Abstract
We have previously demonstrated that cyclic ADP-ribose (cADPR) is a calcium signaling messenger in interleukin 8 (IL-8)-induced lymphokine-activated killer (LAK) cells. In this study we examined the possibility that IL-8 activates CD38 to produce another messenger, nicotinic acid adenine dinucleotide phosphate (NAADP), in LAK cells, and we showed that IL-8 induced NAADP formation after cADPR production. These calcium signaling messengers were not produced when LAK cells prepared from CD38 knock-out mice were treated with IL-8, indicating that the synthesis of both NAADP and cADPR is catalyzed by CD38 in LAK cells. Application of cADPR to LAK cells induced NAADP production, whereas NAADP failed to increase intracellular cADPR levels, confirming that the production of cADPR precedes that of NAADP in IL-8-treated LAK cells. Moreover, NAADP increased intracellular Ca2+ signaling as well as cell migration, which was completely blocked by bafilomycin A1, suggesting that NAADP is generated in lysosome-related organelles after cADPR production. IL-8 or exogenous cADPR, but not NAADP, increased intracellular cAMP levels. cGMP analog, 8-(4-chlorophenylthio)-guanosine 3′,5′-cyclic monophosphate, increased both cADPR and NAADP production, whereas the cAMP analog, 8-(4-chlorophenylthio)-cAMP, increased only NAADP production, suggesting that cAMP is essential for IL-8-induced NAADP formation. Furthermore, activation of Rap1, a downstream molecule of Epac, was required for IL-8-induced NAADP formation in LAK cells. Taken together, our data suggest that IL-8-induced NAADP production is mediated by CD38 activation through the actions of cAMP/Epac/protein kinase A/Rap1 in LAK cells and that NAADP plays a key role in Ca2+ signaling of IL-8-induced LAK cell migration.
Keywords: Receptors/Chemokine, Signal Transduction/Calcium, Signal Transduction/Cyclic Nucleotides, Calcium Intracellular Release, NAD, CD38, Cyclic ADP-ribose, Nicotinic Acid Adenine Dinucleotide Phosphate
Introduction
A type II transmembrane protein, CD38, possesses ADP-ribosyl cyclase (ADPR cyclase)3 and cyclic ADP-ribose hydrolase (cADPR hydrolase) activity (1, 2). These two enzyme activities are involved in the conversion of β-nicotinamide adenine dinucleotide (β-NAD+) first to cADPR and then to ADPR (3–5). The metabolite cADPR is known to increase intracellular Ca2+ concentration, [Ca2+]i, by releasing Ca2+ from intracellular stores or by Ca2+ influx through plasma membrane Ca2+ channels in a variety of cells (6–10). It was shown that CD38 can also synthesize NAADP from NADP in the presence of nicotinic acid by a base exchange reaction in vitro (11). However, it still remains unclear whether the base exchange reaction occurs physiologically as intracellular nicotinic acid concentration is less than the millimolar concentration that is required for the enzymatic synthesis of nicotinic acid adenine dinucleotide phosphate (NAADP) in vitro (12).
NAADP is a potent Ca2+-releasing messenger in a variety of cell types, including mammalian cells (13–15). Although d-myo-inositol 1,4,5-trisphosphate (IP3) and cADPR are firmly established as secondary Ca2+ messengers, receptor-mediated formation of NAADP has been shown in a limited number of cellular systems (16–18). It has been demonstrated that NAADP triggers Ca2+ release from thapsigargin-insensitive Ca2+ stores through the activation of channels distinct from those sensitive to ryanodine and IP3 (19). In sea urchin eggs, NAADP releases Ca2+ from acidic Ca2+ stores, lysosome-related organelles (20). However, NAADP can also release Ca2+ from the endoplasmic reticulum (21–23).
Previously, we have reported that IL-8 stimulated cADPR formation by activation of CD38 via cGMP/protein kinase G and induced an increase of [Ca2+]i and migration of LAK cells (24). In this study we investigated whether NAADP is involved in IL-8-induced Ca2+ signaling and migration of LAK cells. We showed that NAADP plays a key role in IL-8-stimulated long-lasting Ca2+ signaling in cell migration and that IL-8-induced NAADP formation by CD38 is mediated through the actions of cAMP/Epac/PKA/Rap1 after cADPR formation in LAK cells.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
Antibodies were obtained as follows: anti-mouse CD38 monoclonal antibody was from BD Biosciences; anti-Rap1 polyclonal antibody (pAb) was from Upstate Biotechnology (Temecula, CA); anti-TRPM2 pAb was from Abcam (Cambridge, UK); anti-mouse CD38 pAb, anti-Epac pAb, anti-PKA pAb, and horseradish peroxidase-conjugated anti-goat IgG or anti-rabbit IgG were from Santa Cruz Biotechnology (Santa Cruz, CA). Nylon wool was from Polysciences Inc. (Warrington, PA), and human recombinant IL-2 was from Chiron BV (Amsterdam, Netherlands). Xestospongin C, 8-pCPT-cAMP, and 8-pCPT-cGMP were purchased from Calbiochem. 8-pCPT-2′-O-Me-cAMP, N6-benzoyl-cAMP, Rp-8-Br-cAMPS, and N-(p-amylcinnamoyl) anthranilic acid (ACA) were from Biolog Life Science Institute (Bremen, Germany). Human recombinant IL-8, cADPR, NAADP, 8-Br-cADPR, bafilomycin A1, glycylphenylalanine-2-naphthylamide, thapsigargin, SQ22,536 and all other reagents were obtained from Sigma. RPM1 1640 medium, fetal bovine serum, and antibiotics were from Invitrogen. Transwells were purchased from Coring Costar Corp. (Cambridge, MA). The cAMP immunoassay kit was from Assay Designs (Boston, MA).
Animals
CD38 knock-out mice (Cd38−/−; B6.129P2-Cd38tm/Lud) were purchased from The Jackson Laboratory (Bar Harbor, ME). Mice were bred and kept in animal housing facilities at Chonbuk National University Medical School under specific pathogen-free conditions. All experimental animals were used under a protocol approved by the institutional animal care and user committee of the Chonbuk National University Medical School. Standard guidelines for laboratory animal care were followed (25).
Preparation of LAK Cells
LAK cells were prepared as described previously (26). Briefly, spleens of mice were chopped, and then splenocytes were harvested. Red blood cells were lysed by incubation with red blood cell lysis buffer (0.15 m MH4Cl, 10 mm KHCO3, 0.1 mm EDTA (pH 7.2)), and the cells were then washed with serum-free RPMI 1640 twice. The red blood cell-removed cell preparations were incubated on a nylon-wool column at 37 °C for 1 h in a 5% CO2 incubator to remove B lymphocytes and macrophages. Nylon-wool nonadherent cells were collected and incubated at 2 × 106 cells/ml density in a culture media containing 3000 IU/ml IL-2 in a 5% CO2 incubator at 37 °C. The culture media used was RPMI 1640 supplemented with 10% fetal bovine serum, 0.25 μg/ml amphotericin B, 10 units/ml penicillin G, 100 μg/ml streptomycin, 1 mm l-glutamine, 1% nonessential amino acids, and 50 μm 2-mercaptoethanol. After incubation for 4 days, the floating cells were removed, and the adherent cells were cultured in the same culture media containing 3000 IU/ml IL-2. LAK cells induced by IL-2 for 8 days were used throughout the study.
Measurement of Intracellular cADPR Concentration ([cADPR]i)
The level of [cADPR]i was measured using a cyclic enzymatic assay as described previously (27). Briefly, cells were treated with 0.5 ml of 0.6 m perchloric acid under sonication. Precipitates were removed by centrifugation at 20,000 × g for 10 min. Perchloric acid was removed by mixing the aqueous sample with a solution containing three volumes of 1,1,2-trichlorotrifluoroethane to one volume of tri-n-octylamine. After centrifugation at 1500 × g for 10 min, the aqueous layer was collected and neutralized with 20 mm sodium phosphate (pH 8). To remove all contaminating nucleotides, the samples were incubated overnight with the following hydrolytic enzymes at 37 °C: 0.44 unit/ml nucleotide pyrophosphatase, 12.5 units/ml alkaline phosphatase, 0.0625 unit/ml NAD glycohydrolase, and 2.5 mm MgCl2 in 20 mm sodium phosphate buffer (pH 8.0). Enzymes were removed by filtration using Centricon 3 filters. To convert cADPR to β-NAD+, the samples (0.1 ml/tube) were incubated with 50 μl of a cycling reagent containing 0.3 μg/ml Aplysia ADPR cyclase, 30 mm nicotinamide, and 100 mm sodium phosphate (pH 8.0) at room temperature for 30 min. The Aplysia ADPR cyclase was purified as described (28). The samples were further incubated with the cycling reagent (0.1 ml) containing 2% ethanol, 100 μg/ml alcohol dehydrogenase, 20 μm resazurin, 10 μg/ml diaphorase, 10 μm riboflavin 5′-phosphate, 10 mm nicotinamide, 0.1 mg/ml bovine serum albumin (BSA), and 100 mm sodium phosphate (pH 8.0) at room temperature for 2 h. An increase in the resorufin fluorescence was measured at 544-nm excitation and 590-nm emission using a fluorescence plate reader (Molecular Devices Corp., Spectra-Max GEMINI). Various known concentrations of cADPR were also included in the cycling reaction to generate a standard curve.
Measurement of Intracellular NAADP Concentration ([NAADP]i)
The level of [NAADP]i was measured using a cyclic enzymatic assay as described previously (29). Briefly, cells were treated with perchloric acid, and precipitates were removed by centrifugation. Perchloric acid was removed by mixture of 1,1,2-trichlorotrifluoroethane and tri-n-octylamine, and the aqueous layer was collected and neutralized with 20 mm sodium phosphate (pH 8.0) under the exact conditions that were described above. To remove all contaminating nucleotides, the samples were incubated overnight with the following hydrolytic enzymes at 37 °C: 2.5 units/ml apyrase, 0.125 unit/ml NAD glycohydrolase, 2 mm MgCl2, 1 mm NaF, 0.1 mm PPi, and 0.16 mg/ml NMN-adenylyl transferase in 20 mm sodium phosphate buffer (pH 8.0). Enzymes were removed by filtration using Centricon 3 filters. After the hydrolytic treatment, alkaline phosphate (10 units/ml) was added to convert NAADP to NAAD overnight at 37 °C. The alkaline phosphate was removed by filtration using Centricon 3 filters. The samples were further incubated with the cycling reagent (30 μl) containing 2% ethanol, 100 μg/ml alcohol dehydrogenase, 20 μm resazurin, 10 μg/ml diaphorase, 10 μm riboflavin 5′-phosphate, 10 mm nicotinamide, 0.1 mg/ml BSA, and 100 mm sodium phosphate (pH 8.0) at room temperature for 4 h. An increase in the resorufin fluorescence was measured at 544-nm excitation and 590-nm emission using a fluorescence plate reader (Spectra-Max GEMINI). Various known concentrations of NAADP were also included in the cycling reaction to generate a standard curve.
Measurement of cADPR and NAADP Transport
Time courses of cADPR and NAADP influx into LAK cells were measured by a rapid oil-stop procedure as described previously (30, 31). The influx assay was initiated by the rapid addition of cADPR or NAADP to a 100-μl portion of cell suspension (1 × 106 cells) layered over 100 μl of oil (silicone oil/paraffin oil, 80:20) and ended by pelleting the cells at 16,000 × g for 30 s. Transported cADPR and NAADP into pelleted cells were measured using a cyclic enzymatic assay as described (27, 29).
Measurement of [Ca2+]i
Cells were washed with Hanks' balanced salt solution (2 mm CaCl2, 145 mm NaCl, 5 mm KCl, 1 mm MgCl2, 5 mm d-glucose, 20 mm HEPES (pH 7.3)) containing 1% BSA and incubated for 6 h in the same solution containing 1% BSA. Starved LAK cells were incubated with 5 μm Fluo-3 AM (Molecular Probes, Eugene, OR) in Hanks' balanced salt solution containing 1% BSA at 37 °C for 40 min. The cells were washed three times with Hanks' balanced salt solution. Changes in [Ca2+]i in LAK cells were determined at 488-nm excitation/530-nm emission by air-cooled argon laser system (32). The emitted fluorescence at 530 nm was collected using a photomultiplier. One image was scanned every 6 s for 10 min using a confocal microscope (Nikon, Japan). For the calculation of [Ca2+]i, the method of Tsien et al. (33) was used with the following equation: [Ca2+]i = Kd(F − Fmin)/(Fmax − F), where Kd is 450 nm for Fluo-3, and F is the observed fluorescence levels. Each tracing was calibrated for the maximal intensity (Fmax) by the addition of ionomycin (8 μm) and for the minimal intensity (Fmin) by the addition of EGTA (50 mm) at the end of each measurement.
Immunoprecipitation and Western Blotting
Cells were washed with phosphate-buffered saline and then lysed with an ice-cold lysis buffer (20 mm HEPES (pH 7.2), 1% Triton X-100, 10% glycerol, 100 mm NaCl, 1 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, 50 mm NaF, 1 mm Na3VO4, 10 μg/ml leupeptin, 10 μg/ml pepstatin, and 10 μg/ml aprotinin). After centrifugation at 20,000 × g for 10 min, supernatants were taken. For immunoprecipitation, cell lysate (1 mg) precleared with protein G-agarose was incubated with anti-CD38 mAb overnight at 4 °C and then further incubated with protein G-agarose at 4 °C for 1 h. The immunoprecipitates were washed 4 times with cell lysis buffer and then boiled at 95 °C for 10 min. The immunoprecipitated proteins were subjected to SDS-PAGE in a 10% (CD38 and Epac) and 13% (Rap1) gel. After transferring to nitrocellulose membranes, blots were incubated in blocking buffer (10 mm Tris-HCl (pH 7.6), 150 mm NaCl, 0.05% Tween 20) containing 5% nonfat dry milk for 2 h at room temperature and then with primary antibodies (dilution factors were: CD38, 1:1000; Epac, 1:2000; PKA, 1:2000; Rap1, 1:2000) in the blocking buffer overnight at 4 °C. The blots were rinsed four times with the blocking buffer and incubated with horseradish peroxidase-conjugated anti-goat IgG (1:2500 dilution) or anti-rabbit IgG (1:2500 dilution) in the blocking buffer at room temperature for 1 h. The immunoreactive proteins with respective secondary antibodies were determined using an enhanced chemiluminescence kit (Amersham Biosciences) and exposed to an LAS 1000 Image Reader Lite (Fujifilm, Japan). Protein concentrations were determined using a Bio-Rad protein assay kit, and known concentrations of BSA were used as the standard.
Rap1 Pulldown Assay
Rap1 activity was assayed with a Rap1 activation assay kit (Upstate Biotechnology) according to the manufacturer's protocol. LAK cells starved in serum-free medium overnight were stimulated with various reagents. Stimulation was terminated by adding equal volumes of 2× lysis buffer (100 mm Tris-HCl (pH 7.2), 2% Nonidet P-40, 10% glycerol, 1 m NaCl, 5 mm MgCl2, 2 mm phenylmethylsulfonyl fluoride, 20 mm NaF, 2 mm Na3VO4, 20 μg/ml leupeptin, 20 μg/ml pepstatin, and 20 μg/ml aprotinin). After centrifugation at 20,000 × g for 10 min at 4 °C, supernatant (1 mg) was incubated with 15 μg of agarose-RalGDS-Rap binding domain conjugates for 2 h. Agarose was washed three times with lysis buffer. The precipitates were separated by 13% SDS-PAGE and analyzed by immunoblotting with anti-Rap1 antibody. Fifteen μg from each lysate was used to immunodetect total Rap1.
Knockdown of TRPM2 and Rap1 by Small Interference RNA (siRNA)
Rap1 siRNA, TRPM2 siRNA, and control siRNA were purchased from Santa Cruz Biotechnology. LAK cells (2 × 105) were cultured in an antibiotic-free growth media supplemented with fetal bovine serum. After 24 h, LAK cells were transfected with 60 pmol of siRNA oligonucleotides specific to TRPM2 or Rap1 using transfection reagent without antibiotics according to the instructions of the manufacturer. After 48 h of transfection, cells were prepared for examination.
Measurement of Intracellular cAMP Level
Levels of cAMP were determined by a cAMP immunoassay kit according to the manufacturer's protocol. LAK cells starved in serum-free medium overnight were preincubated with 0.5 mm isobutylmethylxanthine, a phosphodiesterase inhibitor, and challenged with various reagents detailed in figure legends. After incubation for the indicated times, the cells were lysed in 0.1 m HCl to stop the reaction. After centrifugation at 20,000 × g for 10 min at 4 °C, supernatants were collected and acetylated, and then immunoassays were performed. A standard curve of acetylated cAMP was also prepared as described in the manufacturer's protocol.
Determination of Cell Migration
Cell migration was determined as described previously (34). In brief, cells were washed with RPMI 1640, scraped using a policeman, and washed with RPM1 1640. Transwells with 8-μm pore-size polycarbonate filters were used. Lower chambers contained 500 μl of RPMI 1640 containing 1% BSA. LAK cells (4 × 105) in 100 μl of RPM1 1640 containing various agents (specified in the figure legend) were placed in the upper chamber. The chambers were incubated in a 5% CO2 incubator at 37 °C for 2 h. After removing the unattached cells, the filters were removed, fixed with ice-cold 100% methanol, and stained with 15% Wright-Giemsa stain for 7 min. The cells were counted under a phase-contrast microscope.
Statistical Analysis
Data represent the means ± S.E of at least three separate experiments. Statistical analysis was performed using Student's t test. A value of p < 0.05 was considered significant.
RESULTS
Treatment of LAK Cells with IL-8 Induces the Production of NAADP by CD38
We previously demonstrated that IL-8-mediated cADPR production through activation of CD38 is essential for the migration of LAK cells (24). In this study we examined whether NAADP is produced in IL-8-treated LAK cells. As shown in Fig. 1, treatment of LAK cells with IL-8 significantly increased intracellular NAADP levels compared with the control. To further determine whether CD38 is responsible for the production of NAADP or not, we measured NAADP levels in LAK cells prepared from CD38 knock-out mice (Cd38−/−) before and after IL-8 treatment. LAK cells from Cd38−/− failed to form NAADP at all by the treatment with IL-8 (Fig. 1). This result suggests that NAADP is produced via CD38 activation in LAK cells when treated with IL-8.
FIGURE 1.
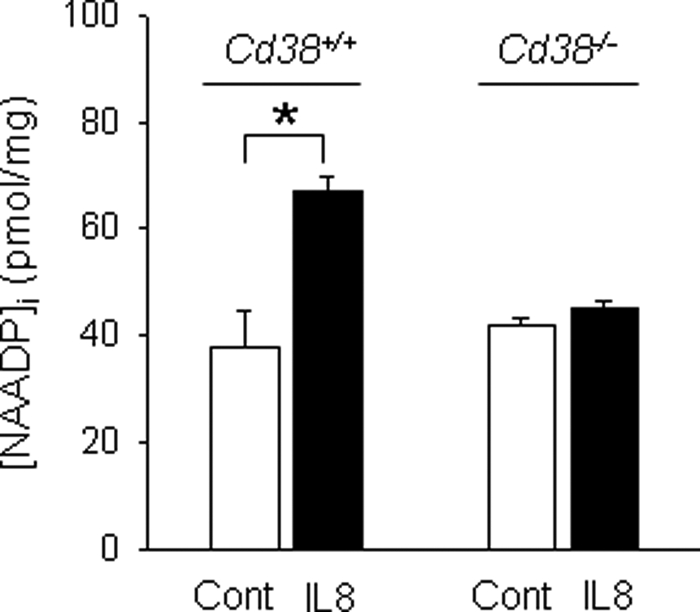
Treatment of LAK cells with IL-8 induces the production of NAADP by CD38. LAK cells prepared from CD38+/+ and Cd38−/− were treated with 10 pm IL-8 for 90 s, and then the concentrations of NAADP formed were measured with the cyclic enzymatic assay. The means ± S.E. of five independent experiments are shown. *, p < 0.05, control (CONT) versus IL-8.
IP3-mediated Ca2+ Increase Is Required for IL-8-mediated cADPR/NAADP Production and Ca2+ Signaling in LAK Cells
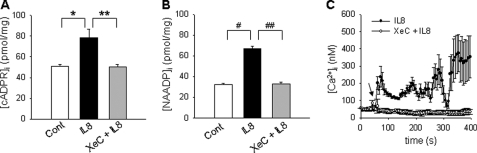
It has been demonstrated that IL-8-mediated cADPR formation was inhibited by xestospongin C (XeC), an IP3 receptor antagonist, in LAK cells (24, Fig. 2A). To study the effect of IP3-mediated Ca2+ increase on IL-8-mediated NAADP production, we measured the levels of NAADP in LAK cells treated with IL-8 after preincubation with XeC. As shown in Fig. 2B, IL-8-mediated formation of NAADP was completely blocked by pretreatment of the cells with XeC. Thus, we next examined the effect of XeC on the IL-8-mediated Ca2+ signal. Pretreatment of LAK cells with XeC completely blocked the IL-8-mediated Ca2+ signal (Fig. 2C). These data suggest that initial Ca2+ increase by IP3 is essential for IL-8-induced cADPR/NAADP production and affects all the Ca2+ signals of IL-8-stimulatory LAK cells.
FIGURE 2.
IP3-mediated Ca2+ increase is required for IL-8-mediated cADPR/NAADP production and Ca2+ signaling in LAK cells. A and B, XeC inhibited IL-8-induced increase of [cADPR]i and [NAADP]i. Levels of cADPR and NAADP were determined after treatment of LAK cells with 10 pm IL-8. LAK cells were preincubated with 2 μm XeC for 15 min before incubation with IL-8. The means ± S.E. of three independent experiments are shown. * and #, p < 0.05, control versus IL-8; ** and ##, p < 0.005, IL-8 versus IL-8 plus XeC. C, IL-8-stimulated increase is shown in [Ca2+]i with (open circle) and without (closed circle) XeC. Arrows indicate the time points of the addition of IL-8.
IL-8 Induces NAADP Production after cADPR Formation
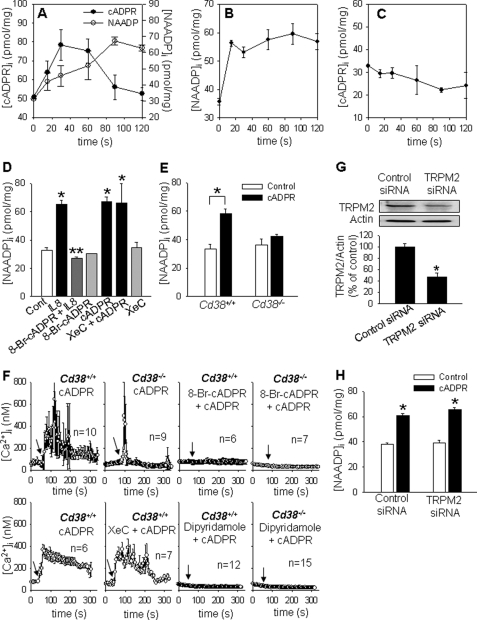
Next, we measured cADPR/NAADP formation at various time intervals after IL-8 treatment of LAK cells. As shown in Fig. 3A, cADPR and NAADP were increased in a time-dependent manner, reaching maximal levels at ∼30 and 90 s, respectively. This result shows a possibility that IL-8-induced cADPR formation precedes NAADP production. To investigate this possibility, we examined whether there is any interdependence between cADPR and NAADP production by taking advantage of transporting cADPR and NAADP to LAK cells (supplemental Fig. S1) like other cells known to be transported (35–37). The addition of exogenous cADPR to LAK cells enhanced intracellular NAADP levels, whereas the addition of NAADP failed to increase intracellular cADPR levels (Fig. 3, B and C), consistent with the above finding that cADPR formation precedes NAADP production. To further confirm the above finding, we examined the effect of 8-Br-cADPR, a cADPR antagonistic analog, on IL-8-mediated NAADP production. As shown in Fig. 3, D–F, 8-Br-cADPR completely inhibited IL-8-induced NAADP formation as well as cADPR-induced Ca2+ signal and NAADP formation, but XeC could not block cADPR-induced Ca2+ signal and NAADP formation because cADPR is downstream of IP3 on IL-8 signaling in LAK cells. Furthermore, cADPR did not induce NAADP formation as well as the long-lasting Ca2+ signal in LAK cells from Cd38−/−, compared with LAK cells from CD38+/+ (Fig. 3, E and F). We next examined the effect of dipyridamole, an inhibitor of transporters for cADPR and NAADP on cADPR- and NAADP-induced [Ca2+]i changes, in CD38+/+ or Cd38−/− LAK cells. As shown in Fig. 3F and supplemental Fig. S2, A and B, cADPR- as well as NAADP-induced [Ca2+]i increase was completely inhibited by dipyridamole. To further examine whether exogenous cADPR addition triggers NAADP production via the Ca2+-permeable cation channel, such as the TRPM2 channel (38), we measured NAADP formation by the addition of cADPR in TRPM2 knockdown LAK cells (Fig. 3G). As presented in Fig. 3H, cADPR-induced NAADP production was not abrogated in TRPM2 knockdown LAK cells, suggesting that exogenous cADPR acts on intracellular Ca2+ stores but not on the TRPM2 channel in LAK cells, triggering the pathway that results in NAADP formation. Consistent with this observation, ACA, a specific TRPM2 inhibitor, failed to inhibit IL-8-induced cADPR formation as well as NAADP formation (supplemental Fig. S3, A and B). Taken together, these data indicate that IL-8 sequentially induces the formation of cADPR and NAADP in LAK cells through CD38 and suggests that cADPR initially induces a transient Ca2+ signal, which in turn triggers NAADP production, resulting in a long-lasting Ca2+ signal.
FIGURE 3.
IL-8 induces NAADP production after cADPR formation. A, LAK cells were treated with 10 pm IL-8 for the indicated times, and then cADPR (closed circle) and NAADP (open circle) concentrations were measured. The means ± S.E. of three independent experiments are shown. B, after LAK cells were treated with 200 μm cADPR for the indicated times, the levels of NAADP were measured. The means ± S.E. of three independent experiments are shown. C, cADPR levels were measured in LAK cells treated with 50 nm NAADP for the indicated times. The means ± S.E. of three independent experiments are shown. D, IL-8-induced NAADP formation was blocked by 8-Br-cADPR. LAK cells were preincubated with 100 μm 8-Br-cADPR for 15 min before incubation with 10 pm IL-8 for 90 s. XeC (2 μm) was preincubated for 15 min before the treatment with cADPR (200 μm) for 30 s. The means ± S.E. of three independent experiments are shown. *, p < 0.05, control versus IL-8 or cADPR or cADPR plus XeC; **, p < 0.005, IL-8 versus IL-8 plus 8-Br-cADPR. Cont, control. E, NAADP levels of LAK cells, which were prepared from CD38+/+ and Cd38−/−, were measured after treatment with 200 μm cADPR for 30 s. The means ± S.E. of three independent experiments are shown. *, p < 0.05, control versus cADPR. F, cADPR-stimulated increase in [Ca2+]i was measured in LAK cells prepared from wild type and CD38 knock-out mice. Arrows indicate the time points of the addition of 200 μm cADPR. Before treatment with cADPR, 8-Br-cADPR (200 μm), XeC (2 μm), or dipyridamole (100 μm) was preincubated at 37 °C for 15 min. n indicates the number of cells examined for Ca2+ measurement. G, TRPM2 knockdown using siRNA is shown. A total of 60 pmol of siRNA was transfected into 5 × 105 cells using a transfection reagent. After 48 h of transfection, the cells were subjected to immunoblotting with anti-TRPM2 pAb (upper panel) and the densitometric changes of TRPM2 expression (lower panel) are shown. *, p < 0.05, control siRNA versus TRPM2 siRNA. H, NAADP formation by cADPR was not inhibited in TRPM2 knockdown LAK cells. NAADP formed was measured after the treatment with cADPR (200 μm) for 30 s in LAK cells. The means ± S.E. of three independent experiments are shown. *, p < 0.05, control versus cADPR.
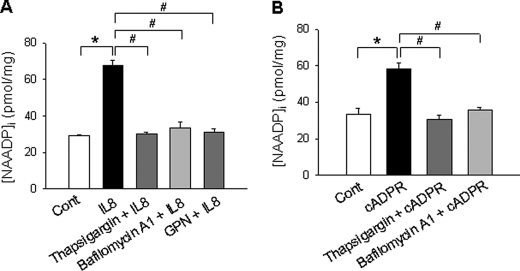
IL-8-induced NAADP Production Occurs in Acidic Organelles and Is Mediated by Ca2+ Release from Thapsigargin-sensitive Stores by cADPR
To determine the role of Ca2+ stores in IL-8-induced NAADP formation, we measured NAADP formation induced by IL-8 or cADPR in LAK cells in the presence of thapsigargin, a sarcoplasmic/endoplasmic reticulum Ca2+-ATPase (SERCA) inhibitor. Fig. 4 shows that thapsigargin significantly blocked IL-8- or cADPR-induced NAADP formation, indicating that Ca2+ release from thapsigargin-sensitive stores by cADPR is essential for NAADP production when the cells were treated with IL-8. Next, we tested whether lysosome-related organelles are involved in NAADP formation, as previous studies suggested that NAADP is formed in the acidic organelles. Therefore, we used two different lysosome-disturbing reagents, bafilomycin A1 and glycylphenylalanine-2-naphthylamide. As shown in Fig. 4, bafilomycin A1 and glycylphenylalanine-2-naphthylamide significantly blocked IL-8- or cADPR-induced NAADP formation, suggesting that lysosome-related organelles are the site of NAADP production induced by IL-8.
FIGURE 4.
IL-8-inducd NAADP formation is mediated by Ca2+ release from thapsigargin sensitive store by cADPR. A, LAK cells pretreated with bafilomycin A1 (1 μm), thapsigargin (10 μm), or glycylphenylalanine-2-naphthylamide (GPN, 50 μm) for 20 min were incubated with 10 pm IL-8 at 37 °C for 90 s. Then the levels of NAADP were measured as described under “Experimental Procedures.” The means ± S.E. of three independent experiments are shown. *, p < 0.05, control (Cont) versus IL-8; #, p < 0.005, IL-8 versus IL-8 plus thapsigargin or IL-8 versus IL-8 plus bafilomycin A1 or IL-8 versus IL-8 plus glycylphenylalanine-2-naphthylamide. B, LAK cells pretreated with bafilomycin A1(1 μm) or thapsigargin (10 μm) for 20 min were incubated with 200 μm cADPR. After incubation for 30 s, NAADP levels were measured as described under “Experimental Procedures.” The means ± S.E. of three independent experiments are shown. *, p < 0.05, control versus cADPR; #, p < 0.005, cADPR versus cADPR plus thapsigargin or cADPR versus cADPR plus bafilomycin A1.
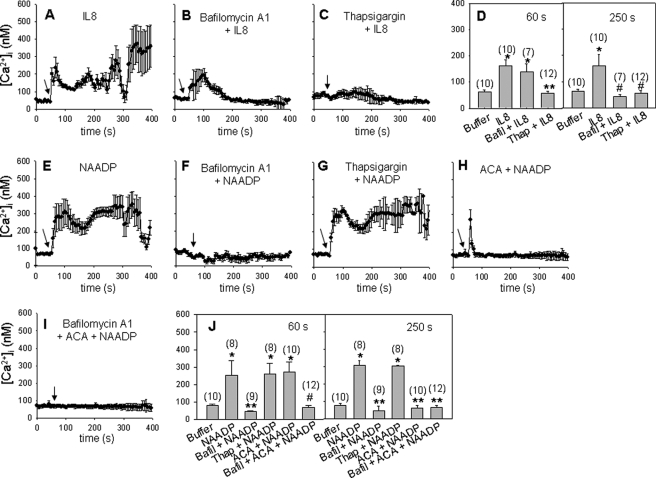
NAADP Releases Ca2+ from Lysosome-like Acidic Organelles in IL-8-induced Ca2+ Signaling
Pretreatment of LAK cells with bafilomycin A1 blocked IL-8-mediated sustained Ca2+ signaling (Fig. 5, A, B, and D), indicating that the late phase of sustained Ca2+ signal is mediated by NAADP through Ca2+ release from lysosome-related acidic organelles. Pretreatment of LAK cells with thapsigargin completely blocked IL-8-mediated Ca2+ signaling (Fig. 5, C and D), consistent with the above findings that initial Ca2+ signaling was essential for NAADP production. These findings were further confirmed by observing that NAADP-induced long-lasting Ca2+ signal was completely blocked by bafilomycin A1 but not by thapsigargin (Fig. 5, E–G and J). To rule out any possibility of the effect of exogenously added NAADP on purinoreceptors, we tested the effects of equimolar concentrations of NAD or NADP under the same condition. However, we could not find any significant signals with high concentration of the nucleotides (data not shown). It was reported that NAADP-mediated Ca2+ influx is triggered from the TRPM2 channel (38). To determine the role of TRPM2 on IL-8-induced Ca2+ signaling, we measured the change in [Ca2+]i in the presence of ACA, a specific TRPM2 channel blocker. As shown in Fig. 5, H and J, ACA prevented an NAADP-mediated sustained Ca2+ increase, but not NAADP-mediated initial Ca2+ increase. Similarly, the late phase of IL-8-induced Ca2+ signal (TRPM2 channel-mediated Ca2+influx) was blocked by the treatment with ACA but not the initial Ca2+ signal (Ca2+ mobilization) (supplemental Fig. S3C). On the other hand, bafilomycin A1 completely blocked NAADP-mediated initial Ca2+ release from acidic organelles and sustained Ca2+ influx in the presence of ACA (Fig. 5, I and J). These results indicate that NAADP-mediated Ca2+ release from acidic organelles triggers Ca2+ influx through the TRPM2 channel in IL-8-induced Ca2+ signaling.
FIGURE 5.
NAADP releases Ca2+ from lysosome-like acidic organelles in IL-8-induced Ca2+ signaling. A–I, LAK cells were loaded with Fluo-3, and the changes in Ca2+ level were measured. The time points of 10 pm IL-8 or 50 nm NAADP additions are indicated by the arrows. Cells were preincubated with 1 μm bafilomycin A1 (B) or 10 μm thapsigargin (C) for 20 min before IL-8 treatment. D, shown is a direct comparison of mean [Ca2+]i during increases of [Ca2+]i. The data shown are analyzed at 60 s. *, p < 0.05, buffer versus IL-8 or IL-8 plus bafilomycin A1 (Balfil); **, p < 0.001, IL-8 versus IL-8 plus thapsigargin (Thap). The data shown are analyzed at 250 s. *, p < 0.05, buffer versus IL-8; #, p < 0.01, IL-8 versus IL-8 plus bafilomycin A1 or IL-8 plus thapsigargin. Also, NAADP was applied after the cells were preincubated with 1 μm bafilomycin A1 (F) or 10 μm thapsigargin (G) or 20 μm ACA (H) for 20 min. J, shown is a direct comparison of mean [Ca2+]i during increases of [Ca2+]i. The data shown are analyzed at 60 s. *, p < 0.05, buffer versus NAADP or NAADP plus thapsigargin or NAADP plus ACA; **, p < 0.005, NAADP versus NAADP plus bafilomycin A1; #, p < 0.001, ACA plus NAADP versus ACA plus NAADP plus bafilomycin A1. The data shown are analyzed at 250 s. *, p < 0.05, buffer versus NAADP or NAADP plus thapsigargin; **, p < 0.001, NAADP versus NAADP plus bafilomycin A1 or NAADP plus ACA or NAADP plus ACA plus bafilomycin A1. Cell numbers are presented in the parentheses. Data are the mean ± S.E.
NAADP-mediated Signal Is Essential for IL-8-induced Migratory Activity of LAK Cells
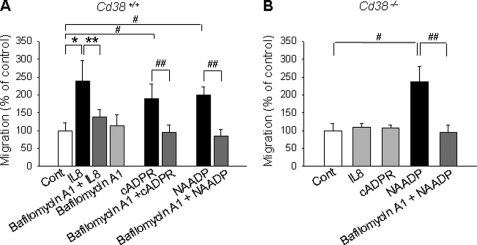
To examine whether NAADP plays any role in IL-8-mediated cell migration, we evaluated the effect of NAADP on the migratory activity of LAK cells. IL-8-mediated migration of LAK cells was blocked by pretreatment with bafilomycin A1 and ACA (Fig. 6A, supplemental Fig. S3D). Moreover, the addition of exogenous cADPR and NAADP to LAK cells stimulated cell migration, which was completely blocked by pretreatment of the cells with bafilomycin A1. In addition, cADPR- or NAADP-induced cell migration was significantly inhibited by dipyridamole (supplemental Fig. S2, C and D). Treatment of LAK cells that were isolated from Cd38−/− with IL-8 and cADPR did not induce cell migration (Fig. 6B), whereas treatment of LAK cells from Cd38−/− with NAADP induced cell migration. Furthermore, bafilomycin A1 completely blocked NAADP-induced migration of LAK cells, which were isolated from Cd38−/−. These results indicate that NAADP is produced in lysosome-like acidic organelles by CD38 and that the NAADP-mediated Ca2+ signal plays an important role in IL-8-mediated migration of LAK cells.
FIGURE 6.
NAADP-mediated signal is essential for IL-8-induced migratory activity of LAK cells. LAK cells, prepared from CD38+/+ (A) and Cd38−/− (B), were preincubated with 1 μm bafilomycin A1 for 20 min, and then the cells were incubated with 10 pm IL-8 or 200 μm cADPR or 50 nm NAADP. After incubation for 2 h, cell migration was assayed as described under “Experimental Procedures.” The means ± S.E. of three independent experiments are shown. *, p < 0.05, control (Cont) versus IL-8; **, p < 0.01, IL-8 versus IL-8 plus bafilomycin A1; #, p < 0.005, control versus cADPR or NAADP; ##, p < 0.05, cADPR versus cADPR plus bafilomycin A1 or NAADP versus NAADP plus bafilomycin A1.
cAMP and cGMP Are Mediators for Differential Production of cADPR and NAADP in IL-8-treated LAK Cells
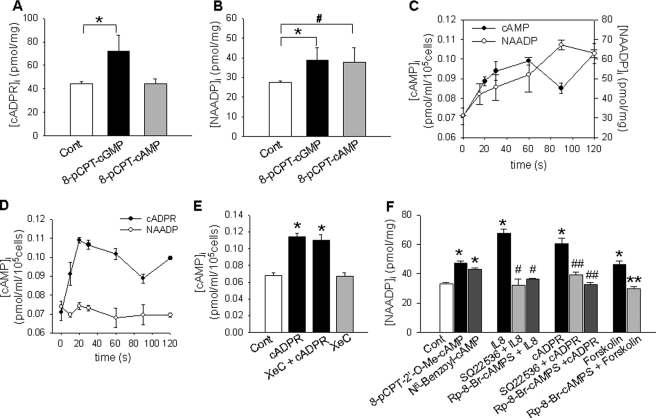
It has been reported that cAMP and cGMP differentially mediate the production of cADPR and NAADP in sea urchin eggs (39), and we previously demonstrated that cGMP regulated the production of cADPR through CD38 internalization, which was induced by an increase of the association CD38 with phosphorylated nonmuscle myosin IIA by protein kinase G (24, 40). Therefore, we examined whether cAMP and cGMP differentially mediate the production of cADPR and NAADP by CD38 in LAK cells. As presented in Fig. 7, A and B, 8-pCPT-cGMP induced the production of NAADP as well as cADPR, whereas 8-pCPT-cAMP induced NAADP production, but not cADPR production, suggesting that the production of NAADP by cAMP is triggered by the action of cADPR. Forskolin, an adenylyl cyclase activator, increased the level of NAADP but not cADPR (data not shown). Collectively, these data indicate that cGMP and cAMP differentially mediate the production of cADPR and NAADP by CD38 in IL-8-treated LAK cells, respectively. Because the above data showed that NAADP production was triggered by the action of cADPR and that the cAMP analog directly induced NAADP production, we asked the question of whether IL-8-stimulated cADPR formation induces NAADP production via cAMP formation. As presented in Fig. 7C, cAMP increase by IL-8 precedes NAADP production. Moreover, cADPR itself induced cAMP formation in a time-dependent manner, which was not blocked by XeC pretreatment, indicating that cADPR induces NAADP production through cAMP (Fig. 7, D and E). To further examine which molecules are involved in cAMP-induced NAADP production, we utilized specific activators of PKA and Epac. Treatment of LAK cells with 8-pCPT-2′-O-Me-cAMP (Epac activator) and N6-benzoyl-cAMP (PKA activator) increased the production of NAADP (Fig. 7F). Moreover, SQ22,536, an adenylate cyclase inhibitor, and Rp-8-Br-cAMPS, a specific PKA inhibitor, significantly inhibited the IL-8-, cADPR-, or forskolin-induced NAADP formation (Fig. 7F). These results indicate that these two cAMP-sensitive molecules play a role in cAMP-induced NAADP production.
FIGURE 7.
NAADP formation is mediated by cAMP/Epac/PKA. A and B, cADPR formation is mediated by cGMP, whereas NAADP formation is regulated by cAMP and cGMP. LAK cells were treated with a cell-permeable cGMP analog, 8-pCPT-cGMP (100 μm), or a cell permeable cAMP analog, 8-pCPT-cAMP (100 μm), for 15 s. Then cADPR production and NAADP production were measured as described under “Experimental Procedures.” The means ± S.E. of three independent experiments are shown. *, p < 0.05, control (Cont) versus 8-pCPT-cGMP; #, p < 0.001, control versus 8-pCPT-cAMP. C, IL-8-induced cAMP increase precedes NAADP formation. Levels of cAMP and NAADP were determined after LAK cells were treated with 10 pm IL-8 for the indicated times. The means ± S.E. of three independent experiments are shown. D, cADPR but not NAADP induces cAMP increase. Levels of cAMP were determined after LAK cells were treated with 200 μm cADPR or 50 nm NAADP for the indicated times. The means ± S.E. of three independent experiments are shown. E, shown is the effect of XeC on cADPR-induced cAMP increase. Levels of cAMP were determined after the LAK cells were treated with 200 μm cADPR for 30 s. XeC (2 μm) was preincubated for 15 min. The means ± S.E. of three independent experiments are shown. *, p < 0.05, control versus cADPR or cADPR plus XeC. F, levels of NAADP were determined after the treatment of LAK cells with 100 μm 8-pCPT-2′-O-Me-cAMP (Epac activator), 100 μm N6-benzoyl-cAMP (PKA activator), 10 pm IL-8, 200 μm cADPR, or 1 μm forskolin. 8-pCPT-2′-O-Me-cAMP, N6-benzoyl-cAMP, cADPR, and forskolin were treated for 30 s, and IL-8 was treated for 90 s. Rp-8-Br-cAMPS (100 μm) and SQ22,536 (250 μm) were preincubated for 30 min. The levels of NAADP were measured as described under “Experimental Procedures.” The means ± S.E. of three independent experiments are shown. *, p < 0.05, control versus 8-pCPT-2′-O-Me-cAMP or N6-benzoyl-cAMP or IL-8 or cADPR or forskolin; #, p < 0.005, IL-8 versus IL-8 plus SQ22,536 or IL-8 plus Rp-8-Br-cAMPS; ##, p < 0.01, cADPR versus cADPR plus SQ22,536 or cADPR plus Rp-8-Br-cAMPS; **, p < 0.05, forskolin versus forskolin plus Rp-8-Br-cAMPS.
IL-8 Induced Rap1 Activation through Association of CD38 with Epac, PKA, and Rap1
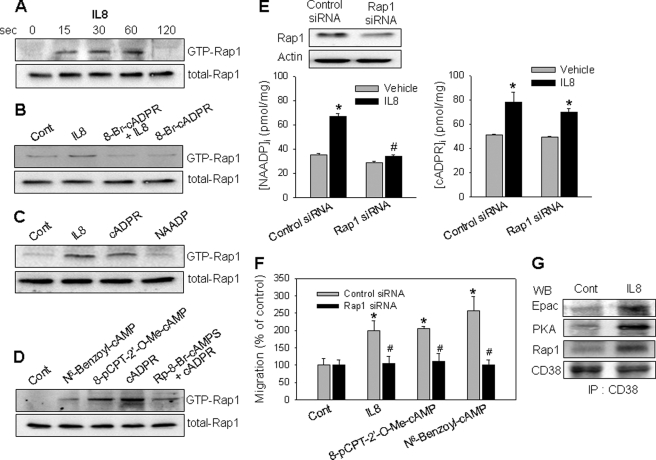
Because NAADP formation by CD38 is regulated by cAMP/PKA in IL-8-induced LAK cells, we next examined whether PKA phosphorylates CD38 directly. However, the phosphorylation level of CD38 was not increased by the treatment of LAK cells with IL-8 or PKA activator (data not shown). It has been reported that Rap1 may be activated by either Epac or PKA (41, 42). Because both Epac and PKA are involved in IL-8-induced NAADP formation as described above, we examined whether Rap1 is involved in the IL-8-induced NAADP formation in LAK cells. As shown in Fig. 8A, IL-8 induced Rap1 activation with a maximum effect at 1 min. Pretreatment of LAK cells with 8-Br-cADPR abolished the IL-8-induced Rap1 activation (Fig. 8B). Treatment of the cells with exogenous cADPR, but not NAADP, activated Rap1 that was comparable with that induced by IL-8 (Fig. 8C). In addition, treatment of LAK cells with an Epac activator and PKA activator increased the activation of Rap1 (Fig. 8D). However, a PKA inhibitor significantly inhibited cADPR-induced Rap1 activation (Fig. 8D). These results indicate that Rap1 activation through Epac and PKA is required for NAADP production in IL-8-stimulated LAK cells. The role of Rap1 in IL-8-induced NAADP formation was further examined by knockdown of Rap1 using siRNA. Rap1 siRNA-transfected cells showed a significantly reduced expression of Rap1 (Fig. 8E). The Rap1 knockdown abrogated the IL-8-induced NAADP formation but not cADPR production (Fig. 8E). The Rap1 knockdown LAK cells did not induce cell migration by the treatment with IL-8, Epac activator, or PKA activator (Fig. 8F). We also analyzed whether Rap1 is associated with Epac, PKA, and CD38 in IL-8-stimulated LAK cells. Indeed, IL-8 induced association of Epac, PKA, and Rap1 with CD38 (Fig. 8G). These results suggest that Epac, PKA, and Rap1 have a critical role in IL-8-induced NAADP production through association with CD38.
FIGURE 8.
IL-8 induces Rap1 activation through association of CD38 with Epac, PKA, and Rap1. A, IL-8 activates Rap1. LAK cells were treated with 10 pm IL-8 for a specified time, after which a pulldown assay was used to detect the active form of Rap1. B, 8-Br-cADPR blocks IL-8-induced Rap1 activation. The cells were preincubated with 8-Br-cADPR (100 μm) for 20 min before treatment with 10 pm IL-8 for 60 s. C, cADPR but not NAADP activates Rap1. LAK cells were treated with 200 μm cADPR or 50 nm NAADP for 30 s. D, cADPR-induced Rap1 activation was inhibited by a PKA inhibitor. Rap1 activation was determined after the treatment of LAK cells with 100 μm N6-benzoyl-cAMP, 100 μm 8-pCPT-2′-O-Me-cAMP, or 200 μm cADPR for 30 s. The cells were preincubated with Rp-8-Br-cAMPS (100 μm) for 30 min before treatment with 200 μm cADPR. E, Rap1 knockdown in LAK cells exhibits a reduced NAADP formation by IL-8 but not cADPR formation. A total of 60 pmol of siRNA was transfected into 5 × 105 cells using a transfection reagent. After 48 h of transfection, the cells were subjected to immunoblotting with anti-Rap1 pAb (upper panel) and measurement of NAADP/cADPR formation (lower panel) as described under “Experimental Procedures.” The means ± S.E. of three independent experiments are shown. *, p < 0.05, control (Cont) versus IL-8; #, p < 0.01, IL-8 in control siRNA versus IL-8 in Rap1 siRNA. F, IL-8/Epac/PKA-mediated cell migration is blocked in Rap1 knockdown LAK cells. Cells were treated with 10 pm IL-8, 100 μm 8-pCPT-2′-O-Me-cAMP, or 100 μm N6-benzoyl-cAMP. After incubation for 2 h, cell migration was assayed as described under “Experimental Procedures.” The means ± S.E. of three independent experiments are shown. *, p < 0.05, control versus IL-8 or 8-pCPT-2′-O-Me-cAMP or N6-benzoyl-cAMP; **, p < 0.01, IL-8 in control siRNA versus IL-8 in Rap1 siRNA or 8-pCPT-2′-O-Me-cAMP in control siRNA versus 8-pCPT-2′-O-Me-cAMP in Rap1 siRNA or N6-benzoyl-cAMP in control siRNA versus N6-benzoyl-cAMP in Rap1 siRNA. G, IL-8 induces the association of CD38 with Epac, PKA, and Rap1. LAK cells were treated with IL-8 for 30 s. The cells were extracted with a lysis buffer and then subjected to immunoprecipitation (IP) using anti-CD38 mAb. The immunoprecipitated proteins were analyzed by immunoblotting (WB) with anti-CD38 pAb, anti-Epac pAb, anti-PKA pAb, or anti-Rap1 pAb.
cAMP/Epac/PKA Mediates IL-8-stimulated Migratory Activity of LAK Cells
Next, we further examined the effect of Epac and PKA on the migratory activity of LAK cells. As shown in Fig. 9, LAK cell migration was significantly increased by IL-8, forskolin, or cADPR. On the other hand, the IL-8-, forskolin-, or cADPR-induced cell migration was blocked by pretreatment of the cells with a PKA inhibitor. This result indicates that cAMP/Epac/PKA system plays an essential role in IL-8-mediated migration of LAK cells.
FIGURE 9.
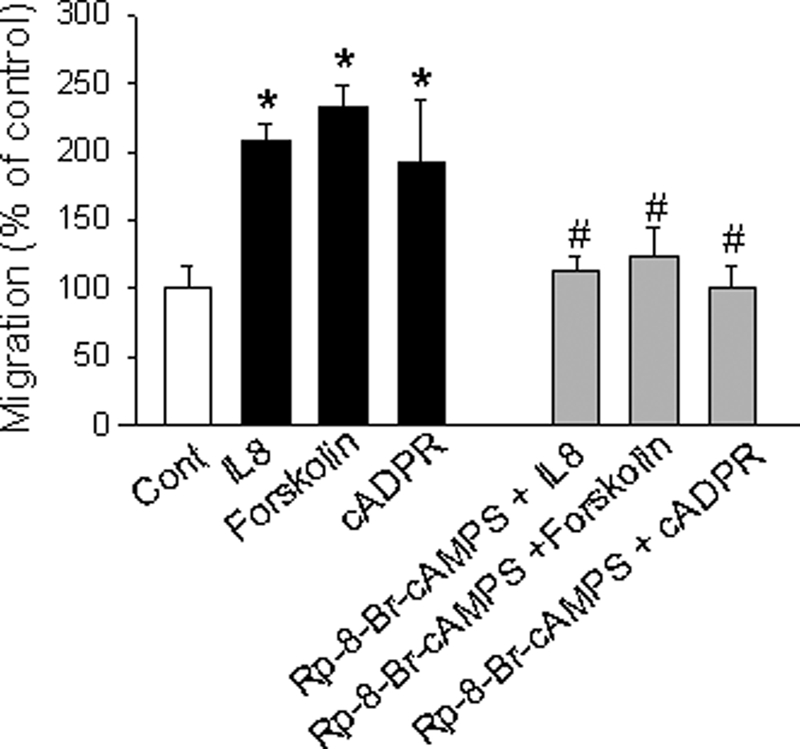
Epac and PKA mediate IL-8-induced migratory activity of LAK cells. LAK cells were treated with 10 pm IL-8, 1 μm forskolin, or 200 μm cADPR. Cells were preincubated with 100 μm Rp-8-Br-cAMPS, a PKA inhibitor, for 20 min. After incubation of cells with various reagents at 37 °C for 2 h, the cells in the top chamber were removed, and the cells adhered to the bottom of the filters were stained with Wright-Giemsa. Cells were counted under a phase-contrast microscope. The means ± S.E. of three independent experiments are shown. *, p < 0.05, control versus IL-8 or forskolin or cADPR; #, p < 0.005, IL-8 versus IL-8 plus Rp-8-Br-cAMPS or forskolin versus forskolin plus Rp-8-Br-cAMPS or cADPR versus cADPR plus Rp-8-Br-cAMPS.
DISCUSSION
The molecular mechanism of CD38 activation has not been fully elucidated, although activation of G-protein coupled receptors and T cell receptor has been shown to increase cADPR production via CD38/ADPR-cyclases (9, 43–47). We have demonstrated that IL-8-induced CD38 activation in LAK cells is mediated by sequential signaling involving an IL-8 receptor, IP3-mediated Ca2+ increase, and activation of protein kinase G (24). We have also demonstrated that protein kinase G-mediated phosphorylation of nonmuscle myosin IIA and association of phospho-nonmuscle myosin IIA with CD38 via Lck are essential for internalization and activation of CD38 (40). In this study we further explored IL-8-induced signaling in LAK cells by asking whether another important Ca2+ messenger, NAADP, is formed by CD38 activation. We showed that IL-8 induced NAADP formation in acidic organelles after cADPR production and that NAADP induced Ca2+ mobilization from acidic stores, contributing to IL-8-stimulated long-lasting Ca2+ signal. Our results also indicated a role of cAMP-sensitive molecules in the communication between thapsigargin-sensitive Ca2+ stores and acidic organelles. These results have revealed for the first time that CD38 produces two messengers, cADPR and NAADP, for Ca2+ signaling in IL-8-stimulatory LAK cell migration. We previously reported that the IL-8-dependent influx of extracellular calcium might be due to gating of unspecified calcium influx channels by cADPR (24). However, from the extended study, we could elucidate that NAADP, downstream of cADPR, is a direct player for Ca2+ influx, which is mediated by TRPM2 channels. Further studies remain to clarify how NAADP-induced Ca2+ release from acidic stores triggers TRPM2 channels for Ca2+ influx.
NAADP is known as an important second messenger in glucose-stimulated pancreatic β cells (16), cholecystokinin-mediated Ca2+ signaling in pancreatic acinar cells (17), and histamine-induced myometrial cells (48). Moreover, NAADP has been shown to be produced by CD38 via a base-exchange reaction (11). However, Soares et al. (48) reported that in vivo NAADP formation does not need CD38 or a base-exchange reaction in myometrial cells, and an existence of enzymes capable of generating NAADP and/or cADPR beside CD38 has recently been reported in various tissues (49). Our present data that IL-8-induced production of NAADP and cADPR was completely ablated in Cd38−/− LAK cells (Fig. 1), indicating that CD38 is an authentic NAADP-producing enzyme in LAK cells. Furthermore, it is of interest to observe that the location of NAADP production by CD38 appears to be distinct from that of cADPR synthesis; cADPR production is predominantly in early endosomes (45), whereas NAADP is produced mainly in lysosome-related acidic organelles (Ref. 38 and Fig. 4). Interestingly, CD38 exists in lysosomes as well as plasma membrane in unstimulated conditions of LAK cells (supplemental Fig. S4) and is constitutively expressed in lysosomes, as indicated by the little change in CD38 activity before and after IL-8 treatment (supplemental Fig. S5). Comparing the amounts of immunoreactive CD38 in lysosomes and plasma membrane fractions from LAK cells, CD38 in plasma membrane shows much lower ADP-ribosyl cyclase activity than that in lysosomes (supplemental Fig. S5), although the exact mechanism has yet to be elucidated. Therefore, as we previously observed that IL-8-induced cADPR formation requires internalization of CD38 from the plasma membrane (40), but NAADP production is mediated by CD38 expressed in lysosomes via Ca2+ release from the endoplasmic reticulum and cAMP/Epac/PKA/Rap 1 signaling.
In the study of sea urchin egg homogenates, soluble and membrane-bound ADP-ribosyl cyclases were identified to produce cADPR (50). The soluble cyclase was stimulated by cGMP, but not by cAMP, whereas the membrane-bound form was independent of cGMP. Both the soluble and membrane fractions also catalyzed the synthesis of NAADP through a base-exchange reaction that was independent of cGMP. Studies with sea urchin egg have indicated that cAMP-induced accumulation of NAADP and also cGMP contributed to the production of cADPR (39). Similarly, in our present study the formation of NAADP in IL-8-treated LAK cells was shown to be selectively regulated by adenylyl cyclase/cAMP, whereas cADPR formation was mediated in a cGMP-dependent manner. It is quite surprising, therefore, to find a similarity in the regulation of Ca2+ signaling between evolutionarily remote organisms. As for the mechanism by which cADPR-mediated Ca2+ increase activates adenylyl cyclase, it would be interesting to examine whether a Ca2+-regulated adenylyl cyclase is involved in the IL-8-induced signaling.
Rap1 is a member of the Ras superfamily of small GTP-binding proteins and is known to be involved in cell adhesion, gap junction formation, proliferation, and differentiation (51, 52). Rap activation is important for lymphocyte migration in vivo (53, 54). The Rap-dependent signaling pathway activated by Epac is known to be cell-specific (55, 56), and there is evidence for Rap-mediated action of Epac to stimulate mitogen-activated protein kinases (p38, extracellular signal-regulated kinase 1/2) in neurons, endocrine cells, and T-cells (57–59). Rap1 was reported to be activated by PKA through the phosphorylation of Rap1GAP (60), consistent with our finding that treatment of LAK cells with IL-8 or PKA activator did not show Rap1 phosphorylation. Our present data shows that IL-8 induced activation of Rap1, which was blocked by pretreatment with 8-Br-cADPR. Furthermore, treatment of LAK cells with exogenous cADPR induced activation of Rap1, whereas the addition of NAADP did not, suggesting that Rap1 activation is upstream of NAADP formation. Rap1 knockdown experiments also revealed that Rap1 is involved in only NAADP production but not cADPR production. It is quite interesting from an enzymology point of view to find a means of selectively regulating the multifunctionality of CD38 enzymatic activities through Rap1. Consistent with findings of IL-8-dependent association of Rap1 with Epac, PKA, and CD38, which are essential components for NAADP production, our preliminary finding that Rap1 is predominantly localized in lysosome-containing fractions from LAK cells (supplemental Fig. S6) suggests that Rap1 regulates CD38 for NAADP production in IL-8-induced LAK cells.
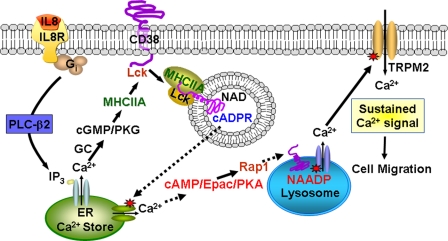
In conclusion, our results revealed that IL-8 elevates [Ca2+]i through cooperative actions of three different Ca2+ signal messengers: IP3, cADPR, and NAADP (Fig. 10). We found that NAADP-mediated Ca2+ signaling acts as the final determinant for IL-8-mediated long-lasting Ca2+ signals, which are essential for cell migration. Our present study also showed that CD38 produces NAADP and cADPR and that cGMP and cAMP may act as key modulators between NAADP- and cADPR-induced Ca2+ signals.
FIGURE 10.
Schematic pathway of IL-8-mediated NAADP formation by CD38 in LAK cells. Ligation of IL-8 receptor (IL8R) stimulates phospholipase Cβ2 (PLC-β2) via Gi, resulting in production of IP3 and diacylglycerol. IP3 binds to the receptor releasing Ca2+ from endoplasmic reticulum (ER) Ca2+ stores. IP3-mediated increase of intracellular Ca2+ levels results in elevation of cGMP levels via guanylyl cyclase (GC), thus activating protein kinase G (PKG) (24). Phosphorylated nonmuscle myosin IIA (MHCIIA) by protein kinase G is recruited in the CD38-Lck complex, inducing the internalization of CD38 and cADPR production (40). The cADPR produced by CD38 in the endocytic vesicles is transported via the promiscuous nucleoside/cADPR transporters, as previously described (35), or by the internalized CD38 itself, as previously proposed (61). cADPR-mediated Ca2+ release induces NAADP production by Rap1 activation via cAMP/Epac/PKA, resulting in release of Ca2+ from lysosome-related acidic organelles. NAADP-mediated Ca2+ release regulates the Ca2+ influx through TRPM2 channel in LAK cells, resulting in cell migration of LAK cells.
Supplementary Material
This work was supported by Korea Science and Engineering Foundation Grant R0A-2007-000-20121-0 (to U.-H. K.) and by Korea Research Foundation Grant KRF-2008-359-E00004 (to S.-Y. R.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Methods and Figs. S1–S6.
- ADPR
- ADP-ribosyl
- cADPR
- cyclic ADPR
- IL-8
- interleukin 8
- LAK
- lymphokine-activated killer
- NAADP
- adenine dinucleotide phosphate
- 8-pCPT
- 8-(4-chlorophenylthio)
- IP3
- d-myo-inositol 1,4,5-trisphosphate
- pAb
- polyclonal antibody
- ACA
- N-(p-amylcinnamoyl) anthranilic acid
- BSA
- bovine serum albumin
- siRNA
- small Interference RNA
- XeC
- xestospongin C
- Rp-8-Br-cAMPS
- 8-bromoadenosine 3′,5′-cyclic monophosphorothicate, Rp-isomer.
REFERENCES
- 1.Lund F., Solvason N., Grimaldi J. C., Parkhouse R. M., Howard M. (1995) Immunol. Today 16, 469–473 [DOI] [PubMed] [Google Scholar]
- 2.Mehta K., Shahid U., Malavasi F. (1996) FASEB J. 10, 1408–1417 [DOI] [PubMed] [Google Scholar]
- 3.Kim H., Jacobson E. L., Jacobson M. K. (1993) Science 261, 1330–1333 [DOI] [PubMed] [Google Scholar]
- 4.Howard M., Grimaldi J. C., Bazan J. F., Lund F. E., Santos-Argumedo L., Parkhouse R. M., Walseth T. F., Lee H. C. (1993) Science 262, 1056–1059 [DOI] [PubMed] [Google Scholar]
- 5.Zocchi E., Franco L., Guida L., Benatti U., Bargellesi A., Malavasi F., Lee H. C., De Flora A. (1993) Biochem. Biophys. Res. Commun. 196, 1459–1465 [DOI] [PubMed] [Google Scholar]
- 6.Lee H. C. (2004) Curr. Mol. Med. 4, 227–237 [DOI] [PubMed] [Google Scholar]
- 7.Galione A., Lee H. C., Busa W. B. (1991) Science 253, 1143–1146 [DOI] [PubMed] [Google Scholar]
- 8.Lee H. C., Aarhus R., Graeff R. M. (1995) J. Biol. Chem. 270, 9060–9066 [DOI] [PubMed] [Google Scholar]
- 9.Guse A. H., da Silva C. P., Berg I., Skapenko A. L., Weber K., Heyer P., Hohenegger M., Ashamu G. A., Schulze-Koops H., Potter B. V., Mayr G. W. (1999) Nature 398, 70–73 [DOI] [PubMed] [Google Scholar]
- 10.Partida-Sánchez S., Cockayne D. A., Monard S., Jacobson E. L., Oppenheimer N., Garvy B., Kusser K., Goodrich S., Howard M., Harmsen A., Randall T. D., Lund F. E. (2001) Nat. Med. 7, 1209–1216 [DOI] [PubMed] [Google Scholar]
- 11.Aarhus R., Graeff R. M., Dickey D. M., Walseth T. F., Lee H. C. (1995) J. Biol. Chem. 270, 30327–30333 [DOI] [PubMed] [Google Scholar]
- 12.Rongvaux A., Andris F., Van Gool F., Leo O. (2003) Bioessays 25, 683–690 [DOI] [PubMed] [Google Scholar]
- 13.Cancela J. M., Churchill G. C., Galione A. (1999) Nature 398, 74–76 [DOI] [PubMed] [Google Scholar]
- 14.Berg I., Potter B. V., Mayr G. W., Guse A. H. (2000) J. Cell Biol. 150, 581–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harmer A. R., Gallacher D. V., Smith P. M. (2001) Biochem. J. 353, 555–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masgrau R., Churchill G. C., Morgan A. J., Ashcroft S. J., Galione A. (2003) Curr. Biol. 13, 247–251 [DOI] [PubMed] [Google Scholar]
- 17.Yamasaki M., Thomas J. M., Churchill G. C., Garnham C., Lewis A. M., Cancela J. M., Patel S., Galione A. (2005) Curr. Biol. 15, 874–878 [DOI] [PubMed] [Google Scholar]
- 18.Gasser A., Bruhn S., Guse A. H. (2006) J. Biol. Chem. 281, 16906–16913 [DOI] [PubMed] [Google Scholar]
- 19.Genazzani A. A., Galione A. (1996) Biochem. J. 315, 721–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Churchill G. C., Okada Y., Thomas J. M., Genazzani A. A., Patel S., Galione A. (2002) Cell 111, 703–708 [DOI] [PubMed] [Google Scholar]
- 21.Hohenegger M., Suko J., Gscheidlinger R., Drobny H., Zidar A. (2002) Biochem. J. 367, 423–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerasimenko J. V., Sherwood M., Tepikin A. V., Petersen O. H., Gerasimenko O. V. (2006) J. Cell Sci. 119, 226–238 [DOI] [PubMed] [Google Scholar]
- 23.Steen M., Kirchberger T., Guse A. H. (2007) J. Biol. Chem. 282, 18864–18871 [DOI] [PubMed] [Google Scholar]
- 24.Rah S. Y., Park K. H., Han M. K., Im M. J., Kim U. H. (2005) J. Biol. Chem. 280, 2888–2895 [DOI] [PubMed] [Google Scholar]
- 25.Institute of Laboratory Animal Research, Commission of Life Sciences, National Resesrch Council (1996) Guide for the Care and Use of Laboratory Animals, pp. 8–20, National Academy Press, Washington, DC [Google Scholar]
- 26.Basse P., Herberman R. B., Nannmark U., Johansson B. R., Hokland M., Wasserman K., Goldfarb R. H. (1991) J. Exp. Med. 174, 479–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Graeff R., Lee H. C. (2002) Biochem. J. 361, 379–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee H. C., Aarhus R. (1991) Cell Regul. 2, 203–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Graeff R., Lee H. C. (2002) Biochem. J. 367, 163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dagnino L., Bennett L. L., Jr., Paterson A. R. P. (1991) J. Biol. Chem. 266, 6308–6311 [PubMed] [Google Scholar]
- 31.Kim U. H., Han M. K., Park B. H., Kim H. R., An N. H. (1993) Biochim. Biophys. Acta 1178, 121–126 [DOI] [PubMed] [Google Scholar]
- 32.Xie G. H., Rah S. Y., Yi K. S., Han M. K., Chae S. W., Im M. J., Kim U. H. (2003) Biochem. Biophys. Res. Commun. 307, 713–718 [DOI] [PubMed] [Google Scholar]
- 33.Tsien R. Y., Pozzan T., Rink T. J. (1982) Nature 295, 68–71 [DOI] [PubMed] [Google Scholar]
- 34.Sebok K., Woodside D., al-Aoukaty A., Ho A. D., Gluck S., Maghazachi A. A. (1993) J. Immunol. 150, 1524–1534 [PubMed] [Google Scholar]
- 35.Guida L., Bruzzone S., Sturla L., Franco L., Zocchi E., De Flora A. (2002) J. Biol. Chem. 277, 47097–47105 [DOI] [PubMed] [Google Scholar]
- 36.Podestà M., Benvenuto F., Pitto A., Figari O., Bacigalupo A., Bruzzone S., Guida L., Franco L., Paleari L., Bodrato N., Usai C., De Flora A., Zocchi E. (2005) J. Biol. Chem. 280, 5343–5349 [DOI] [PubMed] [Google Scholar]
- 37.Billington R. A., Bellomo E. A., Floriddia E. M., Erriquez J., Distasi C., Genazzani A. A. (2006) FASEB J. 20, 521–523 [DOI] [PubMed] [Google Scholar]
- 38.Lange I., Penner R., Fleig A., Beck A. (2008) Cell Calcium 44, 604–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson H. L., Galione A. (1998) Biochem. J. 331, 837–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rah S. Y., Park K. H., Nam T. S., Kim S. J., Kim H., Im M. J., Kim U. H. (2007) J. Biol. Chem. 282, 5653–5660 [DOI] [PubMed] [Google Scholar]
- 41.Li J., O'Connor K. L., Cheng X., Mei F. C., Uchida T., Townsend C. M., Jr., Evers B. M. (2007) Mol. Endocrinol. 21, 159–171 [DOI] [PubMed] [Google Scholar]
- 42.Shimomura H., Imai A., Nashida T. (2004) Arch. Biochem. Biophys. 431, 124–128 [DOI] [PubMed] [Google Scholar]
- 43.Higashida H., Egorova A., Higashida C., Zhong Z. G., Yokoyama S., Noda M., Zhang J. S. (1999) J. Biol. Chem. 274, 33348–33354 [DOI] [PubMed] [Google Scholar]
- 44.Higashida H., Yokoyama S., Hashii M., Taketo M., Higashida M., Takayasu T., Ohshima T., Takasawa S., Okamoto H., Noda M. (1997) J. Biol. Chem. 272, 31272–31277 [DOI] [PubMed] [Google Scholar]
- 45.Kim B. J., Park K. H., Yim C. Y., Takasawa S., Okamoto H., Im M. J., Kim U. H. (2008) Diabetes 57, 868–878 [DOI] [PubMed] [Google Scholar]
- 46.Gul R., Kim S. Y., Park K. H., Kim B. J., Kim S. J., Im M. J., Kim U. H. (2008) Am. J. Physiol. Heart Circ. Physiol. 295, H77–H88 [DOI] [PubMed] [Google Scholar]
- 47.Kim S. Y., Gul R., Rah S. Y., Kim S. H., Park S. K., Im M. J., Kwon H. J., Kim U. H. (2008) Am. J. Physiol. Renal Physiol. 294, F982–F989 [DOI] [PubMed] [Google Scholar]
- 48.Soares S., Thompson M., White T., Isbell A., Yamasaki M., Prakash Y., Lund F. E., Galione A., Chini E. N. (2007) Am. J. Physiol. Cell Physiol. 292, C227–C239 [DOI] [PubMed] [Google Scholar]
- 49.Churamani D., Boulware M. J., Geach T. J., Martin A. C., Moy G. W., Su Y. H., Vacquier V. D., Marchant J. S., Dale L., Patel S. (2007) PLoS ONE. 2, e797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Graeff R. M., Franco L., De Flora A., Lee H. C. (1998) J. Biol. Chem. 273, 118–125 [DOI] [PubMed] [Google Scholar]
- 51.Bos J. L. (2005) Curr. Opin. Cell Biol. 17, 123–128 [DOI] [PubMed] [Google Scholar]
- 52.Somekawa S., Fukuhara S., Nakaoka Y., Fujita H., Saito Y., Mochizuki N. (2005) Circ. Res. 97, 655–662 [DOI] [PubMed] [Google Scholar]
- 53.McLeod S. J., Li A. H., Lee R. L., Burgess A. E., Gold M. R. (2002) J. Immunol. 169, 1365–1371 [DOI] [PubMed] [Google Scholar]
- 54.Chu H., Awasthi A., White G. C., 2nd, Chrzanowska-Wodnicka M., Malarkannan S. (2008) J. Immunol. 181, 3373–3383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sabbatini M. E., Chen X., Ernst S. A., Williams J. A. (2008) J. Biol. Chem. 283, 23884–23894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Crittenden J. R., Bergmeier W., Zhang Y., Piffath C. L., Liang Y., Wagner D. D., Housman D. E., Graybiel A. M. (2004) Nat. Med. 10, 982–986 [DOI] [PubMed] [Google Scholar]
- 57.Guo F. F., Kumahara E., Saffen D. (2001) J. Biol. Chem. 276, 25568–25581 [DOI] [PubMed] [Google Scholar]
- 58.Quilliam L. A., Mueller H., Bohl B. P., Prossnitz V., Sklar L. A., Der C. J., Bokoch G. M. (1991) J. Immunol. 147, 1628–1635 [PubMed] [Google Scholar]
- 59.Tsygankova O. M., Saavedra A., Rebhun J. F., Quilliam L. A., Meinkoth J. L. (2001) Mol. Cell. Biol. 21, 1921–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McAvoy T., Zhou M. M., Greengard P., Nairn A. C. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 3531–3536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Franco L., Guida L., Bruzzone S., Zocchi E., Usai C., De Flora A. (1998) FASEB J. 12, 1507–1520 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.