the 5′ end of the translocating strand (Fig. 1). After annealing the appropriate strands, partial duplex DNAs were formed in which a 24 base-paired duplex was adjacent to a 6-nucleotide mismatched region followed by a 20-mer poly(dA) tail, ending in a 3′-OH. To form the site-specific DPC, an AP site was created by reaction with uracil DNA glycosylase followed by a reaction with T4-pdg in the presence of NaBH4 to reduce the covalent imine intermediate between the C1′ of the deoxyribose and the α-amino group of T4-pdg (29, 30). This reaction is very efficient and results in 90–95% covalently bound product (Figs. 2A and 3A, the first lanes designated to DPC-containing DNA). dU-containing DNAs were used as controls in this study. Independent experiments were performed to confirm that UvrD unwinding activity was comparable for both uracil DNA glycosylase-treated and untreated DNA substrates (supplemental Fig. S4); these observations exclude the possibility that any of the differences observed between control (non-damaged) and DPC-containing DNA were caused by minor amounts of contaminating AP-containing DNA.
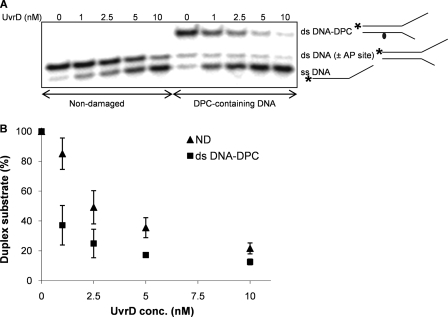
FIGURE 2.
UvrD helicase activity on a DNA substrate containing DPC in the complementary strand. A, UvrD helicase activity on non-damaged duplex (50C-30C) and DNA substrate containing a DPC in the complementary strand is shown. The 32P-labeled 50-mer oligodeoxynucleotide (50C) was annealed to the complementary 30-mer oligodeoxynucleotide (30C), and T4-pdg was covalently linked to the 30-mer strand. Reaction mixtures containing 1 nm concentrations of the indicated duplex DNA substrate and specified concentrations of UvrD were incubated at room temperature for 5 min under standard conditions. The asterisk indicates the 5′ of the 32P-labeled strand of the duplex substrate. B, quantitation of the experiments in A. Percentage duplex substrate is graphically represented as a function of UvrD concentration. The error bars indicate the S.D. derived from three independent helicase reactions. The abbreviations ND, ss DNA, ds DNA (± AP site), and ds DNA-DPC correspond to the non-damaged, single-stranded DNA, double-stranded DNA with or without AP site, and double-stranded DNA containing a DPC, respectively.
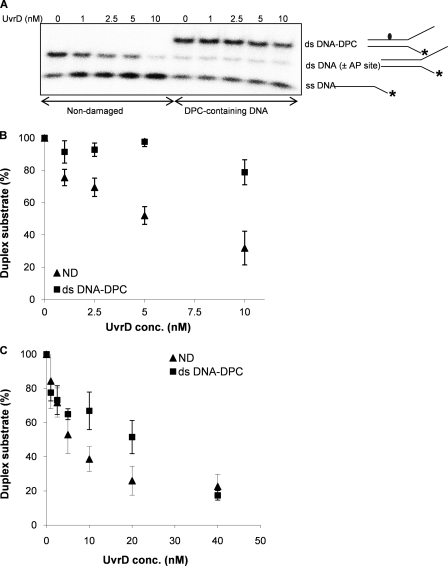
FIGURE 3.
UvrD helicase activity on a DNA substrate containing DPC in the translocating strand. A, UvrD helicase activity on the non-damaged (50T-30T) and the damaged duplex substrate containing a DPC in the translocating strand. The 32P-labeled 30-mer oligodeoxynucleotide (30T) was annealed to the complementary 50-mer oligodeoxynucleotide (50T), and T4-pdg was covalently linked to the 50-mer strand. Reaction mixtures containing 1 nm concentrations of the indicated duplex DNA substrate and specified concentrations of UvrD were incubated at room temperature for 5 min under standard conditions. The asterisk indicates the 5′ of the 32P-labeled strand of the duplex substrate. B, quantitation of the experiments in A. Percentage duplex substrate is graphically represented as a function of UvrD concentration used. The error bars indicate the S.D. derived from three independent helicase reactions. C, reactions with UvrD helicase at higher concentrations (0–40 nm) and a longer incubation time (15 min). Standard helicase reactions were run with the non-damaged duplex (50T-30T) and damaged substrate containing a DPC in the translocating strand. The percentage of duplex substrate is graphically represented as a function of the UvrD concentration. The error bars indicate the S.D. derived from three independent helicase reactions. The abbreviations ND, ss DNA, ds DNA (±AP site), and ds DNA-DPC correspond to the non-damaged, single-stranded DNA, double-stranded DNA with or without AP site, and double-stranded DNA containing a DPC, respectively.
Having generated these substrates, the effect of a DPC on UvrD-catalyzed DNA unwinding was evaluated when the lesion was located in the non-translocating strand (Fig. 2). Data in panel A demonstrate that UvrD was capable of unwinding DPC-containing DNA without any hindrance. Because the co-crystal structure of T4-pdg covalently bound to an AP site reveals that the protein bends the DNA to ∼70° and makes numerous contacts over ∼6 base pairs with both the strand to which it is bound and the complementary strand (38), the lack of any discernable inhibition of UvrD was somewhat unexpected. As shown in Fig. 2B, when the concentration dependence of unwinding was statistically analyzed for damage-containing and control DNAs, no significant difference was observed (p = 0.42). However, at the lowest concentrations of UvrD, the extent of strand separation was slightly higher in the presence of the DPC. This observation suggests that the DNA conformational change imposed by the DPC in the translocating strand may facilitate the UvrD-mediated DNA-unwinding process. This increased efficiency of unwinding in the presence of a damage on the complementary strand has also been observed for Bloom and Werner helicases when assayed on thymine glycol containing DNAs (25).
Conversely, when the covalently linked protein was positioned in the same strand on which the translocation should occur, the presence of the covalently linked T4-pdg significantly inhibited translocation (p < 0.001) as evidenced by little or no single-stranded DNA product (Fig. 3A). The very modest increase in the percent of single-stranded DNA can be attributed to spontaneous melting during the course of the reaction. Even at 10-fold molar excess of the enzyme over the DNA substrate, UvrD only unwound 20% of the damaged substrate relative to 75% unwinding achieved for the non-damaged substrate (Fig. 3B). As expected, UvrD was highly efficient at strand displacement on the control substrate.
Given that UvrD is induced upon SOS response (39), we assessed UvrD activity on the same substrate at higher UvrD concentrations (20- and 40-fold molar excess of the enzyme over the DNA substrate) and increased incubation times (15 min). Fig. 3C shows that upon increasing the concentration and reaction time, comparable unwinding (80%) could be achieved for both the non-damaged and DPC-containing substrate. Thus, the possibility cannot be excluded that UvrD would separate DNA strands in a physiological environment, even when the DPC is in the translocating strand.
Accessory factors, such as single-strand binding protein (SSB), have been shown to modulate the activity of DNA helicases. The effect of SSB on unwinding of non-damaged and DPC-containing DNA substrates by UvrD was tested. When helicase reactions were carried out in the presence of varying amounts of SSB protein (0.625–20 nm), similar inhibition of UvrD activity was observed on both the non-damaged and damaged substrate in the presence of SSB protein (data not shown). Our results are consistent with most of the previously published work, demonstrating an inhibition of UvrD helicase activity in the presence of SSB protein (40, 41).
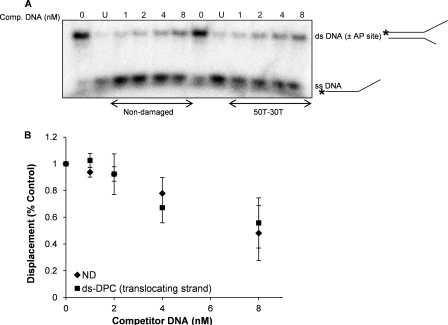
It has previously been shown that various helicases can be sequestered on DNA substrates carrying DNA adducts (42). Because UvrD is significantly inhibited by the DPC located in the translocating strand, we hypothesized that UvrD may be sequestered on this substrate. To test this hypothesis, UvrD was incubated in the presence of either unlabeled control or DPC-containing DNA followed by the addition of a 32P-labeled control non-damaged substrate. As shown in Fig. 4, A and B, preincubation with either control or DPC-containing DNAs (up to 8-fold molar excess) did not differentially affect UvrD unwinding activity on 32P-labeled DNA. Thus, despite a strong inhibition of UvrD helicase activity by a single DPC adduct in the translocating strand, the damaged partial DNA duplexes (double-stranded DPC) did not trap UvrD to any greater extent than the non-damaged duplex molecule. This observation suggests that there is no difference in dissociation of UvrD from the damaged DNA versus non-damaged DNA substrate. Similar results were obtained when the sequestration assay was performed with the substrate containing DPC in the non-translocating strand (data not shown).
FIGURE 4.
UvrD dissociates from the DNA substrate containing a DPC in the translocating strand. A, reactions for sequestration assays were initiated under conditions similar to that described for the helicase reactions in the presence of specified concentrations (1–8 nm) of unlabeled non-damaged (50T-30T) or damaged competitor substrate. After helicase reactions with the competitor DNA, the 32P-labeled tracker substrate was added to the reactions and incubated for 5 min at room temperature. B, quantitation of the experiments in A. The error bars indicate the S.D. derived from three independent sequestration experiments. The abbreviations ND, ss DNA, ds DNA (±AP site), and ds-DPC correspond to the non-damaged, single-stranded DNA, double-stranded DNA with or without AP site, and double-stranded DNA containing a DPC, respectively.
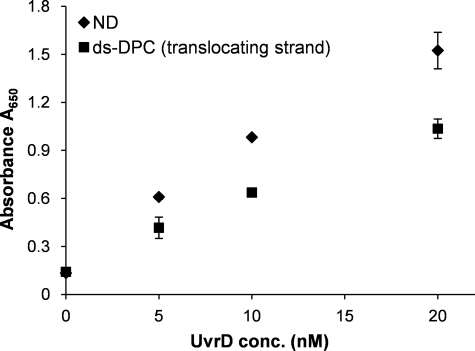
The unwinding reaction of UvrD is fueled by energy derived from ATP hydrolysis (43). The observed inhibitory effect of the DPC lesion in the translocating strand raised the possibility that the ATPase activity of UvrD might also be inhibited on the damaged DNA substrate. To address this question, ATP hydrolysis was measured as a function of UvrD concentration in the presence of non-damaged and DPC-containing substrate (translocating strand) (Fig. 5). Relative to the non-damaged substrate, ATP hydrolysis on the DPC substrate was reduced ∼2-fold (p < 0.001). Because the duplex region in the DPC-containing substrate is approximately half that of the non-damaged substrate, the reduction in ATP hydrolysis likely reflects such a difference. Collectively with the observation that UvrD is not sequestered at the DPC-containing substrate, these data suggest that the apparent reduction in ATP hydrolysis could be because of premature helicase dissociation from the DNA substrate rather than specific inhibition of the hydrolysis.
FIGURE 5.
UvrD ATPase activity is affected in the presence of DPC-containing DNA substrate. Reactions for ATPase assay containing non-damaged (50T-30T) or damaged duplex substrates were prepared under conditions similar to that described for helicase reactions. After a 5-min incubation of DNA substrate (2 nm) and specified concentrations of UvrD enzyme, ATP-containing solution (0.5 mm) was added, and reactions were incubated at room temperature for additional 5 min before terminating the reactions. A650 values for controls containing buffer alone, UvrD enzyme alone, and DNA substrate alone were found to be comparable. The error bars indicate the S.D. derived from three independent ATPase reactions. ND, non-damaged; ds-DPC double-stranded DNA containing a DPC.
UvrD Is Important for the Replication of Plasmid DNAs Containing DPCs
Having observed efficient unwinding by UvrD when the site-specific DPC was located in the non-translocating strand, we next sought to investigate whether UvrD is involved in replication of plasmid DNA containing DPCs. The experimental strategy to test for this possibility is outlined in Fig. 6A. Specifically, we compared the relative efficiencies by which the wild-type and ΔuvrD E. coli strains could replicate DPC-containing plasmids. The status of the deletion in uvrD was confirmed by the absence or presence of the PCR amplification product from the bacterial strain JW3786–5 as compared with a wild-type strain, BW25113 (supplemental Fig. S1). First, the pMS2 plasmid was UV-irradiated to introduce ∼8 CPDs per plasmid molecule. Subsequent treatment with T4-pdg in the presence of NaBH4 converted all CPDs into DPCs (see the inset panel, Fig. 6A). The efficiency of this reaction is quantitative and does not result in the loss of supercoiled density of the plasmid as the phosphodiester bond is never broken. In these experiments non-damaged pBR322 was used as the reference plasmid to normalize for possible interstrain differences in the ability to replicate the plasmid DNA. The reference pBR322 and damaged pMS2-DPC were mixed and transformed into the wild-type and ΔuvrD cells. The successful transformants were selected for amp resistance and subsequently selected for tet resistance, thus allowing identification of pMS2- and pBR322-containing transformants, respectively. The data in Fig. 6B show that when DPC-containing pMS2 plasmid was replicated in the ΔuvrD strain, there was a significant reduction in the number of pMS2 transformants relative to the wild-type E. coli with a near absence of the pMS2-containing clones in the total amp resistant population. This small percentage may reflect the rare undamaged pMS2 molecule within the population.
FIGURE 6.
Relative colony-forming ability of E. coli strains after transformation with DPC containing plasmids. A, a flow chart diagram shows sequential steps involved in the generation of DPC-containing plasmids. The ds pMS2 plasmid (ampr) was irradiated with UV-C, and T4-pdg was trapped at the UV-induced cyclobutane pyrimidine dimer sites. The non-damaged plasmid pBR322 (ampr and tetr) was mixed with either the UV-irradiated (triangle) or DPC containing (star) pMS2 plasmid, and the resultant mixture was used to transform the wild-type and ΔuvrD E. coli strains followed by selecting the transformants for amp resistance. Randomly selected amp resistant clones were further assessed for tet resistance to discriminate between the clones carrying pMS2 versus pBR322 plasmid. An aliquot of UV-irradiated pMS2 vector was collected before and after trapping DPC and analyzed on a 1% agarose gel. Efficiency to replicate pMS2 plasmid containing DPCs (B) or UV-induced lesions (C) is shown. The relative colony forming ability of E. coli strains was assessed after transformation with a mixture of non-damaged pBR322 and pMS2 containing randomly integrated DPCs or UV-induced lesions. For both the wild-type and ΔuvrD strains, the percentage of pMS2-DPCs transformants was calculated relative to pBR322 transformants. The apparent transformation efficiency with the reference plasmid pBR322 was comparable for all the strains tested. The data collected from two independent experiments are shown.
In addition to CPDs, UV induces other lesions, such as (6–4)-photoproducts (44). Thus, it was important to rule out the possibility that these lesions, but not DPCs, were responsible for the low efficiency of the ΔuvrD strain to replicate DPC-containing plasmids. To test this possibility, we transformed the wild-type and ΔuvrD strains with a mixture of non-damaged pBR322 and UV-irradiated/NaBH4-treated pMS2 plasmids. These data revealed that the ΔuvrD strain could replicate damage-containing pMS2 with an efficiency only 2-fold reduced relative to the wild-type (Fig. 6C). These results suggest that the remarkably low efficiency of the ΔuvrD strain to replicate the DPC-containing pMS2 could be attributed to DPCs and not other UV-induced DNA lesions. Thus, it can be concluded that UvrD is important for efficient processing of DPCs to allow replication of DPC-containing plasmids.
Sensitivity of the ΔUvrD Strain to the DPC-inducing Agent, Formaldehyde
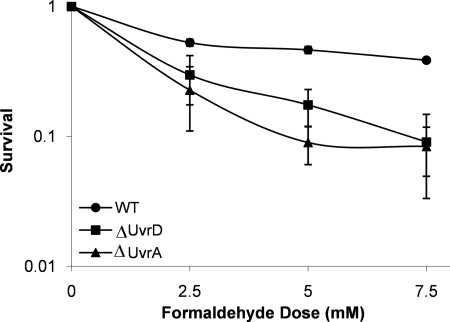
Although the previous replication assay demonstrated a critical role for UvrD in replication of the DPC-containing DNAs, experiments were designed to test the cytotoxic effect of deletion of this gene to the DPC-inducing agent, formaldehyde. Isogenic wild-type and ΔuvrD E. coli strains were constructed and challenged by exposing the cells to the DPC-inducing agent, formaldehyde. Fig. 7 shows that loss of uvrD resulted in an increased cytotoxicity (p < 0.001) relative to the wild type with a D10 (concentration of formaldehyde causing 90% cell death) of ∼5 mm formaldehyde, a result that is consistent with a previous study done in a different strain background (45). These data confirm that UvrD is critical for limiting DPC-induced cytotoxicity.
FIGURE 7.
Cell survival after exposure to the DPC inducing agent, formaldehyde. Exponentially growing wild-type (circles), ΔuvrD (squares), and ΔuvrA (triangles) strains were exposed to formaldehyde at the indicated concentrations for 30 min at 37 °C. The mean (symbol) and S.D. (error bar) from at least three or more independent experiments are shown. WT, wild type.
Two independent studies (37, 45) have also demonstrated that uvrA mutants are sensitive to formaldehyde. One of these studies examined the formaldehyde sensitivity of both ΔuvrD and ΔuvrA strains, but these strains were generated from two different parental backgrounds. Thus, to assess how comparable formaldehyde sensitivity was for our ΔuvrA and ΔuvrD strains, it was necessary to construct an isogenic ΔuvrA strain (see “Experimental Procedures”). Deletion of the uvrA gene was confirmed by PCR (supplemental Fig. S2). Survival assays were performed with isogenic wild-type, ΔuvrA, and ΔuvrD strains and demonstrated that the formaldehyde sensitivities of both deletion strains were comparable (Fig. 7), suggesting that the contribution of both proteins is equally important in protecting against formaldehyde-induced DNA lesions.
DISCUSSION
DPCs can be produced by exposure to a variety of chemical and physical agents, including formaldehyde, transplatin, and ionizing radiation (46–48). The biochemical pathways by which these lesions are repaired and/or tolerated are not well understood; however, the importance of DNA helicases for cellular tolerance after formaldehyde exposure has recently been demonstrated (24, 45).
The DPCs, if not reversed or removed, may cause pausing of replication, presumably by blocking the actions of the replicative polymerases and helicases, the core components of the replication machinery. For example, treatment with a cytidine analog, azacytidine C, results in covalently linked methyltransferase-DNA adducts that cause blockage of DNA replication in vivo (49). This blockage may be overcome by the action of specialized, translesion synthesis DNA polymerases that have been shown to be required for bypassing DNA-peptide cross-links, the potential intermediates generated during processing of DPCs (50). A similar role can be predicted for the helicases, rationalizing for the existence of several specialized helicases that participate in processing of blocked replication forks via different repair processes in a lesion-specific manner.
In the human reconstituted system, there is strong evidence that the repair of DNA-peptide cross-links is initiated by NER (51). However, in prokaryotic organisms it has been shown that NER can initiate repair of both DNA-peptide and small DNA-protein cross-links (26, 37, 52). Minko et al. (26) demonstrated the ability of the UvrABC complex to make a dual incision at the eighth phosphodiester bond 5′ and fourth phosphodiester bond 3′ to a DPC on a DNA substrate in which T4-pdg was covalently linked to a synthetic oligodeoxynucleotide via a site-specific AP site. Upon the addition of purified UvrABC, a 12-mer oligodeoxynucleotide containing the intact T4-pdg was produced, with the efficiency of the incision approximately equal to that measured for a benzo[a]pyrene diolepoxide lesion linked through N2-guanine (26, 52).
However, UvrABC-mediated dual incisions around the lesion are only the first step leading to the complete restoration of undamaged duplex DNA. The following step in prokaryotic NER repair is hypothesized to be UvrD helicase-catalyzed strand displacement to facilitate the removal of the incised strand (53). Previously, a model had been proposed for a post-incision role for UvrD in E. coli NER (54). This model suggested that UvrD would be loaded on the 3′ end of the incised strand and would facilitate release of the 12-mer containing the damage.
The progression of UvrD was severely blocked in the presence of a DPC in the translocating strand. The sequestration studies revealed similar dissociation of UvrD for both the non-damaged substrate and the substrate containing a DPC in the translocating strand, supporting a model where UvrD translocates up to the site of the DPC but, due to strong blockage by the DPC, is dissociated from the DNA. Collectively, these data suggest that UvrD would be unsuccessful in displacing the DPC-containing strand if it is loaded on the 3′ end. Furthermore, because the molecular footprint of T4-pdg extends very close to the site of the 3′ incision, the steric hindrance of T4-pdg near the 3′-OH would likely block loading of UvrD (38).
The DPC-mediated blockage of UvrD was alleviated when the UvrD concentration was raised by 20–40-fold molar excess over the DNA substrate. Considering that high levels of UvrD can accumulate in a full SOS response (2–6 × 104 molecules per cell) (55), it is plausible that the SOS induction of UvrD protein is a prerequisite for overcoming the barrier posed by bulky adducts such as DPCs. In support of this, it has been previously shown using non-damaged DNA substrates that although UvrD preferentially utilizes the 3′ single-stranded end for loading (56), at high concentrations it can initiate unwinding of blunt-ended DNA substrates at nicked sites (57, 58), forked junctions, and free single-stranded 5′ ends (59). Similar results were obtained for DNA substrates containing a benzo[a]pyrene adduct such that a partial rescue of UvrD inhibition was achieved utilizing the blunt end of the DNA substrate in the presence of high concentrations of UvrD (60). Qualitatively similar results have been reported for FancJ helicase activity on a thymine glycol containing DNA (25). Thus, it is possible that UvrD functions on DPC-containing substrates in a similar manner.
The impediment of UvrD upon colliding with adducts is not a generalized phenomenon, and lesions such as thymine glycol that cause a significant distortion of the DNA helix (61) do not impede UvrD helicase activity irrespective of the lesion being placed on either the translocating or non-translocating strand (25). Therefore, it may be that it is the bulk of adduct, not the distortion of DNA, that limits the unwinding activity of UvrD.
Our in vivo studies provide strong evidence for the involvement of UvrD in the processing of DPC lesions. These data clearly demonstrate an essential role for UvrD in replication of plasmid DNAs containing multiple DPCs. We propose that a potential initial role for UvrD may be as a sensor of DNA damage in association with its role in the polymerase III replication complex. After the damage sensing event, the mechanism(s) by which these lesions are repaired or tolerated may include NER, homologous recombination, and possibly translesion synthesis pathways. Because UvrD is known to modulate both NER and homologous recombination mechanisms, it is unclear which pathway dominates to yield lesion-free plasmid DNAs. Due to the extremely poor ability of ΔuvrD cells to replicate DPC-containing plasmids, an analysis of UvrD epistasis with NER or homologous recombination mutants using the plasmid-based assay is technically not feasible. Studies are in progress to delineate the role of UvrD in the various pathways for the repair and/or tolerance of DPCs.
In summary, our data reveal a novel and unique role for UvrD in the processing of DPC lesions. However, considering the versatile nature of the UvrD protein to participate in multiple biological processes, it remains to be established whether UvrD participates in the processing of DPCs in an NER-dependent or independent manner.
Supplementary Material
Acknowledgments
We are thankful to T. M. Lohman (Washington University School of Medicine, St. Louis, MO) for helpful discussions and for providing the purified UvrD protein. We also thank Justin Courcelle (Portland State University, Portland, OR) for kindly providing the bacterial strain (DY329) and P1 phage and the Courcelle laboratory for assistance in making the uvrA deletion strain. We also thank Michael R. Lasarev (Oregon Health & Science University, Portland, OR) for helping with the statistical analyses of the data.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 CA106858 (to A. K. M. and R. S. L.) and PO1 ES05355 (to R. S. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- NER
- nucleotide excision repair
- AP
- apurinic/apyrimidinic
- CPD
- cyclobutane pyrimidine dimer
- DPC
- DNA-protein cross-link
- SSB
- single-strand binding protein
- T4-pdg
- T4 pyrimidine dimer glycosylase/AP site lyase
- cam
- chloramphenicol
- amp
- ampicillin
- tet
- tetracycline
- kan
- kanamycin.
REFERENCES
- 1.Matson S. W. (1986) J. Biol. Chem. 261, 10169–10175 [PubMed] [Google Scholar]
- 2.Arthur H. M., Lloyd R. G. (1980) Mol. Gen. Genet. 180, 185–191 [DOI] [PubMed] [Google Scholar]
- 3.Ossanna N., Mount D. W. (1989) J. Bacteriol. 171, 303–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mendonca V. M., Kaiser-Rogers K., Matson S. W. (1993) J. Bacteriol. 175, 4641–4651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ogawa H., Shimada K., Tomizawa J. (1968) Mol. Gen. Genet. 101, 227–244 [DOI] [PubMed] [Google Scholar]
- 6.Atkinson J., Guy C. P., Cadman C. J., Moolenaar G. F., Goosen N., McGlynn P. (2009) J. Biol. Chem. 284, 9612–9623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hall M. C., Jordan J. R., Matson S. W. (1998) EMBO J. 17, 1535–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamaguchi M., Dao V., Modrich P. (1998) J. Biol. Chem. 273, 9197–9201 [DOI] [PubMed] [Google Scholar]
- 9.Lahue R. S., Au K. G., Modrich P. (1989) Science 245, 160–164 [DOI] [PubMed] [Google Scholar]
- 10.Joyce C. M., Grindley N. D. (1984) J. Bacteriol. 158, 636–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moolenaar G. F., Moorman C., Goosen N. (2000) J. Bacteriol. 182, 5706–5714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bidnenko V., Lestini R., Michel B. (2006) Mol Microbiol 62, 382–396 [DOI] [PubMed] [Google Scholar]
- 13.Florés M. J., Sanchez N., Michel B. (2005) Mol Microbiol 57, 1664–1675 [DOI] [PubMed] [Google Scholar]
- 14.Veaute X., Delmas S., Selva M., Jeusset J., Le Cam E., Matic I., Fabre F., Petit M. A. (2005) EMBO J. 24, 180–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang G., Deng E., Baugh L., Kushner S. R. (1998) J. Bacteriol. 180, 377–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Centore R. C., Leeson M. C., Sandler S. J. (2009) J. Bacteriol. 191, 1429–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Husain I., Van Houten B., Thomas D. C., Abdel-Monem M., Sancar A. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 6774–6778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumura K., Sekiguchi M., Steinum A. L., Seeberg E. (1985) Nucleic Acids Res. 13, 1483–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caron P. R., Kushner S. R., Grossman L. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 4925–4929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Houten B., Gamper H., Hearst J. E., Sancar A. (1988) J. Biol. Chem. 263, 16553–16560 [PubMed] [Google Scholar]
- 21.Carlson K. M., Smith K. C. (1981) Mutat. Res. 84, 257–262 [DOI] [PubMed] [Google Scholar]
- 22.Kuemmerle N. B., Ley R. D., Masker W. E. (1982) Mutat. Res. 94, 285–297 [DOI] [PubMed] [Google Scholar]
- 23.Khodursky A. B., Cozzarelli N. R. (1998) J. Biol. Chem. 273, 27668–27677 [DOI] [PubMed] [Google Scholar]
- 24.de Graaf B., Clore A., McCullough A. K. (2009) DNA Repair 8, 1207–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suhasini A. N., Sommers J. A., Mason A. C., Voloshin O. N., Camerini-Otero R. D., Wold M. S., Brosh R. M., Jr. (2009) J. Biol. Chem. 284, 18458–18470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Minko I. G., Zou Y., Lloyd R. S. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 1905–1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Runyon G. T., Wong I., Lohman T. M. (1993) Biochemistry 32, 602–612 [DOI] [PubMed] [Google Scholar]
- 28.Yu D., Ellis H. M., Lee E. C., Jenkins N. A., Copeland N. G., Court D. L. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 5978–5983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dodson M. L., Schrock R. D., 3rd, Lloyd R. S. (1993) Biochemistry 32, 8284–8290 [DOI] [PubMed] [Google Scholar]
- 30.Schrock R. D., 3rd, Lloyd R. S. (1993) J. Biol. Chem. 268, 880–886 [PubMed] [Google Scholar]
- 31.Moriya M. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 1122–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lloyd R. S., Haidle C. W., Hewitt R. R. (1978) Cancer Res. 38, 3191–3196 [PubMed] [Google Scholar]
- 33.Kanuri M., Minko I. G., Nechev L. V., Harris T. M., Harris C. M., Lloyd R. S. (2002) J. Biol. Chem. 277, 18257–18265 [DOI] [PubMed] [Google Scholar]
- 34.Venables W. N., Ripley B. D. (2002) Modern Applied Statistics with S, 4th Ed., Springer-Verlag, New York [Google Scholar]
- 35.Sung J. S., Demple B. (2006) Methods Enzymol. 408, 48–64 [DOI] [PubMed] [Google Scholar]
- 36.McCullough A. K., Sanchez A., Dodson M. L., Marapaka P., Taylor J. S., Lloyd R. S. (2001) Biochemistry 40, 561–568 [DOI] [PubMed] [Google Scholar]
- 37.Nakano T., Morishita S., Katafuchi A., Matsubara M., Horikawa Y., Terato H., Salem A. M., Izumi S., Pack S. P., Makino K., Ide H. (2007) Mol. Cell 28, 147–158 [DOI] [PubMed] [Google Scholar]
- 38.Golan G., Zharkov D. O., Grollman A. P., Dodson M. L., McCullough A. K., Lloyd R. S., Shoham G. (2006) J. Mol. Biol. 362, 241–258 [DOI] [PubMed] [Google Scholar]
- 39.Siegel E. C. (1983) Mol. Gen. Genet. 191, 397–400 [DOI] [PubMed] [Google Scholar]
- 40.Shen J. C., Gray M. D., Oshima J., Loeb L. A. (1998) Nucleic Acids Res. 26, 2879–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abdel-Monem M., Dürwald H., Hoffmann-Berling H. (1977) Eur. J. Biochem. 79, 39–45 [DOI] [PubMed] [Google Scholar]
- 42.Villani G., Tanguy Le Gac N. (2000) J. Biol. Chem. 275, 33185–33188 [DOI] [PubMed] [Google Scholar]
- 43.Abdel-Monem M., Chanal M. C., Hoffmann-Berling H. (1977) Eur. J. Biochem. 79, 33–38 [DOI] [PubMed] [Google Scholar]
- 44.Douki T., Reynaud-Angelin A., Cadet J., Sage E. (2003) Biochemistry 42, 9221–9226 [DOI] [PubMed] [Google Scholar]
- 45.Salem A. M., Nakano T., Takuwa M., Matoba N., Tsuboi T., Terato H., Yamamoto K., Yamada M., Nohmi T., Ide H. (2009) J. Bacteriol. 191, 5657–5668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zwelling L. A., Anderson T., Kohn K. W. (1979) Cancer Res. 39, 365–369 [PubMed] [Google Scholar]
- 47.Olinski R., Wedrychowski A., Schmidt W. N., Briggs R. C., Hnilica L. S. (1987) Cancer Res. 47, 201–205 [PubMed] [Google Scholar]
- 48.Olinski R., Briggs R. C., Hnilica L. S. (1981) Radiat Res 86, 102–114 [PubMed] [Google Scholar]
- 49.Kuo H. K., Griffith J. D., Kreuzer K. N. (2007) Cancer Res. 67, 8248–8254 [DOI] [PubMed] [Google Scholar]
- 50.Minko I. G., Yamanaka K., Kozekov I. D., Kozekova A., Indiani C., O'Donnell M. E., Jiang Q., Goodman M. F., Rizzo C. J., Lloyd R. S. (2008) Chem. Res. Toxicol. 21, 1983–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reardon J. T., Sancar A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 4056–4061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Minko I. G., Kurtz A. J., Croteau D. L., Van Houten B., Harris T. M., Lloyd R. S. (2005) Biochemistry 44, 3000–3009 [DOI] [PubMed] [Google Scholar]
- 53.Petit C., Sancar A. (1999) Biochimie 81, 15–25 [DOI] [PubMed] [Google Scholar]
- 54.Batty D. P., Wood R. D. (2000) Gene 241, 193–204 [DOI] [PubMed] [Google Scholar]
- 55.Kuzminov A. (1999) Microbiol. Mol. Biol. Rev. 63, 751–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ali J. A., Maluf N. K., Lohman T. M. (1999) J. Mol. Biol. 293, 815–834 [DOI] [PubMed] [Google Scholar]
- 57.Runyon G. T., Bear D. G., Lohman T. M. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 6383–6387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Runyon G. T., Lohman T. M. (1989) J. Biol. Chem. 264, 17502–17512 [PubMed] [Google Scholar]
- 59.Cadman C. J., Matson S. W., McGlynn P. (2006) J. Mol. Biol. 362, 18–25 [DOI] [PubMed] [Google Scholar]
- 60.Choudhary S., Doherty K. M., Handy C. J., Sayer J. M., Yagi H., Jerina D. M., Brosh R. M., Jr. (2006) J. Biol. Chem. 281, 6000–6009 [DOI] [PubMed] [Google Scholar]
- 61.Wallace S. S. (2002) Free Radic. Biol. Med. 33, 1–14 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.