Abstract
A central step to high fidelity protein synthesis is selection of the proper start codon. Recent structural, biochemical, and genetic analyses have provided molecular insights into the coordinated activities of the initiation factors in start codon selection. A molecular model is emerging in which start codon recognition is linked to dynamic reorganization of factors on the ribosome and structural changes in the ribosome itself.
Keywords: Eukaryote, Protein Synthesis, Translation, Translation Control, Translation Initiation Factors, Gcd-, Sui-, eIF1, eIF1A, eIF2
Overview of the Eukaryotic Translation Initiation Pathway
Assembly of an 80 S ribosome at the start codon of an mRNA is facilitated by translation initiation factors that function in a stepwise manner, rearranging both interfactor and factor-ribosome contacts at each step. In this review, we will highlight recent advances in our understanding of the structure-function properties of the initiation factors that function on the ribosome to promote assembly of the 43 S preinitiation complex and govern start site selection. Translation initiation (Fig. 1) (reviewed in Ref. 1) begins with formation of TC3 between initiator Met-tRNAi and the GTP-bound form of eIF2. The TC then associates with the small (40 S) ribosomal subunit. Binding of eIF1 and eIF1A alters the conformation of the 40 S subunit and promotes TC loading, which is also aided by eIF3. In a reaction facilitated by the eIF4 family of factors as well as by eIF3, the 43 S PIC (40 S + eIF1, eIF1A, TC, eIF3) binds to an mRNA near the 5′ cap and scans in a 3′ direction in search of a start codon. Upon start codon recognition, eIF2 completes hydrolysis of its bound GTP in a reaction promoted by eIF5. Base pairing between the start codon on the mRNA and the anticodon loop of Met-tRNAi in the 43 S complex triggers eIF1 release from its ribosomal binding site and dissociation of Pi from eIF2 to form eIF2·GDP, which is now unstably associated with the 40 S subunit. In a second GTP-dependent reaction, the factor eIF5B promotes joining of the large (60 S) ribosomal subunit to the 43 S complex. Hydrolysis of GTP by eIF5B following subunit joining enables eIF5B and eIF1A to dissociate from the 80 S initiation complex, leaving Met-tRNAi in the P site base paired to the start codon. The ribosome is now poised to enter the elongation phase of protein synthesis.
FIGURE 1.
Translation initiation pathway. The scheme of 80 S complex assembly is shown as described in the text. Note that hydrolysis of GTP on eIF2 to GDP·Pi initiates prior to mRNA binding. In addition, following start codon selection and Pi release, the PIC transitions from the open (scanning-competent) to closed (scanning-arrested) state. Factors involved in mRNA binding to the PIC are not shown for clarity.
eIF2
Although the structure of the eIF2 complex, consisting of α, β, and γ subunits, has not yet been determined, structural studies of individual subunits as well as of the corresponding archaeal factor aIF2, in conjunction with in vivo and in vitro analyses, have recently shed light on the structure-function properties of the factor. The eIF2α subunit domain structure is conserved between eukaryotes and Archaea (Fig. 2) (2, 3); however, an N-terminal extension makes the eukaryotic β subunit twice the length of the archaeal protein. This extension contains three lysine-rich segments (K-boxes) consisting of 6–8 consecutive lysine residues (Fig. 2A). The K-boxes in eIF2β mediate the binding of eIF2 to both its GAP, eIF5, and the catalytic subunit of its guanine nucleotide exchange factor, eIF2Bϵ (4). The γ subunit of eIF2 contains three domains (Fig. 2) and shows striking similarity to the structure of EF-Tu/eEF1A, the GTPase that brings aminoacyl-tRNAs onto the ribosome during elongation.
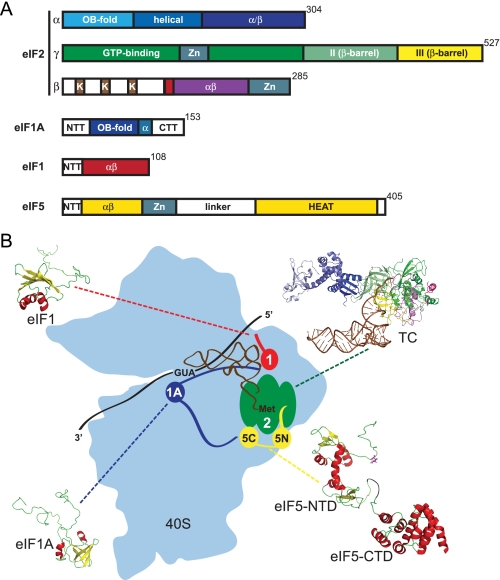
FIGURE 2.
Architecture of the PIC. A, schematic depictions of the key PIC factors from yeast. eIF2 is composed of α, β, and γ subunits. eIF2α consists of an OB-fold, an α-helical domain, and an α/β domain; the N-terminal half of eIF2β contains three Lys-rich (K) segments followed by a short unfolded domain (that adopts a helical structure when in the eIF2 complex, red), a core αβ domain, and a C-terminal zinc finger (gray) domain (9, 48, 49). eIF2γ has three domains: an N-terminal GTP-binding (G) domain and two β-barrel domains II and III; a zinc-binding knuckle is present within the G-domain (6, 7). eIF1A consists of a core OB-fold domain and long unstructured N- and C-terminal tails (NTT and CTT). eIF1 contains an unstructured N-terminal tail linked to an αβ core similar to eIF2β and the eIF5 NTD. eIF5 contains N-terminal αβ and zinc finger domains connected by an unstructured linker to the C-terminal HEAT (Huntington, elongation factor 3, PR65/A, TOR) domain. B, structures of human eIF1 (Protein Data Bank (PDB) code 2IF1), human eIF1A (1D7Q), eIF5 (human NTD, 2G2K; yeast CTD, 2FUL), and a composite archaeal TC are displayed around a schematic depicting the PIC constituents bound to the 40 S ribosomal subunit (light blue). The TC composite structure consists of archaeal aIF2 (3CW2; α, domain I, light blue; domain II, slate blue; domain III, blue; β, N-terminal helix, pink; domain II, violet; domain III, purple; and γ, G-domain, green; domain II, pale green; domain III, yellow) and yeast tRNAi (1YFG, brown). The position of tRNAi was modeled by superimposing the structures of aIF2 (3CW2 and 2QMU) with the structure of EF-Tu·GDPNP·Phe-tRNAPhe (1TTT). The structures of eIF1, eIF1A, and eIF5 are displayed with α-helices in red and β-strands in yellow. The Arg-15 residue in eIF5 that is required for GAP activity is colored magenta.
In EF-Tu, domains II and III move relative to the G-domain in response to GTP versus GDP binding. In the GDP state, domains II and III are remote from the G-domain, whereas in the presence of GTP, domains II and III move toward the G-domain to form the aminoacyl-tRNA-binding pocket (5). In the structures of aIF2γ, the protein is in the closed state in the presence of GTP, GDP, or no nucleotide (6, 7). It seems that rather than large conformational changes, modest reorientations in the G-domain govern Met-tRNAi binding by aIF2. Consistently, only a modest change in Met-tRNAi affinity was detected between eIF2·GTP and eIF2·GDP (15-fold) in contrast to the very large change in aminoacyl-tRNA affinity between EF-Tu in its two nucleotide states (8) (see below).
The eIF2γ subunit forms the keystone of the eIF2αβγ heterotrimeric complex. In the aIF2αβγ complex structure, domain III of aIF2α contacts a loop on domain II of aIF2γ (9). Consistently, mutations that alter conserved surface residues in domain II of aIF2γ impair aIF2α binding in vitro. Importantly, the growth defect associated with the corresponding mutations in yeast eIF2γ were suppressed by overexpression of eIF2α (6). The binding configuration of aIF2α to aIF2γ is the same in the aIF2αγ heterodimer and the aIF2αβγ complex, consistent with the notion that aIF2α and aIF2β bind independently to aIF2γ (3, 9, 10).
Ternary Complex Formation
No structures of a/eIF2 with bound Met-tRNAi have yet been reported. Despite the lack of this important structure, significant insights into the mechanism of Met-tRNAi binding by eIF2 have been obtained. In the “on” GTP-bound state, eIF2 binds Met-tRNAi with a Kd of ∼10 nm, 15-fold more tightly than the “off” GDP-bound state (8). This GTP/GDP switch involves toggling on and off of an interaction between eIF2 and the methionine on the tRNAi. Recombinant aIF2 also binds Met-tRNAi in a GTP-dependent manner (11). As might be expected based on its structural similarity to EF-Tu, the isolated aIF2γ subunit binds Met-tRNAi, albeit weakly (Kd ∼ 5 μm) (11). Neither aIF2α nor aIF2β binds Met-tRNAi, and the aIF2βγ complex binds Met-tRNAi with similar affinity as isolated aIF2γ. In contrast, the aIF2αγ complex binds Met-tRNAi with a Kd of ∼40 nm, similar to intact aIF2. Interestingly, this increased Met-tRNAi binding affinity is observed when only the C-terminal domain III of aIF2α is in complex with aIF2γ. As isolated domain III from neither aIF2α nor eIF2α showed significant Met-tRNAi binding, it seems that aIF2α allosterically enhances the Met-tRNAi binding by aIF2γ. Consistent with this idea, deletion of the aIF2α-binding loop in domain II of aIF2γ impairs Met-tRNAi binding to isolated aIF2γ (11). At odds with these findings, a yeast eIF2βγ complex isolated from a strain lacking eIF2α bound Met-tRNAi with only 5-fold lower affinity than intact eIF2 (12). Moreover, as the growth rate of the eIF2α-less strain (expressing a mutant form of eIF2γ and overexpressing tRNAi) is <2-fold slower than a wild-type strain (13), it seems that eIF2α does not play a crucial role in translation initiation in vivo aside from its role in regulation.
Although it was previously thought that GTP hydrolysis on eIF2 was triggered upon base pairing between the anticodon loop of Met-tRNAi in the 43 S complex and a start codon in the mRNA, more recent work has established that some of the GTP in the ternary complex is hydrolyzed to GDP+Pi in the PIC prior to start codon recognition (14) (Fig. 1). Release of Pi, rather than GTP hydrolysis, is the step that appears to be strongly controlled by start codon recognition. As will be discussed below, dissociation of eIF1 from the 43 S complex upon start codon recognition is the event that triggers release of Pi from eIF2.
The structures of various aIF2 complexes provide a rationale for the Pi-regulated binding of Met-tRNAi to eIF2. In aIF2γ·GDP structures, the tRNA-binding pocket is not formed (6, 7); however, in the aIF2αγ·GDPNP structure (3), a cleft for binding the methionine and terminal A76 of Met-tRNAi is observed (3). Surprisingly, when aIF2αβγ was crystallized in the presence of GDP, a similar Met-tRNAi-binding cleft was observed (9). Careful examination of the structure suggested the presence of GDP+Pi, rather than GDP, in the nucleotide-binding pocket. As the Met-tRNAi-binding cleft is in the open conformation in aIF2αβγ·GDP+Pi, this structure is compatible with the notion that Pi release and the resultant reconfiguration of eIF2 to the GDP conformation are necessary for release of eIF2 from Met-tRNAi in the ribosomal P site.
eIF2 Structure-Function Insights from Yeast Genetics
Genetic analyses in yeast have provided novel insights into the structure and function of the initiation factors. Mutations that impair TC formation or TC binding to the ribosome affect translational control of the GCN4 mRNA and produce a Gcd− phenotype (15). A second assay system monitoring His4p production in yeast from an mRNA lacking its normal start codon allows the fidelity of start codon recognition to be assessed. Sui− mutations enhance initiation at an in-frame UUG codon allowing His4p synthesis from the mutant mRNA (16).
Both Gcd− and Sui− mutations have been identified in all three subunits of eIF2. For example, Gcd− mutations in eIF2γ that affect guanine nucleotide-binding residues (16–18), the predicted methionine-binding pocket (6), and residues proposed to interact with the body of the tRNA (3) have been isolated and characterized. In all cases, phenotypes associated with the mutations are suppressed by overexpressing tRNAi, as predicted for amino acid changes that affect TC formation. Mutations in eIF2 causing Sui− phenotypes could operate by increasing spurious Pi or Met-tRNAi release from the factor. However, as the Sui− phenotypes do not simply correlate with the Met-tRNAi binding affinities of the mutant factors, it has been proposed that subtle alterations in the conformation of Met-tRNAi on the 40 S subunit affect base pairing between the anticodon and base triplets in the scanned mRNA, thereby impairing the fidelity of start codon recognition (17).
eIF1 and eIF1A
eIFs 1 and 1A are small initiation factors that bind directly to the 40 S ribosomal subunit and play central roles in both TC recruitment and the identification of the start codon (19). Together, eIF1 and eIF1A induce a conformational change in the 40 S subunit that promotes rapid binding of the TC (20). This open complex (Fig. 1), in which the mRNA entry channel latch is unlocked and a connection is formed between the head and shoulder of the subunit, is proposed to be competent for scanning the mRNA. eIF1 accelerates the rate of TC binding to the 40 S subunit as well as its release, whereas eIF1A accelerates the rate of TC binding but slows its release. These data suggest that eIF1 primarily acts to induce the formation of the open state of the 40 S subunit along with eIF1A, whereas eIF1A both performs this function and also interacts with the TC to stabilize its binding to the PIC. As expected based on the important roles these factors play in TC loading, a variety of mutations have been found in each that produce Gcd− phenotypes (e.g. Refs. 21–23).
eIF1A is made up of a central OB-fold domain that binds to the A site of the 40 S subunit and two long, unstructured N- and C-terminal tails (24, 25) (Fig. 2). These tails, which are not found in the orthologous bacterial factor, IF1, have been shown to be intimately involved in TC loading onto the 40 S subunit and in start codon recognition (19, 26). Prior to start codon recognition by the PIC, the tails of eIF1A are located in or near the P site of the 40 S subunit, which likely prevents the initiator tRNA from fully entering it (25, 26). When the start codon is encountered, the tRNA moves fully into the P site, and the C-terminal tail moves out of it (25, 26). It has been shown that eIF1A interacts strongly with eIF5, either directly or indirectly, upon start codon recognition (27), and the movement of the C-terminal tail of eIF1A may be involved in mediating this new interaction.
Mutations in both the N-terminal and the C-terminal tails have been found that affect the fidelity of start codon recognition. Amino acid changes in the N-terminal tail suppress Sui− mutations elsewhere in the protein or in other factors, as do changes in a region at the very N-terminal edge of the unstructured C-terminal tail. These two regions are proposed to pack on an α-helical segment of the factor, mutation of which also confers an Ssu− (suppressor of Sui−) phenotype. The structure formed by these three regions of eIF1A forms a sort of “brake” that inhibits scanning by the PIC. Disrupting this scanning inhibitor reduces the ability of the complex to stop and enter the closed, post-start codon recognition state and hence suppresses Sui− phenotypes. In contrast, mutations in two other regions of the C-terminal tail, termed SE 1 and 2, produce Sui− phenotypes. The data suggest that SE1 and SE2 promote formation of the open, scanning-competent state of the PIC, in which the initiator tRNA is not fully engaged in the P site. Movement of these elements out of the P site upon initial start codon recognition is proposed to allow proper engagement of the tRNA (25, 26).
The core (αβ) domain of eIF1 is similar to several ribosomal proteins, as well as to eIF2β and the N-terminal domain of eIF5 (Fig. 2A) (28). Footprinting studies have shown that eIF1 binds near the P site but not in a position that allows it to directly monitor codon/anticodon pairing between the mRNA and initiator tRNA (29).
Work in vitro and in vivo has shown that eIF1 is the key switch in triggering downstream events in response to the PICs encountering a start codon (21, 30). Initial start codon recognition results in a rapid conformational change in the PIC that moves the C termini of eIF1 and eIF1A away from each other. This is followed by release of eIF1 from its binding site within the PIC. Dissociation of eIF1 triggers Pi release from eIF2·GDP+Pi (14), as well as conversion from the open conformation of the PIC to the closed one (23) (Fig. 1). This latter finding is consistent with the fact that both eIF1 and eIF1A are required for stable formation of the open state of the yeast 40 S subunit, which suggested that release of eIF1 from the PIC should result in complex closure (20). The presence or absence of eIF1 within the PIC is thus the key control point that determines whether the complex continues searching for the start codon or halts and commits to downstream events in the initiation process. Consistent with this, a number of mutations have been found in eIF1 that reduce the affinity of the factor for the PIC, increasing aberrant release and thus producing strong Sui− phenotypes (21, 23, 31).
Both eIF1 and eIF1A have been reported to interact with eIF2. In solution, eIF1A interacts with eIF2 via the unstructured N-terminal tail of eIF1A, which also mediates an interaction with eIF3 (32). Pulldown experiments have indicated that eIF1 interacts in solution with eIF2β. This interaction appears to be mediated by both the unstructured N terminus of eIF1 and a basic region in its structured domain (28). These interactions may provide communication links between eIFs 1 and 1A and eIF2, allowing signaling within the PIC in response to start codon recognition.
eIF5
eIF5 acts as a GAP for eIF2 (33, 34), increasing the rate of GTP hydrolysis within the PIC by over 6 orders of magnitude (14). However, several lines of evidence indicate that eIF5 plays a more direct role in the mechanism of start codon recognition than simply acting as a constitutive GAP. For example, although Archaea have an ortholog of eIF2, they lack any detectable eIF5 ortholog, suggesting that this factor evolved in response to a eukaryote-specific requirement. As Archaea utilize either 5′-proximal AUGs or Shine-Dalgarno sequences to locate start codons, whereas eukaryotes employ an entirely different mechanism, eIF5 may play an important role in the eukaryotic scanning mechanism of start codon selection. In addition, mutations in eIF5 have been isolated that alter the fidelity of start codon recognition. Strikingly, the Sui− mutation G31R is codon-specific in its effect; it efficiently enhances use of UUG codons as start sites but does not enhance use of GUG, CUG, or AUU codons (16). This result suggests that the mutation alters the ability of eIF5 to sense and respond to AUG codons, endowing it with specific ability to also respond to UUG but not other near cognate codons. If eIF5 acted simply as a constitutive GAP, rather than playing a more direct role in start codon recognition, it is unclear how a mutation could lead to this sort of specificity given that GUG and CUG codons are inherently as efficient as UUG codons for initiation in Saccharomyces cerevisiae (35).
A number of recent in vitro studies support a direct role for eIF5 in detecting and responding to the start codon. eIF1A and eIF5 interact strongly within the PIC upon start codon recognition (27). With wild-type factors, this interaction is stronger with an AUG codon than a UUG codon. Remarkably, this specificity is switched when wild-type eIF5 is replaced with the Sui− G31R version of the factor, mimicking the codon specificity observed in vivo. More recently, it was shown that eIF5 antagonizes eIF1 binding to the PIC, suggesting that these two factors also communicate with each other and that release of eIF1 upon start codon recognition may allow eIF5 to alter its position within the complex, possibly an event that allows interaction with eIF1A and/or Pi release by eIF2 (23).
Structures of the N- and C-terminal domains of eIF5 have been determined by NMR and x-ray crystallography, respectively (36–38) (Fig. 2). A variety of studies have indicated that eIF5 interacts directly with eIF1, eIF2, eIF3, and eIF4G. The interaction with eIF2 is mediated by both the NTD and the CTD of eIF5. The NTD of eIF5 interacts with the G-domain of eIF2γ (39), and the CTD of eIF5 interacts with the K-boxes in the N-terminal region of eIF2β (4, 40, 41). eIFs 1, 3, and 4G all appear to interact with the CTD of eIF5, suggesting that one important role of this part of the factor is as a platform for organizing the architecture of the PIC.
The findings that the NTD of eIF5 is structurally similar to eIF1 (37) and that eIF5 and eIF1 antagonize each other's binding to the PIC (23) have suggested a model to explain part of the mechanism of action of eIF5 during start codon recognition. In this proposal, eIF1 occupies a binding site within the PIC in competition with part of eIF5 (e.g. its NTD). When eIF1 is released following start codon recognition, eIF5 can move into this site. This change in the position of eIF5 could then allow Pi release, interaction with eIF1A, and stabilization of the closed state of the complex. It is appealing to think that the domain that moves is the NTD, which is the domain containing the GAP activity and thus might also be expected to serve as the physical gate that prevents premature Pi release. Its structural similarity to eIF1 is also consistent with the idea that the NTD of eIF5 and eIF1 can bind to the same region within the PIC (37). However, given the interaction between eIF1 and the CTD of eIF5 observed in solution, it is also possible that these events are mediated by the proximity of the eIF5 CTD and eIF1 within the PIC (28). Either way, in this model, a key function of eIF5 is to act as the gate that prevents Pi release until start codon recognition has taken place. If this is the case, having eIF5 act as a GAP for eIF2 makes sense as it prevents GTP hydrolysis until the Pi gate is in place. Premature GTP hydrolysis and Pi release could result in selection of aberrant start sites and the production of miscoded proteins.
Perspectives
Over the past 5 years, it has become clear that the eukaryotic translation preinitiation complex is a dynamic version of a three-dimensional jigsaw puzzle. Each component makes a number of interactions with other components, and these interactions change as the initiation process proceeds.
In addition to the expected conformational changes in the 40 S ribosomal subunit itself, it is becoming increasingly clear that the movements of the factors, and the domains within them, are critical to the mechanics of the process. Competition between components for a single binding site, which allows the movement or release of one factor to trigger movement of another, may be an emerging theme. In addition, many factors have long, unstructured regions that are increasingly appearing to play critical roles in various stages of initiation. These regions may allow efficient communication over long distances, as well as provide factors with the ability to readily change binding partners. For example, the C-terminal tail of eIF1A plays a role in TC recruitment to the 40 S subunit. It moves out of the P site upon start codon recognition, possibly an event that is involved in triggering a new interaction with eIF5, and then interacts with eIF5B at the very end of initiation to stimulate subunit joining (42, 43).
At the heart of start codon recognition is the formation of 3 bp between the anticodon of the initiator tRNA and the mRNA. This is a simple molecular interaction, yet it triggers a series of complicated rearrangements that commit the complex to completing the initiation process from the selected point on the mRNA. How this signal is transmitted and amplified into the events described above is not yet clear. Recent work has indicated that like decoding in the A site during elongation, recognition of the start codon relies only on formation of 3 bp, not on the specific sequence of those pairs, suggesting that the PIC monitors duplex formation in the P site in some way (35). However, the structures of the P site of bacterial elongation complexes do not provide any obvious candidates for ribosomal sensors analogous to A1492, A1493, and G530, the bases in the A site that swing out to recognize formation of a duplex between the incoming tRNA and the mRNA codon (44). It is possible that the structure of the P site is altered in the eukaryotic initiation complex in such a way as to allow it to directly recognize base pairing. Alternatively, the initiation factors themselves may be the sensors of base pair formation. The movement of the C-terminal tail of eIF1A out of the P site upon initial start codon recognition is likely part of this sensor system. eIF1, eIF2, and eIF5 may sense formation of the codon/anticodon duplex either directly or, perhaps more likely given the crowded nature of the P site, indirectly. Indirect sensing of base pairing might be mediated by changes in the conformation or position of the initiator tRNA body (45, 46), similar to the active role proposed for tRNAs during decoding in the A site (47). These changes in the conformation or position of the initiator tRNA could be sensed by eIFs 1, 1A, 2, or 5, triggering downstream events including eIF1 and Pi release from the PIC.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant GM62128 (to J. R. L.) and by the Intramural Research Program of the NICHD, National Institutes of Health (to T. E. D.). This work was also supported by a grant from the Human Frontier Science Program (to J. R. L.). This is the third article in the Thematic Minireview Series on Protein Synthesis. This minireview will be reprinted in the 2010 Minireview Compendium, which will be available in January, 2011.
- TC
- ternary complex
- aIF
- archaeal initiation factor
- eIF
- eukaryotic initiation factor
- GDPNP
- guanosine 5′-βγ-imidodiphosphate
- CTD
- C-terminal domain
- NTD
- N-terminal domain
- Gcd−
- general control derepressed
- Sui−
- suppressor of initiation
- Met-tRNAi
- methionyl initiator tRNA
- PIC
- preinitiation complex
- GAP
- GTPase-activating protein
- SE
- scanning enhancer.
REFERENCES
- 1.Sonenberg N., Hinnebusch A. G. (2009) Cell 136, 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ito T., Marintchev A., Wagner G. (2004) Structure 12, 1693–1704 [DOI] [PubMed] [Google Scholar]
- 3.Yatime L., Mechulam Y., Blanquet S., Schmitt E. (2006) Structure 14, 119–128 [DOI] [PubMed] [Google Scholar]
- 4.Asano K., Krishnamoorthy T., Phan L., Pavitt G. D., Hinnebusch A. G. (1999) EMBO J. 18, 1673–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andersen G. R., Nyborg J. (2001) Cold Spring Harbor Symp. Quant. Biol. 66, 425–437 [DOI] [PubMed] [Google Scholar]
- 6.Roll-Mecak A., Alone P., Cao C., Dever T. E., Burley S. K. (2004) J. Biol. Chem. 279, 10634–10642 [DOI] [PubMed] [Google Scholar]
- 7.Schmitt E., Blanquet S., Mechulam Y. (2002) EMBO J. 21, 1821–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kapp L. D., Lorsch J. R. (2004) J. Mol. Biol. 335, 923–936 [DOI] [PubMed] [Google Scholar]
- 9.Yatime L., Mechulam Y., Blanquet S., Schmitt E. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 18445–18450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stolboushkina E., Nikonov S., Nikulin A., Bläsi U., Manstein D. J., Fedorov R., Garber M., Nikonov O. (2008) J. Mol. Biol. 382, 680–691 [DOI] [PubMed] [Google Scholar]
- 11.Yatime L., Schmitt E., Blanquet S., Mechulam Y. (2004) J. Biol. Chem. 279, 15984–15993 [DOI] [PubMed] [Google Scholar]
- 12.Nika J., Rippel S., Hannig E. M. (2001) J. Biol. Chem. 276, 1051–1056 [DOI] [PubMed] [Google Scholar]
- 13.Erickson F. L., Nika J., Rippel S., Hannig E. M. (2001) Genetics 158, 123–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Algire M. A., Maag D., Lorsch J. R. (2005) Mol. Cell 20, 251–262 [DOI] [PubMed] [Google Scholar]
- 15.Hinnebusch A. G. (2005) Annu. Rev. Microbiol. 59, 407–450 [DOI] [PubMed] [Google Scholar]
- 16.Huang H. K., Yoon H., Hannig E. M., Donahue T. F. (1997) Genes Dev. 11, 2396–2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alone P. V., Cao C., Dever T. E. (2008) Mol. Cell. Biol. 28, 6877–6888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Erickson F. L., Hannig E. M. (1996) EMBO J. 15, 6311–6320 [PMC free article] [PubMed] [Google Scholar]
- 19.Mitchell S. F., Lorsch J. R. (2008) J. Biol. Chem. 283, 27345–27349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Passmore L. A., Schmeing T. M., Maag D., Applefield D. J., Acker M. G., Algire M. A., Lorsch J. R., Ramakrishnan V. (2007) Mol. Cell 26, 41–50 [DOI] [PubMed] [Google Scholar]
- 21.Cheung Y. N., Maag D., Mitchell S. F., Fekete C. A., Algire M. A., Takacs J. E., Shirokikh N., Pestova T., Lorsch J. R., Hinnebusch A. G. (2007) Genes Dev. 21, 1217–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fekete C. A., Mitchell S. F., Cherkasova V. A., Applefield D., Algire M. A., Maag D., Saini A. K., Lorsch J. R., Hinnebusch A. G. (2007) EMBO J. 26, 1602–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nanda J. S., Cheung Y. N., Takacs J. E., Martin-Marcos P., Saini A. K., Hinnebusch A. G., Lorsch J. R. (2009) J. Mol. Biol. 394, 268–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Battiste J. L., Pestova T. V., Hellen C. U., Wagner G. (2000) Mol. Cell 5, 109–119 [DOI] [PubMed] [Google Scholar]
- 25.Yu Y., Marintchev A., Kolupaeva V. G., Unbehaun A., Veryasova T., Lai S. C., Hong P., Wagner G., Hellen C. U., Pestova T. V. (2009) Nucleic Acids Res. 37, 5167–5182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saini A. K., Nanda J. S., Lorsch J. R., Hinnebusch A. G. (2010) Genes Dev. 24, 97–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maag D., Algire M. A., Lorsch J. R. (2006) J. Mol. Biol. 356, 724–737 [DOI] [PubMed] [Google Scholar]
- 28.Reibarkh M., Yamamoto Y., Singh C. R., del Rio F., Fahmy A., Lee B., Luna R. E., Ii M., Wagner G., Asano K. (2008) J. Biol. Chem. 283, 1094–1103 [DOI] [PubMed] [Google Scholar]
- 29.Lomakin I. B., Kolupaeva V. G., Marintchev A., Wagner G., Pestova T. V. (2003) Genes Dev. 17, 2786–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maag D., Fekete C. A., Gryczynski Z., Lorsch J. R. (2005) Mol. Cell 17, 265–275 [DOI] [PubMed] [Google Scholar]
- 31.Yoon H. J., Donahue T. F. (1992) Mol. Cell. Biol. 12, 248–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olsen D. S., Savner E. M., Mathew A., Zhang F., Krishnamoorthy T., Phan L., Hinnebusch A. G. (2003) EMBO J. 22, 193–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Das S., Ghosh R., Maitra U. (2001) J. Biol. Chem. 276, 6720–6726 [DOI] [PubMed] [Google Scholar]
- 34.Paulin F. E., Campbell L. E., O'Brien K., Loughlin J., Proud C. G. (2001) Curr. Biol. 11, 55–59 [DOI] [PubMed] [Google Scholar]
- 35.Kolitz S. E., Takacs J. E., Lorsch J. R. (2009) RNA 15, 138–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bieniossek C., Schütz P., Bumann M., Limacher A., Uson I., Baumann U. (2006) J. Mol. Biol. 360, 457–465 [DOI] [PubMed] [Google Scholar]
- 37.Conte M. R., Kelly G., Babon J., Sanfelice D., Youell J., Smerdon S. J., Proud C. G. (2006) Biochemistry 45, 4550–4558 [DOI] [PubMed] [Google Scholar]
- 38.Wei Z., Xue Y., Xu H., Gong W. (2006) J. Mol. Biol. 359, 1–9 [DOI] [PubMed] [Google Scholar]
- 39.Alone P. V., Dever T. E. (2006) J. Biol. Chem. 281, 12636–12644 [DOI] [PubMed] [Google Scholar]
- 40.Das S., Maitra U. (2000) Mol. Cell. Biol. 20, 3942–3950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamamoto Y., Singh C. R., Marintchev A., Hall N. S., Hannig E. M., Wagner G., Asano K. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 16164–16169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Acker M. G., Shin B. S., Dever T. E., Lorsch J. R. (2006) J. Biol. Chem. 281, 8469–8475 [DOI] [PubMed] [Google Scholar]
- 43.Marintchev A., Kolupaeva V. G., Pestova T. V., Wagner G. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 1535–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmeing T. M., Ramakrishnan V. (2009) Nature 461, 1234–1242 [DOI] [PubMed] [Google Scholar]
- 45.Kapp L. D., Kolitz S. E., Lorsch J. R. (2006) RNA 12, 751–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kolitz S. E., Lorsch J. R. (2010) FEBS Lett. 584, 396–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cochella L., Green R. (2005) Science 308, 1178–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gutiérrez P., Osborne M. J., Siddiqui N., Trempe J. F., Arrowsmith C., Gehring K. (2004) Protein Sci. 13, 659–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sokabe M., Yao M., Sakai N., Toya S., Tanaka I. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 13016–13021 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.