Abstract
The transcriptional co-activator PGC-1α and the NAD+-dependent deacetylase SIRT1 are considered important inducers of mitochondrial biogenesis because in the nucleus they regulate transcription of nucleus-encoded mitochondrial genes. We demonstrate that PGC-1α and SIRT1 are also present inside mitochondria and are in close proximity to mtDNA. They interact with mitochondrial transcription factor A (TFAM) as assessed by confocal microscopy analysis and by blue native-PAGE. Nucleoid purification allowed us to identify SIRT1 and PGC-1α as proteins associated with native and cross-linked nucleoids, respectively. After mtDNA immunoprecipitation analysis, carried out on mitochondrial extracts, we found that PGC-1α is present on the same D-loop region recognized by TFAM. Finally, by oligonucleotide pulldown assay, we found PGC-1α and SIRT1 associated with the TFAM consensus sequence (human mitochondrial transcription factor-binding site H). The results obtained suggest that in mitochondria PGC-1α and SIRT1 may function as their nuclear counterparts and represent the genuine factors mediating the cross-talk between nuclear and mitochondrial genome. Finally, this work adds new knowledge on the function of SIRT1 and PGC-1α and highlights the direct involvement of such proteins in regulation of mitochondrial biogenesis.
Keywords: Bioenergetics, Subcellular Organelles/Mitochondria, Mitochondrial DNA, Mitochondrial Metabolism, SIRT
Introduction
The mitochondrial proteome includes ∼1500 proteins among which only 13 are expressed by the mitochondrial genome (1). Accordingly, abundance, morphology, and functional properties of mitochondria are finely controlled at the nuclear genome, where an interconnected network of transcription factors regulates the expression of mitochondrial proteins, including those that control replication and transcription of the mitochondrial genome. The same transcriptional network senses alterations of energetic homeostasis of the cells and is able to adapt mitochondrial function and biogenesis in response to nutrient availability and energy demand (2, 3).
In the last few years, the NAD+-dependent protein deacetylase sirtuin 1 (SIRT1) is emerging as a crucial regulator of mitochondrial biogenesis (4). Mammalian SIRT1 is homologous to the yeast silent information regulator 2 (Sir2) and belongs to class III histone deacetylates (HDACIII) that include other sirtuins (SIRT2–7) with specific subcellular localization and protein substrates. The dependence of SIRT1 on NAD+ links its enzymatic activity directly to the energy status of the cell, thus being activated in conditions of nutrient deprivation such as fasting and caloric restriction (5). Besides its role in modification of chromatin and silencing of transcription, by heterochromatin formation through histones modification, SIRT1 targets a wide range of transcriptional factors, including protein 53 (p53) and forkhead box O (FoxO) (6, 7). However, by regulating peroxisome proliferator-activated receptor γ co-activator 1α (PGC-1α), through a functional protein-protein interaction, SIRT1 influences the activity of one of the most versatile metabolic transcriptional co-activators of genes involved in energy metabolism (5, 8, 9). In particular, PGC-1α represents an upstream inducer of genes of mitochondrial metabolism by positively affecting the activity of some hormone nuclear receptors (peroxisome proliferator-activated receptor γ and estrogen-related receptor α) and nuclear transcription factors (NRF-1,2) (10). NRF-1 is a downstream effector of SIRT1/PGC-1α and activates the expression of OxPhos components, mitochondrial transporters, and ribosomal proteins. Additionally, NRF-1 regulates the activation of the Tfam, Tfb1m, and Tfb2m promoters and indirectly affects the expression of Cox genes, Glut4 and PGC-1α itself (1). Importantly, the coordination of the two genomes seems to be exclusively achieved by the nucleus-encoded proteins TFAM,2 TFB1M, and TFB2M, among which the mitochondrial transcription factor A seems to play a central role being essential for transcription, replication, and maintenance of mtDNA (11, 12).
mtDNA is packaged into the matrix in DNA-protein complexes, termed nucleoids, containing several copies of mtDNA and several different proteins. The molecular structure of nucleoids is largely unknown, but a recent model presented by Bogenhagen et al. (13) proposes that nucleoids are a “layered” structure formed by a central core and a peripheral zone. The first region contains proteins involved in mtDNA transcription and replication including TWINKLE, mitochondrial single-stranded DNA-binding protein, mitochondrial DNA polymerase γ, and TFAM, whereas the second region is fundamental for complex assembly and translation. Numerous sets of proteins have been identified in nucleoids, including several metabolic proteins, chaperons, and antioxidant enzymes (11, 13–16).
SIRT1 and PGC-1α have been recognized to have a primary role in mitochondrial biogenesis/metabolism, and they were found mainly located inside the nuclear compartment. However, their possible direct involvement in the regulation of expression of mitochondrion-encoded genes has not yet been investigated. In this study, we found that SIRT1 and PGC-1α are localized inside mitochondria, are associated with nucleoids, and form a multiprotein complex with TFAM, suggesting their possible involvement in regulation of mitochondrial biogenesis and metabolism.
EXPERIMENTAL PROCEDURES
Cell Cultures and Transfection
Human SH-SY5Y neuroblastoma, HeLa cervix carcinoma cells, and human embryonic kidney cells (HEK293) were purchased from the European Collection of Cell Cultures (Salisbury, UK) and cultured according to the manufacturer's instructions. Cells were maintained in culture medium supplemented with 10% fetal calf serum, 2 mm glutamine, 100 units/ml penicillin/streptomycin (Lonza Sales, Switzerland) and maintained at 37 °C in an atmosphere of 5% CO2 in air.
SH-SY5Y neuroblastoma cells were transiently transfected with the Addgene plasmid pSV-PGC1 (Addgene, Cambridge, MA) (17) by electroporation using a Gene Pulser Xcell system (Bio-Rad), according to the manufacturer's instructions, and were immediately seeded into fresh medium. Transfection efficiency was estimated by co-transfecting the cells with pMAX-FP-GreenC vector (Lonza, Basel, Switzerland). Only experiments that gave >80% transfection efficiency were considered. SH-SY5Y cells were transfected with a 21-nucleotide siRNA duplex (siPGC-1α) directed against the following human PGC-1α target sequence (18): 5′-AAGACCAGCCUCUUUGCCCAG-3′. Transfection with a scramble siRNA duplex (siscr), with no homology to other human mRNAs, was used as control. All the experiments were performed on untransfected cells as additional control. Because no differences were found between siscr and untransfected cells, only siscr were reported. Cells were transfected by electroporation as described previously (19), and transfection efficiency of siRNA was evaluated by co-transfecting siRNAs with nonspecific rhodamine-conjugated oligonucleotides. Only experiments that gave transfection efficiency of >80% were considered.
Isolation of Mitochondria from HeLa Cells and Mice Organs
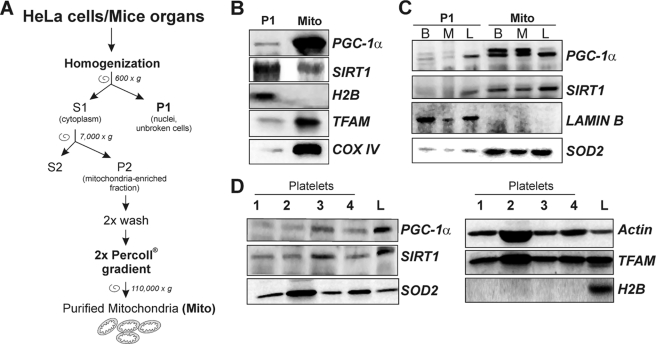
Crude mitochondria from HeLa cells were obtained according to Sun et al. (20). Isolation of mitochondria from mouse brain, muscle, and liver was performed according to the protocol from Frezza et al. (21). Enriched fractions of mitochondria from HeLa cells and mice organs were purified on Percoll® (Sigma) gradient according to the protocol from Pellon-Maison et al. (22). To eliminate the possible presence of nuclear DNA contaminants, Percoll®-purified mitochondria were incubated for 1 h at 4 °C in the presence of DNase I (20 units/mg), and reaction was then stopped by addition of 5 mm EDTA, pH 8.0. A scheme of mitochondria purification is shown in Fig. 2A. All experiments were made to minimize animal suffering and to reduce the number of mice used, in accordance with the European Community Council Directive of November 24, 1986 (86/609/EEC).
FIGURE 2.
Detection of mitochondrial SIRT1 and PGC-1α in mitochondria purified from HeLa cells and mouse organs and in human platelets. A, scheme for purification of mitochondria from HeLa and mouse organs. B, 10 μg of proteins obtained from HeLa purified mitochondria (Mito) and nuclei-enriched pellets (P1) were subjected to SDS-PAGE followed by Western blot analysis. The possible presence of nuclear protein contaminants was measured by incubating nitrocellulose membrane with rabbit anti-H2B. The effective mitochondria isolation was assessed by staining with rabbit anti-TFAM and mouse anti-cytochrome c oxidase sub IV (COX IV). PGC-1α and SIRT1 were detected by using rabbit Santa Cruz Biotechnology antibodies. C, 10 μg of proteins obtained from nuclei-enriched pellets (P1) and mitochondria (Mito) isolated from mice brain (B), skeletal muscle (M), and liver (L) were subjected to SDS-PAGE followed by Western blot analysis. The possible presence of nuclear protein contaminants was measured by incubating nitrocellulose membrane with rabbit anti-lamin B. The effective mitochondrial isolation was assessed by staining with rabbit anti-SOD2. PGC-1α and SIRT1 were detected by using Santa Cruz Biotechnology antibodies. D, human platelets were purified from peripheral blood, and 20 μg of protein extracts were subjected to SDS-PAGE followed by Western blot analysis. The possible presence of nuclear proteins was measured by incubating the nitrocellulose membrane with rabbit anti-H2B. PGC-1α and SIRT1 were detected by using Santa Cruz Biotechnology antibodies. Immunoblots reported are from one experiment representative of at least three that gave similar results.
Isolation of Native and Cross-linked Nucleoids
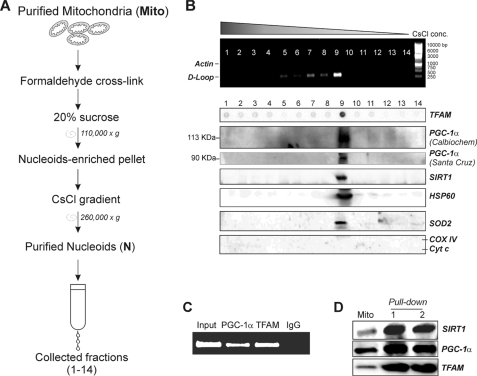
Native nucleoids were purified on a sucrose gradient according to Kaufman et al. (15) with the following modification: before centrifugation on a sucrose gradient, isolated mitochondria were homogenized with a glass Dounce potter. Cross-linked nucleoids were isolated on a CsCl gradient according to Garrido et al. (23) with some modifications. In brief, isolated mitochondria were cross-linked with 1% formaldehyde and homogenized in a glass Dounce potter, and nucleoid-enriched pellets were recovered by 20% sucrose centrifugation. The pellet was treated with RNase I for 1 h and charged on CsCl (RI = 1.365) in TE buffer with 1% Sarkosyl. Gradients were formed by centrifugation at 260,000 × g overnight in a Beckman SW41Ti rotor. The gradient was divided into 14 fractions that were collected from the bottom of the tube. A scheme of the procedure is shown in Fig. 4.
FIGURE 4.
Assay of the presence of SIRT1 and PGC-1α in cross-linked mitochondrial nucleoids. A, scheme for purification of nucleoids (N) from mouse liver mitochondria. Fractions (1–14) were finally collected by pricking the bottom of the tube. B, presence of mtDNA in the collected fractions was assayed performing PCR analysis of D-loop region. The presence of genomic DNA was excluded by performing PCR analysis of actin gene. After cross-linking reversion, fractions were subjected to SDS-PAGE. Western blot analysis with mouse anti-cytochrome c oxidase subunit IV (COX IV) and mouse anti-cytochrome c (Cyt c) was carried out to determine the possible presence of protein contaminants. SIRT1, HSP60, TFAM, and SOD2 were detected by using specific rabbit antibodies. PGC-1α was detected by using both the Santa Cruz Biotechnology and the Calbiochem antibodies. C, cross-linked mitochondria from mouse liver were sonicated and immunoprecipitated with goat anti-TFAM or goat anti-PGC-1α antibody (Santa Cruz Biotechnology). DNA was extracted, and PCR analysis of the region +15,600/+15,868 of mtDNA was successively carried out. Input and immunoprecipitation with a control IgG were used as positive and negative controls. D, mitochondrial protein extracts from HEK293 cells were analyzed by oligonucleotide pulldown assay using TFAM biotinylated consensus oligonucleotide and Western blot analysis using Santa Cruz Biotechnology anti-TFAM, anti-SIRT1, and anti-PGC-1α antibody. Mito, total mitochondrial lysates; lanes 1 and 2, two independent oligonucleotide pulldown experiments. Immunoblots reported are from one experiment representative of at least three that gave similar results.
Platelet Isolation
Peripheral human blood from four healthy donors was layered on top of Histopaque 1077 (Sigma), and cells were separated by centrifugation according to the manufacturer's instructions. The ring containing leukocytes and platelets was collected and centrifuged at 800 rpm for 10 min. The supernatant containing platelets was centrifuged three times at 800 rpm for 10 min to eliminate leukocytes (pellet). Platelets were then pelleted by centrifugation at 1500 rpm for 10 min. The presence of leukocyte contamination was excluded by visualization through an optical microscope.
Western Blotting and Dot Blot
Isolated mitochondria or platelets were lysed in RIPA buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 12 mm deoxycholic acid, 0.5% Nonidet P-40) containing protease inhibitor mixture (Sigma). Fractions from sucrose gradient were first dialyzed against TE buffer. Samples from CsCl gradient were incubated at 65 °C overnight in the presence of 1% SDS for cross-linking reversion. Protein samples were used for SDS-PAGE followed by Western blotting. Nitrocellulose membranes were stained with the following: mouse cytochrome c oxidase sub-IV mAb (1:500) (Molecular Probes); rabbit SOD2 pAb (1:2000) (Upstate Biotechnology, Inc.); mouse PGC-1α mAb (1:1000) (Calbiochem); mouse SIRT1 mAb (1:1000) (Cell Signaling); rabbit HSP60 pAb (1:2000); rabbit H2B pAb (1:1000); rabbit PGC-1α pAb (clone H-300) (1:1000); rabbit SIRT1 pAb (clone H-300) (1:1000); rabbit Lamin B pAb (1:500) (Santa Cruz Biotechnology); and rabbit TFAM pAb (1:1000) (kind gift from Prof. Dan Bogenhagen, Stony Brook University, Stony Brook NY). Afterward, the membranes were incubated with the appropriate horseradish peroxidase-conjugate secondary antibody (Bio-Rad), and immunoreactive bands were detected by a Fluorchem Imaging system (Alpha Innotech Corp.).
PGC-1α was detected by Western blot with the following two alternative antibodies: mouse mAb from Calbiochem and rabbit pAb from Santa Cruz Biotechnology. The majority of the experiments was performed with the antibody from Santa Cruz Biotechnology as it displayed a more efficient cross-reactivity toward mouse PGC-1α (see below).
For dot blot, 25 μl of fractions derived from sucrose gradients were spotted on nitrocellulose membrane using a Bio-Rad Dot Blot Apparatus (Bio-Rad) according to the manufacturer's instruction. Nitrocellulose membrane was stained with goat TFAM pAb (clone E-16) (1:500) (Santa Cruz Biotechnology).
DNA Extraction and PCR
The analysis of mtDNA after CsCl and sucrose gradient was performed as follows. 50 μl of each fraction obtained by the CsCl gradient was diluted once with TE buffer, extracted two times with phenol/chloroform/isoamyl alcohol, 25:24:1 (Sigma), and precipitated overnight with 0.1 volume of 3 m sodium acetate and 2 volumes of 2-propanol. Precipitates were washed with 75% ethanol and resuspended in 10 μl of TE buffer. Alternatively, after dialysis 50 μl of the fractions obtained from sucrose gradient were used for DNA extraction. Primers (Sigma) (forward, 5′-TCA GAC ATC T GG TTC TTA CT T CAG-3′, and reverse, 5′-CAT GAA TAA TTA GCC TTA GGT GAT-3′) gave an amplicon of 263 bp from the D-loop region of mouse mtDNA (+15,783/+16,045). PCR cycle was performed as follows: 95 °C for 2 min, 25 cycles (95 °C for 30 s; 59 °C for 30 s; 72 °C for 1 min), with final extension at 72 °C for 5 min. PCR product was loaded on 1.2% agarose gel electrophoresis.
Reverse Transcription-PCR Analysis
Total RNAs were prepared using TRIzol® reagent (Invitrogen) according to the manufacturer's instructions and used for the detection of PGC-1α mRNAs in semi-quantitative reverse transcription-PCR. GAPDH was used as internal control. Briefly, 3 μg of total RNA was retrotranscripted in the presence of random primers and Moloney murine leukemia virus according to the manufacturer's instructions. PCRs of PGC-1α and GAPDH cDNA (30 cycles of 15 s melting at 95 °C, 30 s annealing at 56 °C, and 30 s of extension at 72 °C) were performed with Platinum TaqDNA polymerase (Invitrogen) using the primers listed below: PGC-1α forward, 5′-AATTCACAATCACAGGATCAGAACA-3′, and PGC-1α reverse, 5′-ACTTAAGGTGCGTTCAATAGTCTT-3′; GAPDH forward, 5′-CTCCTCCACCTTTGACGCTG-3′, and GAPDH reverse, 5′-CCACCCTGTTGCTGTAGCCA-3′. The products obtained were of 465 bp for PGC-1α and 100 bp for GAPDH.
mtDNA Immunoprecipitation
mtDNA immunoprecipitation was performed according to Kucej et al. (24) with some modifications. Briefly, after Percoll® gradient purification and DNase I treatment, mitochondria from mouse liver were washed and collected. Mitochondria were cross-linked with 1% formaldehyde at 4 °C and shared on ice using a Branson Sonifier 250 (Branson Ultrasonix Corp.) (output 20%, 10 times for 25 s, with a 20-s pause each time). The fragmentation of mtDNA (about 500 bp) was verified by agarose gel electrophoresis after rapid cross-linking reversion (95 °C for 30 min). Samples were then pre-cleared with protein A/G-agarose (Santa Cruz Biotechnology) for 1 h at 4 °C, and 500 μg/ml proteins were immunoprecipitated overnight at 4 °C using 4 μg of goat anti-PGC-1α or goat anti-TFAM (Santa Cruz Biotechnology) as positive control. As negative control, immunoprecipitation was performed with control goat IgG. Immunocomplexes were precipitated for 2 h at 4 °C using 20 μl of protein A/G-agarose in the presence of 5 μg of shared salmon sperm DNA (Sigma) to reduce unspecific DNA-bead interactions. Immunoprecipitates were washed under stringent conditions (50 mm Tris-HCl, pH 7.4, 500 mm NaCl, 2 mm EDTA) and incubated overnight at 65 °C in the presence of 1% SDS for cross-linking reversion. DNA was extracted from supernatants, and 50 ng of the obtained mtDNA was used for PCR experiments. Primers (Sigma) (forward, 5′-GGA CAT ATC TGT GTT ATC TGA CAT-3′, and reverse, 5′-CCG ATA CCA TCG AGA TGT CTT A-3′) (+15,600/15,868) gave an Amplicon of 269 bp from the mouse mtDNA D-loop region. PCR was performed as described above.
Oligonucleotide Pulldown Assay
The assay was performed essentially as described previously by Westerheide et al. (25). Mitochondria were purified from HEK293 cells and lysed in cold lysis buffer (25 mm HEPES, pH 7.4, 100 mm NaCl, 5 mm EDTA, 20 mm β-glycerophosphate, 20 mm p-nitrophenyl phosphate, 0.5% Triton X-100, 20 mm 100 μm sodium orthovanadate) containing protease inhibitors. Protein extracts were incubated with 1 μg of human TFAM consensus sequence human mitochondrial transcription factor-binding site H (MT-TFH) biotinylated at 5′ (5′-CGCTGCTAACCCCATACCCCGA-3′ and 5′-TCGGGGTATGGGGTTAGCAGCG-3′), and proteins were allowed to bind the oligonucleotide for 30 min at room temperature. The oligonucleotides were precipitated with UltraLink streptavidin beads (Pierce) for 1 h at 4 °C. Bound fractions were washed three times with wash buffer (20 mm Tris-HCl, pH 7.5, 1 mm EDTA, 10% glycerol, 0.1% Triton X-100), eluted with denaturing buffer, and analyzed by Western blotting.
Blue Native-PAGE (BN-PAGE)
6–20% BN-PAGE and second dimension 10% SDS-PAGE were carried out on proteins from purified liver mitochondria according to Camacho-Carvajal et al. (26). This technique allows finely separating very high molecular weight multiprotein complexes.
Microscopy Analyses
Cells were seeded directly on glass coverslips and after 24 h were fixed with 4% paraformaldehyde and permeabilized by incubation with 0.2% Triton X-100. For analysis of mitochondrial localization of SIRT1 and PGC-1α and SIRT1-PGC-1α interaction, SH-SY5Y cells were incubated with mouse cytochrome c mAb (1:200) and rabbit SIRT1 pAb (1:100) or rabbit PGC-1α pAb (1:100) or goat PGC-1α pAb (clone K-15) (Santa Cruz Biotechnology). After staining with the appropriate AlexaFluor®-conjugated secondary antibodies (1:1000) (Invitrogen), fluorescence images were acquired by Olympus IX 70 equipped with Nanomover® and Softworx DeltaVision (Applied Precision Inc.) with a U-PLAN-APO 60× objective. Incubation with Hoechst 33342 (Invitrogen) was carried out to visualize nuclei. The images presented were captured under constant exposure time, gain, and offset. To improve the contrast and resolution of digital images captured in the microscope, they were elaborated by deconvolution software.
For co-localization of PGC-1α and SIRT1 with TFAM, HeLa cells were incubated with goat TFAM pAb (1:100) and rabbit PGC-1α pAb (1:100) or rabbit SIRT1 pAb (1:100) (Santa Cruz Biotechnology, CA). Images were visualized with an Olympus Fluoview 1000 confocal laser scanning system. In addition, Pearson's correlation coefficient, R(r), was calculated for fluorescence intensities of both the confocal channels. This coefficient describes the correlation between the intensity distribution, or pattern overlap, in two channels in terms of a least squares fit. This value can be between −1 and 1, and R = 1 indicates complete correlation between the two channels. R(r) values higher than 0.5 (50% of co-localization) were considered significant.
For identification of the association of PGC-1α and SIRT1 with mtDNA, HeLa cells were metabolically labeled with BrdUrd for 6 h and stained with anti-BrdUrd antibody (Promega, UK) according to Garrido et al. (23). Protein concentration was determined by the method of Lowry et al. (27).
Statistical Analysis
The results are presented as means ± S.D. Statistical evaluation was conducted by Student's t test. Differences were considered to be significant at p < 0.05.
RESULTS
Validation of the Antibodies Used for Detection of PGC-1α
In this study, we utilized different antibodies against PGC-1α for accuracy in the detection of the protein. In particular, we checked the efficacy of the anti-PGC-1α mouse monoclonal antibody (from Calbiochem) and the rabbit polyclonal antibody (from Santa Cruz Biotechnology) raised against amino acids 1–120 and 1–300, respectively. The results reported in supplemental Fig. 1A show that in total extracts of SH-SY5Y neuroblastoma cells, a band at 113 kDa was detected by the Calbiochem antibody, and on the same sample, a prominent band at 90 kDa was detected by the Santa Cruz Biotechnology antibody. These different molecular weights for PGC-1α are in line with those reported by the manufacturers and most probably are due to diverse forms of the protein, which could be profoundly modified by post-transcriptional processes, including the splicing and/or post-translational processes (e.g. acetylated, phosphorylated, etc.) (5, 28–30). However, to be more convinced that the two bands correspond to PGC-1α, we used two different approaches. On the one hand, we overexpressed the mouse PGC-1α using a pSV-PGC1 vector in SH-SY5Y cells. Western blot analysis revealed that the band both at 90 and 113 kDa was efficiently increased indicating that the antibodies used really recognized different forms of PGC-1α (supplemental Fig. 1B). On the other hand, we down-regulated PGC-1α by transiently transfecting SH-SY5Y cells with siRNA against PGC-1α (siPGC-1α). As negative control, we used a scrambled sequence matching no intracellular mRNA (siscr). The supplemental. Fig. 1C shows that PGC-1α mRNA content was efficiently down-regulated, and Western blot analyses revealed that both the 90- and the 113-kDa bands were down-regulated, confirming that they correspond to PGC-1α.
PGC-1α and SIRT1 Are Localized within Mitochondria
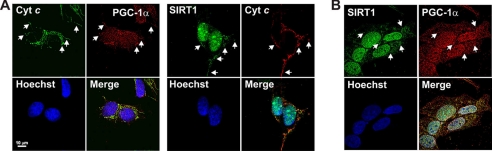
SIRT1 and PGC-1α are considered nuclear proteins, and to date only their effect on transcription of nucleus-encoded mitochondrial target genes has been documented (31). We explored whether SIRT1 and PGC-1α could be localized within mitochondria by analyzing their distribution in paraformaldehyde-fixed SH-SY5Y cells thorough an “Olympus Delta Vision” epifluorescence microscope equipped with a Nanomover®. These cells were selected on the basis of their well defined mitochondrial network. As observed in the images elaborated by deconvolution software and reported in the Fig. 1A, PGC-1α and SIRT1 fluorescence matches with that of Hoechst 33342 according to their well known nuclear localizations. Surprisingly, these proteins also exhibit a heterogeneous distribution in the cytoplasmic milieu that nicely coincides with mitochondrial lattice as evidenced by cytochrome c counterstaining. We also verified whether the well known PGC-1α-SIRT1 interaction was operative inside the mitochondria. The images illustrated in Fig. 1B show that, in SH-SY5Y cells, fluorescence signals derived from PGC-1α and SIRT1 significantly overlapped both at the cytoplasmic and nuclear levels. It is important to point out that an alternative goat anti-PGC-1α was used in Fig. 1B. Such antibody equally evidenced a distribution of PGC-1α outside the nucleus, suggesting that the cytoplasmic fluorescent signal reported in Fig. 1A was not due to a nonspecific staining.
FIGURE 1.
Detection of mitochondrial SIRT1 and PGC-1α by immunofluorescence microscopy in SH-SY5Y cells. A, mitochondrial network and nuclei were evidenced by incubation with mouse anti-cytochrome c and Hoechst 33342, respectively. PGC-1α or SIRT1 was stained by using rabbit antibodies (Santa Cruz Biotechnology). B, PGC-1α and SIRT1 were stained by using goat anti-PGC-1α (Santa Cruz Biotechnology) and rabbit anti-SIRT1. Cells were analyzed by an Olympus Delta vision deconvolution fluorescent microscope. The yellow-orange color obtained by image merge indicates the regions where the green and red signals superimpose. White arrows indicate some regions of co-localization signals. Scale bar, 10 μm.
To confirm the mitochondrial localization of PGC-1α and SIRT1, we carried out Western blot analysis of both PGC-1α and SIRT1 on purified mitochondria from HeLa cells and mouse muscle, brain, and liver. Fig. 2A shows the procedure of mitochondria purification. In brief, HeLa cells or mice organs were homogenized in a Teflon potter and centrifuged to separate cytoplasm (S1) from nuclei and unbroken cells (P1). S1 was then centrifuged to collect mitochondria-enriched fraction (P2). Finally, P2 was layered on top of 30% Percoll® solution, and mitochondria were separated from associated microvesicles through a Percoll® self-forming gradient according to the method of Pellon-Maison et al. (22). Although isolated mitochondria were treated with DNase I, the possible contamination of mitochondria with nuclei components was excluded by carrying out Western blot analysis of two of the most abundant nuclear proteins, i.e. H2B or lamin B for HeLa and mouse organs, respectively (Fig. 2, B and C). High amounts of mitochondrial proteins, such as subunit IV of cytochrome c oxidase (COX IV), SOD2, and TFAM, were observed in mitochondria from HeLa cells and the screened mice organs, suggesting that these organelles were efficiently isolated. Immunoblot analysis was then carried out to confirm the presence of PGC-1α and SIRT1 inside the mitochondria using rabbit anti-SIRT1 and anti-PGC-1α from Santa Cruz Biotechnology. Protein extracts of purified mitochondria from HeLa cells, mouse brain, muscle, and liver displayed the presence of these proteins (Fig. 2, B and C). SIRT1 was also detected by using an alternative antibody from Calbiochem that recognized the same protein band (data not shown). Moreover, the results reported in supplemental Fig. 1D show that in mitochondria from mouse liver both the bands of 113 and 90 kDa were detected by using the two different anti-PGC-1α antibodies (Santa Cruz Biotechnology and Calbiochem, as described previously). Moreover, the Calbiochem antibody cross-reacts with additional proteins in the mitochondria of mouse liver, one of which could be ascribed to the 90-kDa PGC-1α (supplemental Fig. 1D). The 113- and the 90 kDa-bands were also obtained on isolated mitochondria from HeLa cells supplemental Fig. 1E, confirming that the same electrophoretic pattern of PGC-1α was present in total extracts and mitochondria.
We then looked at the possible presence of PGC-1α and SIRT1 in purified human platelets that are known to be cells completely void of nuclei, and thus the presence of the nuclear PGC-1α and SIRT1 could be excluded. The immunoblots reported in Fig. 2D demonstrate that purification of human platelets was properly achieved without any presence of leukocyte contamination. In fact, platelet protein extracts do not contain any nuclear components, as demonstrated by the absence of H2B. On the contrary, mitochondrial protein such as TFAM and SOD2 are highly expressed. Fig. 2D also shows that SIRT1 and PGC-1α are expressed in platelets derived from different health subjects, indicating that such proteins are normally localized also inside mitochondria.
SIRT1 and PGC-1α Are Present in Mitochondrial Nucleoids
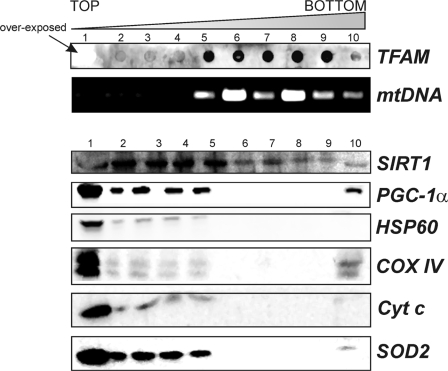
Given the results obtained in the in vitro and in vivo mitochondrial localization of PGC-1α and SIRT1, and the knowledge of the function of these proteins as regulators of gene expression, we speculated on a possible association of these proteins with nucleoids. Actually, nucleoids are functional structures localized in the mitochondrial matrix containing mtDNA packaged with several proteins belonging to mitochondrial transcriptional machinery, including the mitochondrion-specific transcription factor TFAM, and are considered the mitochondrial units of inheritance (13, 23). Isolated liver mitochondria were lysed, and after centrifugation, the nucleoid-enriched pellet was layered on a sucrose gradient according to Garrido et al. (23). After sucrose gradient centrifugation, nucleoid-containing fractions were identified by assessing the content of both TFAM and mtDNA, and the purity was verified by detecting the presence of non-nucleoid mitochondrial protein such as cytochrome c and COX IV by Western blot analysis (Fig. 3). Moreover, other proteins, such as HSP60 and SOD2, with which association with nucleoids is still debated, were also analyzed. Nucleoids were contained in fractions 6–9 because such fractions concomitantly stained positively with TFAM antibody and showed the presence of mtDNA. However, TFAM was mainly localized in fraction 1 according to its presence in mitochondria also as mtDNA-unbound protein. The complete absence of cytochrome c and COX IV in fractions 6–9 excluded the presence of protein contaminants in fractions containing nucleoids. These non-nucleoids proteins were instead particularly abundant in fraction 1 and were present to a minor extent in fractions 2–5 and 10. The same trend was evidenced for SOD2 and HSP60 indicating that they are not likely associated with liver native nucleoids. Being both SOD2 and HSP60 matrix proteins, a possible matrix-protein contamination in the nucleoid containing fractions was finally excluded. Surprisingly, SIRT1 but not PGC-1α was found natively associated with nucleoids. In fact, SIRT1 was found in fractions 6–9.
FIGURE 3.
Assay of the presence of SIRT1 and PGC-1α in mitochondrial native nucleoids. Native nucleoids were obtained from isolated liver mitochondria as described under “Experimental Procedures.” After sucrose gradient, nucleoid-containing fractions were identified by detecting the presence of TFAM by dot blot (using goat anti-TFAM) and mtDNA by PCR analysis of the D-loop region. Western blot analyses with mouse anti-cytochrome c oxidase subunit IV (COX IV), mouse anti-cytochrome c (Cyt c), rabbit anti-SOD2, and rabbit anti-HSP60 were carried out to determine the possible presence of protein contaminants. Fractions 6–9 were considered to contain native nucleoids due to the absence of protein contaminants and to the presence of TFAM and mtDNA. PGC-1α and SIRT1 were detected by using Santa Cruz Biotechnology antibodies. Immunoblots reported are from one experiment representative of at least three that gave similar results.
The absence of PGC-1α in native nucleoids did not exclude that it could be weakly or indirectly associated with mtDNA. For this reason, liver mitochondria were cross-linked with 1% formaldehyde and pelleted through 20% sucrose followed by CsCl gradient (Fig. 4A). The putative nucleoid-containing fraction(s) was identified by assaying the presence of mtDNA and TFAM through PCR analysis of D-loop and dot blot analysis of TFAM protein, respectively (Fig. 4B). The presence of genomic DNA was excluded by performing PCR analysis of actin. Western blot analysis of cytochrome c and COX IV was also performed on each collected fraction to exclude the presence of protein contaminants in nucleoid-containing fractions (Fig. 4B). Cytochrome c and COX IV were not present in any fraction, strongly indicating that the majority of mitochondrial protein contaminants was already eliminated after centrifugation on 20% sucrose (Fig. 4A). As shown in Fig. 4B, among the collected fractions, fraction 9 contains the higher level of both TFAM and mtDNA strongly indicating that in such fractions purified nucleoids were included. Fraction 9 also positively stained for HSP60, which is recognized as protein packaged into cross-linked nucleoids (Fig. 4B) (15). As expected, SIRT1 was present in fraction 9 containing cross-linked nucleoids (Fig. 4B). Moreover, Western blot analysis carried out with rabbit antibody from Santa Cruz Biotechnology indicated that PGC-1α was present in fraction 9, suggesting that also PGC-1α is a component of the nucleoid structure. The same result was obtained by performing Western blot analysis of PGC-1α with the mouse monoclonal antibody from Calbiochem. Also in this case, the two antibodies recognized PGC-1α at different molecular masses (90 and 113 kDa, respectively). The fraction containing purified nucleoids was also loaded in parallel with a mitochondrial lysate to corroborate both the specificity of the antibodies used and the goodness of nucleoid purification (supplemental Fig. 2).
In searching for the role of PGC-1α within nucleoids, we asked whether PGC-1α could interact at the level of the mtDNA D-loop region. To this end, isolated cross-linked mitochondria were immunoprecipitated with anti-PGC-1α, anti-TFAM (positive control), or an IgG isotype (negative control). As expected, PCR analysis of +15,600/+15,868 mtDNA showed that TFAM is highly associated with the D-loop region. No PCR product was detectable in samples immunoprecipitated with the IgG isotype. The presence of D-loop region was identified in samples immunoprecipitated with anti-PGC-1α, suggesting the association of PGC-1α with mtDNA (Fig. 4C). These data were confirmed by performing an oligonucleotide-based pulldown assay in which mitochondrial protein extracts were incubated with a TFAM biotinylated consensus oligonucleotide. After oligonucleotide pulldown, samples were subjected to Western blot analysis. Fig. 4D shows that TFAM protein was efficiently precipitated together with PGC-1α and SIRT1, suggesting that these proteins in some way have the ability to interact with TFAM at the level of DNA.
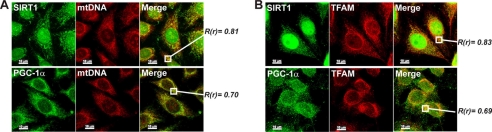
To further demonstrate that SIRT1 and PGC-1α were associated with nucleoids, mtDNA of HeLa cells was metabolically labeled by BrdUrd incorporation and stained with an antibody against BrdUrd. SIRT1 or PGC-1α staining evidenced a high degree of co-localization of mtDNA with these two proteins especially in the perinuclear region, where mitochondria are principally distributed in HeLa cells (Fig. 5A). However, although SIRT1 displayed the expected preponderant nuclear localization, PGC-1α did not, letting us postulate that the lower rate of PGC-1α localization into the nucleus may be a characteristic of this tumor cell line. The extent of co-localization was estimated by calculation of Pearson's correlation coefficient. This analysis gave scores of 0.81 and 0.70 for SIRT1/mtDNA and PGC-1α/mtDNA, respectively. TFAM is a protein that was found to be acetylated on a single lysine residue in rat liver (32). Because TFAM is a transcription factor, it was reasonable to postulate not only a possible modulation by SIRT1 but also by PGC-1α in a similar way to that operating at nuclear levels. Thus, we first evaluated the possible interaction of SIRT1 and/or PGC-1α with TFAM by confocal microscopy. As reported in Fig. 5B, TFAM staining was mainly visualized in the perinuclear region and as an intricate cytoplasmic network according to its mitochondrial localization. We found that fluorescence of SIRT1 and PGC-1α overlapped well with that of TFAM at the mitochondrial level, and calculation of Pearson's correlation coefficient finally demonstrated that they are co-localized with TFAM (0.83 and 0.69, respectively).
FIGURE 5.
Determination of the association of SIRT1 and PGC-1α with mtDNA and TFAM by confocal microscopy in HeLa cells. A, mtDNA was labeled by BrdUrd incorporation followed by methanol-acetone fixing and immunostaining with mouse anti-BrdUrd. SIRT1 or PGC-1α was also immunostained using Santa Cruz Biotechnology antibodies. Cells were then visualized by confocal microscopy, and the level of superimposition of SIRT1 or PGC-1α-derived signals with that of BrdUrd was evaluated by calculation of the Pearson's correlation coefficient (R(r)). Values >0.50 were considered to be significant. B, HeLa cells were fixed with paraformaldehyde and immunostained with goat anti-TFAM and rabbit Santa Cruz Biotechnology anti-PGC-1α or SIRT1. Cells were then visualized by confocal microscopy, and the level of superimposition of SIRT1 or PGC-1α-derived signals with that of TFAM was evaluated by calculation of the Pearson's correlation coefficient (R(r)). Values >0.50 were considered to be significant. Scale bar, 10 μm.
PGC-1α and SIRT1 Interact with TFAM to Form Multiprotein Complexes in Mitochondria
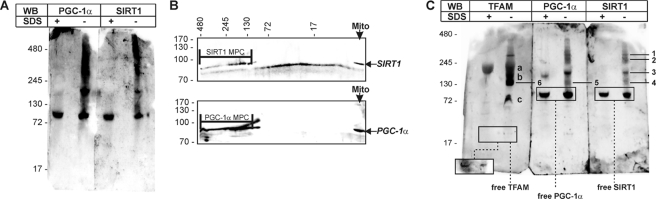
The above reported results prompted us to analyze whether PGC-1α and SIRT1 could participate in mitochondria in the formation of native multiprotein complexes. Purified mitochondria were lysed in a buffer able to preserve native interactions between components of multiprotein complexes and subjected to BN-PAGE followed by Western blot analysis. The same samples were loaded in the presence of SDS in parallel to visualize denatured PGC-1α and SIRT1. The immunoblots reported in Fig. 6A shows that both proteins are present as free factors in mitochondrial lysates and also form protein macro-complexes. A superimposable pattern of anti-SIRT1 and anti-PGC-1α immunoreactive bands was evidenced. It is worth reminding that although mouse SIRT1 and PGC-1α have different molecular masses (90 and 80 kDa, respectively), the BN-PAGE technique used could not allow finely separating these two proteins (under such conditions the difference between the two Rf values should be 0.05 cm). To confirm the specificity of the interactions found in the native samples, we carried out a second dimension denaturing SDS-PAGE followed by Western blot analysis of PGC-1α or SIRT1. A large lane of spots in SDS second dimension corresponding to PGC-1α and SIRT1 proteins was found (Fig. 6B), confirming the existence of several native complexes in which these proteins are engaged.
FIGURE 6.
Identification of mitochondrial multiprotein complexes by BN-PAGE in liver purified mitochondria. A, mitochondrial proteins were extracted from liver purified mitochondria (mito) using a lysis buffer maintaining native interaction in multiprotein complexes. Ten μg of proteins were loaded on blue native gel. The same samples were boiled in the presence of SDS and loaded in parallel with the respective native sample. Western blot (WB) analysis using rabbit Santa Cruz Biotechnology anti-SIRT1 or anti-PGC-1α was then performed. B, after BN-PAGE, a second SDS-PAGE dimension was carried out followed by Western blot analysis using rabbit Santa Cruz Biotechnology anti-PGC-1α or anti-SIRT1. MPC, multiprotein complexes. C, 10 μg of proteins were loaded on blue native gel, and Western blot using a rabbit anti-TFAM was carried out. 1, multiprotein complex containing TFAM, PGC-1α, and SIRT1; 2 and 3, multiprotein complexes containing PGC-1α and SIRT1; 4, multiprotein complex containing TFAM, and SIRT1; 5 multiprotein complex containing TFAM and PGC-1α; 6, multiprotein complex containing TFAM-PGC-1α and TFAM-SIRT1; a–c, other TFAM multiprotein complexes. To visualize free TFAM protein, a major exposure was mandatory due to the lower cross-reactivity of TFAM antibody against native TFAM with respect to TFAM engaged in multiprotein complexes (see inset in TFAM immunoblot). Immunoblots reported are from one experiment representative of four that gave similar results.
To confirm the interaction of TFAM with SIRT1 and PGC-1α evidenced by confocal microscopy, we also carried out Western blot analysis of TFAM in parallel with PGC-1α and SIRT1 after BN-PAGE (Fig. 6C). An immunoreactive band of ∼130 kDa was detected in all the blots, reasonably representing protein complexes containing at least PGC-1α/TFAM and SIRT1/TFAM. The immunoreactive band having the highest molecular mass (>245 kDa) was found in all immunoblots, indicating that SIRT1, PGC-1α, and TFAM are parts of a larger mitochondrial multiprotein complex. A band of molecular mass between 130 and 245 kDa was present exclusively in PGC-1α and SIRT1 but not in TFAM immunoblot, indicating that it could contain the SIRT1-PGC1α protein complex. Other protein interactions exclusive for TFAM were further evidenced.
DISCUSSION
In this study, we demonstrated that SIRT1 and PGC-1α, which have long been considered nuclear proteins participating in regulation of transcription of nucleus-encoded mitochondrial genes, are also distributed inside mitochondria. We demonstrated that SIRT1 and PGC-1α are present in mitochondria of cells of tumor origin, such as HeLa, HEK293, and SH-SY5Y cells, as well as in human platelets (notably nuclei-free cells) and mitochondria from mouse organs. In particular, these proteins have been found in liver, brain, and skeletal muscle mitochondria.
Moreover, the different antibodies used to detect PGC-1α demonstrated that it exists inside the cells at different molecular masses, and in particular, we mainly evidenced two different bands at 90 and 113 kDa, which were both modulated by either overexpression or down-regulation. The diverse molecular mass forms of PGC-1α were in line with those reported by the manufacturers and most probably due to post-transcriptional and/or post-translational processes (28, 29). Work is in progress in our laboratory to identify the modifications responsible for such molecular weights difference.
During mitochondrial biogenesis, SIRT1 participates inside the nucleus in the induction of PGC-1α that is pivotal to orchestrate the activation of a broad set of transcription factors and nuclear hormone receptors enhancing the expression of the nucleus-encoded mitochondrial genes. The presence of SIRT1 and PGC-1α within the mitochondria suggests that these proteins could exert similar functions in this compartment. Among the putative targets of their actions, we focused on TFAM as, being a transcription factor, it can be likely modulated by co-activator(s) and by an acetylation/deacetylation mechanism. Data from Dinardo et al. (32) in part support this assumption as they demonstrated that TFAM is acetylated at a single lysine residue, a modification that is supposed to have a role in regulating the expression of mitochondrial genes or in general the mtDNA maintenance. Also, unpublished results from our laboratory indicate that in liver mitochondria TFAM is present both in acetylated and de-acetylated status.3 To date, neither a deacetylase nor a co-activator has been identified to modulate TFAM activity. The presence of SIRT1 in samples precipitated after incubation with oligonucleotide representing TFAM consensus sequence strongly indicates a possible action of SIRT1 on TFAM. Purification of liver nucleoids allowed us to discover that SIRT1 as well as TFAM are natively associated with nucleoids, whereas PGC-1α is not. These observations led us to speculate that, at least in its nucleoid-associated state, SIRT1 could be the factor responsible for modulation of TFAM acetylation level. We are confident that the presence of SIRT1 in native nucleoids is not due to contamination, because mitochondrial HSP60, which is considered a protein contaminant in native purified nucleoids (13), and other non-nucleoid associated proteins were not detected. Moreover, the cross-reactivity of the SIRT1 antibody (recognizing an ∼90-kDa band) with mitochondrial SIRT3–5 has to be excluded because these family members have very lower molecular weights with respect to SIRT1 as reported in several protein databases (e.g. Swiss Prot and TrEMBL). The evidence that SIRT1 has been recently recognized to be located in cytoplasm of cardiomyocytes (33) strongly supports our findings about the presence of SIRT1 inside mitochondria. Interestingly, our data also indicate that SOD2 is not associated with liver native nucleoids as was instead recently demonstrated for Jurkat cells by Kienhöfer et al. (14). However, in line with our results, the same authors demonstrated that SOD2 could be absent in native nucleoids of other cell types, such as bovine endothelial cells. The results obtained from mitochondria cross-linking confirm that SOD2 is in some way associated with nucleoids and further support the role of this antioxidant enzyme in ensuring mtDNA integrity.
The absence of PGC-1α in natively purified nucleoids is in agreement with its inability to directly bind DNA. In contrast, its presence in cross-linked nucleoids nicely correlates with its function as a transcriptional co-activator also in mitochondria. Results obtained by confocal images confirmed the association of both PGC-1α and SIRT1 with nucleoids, as a very significant Pearson's correlation coefficient of such proteins with mtDNA was observed.
We demonstrate that PGC-1α and SIRT1 are present intramitochondrially as free proteins and also interact with TFAM. In mitochondria, SIRT1 and PGC-1α form several multiprotein complexes among which are included PGC-1α/SIRT1, TFAM/PGC-1α, and TFAM/SIRT1. Moreover, both PGC-1α and SIRT1 reasonably participate with TFAM in the formation of a higher molecular weight multiprotein complex, which could correspond to macro-complexes involved in mtDNA transactions. We also have strong evidence that PGC-1α could positively influence the activity of human TFAM at least on the D-loop region. Not only the oligonucleotide pulldown assay but also data from mtDNA immunoprecipitation analysis demonstrated that TFAM is bound in association with SIRT1 and/or PGC-1α to mtDNA.
The way in which SIRT1 and PGC-1α are included in mitochondria remains obscure, having no known mitochondrial import sequences, and it deserves further investigation. However, an import mechanism similar to that of other mitochondrial proteins, mediated by the translocases of outer and inner mitochondrial membrane, may be operative (3).
The presence of SIRT1 in the mitochondrial compartment is not in contrast with that of mitochondria resident sirtuins (SIRT3–5), which have been uniquely assigned to regulating metabolic mitochondrial enzyme activity (34), and at the same time it reinforces the role of SIRT1 in modulating DNA remodeling and transcription.
All together, our results suggest that SIRT1 and PGC-1α have to be considered not only nuclear but also mitochondrial imported proteins that are associated with nucleoids. The data obtained also indicate that in this submitochondrial region, SIRT1 and PGC-1α could function as regulators of TFAM possibly mediating its deacetylation and mtDNA binding activity. On the basis of such findings, other avenues are now open, For instance, is the acetylation/deacetylation status of PGC-1α and TFAM responsible for their association with nucleoids and for modulating mtDNA transcription? Are mitochondrial SIRT1 and PGC-1α the effective mediators of the cross-talk between cellular metabolism and mitochondrial biogenesis?
Finally, this work strengthens the role of SIRT1/PGC-1α pathway in regulation of mitochondrial biogenesis and metabolism and gives new inputs in the comprehension of the regulatory mechanisms governing mitochondrial genome expression, replication, and maintenance. This unexpected localization also highlights the intriguing perspective that alterations in mitochondrial residents PGC-1α and SIRT1 could be causative of mitochondrial homeostasis impairment underlying pathophysiological processes, including aging, metabolic syndromes, myopathies, and neurodegenerative diseases.
Supplementary Material
Acknowledgments
We gratefully acknowledge Palma Mattioli and Elena Romano (Dept. of Biology, University of Rome Tor Vergata, Rome, Italy) for their assistance in fluorescent and confocal microscopy and images analysis. We thank Dr. Dan Bogenhagen (Stony Brook University, Stony Brook, NY) for the kind gift of polyclonal rabbit TFAM antibody. We also thank Dr. Bruce Spiegelman (Dept. of Medicine, University of Chicago) for providing pSV-PGC-1α vector insert in Addgene deposit plasmid. We thank Dr. Teresa C. Leone for helpful discussions about PGC-1α antibody utilization. We are also grateful to the animal house staff of the University of Rome “Tor Vergata.”
This work was supported in part by grants from Ministero della Salute and MIUR and by Fondazione Roma Research Grant 2008 (to P. V.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
K. Aquilano, P. Vigilanza, S. Baldelli, B. Pagliei, G. Rotilio, and M. R. Ciriolo, unpublished results.
- TFAM
- mitochondrial transcription factor A
- BrdUrd
- 5-bromo-2-deoxyuridine
- BN-PAGE
- blue native-PAGE
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- pAb
- polyclonal antibody
- mAb
- monoclonal antibody
- siRNA
- small interfering RNA
- siscr
- scramble small interference RNA.
REFERENCES
- 1.Hock M. B., Kralli A. (2009) Annu. Rev. Physiol. 71, 177–203 [DOI] [PubMed] [Google Scholar]
- 2.Chow L. S., Greenlund L. J., Asmann Y. W., Short K. R., McCrady S. K., Levine J. A., Nair K. S. (2007) J. Appl. Physiol. 102, 1078–1089 [DOI] [PubMed] [Google Scholar]
- 3.Scarpulla R. C. (2008) Physiol. Rev. 88, 611–638 [DOI] [PubMed] [Google Scholar]
- 4.Guarente L. (2007) Cold Spring Harbor Symp. Quant. Biol. 72, 483–488 [DOI] [PubMed] [Google Scholar]
- 5.Rodgers J. T., Lerin C., Gerhart-Hines Z., Puigserver P. (2008) FEBS Lett. 582, 46–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng H. L., Mostoslavsky R., Saito S., Manis J. P., Gu Y., Patel P., Bronson R., Appella E., Alt F. W., Chua K. F. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 10794–10799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brunet A., Sweeney L. B., Sturgill J. F., Chua K. F., Greer P. L., Lin Y., Tran H., Ross S. E., Mostoslavsky R., Cohen H. Y., Hu L. S., Cheng H. L., Jedrychowski M. P., Gygi S. P., Sinclair D. A., Alt F. W., Greenberg M. E. (2004) Science 303, 2011–2015 [DOI] [PubMed] [Google Scholar]
- 8.Nemoto S., Fergusson M. M., Finkel T. (2005) J. Biol. Chem. 280, 16456–16460 [DOI] [PubMed] [Google Scholar]
- 9.Knutti D., Kralli A. (2001) Trends Endocrinol. Metab. 12, 360–365 [DOI] [PubMed] [Google Scholar]
- 10.Finkel T. (2006) Nature 444, 151–152 [DOI] [PubMed] [Google Scholar]
- 11.Wang Y., Bogenhagen D. F. (2006) J. Biol. Chem. 281, 25791–25802 [DOI] [PubMed] [Google Scholar]
- 12.Kang D., Kim S. H., Hamasaki N. (2007) Mitochondrion 7, 39–44 [DOI] [PubMed] [Google Scholar]
- 13.Bogenhagen D. F., Rousseau D., Burke S. (2008) J. Biol. Chem. 283, 3665–3675 [DOI] [PubMed] [Google Scholar]
- 14.Kienhöfer J., Häussler D. J., Ruckelshausen F., Muessig E., Weber K., Pimentel D., Ullrich V., Bürkle A., Bachschmid M. M. (2009) FASEB J. 23, 2034–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaufman B. A., Newman S. M., Hallberg R. L., Slaughter C. A., Perlman P. S., Butow R. A. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 7772–7777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bogenhagen D. F. (1985) J. Biol. Chem. 260, 6466–6471 [PubMed] [Google Scholar]
- 17.Puigserver P., Wu Z., Park C. W., Graves R., Wright M., Spiegelman B. M. (1998) Cell 92, 829–839 [DOI] [PubMed] [Google Scholar]
- 18.Kawakami Y., Tsuda M., Takahashi S., Taniguchi N., Esteban C. R., Zemmyo M., Furumatsu T., Lotz M., Belmonte J. C., Asahara H. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 2414–2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aquilano K., Vigilanza P., Rotilio G., Ciriolo M. R. (2006) FASEB J. 20, 1683–1685 [DOI] [PubMed] [Google Scholar]
- 20.Sun F. C., Wei S., Li C. W., Chang Y. S., Chao C. C., Lai Y. K. (2006) Biochem. J. 396, 31–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frezza C., Cipolat S., Scorrano L. (2007) Nat. Protoc. 2, 287–295 [DOI] [PubMed] [Google Scholar]
- 22.Pellon-Maison M., Montanaro M. A., Coleman R. A., Gonzalez-Baró M. R. (2007) Biochim. Biophys. Acta 1771, 830–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garrido N., Griparic L., Jokitalo E., Wartiovaara J., van der Bliek A. M., Spelbrink J. N. (2003) Mol. Biol. Cell 14, 1583–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kucej M., Kucejova B., Subramanian R., Chen X. J., Butow R. A. (2008) J. Cell Sci. 121, 1861–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Westerheide S. D., Anckar J., Stevens S. M., Jr., Sistonen L., Morimoto R. I. (2009) Science 323, 1063–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Camacho-Carvajal M. M., Wollscheid B., Aebersold R., Steimle V., Schamel W. W. (2004) Mol. Cell. Proteomics 3, 176–182 [DOI] [PubMed] [Google Scholar]
- 27.Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. (1951) J. Biol. Chem. 193, 265–275 [PubMed] [Google Scholar]
- 28.Chang J. S., Huypens P., Zhang Y., Black C., Kralli A., Gettys T. W. (2010) J. Biol. Chem. 285, 18039–18050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Handschin C., Spiegelman B. M. (2006) Endocr. Rev. 27, 728–735 [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y., Huypens P., Adamson A. W., Chang J. S., Henagan T. M., Boudreau A., Lenard N. R., Burk D., Klein J., Perwitz N., Shin J., Fasshauer M., Kralli A., Gettys T. W. (2009) J. Biol. Chem. 284, 32813–32826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin J. D. (2009) Mol. Endocrinol. 23, 2–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dinardo M. M., Musicco C., Fracasso F., Milella F., Gadaleta M. N., Gadaleta G., Cantatore P. (2003) Biochem. Biophys. Res. Commun. 301, 187–191 [DOI] [PubMed] [Google Scholar]
- 33.Tanno M., Kuno A., Yano T., Miura T., Hisahara S., Ishikawa S., Shimamoto K., Horio Y. (2010) J. Biol. Chem. 285, 8375–8382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schlicker C., Gertz M., Papatheodorou P., Kachholz B., Becker C. F., Steegborn C. (2008) J. Mol. Biol. 382, 790–801 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.