Abstract
Lysosome function is essential to many physiological processes. It has been suggested that deregulation of lysosome function could contribute to cancer. Through a genetic screen in Drosophila, we have discovered that mutations disrupting lysosomal degradation pathway components contribute to tumor development and progression. Loss-of-function mutations in the Class C vacuolar protein sorting (VPS) gene, deep orange (dor), dramatically promote tumor overgrowth and invasion of the RasV12 cells. Knocking down either of the two other components of the Class C VPS complex, carnation (car) and vps16A, also renders RasV12 cells capable for uncontrolled growth and metastatic behavior. Finally, chemical disruption of the lysosomal function by feeding animals with antimalarial drugs, chloroquine or monensin, leads to malignant tumor growth of the RasV12 cells. Taken together, our data provide evidence for a causative role of lysosome dysfunction in tumor growth and invasion and indicate that members of the Class C VPS complex behave as tumor suppressors.
Keywords: Drosophila Genetics, Lysosomes, Ras, Tumor Metastases, Tumor Suppressor, Class C VPS Complex, Tumor Growth, Vps18, deep orange
Introduction
The lysosome plays important roles in many physiological processes and pathological conditions. It has been suggested that deregulation of lysosome function could contribute to tumor development. For example, altered trafficking of lysosomal cathepsins has been found in cultured cancer cells and in malignant tumors in patients (1–3). However, evidence that lysosome dysfunction plays a causative role in tumor development and progression remains elusive.
Proteins responsible for lysosomal delivery and degradation are highly conserved among different eukaryotic species. For example, genetic screens in yeast have identified a group of genes encoding the Class C vacuolar protein sorting (VPS)5 complex, which is involved in the docking and membrane fusion of endosomal vesicles and the lysosome-like yeast vacuole. Mutations in Class C VPS complex genes cause similar phenotypes characterized as accumulation of enlarged multivesicular bodies and defects in sorting cargoes to the vacuole (4, 5). In Drosophila, studies have shown that the components of the Class C VPS complex, e.g. Deep orange (Dor), Carnation (Car), and Vps16A, play a similar role, as do their yeast homologues (6, 7). The Drosophila lysosomal delivery system is also found to be involved in the development of eye pigmentation. Mutations in car and dor cause severe defects in intracellular trafficking to the lysosome and defects in eye color (8). The Class C VPS complex has been shown to be functionally conserved from flies to human (9–15).
To identify genes involved in tumor progression, we have performed a genetic screen for mutations on the X chromosome utilizing a recently established Drosophila metastasis model (16). In the developing eye epithelium, GFP-labeled clones of cells are simultaneously expressing the oncogenic Ras (RasV12) protein and are homozygous for newly induced X-linked mutations. Because clones of RasV12 cells exhibit benign overgrowth, the metastatic phenotype can be easily visualized by the invasion of GFP-positive cells from the primary tumor to other tissues. Here we report the identification of dor, a homologue of yeast vps18, in the screen and uncovered the causative role of lysosomal dysfunction in tumor progression and metastasis by analyses with both mutations and antimalarial drugs.
EXPERIMENTAL PROCEDURES
Fly Strains and Genetics
Flies were raised at 25 °C on standard medium. Duplication and deficiency lines used in mutation mapping were obtained from the Bloomington Stock Center and Kyoto Stock Center. dorC107 and dorD185 were isolated from an ethyl methanesulfonate mutagenesis screen, whereas dor8 was obtained from the Bloomington Stock Center. The DNA fragment for inverted repeats in the carRNAi construct was generated by PCR and cloned into pWIZ (17). The transgenic flies were established using standard procedures (18). UAS-dor, vps16ARNAi, and UAS-GFP-LAMP strains were gifts from Helmut Krämer. The UAS-atg5RNAi and UAS-atg7RNAi strains were kindly provided by Thomas P. Neufeld.
Fluorescence-labeled non-invasive and invasive tumor clones were produced using w, Tub-Gal80, FRT19A; ey-FLP5, Act5C>y+>Gal4, UAS-GFP (19A GFP tester) or w, Tub-Gal80, FRT19A; ey-FLP5, G454/CyO; Act5C>y+>Gal4, UAS-SrcRFP/TM6B. Clones homozygous for dor mutations were generated from dor heterozygous flies by crossing with 19A GFP testers.
The genotypes for the animals described in this study are listed below. In Figs. 1A, 2C, 2D, and 3B, w, sn3, FRT19A/w, Tub-Gal80, FRT19A; ey-FLP5, Act5C>y+>Gal4, UAS-GFP/+. In Figs. 1B, 3D, 3E, and 4, I–L, w, sn3, FRT19A/w, Tub-Gal80, FRT19A; ey-FLP5, Act5C>y+>Gal4, UAS-GFP/UAS-RasV12. In Fig. 1C, SupA186, w, sn3, FRT19A/w, Tub-Gal80, FRT19A; ey-FLP5, Act5C>y+>Gal4, UAS-GFP/UAS-RasV12. In Fig. 1D, EnhA235, w, sn3, FRT19A/w, Tub-Gal80, FRT19A; ey-FLP5, Act5C>y+>Gal4, UAS-GFP/UAS-RasV12. In Fig. 1E, dorC107, w, sn3, FRT19A/w, Tub-Gal80, FRT19A; ey-FLP5, Act5C>y+>Gal4, UAS-GFP/UAS-RasV12. In Fig. 1F, dorD185, w, sn3, FRT19A/w, Tub-Gal80, FRT19A; ey-FLP5, Act5C>y+>Gal4, UAS-GFP/UAS-RasV12. In Fig. 2, E and F, dorC107, w, sn3, FRT19A/w, Tub-Gal80, FRT19A; ey-FLP5, Act5C>y+>Gal4, UAS-GFP/+. In Fig. 2G, dor8, FRT19A/w, Tub-Gal80, FRT19A; ey-FLP5, Act5C>y+>Gal4, UAS-GFP/UAS-RasV12. In Fig. 2H, dor8, FRT19A/w, Tub-Gal80, FRT19A; ey-FLP5, Act5C>y+>Gal4, UAS-GFP/UAS-RasV12; UAS-dor/+. In Fig. 2I, dor8. In Fig. 2J, dor8/Y; Act-Gal4/+; UAS-dor/+. In Fig. 3A (WT)), w1118; Act-Gal4/+. In Fig. 3A (carRNAi), w1118; Act-Gal4/+; carRNAi/+. In Fig. 3C, w, sn3, FRT19A/w, Tub-Gal80, FRT19A; ey-FLP5, Act5C>y+>Gal4, UAS-GFP/+; carRNAi/+. In Fig. 3, F and G, w, sn3, FRT19A/w, Tub-Gal80, FRT19A; ey-FLP5, Act5C>y+>Gal4, UAS-GFP/UAS-RasV12; carRNAi/+. In Fig. 3, H and I, w, sn3, FRT19A/w, Tub-Gal80, FRT19A; ey-FLP5, Act5C>y+>Gal4, UAS-GFP/UAS-RasV12, vps16ARNAi. In Fig. 4, A–D, w, sn3, FRT19A/w, Tub-Gal80, FRT19A; ey-FLP5, G454/UAS-RasV12, UAS-GFP-LAMP; Act5C>y+>Gal4, UAS-SrcRFP/+. In Fig. 4, E–H, dor8, FRT19A/w, Tub-Gal80, FRT19A; ey-FLP5, G454/UAS-RasV12, UAS-GFP-LAMP; Act5C>y+>Gal4, UAS-SrcRFP/+.
FIGURE 1.
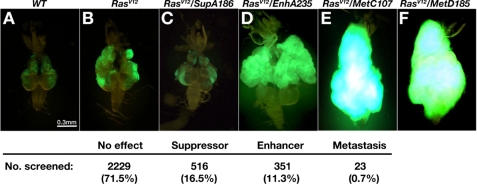
Genetic screen identified mutations that promote tumor growth and metastasis of RasV12 cells. A–F, dorsal views of dissected cephalic complexes from third-instar larvae of mentioned genotypes are shown. The anterior is to the top in all panels; the mouth hook is to the top, and the VNC is pointing down in all panels. ey-FLP-introduced homozygous clones are labeled by GFP (green). The summary of the screen was shown at the bottom. Mutants are grouped into four subcategories based on their tumor progression phenotypes. The screen results are listed as the number of mutant lines and its percentage in each subcategory.
FIGURE 2.
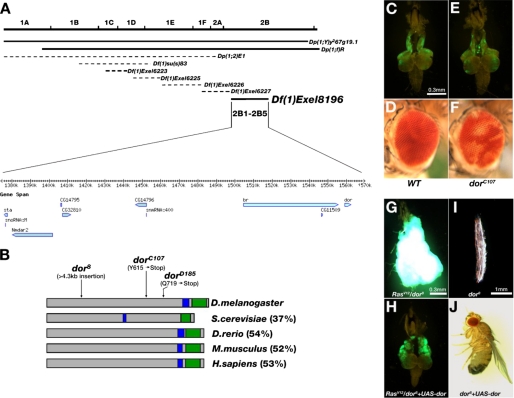
deep orange behaves as a metastasis suppressor. A, genetic mapping using a series of duplication and deficiency lines located the dorC107 mutation in the chromosome region of 2B1-2B5. Solid lines indicate the duplications and deficiencies that rescued or failed to complement with the mutation, respectively. Dotted lines indicate the duplications or deficiencies that failed to rescue or were able to complement the mutation, respectively. The candidate genes located within this region are shown. B, illustration of the Dor protein and its homologues in several species. The coiled-coil domain and the RING-H2 domain are indicated by blue and green boxes, respectively. The percentages of overall similarity between Dor and its homologues are indicated. The molecular lesions of the three dor alleles are also indicated. D.melanogaster, Drosophila melanogaster; S.cerevisiae, Saccharomyces cerevisiae; D.rerio, Danio rerio; M.musculus, Mus musculus; H.sapiens, Homo sapiens. C, E, G, and H, fluorescence images of dissected cephalic complexes showing GFP-labeled homozygous cell clones of indicated relevant genotypes. D and F, micrographs of mosaic compound eyes containing homozygous WT (D) or dorC107 (F) clones labeled by the w mutation. I and J, microscopy images of the three-instar larva (I) and the adult fly (J) with indicated genotypes. Clones homozygous for dorC107 (E) do not exhibit any growth advantage when compared with WT (C). Note the severe pigmentation loss in dorC107 clones (F) when compared with WT (D). G–J, the expression of full-length wild type dor cDNA can fully rescue dor8-caused phenotypes. The excess growth and metastasis of RasV12/dor8 tumors (G) are completely blocked by specific expression of UAS-dor in these tumor clones (H). The image of a viable adult fly indicates that the lethality of dor8 allele (I) is rescued by ubiquitous expression of UAS-dor (J). C and E are in the same magnification. G and H are in the same magnification.
FIGURE 3.
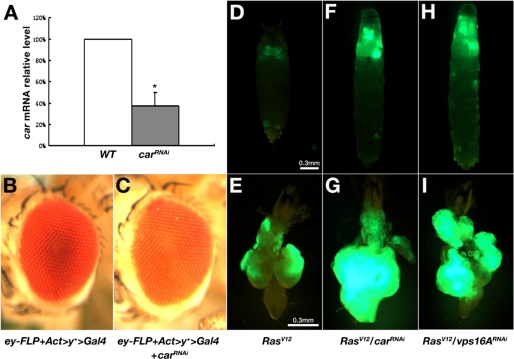
Disruption of the Class C VPS complex promotes tumor growth and metastasis in RasV12 cells. A, quantitative real-time reverse transcription-PCR results indicate that the car mRNA level in third-instar larvae was significantly reduced by RNAi. Actin mRNA levels were used as internal controls. *, p < 0.0001. Error bars indicate S.D. B and C, micrographs of compound eye from control (B) and carRNAi (C) flies, indicating the pigmentation defect in the carRNAi eye. D–I, larvae/pupae and dissected cephalic complexes from third-instar larvae with indicated genotypes. Both carRNAi and vps16ARNAi cause overgrowth and metastasis of GFP-positive RasV12 cells. D, F, and H are in the same magnification. E, G, and I are in the same magnification.
FIGURE 4.
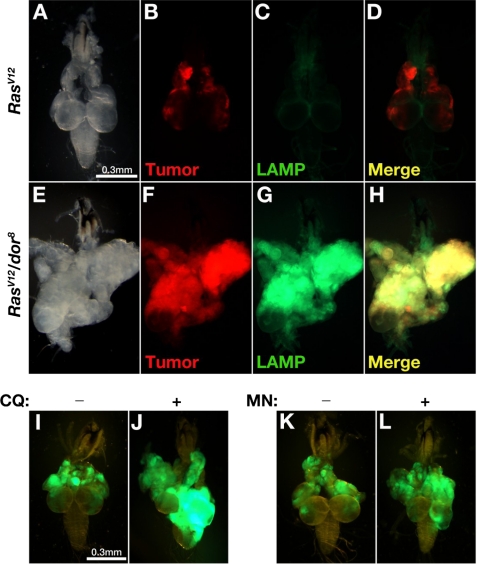
Impaired lysosomal degradation contributes to metastasis. Dissected cephalic complexes from third-instar larvae are shown. A–H, RFP was used for labeling tumor clones to distinguish them from the green signal of the GFP-LAMP fusion protein. I–L, RasV12 tumor cells were labeled by GFP. In RasV12 tumors (A–D), GFP-LAMP fusion proteins were presumably targeted to lysosomes and efficiently degraded. With additional dor8 mutation, the lysosomal degradation of GFP-LAMP proteins was blocked inside tumor tissues with invasive potential (E–H). Inhibition of lysosomal degradation by feeding chloroquine (CQ) (J) or monensin (MN) (L) induced VNC invasion of RasV12 tumor clones (green) when compared with the control samples (I and K).
Metastasis Assays
Assays to detect metastatic behaviors were performed as described previously (16). Briefly, cephalic complexes of wandering third-instar larvae were dissected in phosphate-buffered saline, and the distribution patterns of GFP clones were carefully examined in eye-antennal discs, optical lobes, ventral nerve cords (VNCs), guts, and tracheae. The invasions of GFP clones from their original sites (eye-antennal discs and optical lobes) to VNCs (as well as guts and tracheae) were considered as metastasis events. Metastasis was considered suppressed if no metastasis event was observed or if the majority of animals pupated. All images were captured under Leica MZFLIII microscope.
Chloroquine and Monensin Feeding Assays
10% phosphate-buffered saline and ethanol were used to dissolve chloroquine (Sigma) and monensin (Sigma), respectively. In drug treatment experiments, a single drug solution was mixed into standard fly medium with a final concentration of 1 mg/ml (chloroquine) or 600 μm (monensin). Control medium for each drug was prepared by mixing standard medium with the solvent only. Crosses of parental flies were set up 1 day before they were transferred into vials for drug treatment. The third-instar larvae of the next generation were gathered for the subsequential metastasis assays.
RESULTS
A Genetic Screen for X-linked Mutations That Promote Tumor Overgrowth and Metastasis of RasV12 Cells
The Drosophila X chromosome contains over 13% of the genome and more than 2,200 genes. However, only less than 750 X-linked genes have been phenotypically characterized. This is largely due to the difficulty in establishing lines harboring X-linked lethal mutations, which can only be stocked in females. We have established 3,119 lines with individual ethyl methanesulfonate-mutagenized X chromosomes and discovered that 1,092 lines carry recessive lethal mutations, whereas 78 of the viable lines exhibit visible morphological defects.6 We then examined all of the mutagenized X chromosomes for their ability to promote tumor growth and invasion in cooperation with RasV12. As reported previously, clones of cells expressing RasV12 in the developing eye exhibit moderate benign overgrowth and never invade into the nearby VNC or other tissues (Fig. 1, A and B). When clones of RasV12-expressing cells are also homozygous for a mutation in the metastasis suppressor gene, stardust (sdt), they developed into aggressive tumors with dramatic overgrowth and invasion into VNC (16). When the newly induced X-linked mutations were examined in a similar fashion, 71.5% (2,229) of the mutations did not affect the RasV12 phenotype (Fig. 1). 16.5% (516) of the mutations suppressed RasV12-induced benign overgrowth (Fig. 1C). Furthermore, 11.3% (315) of the mutations enhanced RasV12-induced tumor overgrowth but did not exhibit any invasion phenotype (Fig. 1D). Finally, 0.7% (23) of the mutations, including 22 lethal and 1 viable, triggered both enhanced tumor overgrowth and metastasis (metastasis-promoting mutations).6 Two of the metastasis-promoting mutations belong to the same complementation group, dor, which has an existing null allele, dor8 (also see below). Starting on day 8 after egg laying (AEL), RasV12 tumors with homozygous dor mutation (hereafter referred to as RasV12/dor−) exhibited enhanced overgrowth when compared with RasV12 controls (data not shown). On day 14 AEL, RasV12/dor− tumor cells clearly invaded into VNC with a high frequency (75%, n = 24: data not shown). On day 18 AEL, the dramatically overgrown RasV12/dor− tumors occupied about one-quarter volume of the larvae body (data not shown) and completely enveloped VNC (Fig. 1E, 100%, n = 22; Fig. 1F, 100%, n = 29; Fig. 2G, 100%, n = 38; Table 1). Occasionally, GFP-labeled tumor cells were also found in the gut and the trachea of the RasV12/dor− larvae, indicating secondary tumor formation in distal organs (data not shown). All RasV12/dor− larvae died prior to pupation.
TABLE 1.
Lysosome dysfunction promotes overgrowth and metastasis of RasV12 tumors
| Genotype | Tumor overgrowth | Metastasis ratio | p valuea |
|---|---|---|---|
| WT | − | 0% (n > 200) | NAb |
| RasV12 | + | 0% (n > 200) | NAb |
| RasV12/dor8 | +++ | 100% (n = 38) | 0c |
| RasV12/MetC107 | +++ | 100% (n = 22) | 0c |
| RasV12/MetD185 | +++ | 100% (n = 29) | 0c |
| RasV12/dor8 + UAS-dor | + | 0% (n = 50) | 1c |
| RasV12 + carRNAi | ++ | 29.8% (n = 57) | 0.000001c |
| RasV12 + vps16ARNAi | ++ | 34.9% (n = 43) | 0.000006c |
| RasV12 + 10% phosphate-buffered saline | + | 0% (n = 44) | NAb |
| RasV12 + chloroquine | ++ | 16.1% (n = 56) | 0.0014d |
| RasV12 + ethanol | + | 0% (n = 60) | NAb |
| RasV12 + monensin | ++ | 11.8% (n = 51) | 0.013e |
a p value was calculated using Fisher's exact test.
b NA, not applicable.
c Compared against RasV12.
d Compared against RasV12 + 10% phosphate-buffered saline.
e Compared against RasV12 + monensin. p < 0.05 is considered as a significant difference.
To test whether the dor mutation alone was sufficient to cause tumorigenesis or metastasis, we generated dor− homozygous mutant clones without RasV12 in eye tissues using ey-FLP. When compared with wild type controls (Fig. 2, C and D), clones of dor− cells displayed no visible growth advantages or morphology defects in either larval or adult stages (Fig. 2, E and F). However, severe loss of red pigments was observed in the dor− clones in the adult eye (Fig. 2F), indicating a cell-autonomous pigmentation defect. Taken together, these data indicate that the dor mutation itself is not sufficient to induce tumor growth but can collaborate with the oncogene RasV12 to trigger tumor overgrowth and metastasis.
dor Behaves as a Metastasis Suppressor
To map an X-linked lethal mutation or to perform complementation tests, we must first rescue the male lethality with a chromosome duplication. Hemizygous dor mutant male flies were lethal at the third-instar larvae stage. We examined a collection of chromosome duplications that cover more than 90% of the X chromosome (Bloomington chromosome 1 duplication kit) and successfully rescued the dor male lethality by two duplications with an overlap chromosome region from 1A3 to 2B18 (Dp(1;Y)y267g19.1; Dp(1;f)R) (Fig. 2A). The rescued dor mutant males allowed us to perform complementation tests with other lethal metastasis-promoting mutations, which led to the identification of a complementation group containing two alleles (dorC107 and dorD185; Fig. 1, E and F). Further genetic mapping with strains carrying smaller deficiencies or duplications located dor in a chromosome region between 2B1 and 2B5, containing 10 known or predicted genes (Fig. 2A). Sequencing of the coding regions of these candidate genes identified two nonsense mutations (T to G and C to T) in the dor gene for dorC107 and dorD185, respectively (Fig. 2B). Both mutations were expected to result in premature Dor proteins lacking the functionally critical RING-H2 domain (6). The previously existing dor8 null mutation failed to complement with the new dor alleles and exhibited similar phenotypes including third-instar larval lethality in homozygous mutants (19), eye pigmentation defects in clones of dor− cells (6), and more importantly, enhanced tumor overgrowth and metastasis in clones of RasV12/dor− cells (Fig. 2G). Finally, the wild type dor transgene can rescue both the lethality and the tumor phenotypes of all three alleles (Fig. 2, G–J; Table 1; and data not shown). Therefore, we conclude that the metastasis-promoting mutations are loss-of-function dor alleles and that dor encodes a novel metastasis suppressor.
To test whether the effect of dor on tumor growth and metastasis is context-dependent, we inactivated dor in clones of dMyc-overexpressing cells and did not observe tumor overgrowth or metastasis (dMycover/dor8; supplemental Fig. 1C). Furthermore, we inactivated dor in clones of cells with overexpression of the insulin receptor (InR) and observed enhanced tumor overgrowth but not metastasis (InRover/dor8; supplemental Fig. 1D). These data suggest that the effect of dor on tumor growth and metastasis is context-dependent.
Disruption of the Class C VPS Complex Cooperates with Oncogenic RasV12 in Inducing Metastasis
Three genes have been identified to encode different components of the Class C VPS complex in Drosophila. Mutations of the other two genes, car and vps16A, could lead to similar pigmentation and vesicle-trafficking phenotypes as those exhibited in dor mutants (6, 7). We thus disrupted car or vps16A in the same RasV12 tumor model to examine their potential effects on metastasis.
We used RNAi to knock down the expression of car. Two snap-back car cDNA fragments were placed under the transcriptional control of a UAS promoter to form a carRNAi transgene. When driven by Act-Gal4, the carRNAi transgene efficiently impaired the endogenous expression of car in one of the transgenic lines. Quantitative real-time reverse transcription-PCR using RNA isolated from the third-instar larvae showed that the car mRNA level in carRNAi animals was decreased to 37.6% of that in the wild type controls. As a control, dor expression was not affected (Fig. 3A and data not shown). Transgenic larvae could develop into the pupal stage, but only 4.4% of them (n = 91) survived to adult. Consistent with the function of car in cargo delivery to pigment granules (6, 20), eye-specific RNAi of car induced by ey-FLP and Act>y+>Gal4 resulted in obvious reduction of eye pigments (Fig. 3, B and C). Knockdown of car moderately enhanced the overgrowth of RasV12 tumors on day 18 AEL (Fig. 3F; Table 1). When compared with the non-invasive RasV12 tumors (Fig. 3, D and E, 0%, n = 47), RasV12/carRNAi tumors frequently invaded into the VNC (Fig. 3G, 29.8%, n = 57; Table 1). Knockdown of vps16A in RasV12 cells resulted in similar metastatic phenotypes (RasV12/vps16A RNAi). An established vps16ARNAi allele (7) led to tumor invasion toward VNC in RasV12 cells (Fig. 3, H and I, 34.9%, n = 43; Table 1). The tumor growth- and metastasis-promoting capabilities of carRNAi and vps16ARNAi were weaker than that of dor8, likely attributing to the fact that the RNAi procedure only reduced but did not eliminate the activity of the genes. Nevertheless, these data clearly indicate a tumor growth and metastasis suppression role of the Class C VPS complex genes in cancer progression.
Impaired Lysosomal Degradation Contributed to Metastatic Behavior
Involvement of the Class C VPS complex in lysosomal delivery suggests that the acquired metastatic capability of RasV12 tumor cells may result from defects in lysosomal trafficking. To test this possibility, we examined the lysosomal degradation activity in RasV12/dor− tumor cells with a GFP-LAMP transgene. When expressed, the cytoplasmic tail of human LAMP1 protein is sufficient to direct the fused GFP to the lysosome for degradation (7). When the transgene is expressed in red fluorescent protein (RFP)-labeled clones of RasV12 cells, only basal level GFP signals were detected, indicating effective lysosomal degradation (Fig. 4, A–D). However, massive accumulation of GFP-LAMP proteins were observed in RasV12/dor− tumor cells both in the primary tumors and in the leading edge of invading tumor cells (Fig. 4, E–H), revealing impaired lysosomal degradation.
We also blocked lysosomal degradation in RasV12 tumors by chloroquine or monensin treatment. These two traditional antimalarial drugs have been reported to be able to inhibit lysosomal degradation by raising intralysosomal pH (21, 22). RasV12 larvae were bred on chloroquine- or monensin-containing medium to score the tumor phenotypes. The treatment of either chloroquine or monensin clearly promoted metastasis. On day 18 AEL, 16.1% of the chloroquine-treated RasV12 tumors have prominently invaded into the VNC, although the overgrowth was not notably enhanced (Fig. 4J, n = 56; Table 1). In contrast, the invasive behavior was never observed in control tumors fed with solvent only medium (Fig. 4I, n = 44; Table 1). Monensin treatment resulted in similar phenotypes. 11.8% of the monensin-treated RasV12 animals developed VNC invasion (Fig. 4L, n = 51; Table 1), in comparison with no invasion in the control group (Fig. 4K, n = 60; Table 1). Taken together, these data indicate that lysosomal degradation impairment can cooperate with RasV12 to promote tumor progress and metastasis.
Autophagy, a process involving degradation of cellular components by fusion with the lysosome, has been indicated to play important roles in cancer development (23, 24). However, data suggest that the process could have opposing roles in tumorigenesis (23, 24). beclin1 (atg6) and UVRAG (vps38), two components of the autophagy pathway, have been identified as tumor suppressors (25–27). On the other hand, autophagy has also been implicated for a tumor-promoting role by its contribution to the survival of tumor cells under stress conditions (28). Because the Class C VPS complex is essential in both the endosome-lysosome and the autophagosome-lysosome fusion processes (7, 29, 30), we thus examined whether disruption of the autophagy pathway could contribute to enhanced tumor overgrowth and metastasis. Because atg5 and atg7 are required for autophagy in mammals and Drosophila (31, 32), we used the established atg5 or atg7 RNAi allele (31) to knock down the activity of autophagy in clones of RasV12 cells. Neither enhanced tumor overgrowth nor metastasis was observed (data not shown). Furthermore, in Drosophila, activation of the JNK signaling pathway has been shown to promote the overgrowth and the metastasis of RasV12 cells with disrupted cell polarity (RasV12/scrib−) (33–35). We also disrupted the JNK pathway by expressing a dominant negative form of the Drosophila JNK gene (bskDN) in the RasV12/dor− tumor cells. We found that both enhanced tumor overgrowth and metastasis of the RasV12/dor− tumors were almost completely suppressed (supplemental Fig. 1E). Together, these data suggest that activation of the JNK signaling, rather than disruption of autophagy, is likely the cause of the phenotypes observed in the RasV12/dor− tumors.
DISCUSSION
Deregulation of lysosome functions is linked to many pathological conditions including cancer (36). Dor/Vps18 is one of the essential players in the lysosomal delivery pathway, and its functions have been conserved from yeast to mammals (6, 14, 37, 38). Our study shows that dor functions as a suppressor of tumor growth and metastasis. Although inactivation of dor did not trigger tumorigenesis by itself, it promotes both tumor growth and metastasis in cooperation with oncogenic Ras. Consistent with disruption of the Class C VPS complex, inactivation of either of the two other components of the complex, car and vps16A, resulted in similar tumor growth and metastasis phenotypes. These data indicate that components of the Class C VPS complex behave as metastasis suppressors.
Our data showed that administration of antimalarial drugs, chloroquine or monensin, has a profound effect on promoting tumor progression and metastasis of cells expressing oncogenic Ras. Chloroquine treatment has been reported to have a suppression effect on Myc-induced lymphoma in mice (39). On the other hand, recent studies of chloroquine and quinacrine, another antimalarial drug, have shown promotion of tumor development in rats (40). Perhaps the effect of lysosomal degradation blockage on tumor development and progression is context-dependent. Indeed, we observed that the effect of dor mutations on tumor overgrowth and metastasis is RasV12-dependent. The dor mutation has no effect on dMyc-overexpressing cells and only enhances growth of the InR-overexpressing cells. Our study, taken together with the previous reports, argues for further careful examination of the effect of antimalarial drugs on tumor development and progression.
Autophagy has been indicated to play important roles in cancer development. However, disruption of autophagy is unlikely to be the major cause of the phenotypes that we observed in the RasV12/dor− tumors because inactivation of either of two key components of the autophagy pathway, atg5 and atg7, did not enhance tumor overgrowth or metastasis of the RasV12 cells. On the other hand, disruption of the JNK signaling in the RasV12/dor− tumors blocked both enhanced tumor overgrowth and metastasis. This is consistent with the previous observation that activation of the JNK signaling plays an essential role in tumor growth and progression (33–35).
Cellular homeostasis is tightly controlled in living cells. Normal cells have the ability to modulate the rates of molecular synthesis and degradation in response to different developmental and physiological stimuli. Impaired lysosomal degradation has been detected in cancer and other pathological conditions and is believed to be able to increase the cell mass (41–43). Consistent with this notion, RasV12/dor− tumors exhibited a dramatic overgrowth phenotype. In those studies, however, the role of lysosomal degradation in tumor metastasis is less obvious. Future study is needed to explore whether any processes or signaling events that promote metastasis are affected by lysosomal degradation. Analysis of the downstream events will help to understand how impaired lysosomal degradation contributes to malignant transformation and may indicate a novel direction for therapeutic intervention against metastasis.
Supplementary Material
Acknowledgments
We sincerely thank Helmut Krämer (UT Southwestern Medical Center), Thomas P. Neufeld (University of Minnesota), the Kyoto Stock Center, and the Bloomington Stock Center for providing various fly strains, Fei Gu, Sheng Shi, and Beibei Song for technical assistance, and Beibei Ying, Kejing Deng, Rener Xu, Wufan Tao, Ling Sun, Zhisheng Ye, Chi Zhang, and other members of the Institute of Developmental Biology and Molecular Medicine for discussions.
This work was supported by National Natural Science Foundation of China (Grant 30270694), Chinese Key Projects for Basic Research (973) (Grant 2006CB806702), Hi-tech Research and Development Project (863) (Grant 2007AA022101), 211 and 985 projects of Chinese Ministry of Education, Shanghai Pujiang Program (Grant 05PJ14024), and Shanghai Rising-Star Program (Grant 06QA14006). This work was also supported in part by National Institutes of Health NCI Grant R01 CA069408 (to T. X.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
C. Chi, H. Zhu, and T. Xu, unpublished data.
- VPS
- vacuolar protein sorting
- GFP
- green fluorescent protein
- RFP
- red fluorescent protein
- RNAi
- RNA interference
- VNC
- ventral nerve cord
- AEL
- after egg laying
- JNK
- c-Jun N-terminal kinase
- WT
- wild type
REFERENCES
- 1.Rochefort H., Capony F., Garcia M. (1990) Cancer Metastasis Rev. 9, 321–331 [DOI] [PubMed] [Google Scholar]
- 2.Hirano T., Manabe T., Takeuchi S. (1993) Cancer Lett. 70, 41–44 [DOI] [PubMed] [Google Scholar]
- 3.Brouillet J. P., Dufour F., Lemamy G., Garcia M., Schlup N., Grenier J., Mani J. C., Rochefort H. (1997) Cancer 79, 2132–2136 [PubMed] [Google Scholar]
- 4.Banta L. M., Robinson J. S., Klionsky D. J., Emr S. D. (1988) J. Cell Biol. 107, 1369–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Preston R. A., Manolson M. F., Becherer K., Weidenhammer E., Kirkpatrick D., Wright R., Jones E. W. (1991) Mol. Cell. Biol. 11, 5801–5812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sevrioukov E. A., He J. P., Moghrabi N., Sunio A., Krämer H. (1999) Mol. Cell 4, 479–486 [DOI] [PubMed] [Google Scholar]
- 7.Pulipparacharuvil S., Akbar M. A., Ray S., Sevrioukov E. A., Haberman A. S., Rohrer J., Krämer H. (2005) J. Cell Sci. 118, 3663–3673 [DOI] [PubMed] [Google Scholar]
- 8.Lloyd V., Ramaswami M., Krämer H. (1998) Trends Cell Biol. 8, 257–259 [DOI] [PubMed] [Google Scholar]
- 9.Kim B. Y., Krämer H., Yamamoto A., Kominami E., Kohsaka S., Akazawa C. (2001) J. Biol. Chem. 276, 29393–29402 [DOI] [PubMed] [Google Scholar]
- 10.Huizing M., Didier A., Walenta J., Anikster Y., Gahl W. A., Krämer H. (2001) Gene 264, 241–247 [DOI] [PubMed] [Google Scholar]
- 11.Suzuki T., Oiso N., Gautam R., Novak E. K., Panthier J. J., Suprabha P. G., Vida T., Swank R. T., Spritz R. A. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 1146–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poupon V., Stewart A., Gray S. R., Piper R. C., Luzio J. P. (2003) Mol. Biol. Cell 14, 4015–4027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sadler K. C., Amsterdam A., Soroka C., Boyer J., Hopkins N. (2005) Development 132, 3561–3572 [DOI] [PubMed] [Google Scholar]
- 14.Maldonado E., Hernandez F., Lozano C., Castro M. E., Navarro R. E. (2006) Pigment Cell Res. 19, 315–326 [DOI] [PubMed] [Google Scholar]
- 15.Yu J. F., Fukamachi S., Mitani H., Hori H., Kanamori A. (2006) Pigment Cell Res. 19, 628–634 [DOI] [PubMed] [Google Scholar]
- 16.Pagliarini R. A., Xu T. (2003) Science 302, 1227–1231 [DOI] [PubMed] [Google Scholar]
- 17.Lee Y. S., Carthew R. W. (2003) Methods 30, 322–329 [DOI] [PubMed] [Google Scholar]
- 18.Spradling A. C., Rubin G. M. (1982) Science 218, 341–347 [DOI] [PubMed] [Google Scholar]
- 19.Shestopal S. A., Makunin I. V., Belyaeva E. S., Ashburner M., Zhimulev I. F. (1997) Mol. Gen. Genet. 253, 642–648 [DOI] [PubMed] [Google Scholar]
- 20.Sriram V., Krishnan K. S., Mayor S. (2003) J. Cell Biol. 161, 593–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seglen P. O., Grinde B., Solheim A. E. (1979) Eur. J. Biochem. 95, 215–225 [DOI] [PubMed] [Google Scholar]
- 22.Grinde B. (1983) Exp. Cell Res. 149, 27–35 [DOI] [PubMed] [Google Scholar]
- 23.Kondo Y., Kanzawa T., Sawaya R., Kondo S. (2005) Nat. Rev. Cancer 5, 726–734 [DOI] [PubMed] [Google Scholar]
- 24.Mathew R., Karantza-Wadsworth V., White E. (2007) Nat. Rev. Cancer 7, 961–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aita V. M., Liang X. H., Murty V. V., Pincus D. L., Yu W., Cayanis E., Kalachikov S., Gilliam T. C., Levine B. (1999) Genomics 59, 59–65 [DOI] [PubMed] [Google Scholar]
- 26.Liang X. H., Jackson S., Seaman M., Brown K., Kempkes B., Hibshoosh H., Levine B. (1999) Nature 402, 672–676 [DOI] [PubMed] [Google Scholar]
- 27.Liang C., Feng P., Ku B., Dotan I., Canaani D., Oh B. H., Jung J. U. (2006) Nat. Cell Biol. 8, 688–699 [DOI] [PubMed] [Google Scholar]
- 28.Degenhardt K., Mathew R., Beaudoin B., Bray K., Anderson D., Chen G., Mukherjee C., Shi Y., Gélinas C., Fan Y., Nelson D. A., Jin S., White E. (2006) Cancer Cell 10, 51–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang C., Lee J. S., Inn K. S., Gack M. U., Li Q., Roberts E. A., Vergne I., Deretic V., Feng P., Akazawa C., Jung J. U. (2008) Nat. Cell Biol. 10, 776–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindmo K., Simonsen A., Brech A., Finley K., Rusten T. E., Stenmark H. (2006) Exp. Cell Res. 312, 2018–2027 [DOI] [PubMed] [Google Scholar]
- 31.Scott R. C., Schuldiner O., Neufeld T. P. (2004) Dev. Cell 7, 167–178 [DOI] [PubMed] [Google Scholar]
- 32.Jaeger P. A., Wyss-Coray T. (2009) Mol. Neurodegener. 4, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu M., Pastor-Pareja J. C., Xu T. (2010) Nature 463, 545–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Igaki T., Pastor-Pareja J. C., Aonuma H., Miura M., Xu T. (2009) Dev. Cell 16, 458–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Igaki T., Pagliarini R. A., Xu T. (2006) Curr. Biol. 16, 1139–1146 [DOI] [PubMed] [Google Scholar]
- 36.Polo S., Pece S., Di Fiore P. P. (2004) Curr. Opin. Cell Biol. 16, 156–161 [DOI] [PubMed] [Google Scholar]
- 37.Yogosawa S., Hatakeyama S., Nakayama K. I., Miyoshi H., Kohsaka S., Akazawa C. (2005) J. Biol. Chem. 280, 41619–41627 [DOI] [PubMed] [Google Scholar]
- 38.Rieder S. E., Emr S. D. (1997) Mol. Biol. Cell 8, 2307–2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amaravadi R. K., Yu D., Lum J. J., Bui T., Christophorou M. A., Evan G. I., Thomas-Tikhonenko A., Thompson C. B. (2007) J. Clin. Invest. 117, 326–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dutta P., Karmali R., Pinto J. T., Rivlin R. S. (1994) Cancer Lett. 76, 113–119 [DOI] [PubMed] [Google Scholar]
- 41.Kisen G. O., Tessitore L., Costelli P., Gordon P. B., Schwarze P. E., Baccino F. M., Seglen P. O. (1993) Carcinogenesis 14, 2501–2505 [DOI] [PubMed] [Google Scholar]
- 42.Bradley M. O. (1977) J. Biol. Chem. 252, 5310–5315 [PubMed] [Google Scholar]
- 43.Tessitore L., Bonelli G., Cecchini G., Autelli R., Amenta J. S., Baccino F. M. (1988) Biochem. J. 251, 483–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.