Abstract
The molecular structure of mouse Mucin 21 (Muc21)/epiglycanin is proposed to have 98 tandem repeats of 15 amino acids and three exceptional repeats with 12 or 13 amino acids each, followed by a stem domain, a transmembrane domain, and a cytoplasmic tail. A cDNA of Muc21 having 84 tandem repeats of 15 amino acids was constructed and transfected using a Venus vector into HEK 293T cells. The fluorescent cells, which were considered to express Muc21, were nonadherent. This antiadhesion effect was lessened when constructs with smaller numbers of tandem repeats were used, suggesting that the tandem repeat domain plays a crucial role. Cells expressing Muc21 were significantly less adherent to each other and to extracellular matrix components than control cells. Antibody binding to the cell surface integrin subunits α5, α6, and β1 was reduced in Muc21 transfectants in a tandem repeat-dependent manner, whereas equal amounts of proteins were detected by Western blot analysis. Muc21 was expressed as a large glycoprotein that was highly glycosylated with O-glycans at the cell surface, as detected by flow cytometry, Western blotting, and lectin blotting. Although at least a portion of Muc21 was glycosylated with sialylated glycans, removal of sialic acid did not influence the prevention of adhesion.
Keywords: Cell Adhesion, Glycoconjugate, Glycoprotein, Glycosylation, Tumor Marker, Epiglycanin, Mucin
Introduction
A mucin called epiglycanin was first reported in 1975 (1) as a cell surface glycoprotein expressed by a subline of TA3 cells (2). These cells were isolated as mammary carcinoma cells and originated from a spontaneous tumor in A/HeHa mice in 1949 in the laboratory of Hauschka (3). Two sublines, TA3-Ha (3) and TA3-St (4), were obtained later. TA3-Ha cells could grow in and kill allogeneic mouse strains, rats, and hamsters (5), whereas TA3-St cells grow only in syngeneic hosts. This property has been explained by the presence of epiglycanin on TA3-Ha cells, which was initially identified as a large surface molecule with an extended shape by biochemical and electron microscopic studies (2, 6, 7).
Epiglycanin was the first mucin described to be associated with the malignant behavior of carcinoma cells, and the molecular characteristics of this mucin and its possible functions have been a subject of debate for the last 30 years. When other carcinoma-associated mucins, such as MUC1, were reported to correlate with poor clinical outcomes of patients, epiglycanin was often cited and referred to as an example of a mucin that potentially plays a crucial role in determining the malignant behavior of carcinoma cells. Immune suppression (8) and prevention of cell adhesion (9) have been proposed as putative molecular functions specific to epiglycanin and possibly to other mucins.
However, because of the difficulty of cloning the epiglycanin gene (and mucin genes in general), molecular evidence as to how this mucin determines malignant cellular behavior has been lacking (10). Recently, a novel transmembrane mucin was isolated by representative differential analysis between TA3-Ha and TA3-St cells and designated as Muc21. Muc21 was implicated as the molecular entity of epiglycanin because Muc21 could be detected only in TA3-Ha cells and not in TA3-St cells by Northern blot analysis and reverse transcription-PCR and because there was a high similarity in amino acid composition to purified epiglycanin (11). However, because of the repetitive nature of the gene structure (tandem repeats, TR),2 and because of its large size, the complete Muc21 gene has not been cloned, whereas its human counterpart was (11). Mouse Muc21/epiglycanin is thought to be a highly glycosylated molecule, which makes it likely that its function is dependent on its glycoforms. However, there was no convenient way to examine the glycoforms of Muc21/epiglycanin.
Here, we propose a gene structure for mouse Muc21 and show its expression on cell surfaces using a reconstructed Muc21 cDNA. Striking morphologic changes, including loss of adhesion, were observed in Muc21 transfectants. By expressing various partial gene sequences, it became clear that loss of adhesion is mediated by the TR portion of Muc21.
EXPERIMENTAL PROCEDURES
Cell Culture
Human embryonic kidney (HEK) 293T cells were grown in high glucose Dulbecco's modified Eagle medium (Nissui-pharma) supplemented with l-glutamine and fetal calf serum at 37 °C, 5% CO2.
5′-RACE
5′-RACE was performed with GeneRacer (Invitrogen) according to the manufacturer's instructions. Reverse transcription was performed using a primer that anneals to a sequence in the TR domain: 5′-GCTGTCAACGATACGCTACGTAACGGCATGACAGTGGATGCAGTGGTGGTCAGGGTGGGTGTAGAGCCTGAGCCAGTGCTGGATACAGTGGTGGTCGGG-3′. PCR was performed using the following primers: 5′-CGACTGGAGCACGAGGACACTGA-3′ and 5′-CTGTGGCTATGCTCTTCGTTTCTGATGAAGGATTT-3′ for first round PCR, 5′-GGACACTGACATGGACTGAAGGAGTA-3′ and 5′-GGATTTGAAGTTGTGGTGGAGGAAATATGGGCATAAC-3′ for nested PCR.
Vector Construction
For the Muc21-84TR-IRES2-Venus vector, the N-terminal signal sequences and the C-terminal cytoplasmic domain of Muc21 were amplified using cDNA from mouse tissues as templates and primers (forward, 5′- GCTCTAGACAGTCACAGGGAGGCAGA-3′ and reverse, 5′-GCGAATTCGGCATAACCACTTCCTAAA-3′ for the N-terminal fragment; forward, 5′-TTCCAGCTCTAGCCTGAGTGCCACCC-3′ and reverse, 5′-CCTGGTACCGGGGCATACAGAAGC-3′ for the C-terminal fragment) and sequentially ligated to the pcDNA3.1(−) vector (Invitrogen). A pBS-SalNot vector was digested with BamHI and StuI. This vector was obtained by ligating a BAC clone (RP24-118O5) after digesting with SacI and thus encoded a part of exon II including 84 TR, intron II, and exon III of the Muc21 gene. The fragments thus obtained that had a TR domain were ligated to the previous vector containing the N-terminal signal sequence and the C-terminal cytoplasmic domain. The Muc21-84TR gene in the pcDNA3.1(−)-vector was obtained and further transferred to the pBluescript II SK(+) vector (Stratagene) to obtain restriction enzyme sites. The pBluescript II SK(+)-Muc21-84TR vector was digested with NotI and EcoRV and then ligated into the CSII-EF-MCS-IRES2-Venus vector (Riken BioResource Center). For the Muc21-4TR-IRES2-Venus vector, a DNA fragment that included 4 TR to the C-terminal stop codon was amplified by PCR and ligated to a pcDNA3.1(−) vector having the N-terminal signal sequence of Muc21. The obtained signal-4TR-Muc21 in the pcDNA3.1(−) vector was transferred into a pcDNA3.1(−)/hygro vector (Invitrogen) and then further transferred into the CSII-EF-MCS-IRES2-Venus vector.
For the N-terminal FLAG-tagged Muc21-84TR vector, a pair of oligonucleotides encoding a Kozak sequence, the signal sequence of Muc21, and a FLAG tag (a sense oligonucleotide, 5′-CTAGCAGGCAGAGGAAGATGCGGAGGAGAAGCAGCCTCTGGTGCTGGCTGCTTTTGCAGATCCTGCTTTTAGGAAGTGGTTATGCCGATTACAAGGATGACGACGATAAGGGAGGCGGAGGAGGCGCATCTAGCAAGG-3′ and an antisense oligonucleotide, 5′-AATTCCTTGCTAGATGCGCCTCCTCCGCCTCCCTTATCGTCGTCATCCTTGTAATCGGCATAACCACTTCCTAAAAGCAGGATCTGCAAAAGCAGCCAGCACCAGAGGCTGCTTCTCCTCCGCATCTTCCTCTGCCTG-3′) were annealed and used as N-terminal fragments.
For the N-terminal FLAG-tagged cytoplasmic domain-deficient Muc21 mutant, a DNA fragment encoding only two amino acids (RR) as a C-terminal cytoplasmic domain was prepared by PCR and ligated to BamHI, KpnI-digested N-terminal FLAG-tagged Muc21 with 84 TR in the pcDNA3.1(−) vector. For N-terminal FLAG and C-terminal c-myc-tagged Muc21 with 84 TR vector, a TR domain and a C-terminal domain were inserted into the pcDNA3.1(−) vector and then transferred to the p3XFLAG-myc-CMV-25 expression vector (Sigma). For a vector with N-terminal FLAG and C-terminal c-myc-tagged Muc21 and 29 TR or 19 TR, the N-terminal FLAG and C-terminal c-myc-tagged vector with Muc21 having 84 TR was digested with BglII. The resulting five fragments from the TR domain allowed in-frame translation when each of them was inserted into the above BglII-digested vector. The BglII-digested vector was ligated to itself in the presence of BglII-digested fragments. The resulting vector was digested with BglII to examine the size of the insert. The vector without BglII-digested fragments was assumed to have 19 TR and the vector with a 410-bp BglII fragment to have 29 TR. For the following vectors, each fragment from Muc21 was amplified by PCR, and the PCR fragments were inserted into the p3XFLAG-myc-CMV-25 expression vector: an N-terminal FLAG and C-terminal c-myc-tagged Muc21-4TR vector, an N-terminal FLAG and C-terminal c-myc-tagged Muc21-neck domain vector, and an N-terminal FLAG and C-terminal c-myc-tagged Muc21-SEA (sea-urchin sperm protein, enterokinase, and agrin)-like domain vector.
Transfection
Transfection was performed using TransIT-293 (Takara) according to the manufacturer's instruction. Cells were seeded at 2.5 × 105 cells in 24-well plates 2 h before transfection. DNA and reagent mixtures were added to cells after incubation for 30 min at room temperature. Cells were assayed at 36–48 h after transfection.
Flow Cytometry
Cells were detached from cell culture plates after incubating with 0.02% EDTA in phosphate-buffered saline for 3 min and were stained with one of following antibodies or lectins: Armenian hamster anti-Muc21 mAb 1A4-1, anti-human integrin α3 (mouse IgG1; Immunotech), anti-human integrin α5 (mouse IgG2b, SAM-1; Abcam), anti-human integrin αV (mouse IgG1, P2W7; Sigma), anti-human integrin β1 (mouse IgG1, SG/19; Seikagaku Corporation), mouse IgG (Zymed Laboratories Inc.), anti-human integrin α6 (rat IgG2a, GoH3; Immunotech), or rat IgG (ICN/Cappel). The cells were further stained with biotinylated goat anti-mouse IgG (Sigma) or biotinylated goat anti-rat IgG. Biotinylated peanut (Arachis hypogaea) agglutinin (PNA, 10 μg/ml; Vector Laboratories), biotinylated Vicia villosa agglutinin (VVA)-B4 (10 μg/ml, Vector laboratories), and biotinylated Sambucus seiboldiana agglutinin (SSA, 10 μg/ml; Seikagaku) were also used. Allophycocyanin-conjugated streptavidin (BioLegend) was applied, and then the cells were analyzed by BD FACSAria (BD) at an excitation wavelength 638 nm and emission wavelength 660/20 nm. The fluorescence protein products of Venus gene were also detected at an excitation wavelength 488 nm and emission wavelength 530/30 nm.
Terminal Deoxynucleotidyl Transferase dUTP Nick End Labeling (TUNEL) Assays
Assays were performed with an in situ cell death detection kit (Roche Applied Science) according to the manufacturer's instruction. The floating cells from Muc21 transfectants were collected and incubated with the TUNEL reaction mixture for 60 min at 37 °C after fixation and permeabilization. Fluorescence was detected with flow cytometry. As a negative control, cells were incubated with TUNEL reaction mixture without adding terminal transferase. As a positive control, cells were incubated with DNase I (10 units/100 μl) before reacting with TUNEL reaction mixture.
Quantification of the Floating Cells
Medium from transient transfectants was collected, and cells in the medium were counted after centrifugation. Adherent cells were detached with 0.05% trypsin-EDTA treatment and counted. Total cells were calculated by summing the nonadherent cells and the adherent cells. Transfection efficiency was evaluated by detecting Venus-positive cells or FLAG-positive cells by the use of flow cytometry.
Western Blotting and Lectin Blotting
Transiently transfected 293T cells with mock and Muc21 vectors that had N-terminal FLAG tags were treated in lysis buffer (50 mm Tris-HCl, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100). The cell lysates were immunoprecipitated with an anti-FLAG M2 affinity gel (Sigma) for 3 h at 4 °C. The immunoprecipitated FLAG-tagged Muc21 molecules were electrophoretically separated on 4% polyacrylamide gels in the presence of 0.1% SDS and blotted onto polyvinylidene difluoride membranes (GE Healthcare), and then reacted with biotinylated anti-FLAG antibodies (Sigma) or lectins (2.5 μg/ml) and further reacted with horseradish peroxidase-streptavidin (Zymed Laboratories Inc.). For Western blotting with anti-integrin antibodies, mock and 4 TR transfectants were lysed after transient transfection. For transfectants with 84 TR, only floating cells were collected and lysed with lysis buffer. Fifty micrograms of cell lysates were electrophoretically separated on 7.5% polyacrylamide gels in the presence of 0.1% SDS and blotted onto polyvinylidene difluoride membranes. The membranes were cut in the middle so they could react separately with anti-β-tubulin mAb clone TUB2.1 (expected band at 55 kDa; Sigma) for the lower part and anti-human integrin antibodies for the upper part. Goat anti-human integrin α3 pAb (expected band at 150 kDa; Santa Cruz Biotechnology), mouse anti-human integrin α5 mAb (expected band at 150 kDa; BD Transduction Laboratories), rat anti-human integrin α6 mAb (expected band at 140 kDa, GoH3; Immunotech), mouse anti-human integrin αV mAb (expected band at 160 kDa, P2W7; Sigma), or mouse anti-human integrin β1 mAb (expected band at 115 kDa; Millipore) was used. Horseradish peroxidase-goat anti-mouse IgGs (Zymed Laboratories Inc.), horseradish peroxidase-rabbit anti-goat IgGs (Zymed Laboratories Inc.), or horseradish peroxidase-goat anti-rat IgGs (Zymed Laboratories Inc.) was used as a secondary antibody.
Homotypic Cell Aggregation Assays
Twenty-four-well plates were coated with 500 μl of 1% bovine serum albumin in HCMF (HEPES buffer, Ca2+- and Mg2+-free) buffer (8 g of NaCl, 0.4 g of KCl, 0.12 g of Na2HPO4·12H2O, 1.0 g of glucose, 2.38 g HEPES to 1l, pH 7.4) overnight at 4 °C and rinsed with HCMF buffer twice. Plates were kept on ice until loading cells. Cells were detached by 2 min of 0.05% trypsin-EDTA treatment and were untreated or treated with 5 units/ml sialidase (neuraminidase from Clostridium perfringens (C. welchii); Sigma) in serum-free medium for 30 min at 37 °C. After washing with PBS, cells were resuspended in HCMF buffer containing DNase I (10 μg/ml) and MgCl2 (1 mm final concentration) and then sorted for collection of Venus-positive populations. The sorted cells were resuspended in the same buffer, and 5 × 104 cells in 500 μl/well were added to the previous 24-well plates. After addition of 56 μl of 100 mm CaCl2 to the wells, plates were placed in a gyrating shaker rotating at 80 rpm and incubated for 30 min to 2 h at 37 °C. The number of cell aggregates was counted every 30 min by observation under the microscope. Percent aggregation was determined by (N0 − Nt)/N0 ×100, where Nt is the total particle number (aggregates plus single cells) at incubation time t, and N0 is the total cell number in the cell suspension. More than three visual fields were counted. Sialidase treatment was confirmed by increased binding of treated cells to PNA.
Adhesion Assays
The 96-well plates (Iwaki) were coated with 50 μl of solution/well, each containing one of following molecules; 20 μg/ml fibronectin (Sigma), 20 μg/ml laminin (Sigma), 40 μg/ml collagen IV (Sigma), 50 μg/ml matrigel (BD), 40 μg/ml bovine serum albumin (Sigma) as a negative control, and 0.002% poly-l-lysine (Wako), at 4 °C overnight. After blocking with 1% bovine serum albumin in Dulbecco's phosphate-buffered saline, the plates were washed with Dulbecco's phosphate-buffered saline three times, and 5 × 104 cells were added to each well in triplicate. After 3 h of incubation, the plates were washed with Dulbecco's phosphate-buffered saline three times very carefully, stained with crystal violet, lysed, and measured for absorbance at 550 nm to quantify the attached cells. Additional procedures can be found in supplemental Materials.
RESULTS
Characterization of the Gene Structure of Muc21
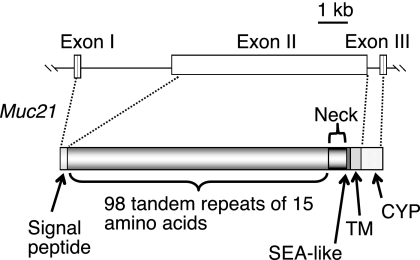
The Muc21 gene was previously identified by representational differential analysis between TA3-Ha and TA3-St cells (11). The gene resides in the major histocompatibility complex class I region of mouse chromosome 17. In the same study, the C-terminal sequences of Muc21 including 10 TR were identified by 3′-RACE using cDNA from TA3-Ha cells. As an effort to clone the full-length cDNA of Muc21, we found a BAC clone (RP24-11805) with a Muc21 gene on one end. The Muc21 gene region was transferred to the pBluescript SK(+) vector to perform sequencing. The results revealed that the Muc21 gene from the BAC clone contained 84 TR and the C-terminal domain. The recently updated NCBI database (NC_000083.5) disclosed TR sequences, including previously unidentified sequences in the middle of the TR domain. Although the predicted gene did not include the complete structure of the Muc21 gene as having 95 TR, the sequences of the 84 TR upstream of the C terminus were exactly the same as the 84 TR from the BAC clone. 5′-RACE based on the DNA sequence data from NCBI resulted in a product that encoded exon I and the first portion of exon II, thus suggesting that intron I is more than 3 kb in length. Exon II encodes 3 amino acids corresponding to a portion of the signal sequence, and the subsequent sequence corresponded to the tandem repeat sequences. The 4th repeat matched with the 95th repeat (counted from the one nearest to the transmembrane domain) revealed by the NCBI database. Therefore, Muc21 was presumed to have 98 TR in its TR domain. Collectively, similar to human Muc21 previously cloned in our laboratory as a type I transmembrane protein, Muc21 consists of 3 exons. Exon I encodes a signal sequence; exon II encodes the end of the signal sequence, a TR domain, a neck domain, an SEA-like sequence, and a transmembrane domain; and exon III encodes a cytoplasmic domain (Fig. 1).
FIGURE 1.
Muc21/epiglycanin is assumed to have 98 TR of 15 amino acids. Muc21 consists of three exons. Exon I encodes a signal sequence; exon II encodes the end of signal sequence, a TR domain, a neck domain, an SEA-like domain, and a transmembrane (TM) domain; and exon III encodes a cytoplasmic (CYP) domain. Exon I was discovered by 5′-RACE, and the full size of the TR domain was estimated by the results of 5′-RACE, NCBI database analysis, and partial sequencing of Muc21 gene from a BAC clone.
All TR were composed of 15 amino acids except the 2nd TR with 13 amino acids and the 9th and the 15th tandem repeats, each having 12 amino acids. Among 98 TR, the 4 C-terminal TR had atypical sequences. These atypical TR differed from the rest of the TR, but still showed a high proportion of serine and threonine. The proposed complete molecular structure of Muc21 is described in Fig. 1.
Construction of the Muc21 Expression Vector and Its Expression in Cells
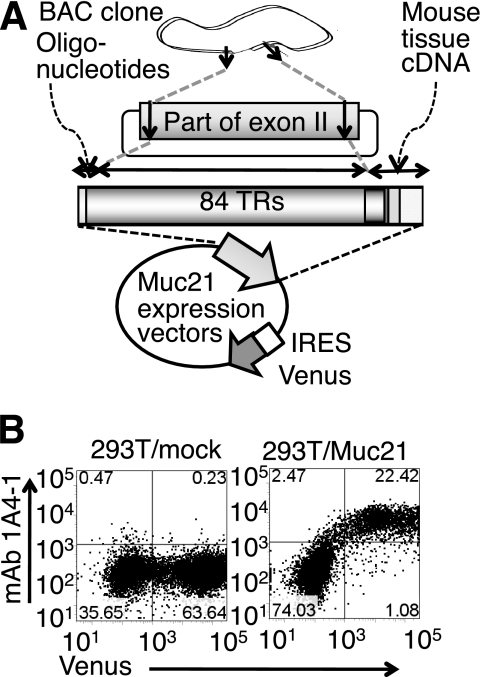
A Muc21 expression vector was constructed by combining three fragments: an N-terminal and a C-terminal fragment from PCR using cDNA from mouse tissues and a fragment having 84 TR from the BAC clone. This artificial Muc21 cDNA contained all elements of the Muc21 gene, except that the N-terminal 14 TR were deleted. This cDNA was incorporated into an IRES-Venus vector, which allows bicistronic gene expression (Fig. 2A). In some experiments shown below, Muc21 with an N-terminal FLAG tag was constructed by synthesizing the N-terminal sequences, including the Kozak sequence, the signal sequence, and the FLAG tag sequence. When the Muc21-IRES2-Venus vector was transfected into 293T cells, Muc21 was detected at the cell surface by Muc21 mAb 1A4-1, and the gene expression was assured by the Venus gene products that make it possible to detect transfected cells (Fig. 2B). The monoclonal antibody 1A4-1 was previously generated in our laboratory and is specific to Muc21. Establishment and specificity of mAb 1A4-1 will be published in a separate paper.
FIGURE 2.
Artificial Muc21/epiglycanin cDNA having 84 TR was constructed and expressed in 293T cells. A, methods of constructing the Muc21 expression vector are summarized. B, cells transiently transfected with the Muc21-IRES2-Venus vector were stained with mAb 1A4-1 and analyzed by flow cytometry using allophycocyanin-conjugated streptavidin as a fluorescence group at an excitation wavelength 638 nm and emission wavelength 660/20 nm.
Expressed Mucin and Its Glycoforms
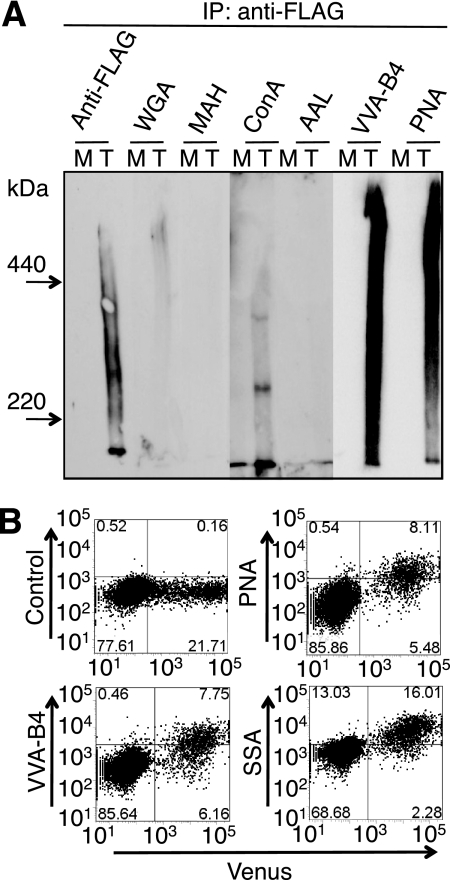
When cell lysates were prepared after transient transfection of the N-terminal FLAG-tagged Muc21 vector and immunoprecipitated with anti-FLAG antibody, mucin-like large highly glycosylated glycoproteins were detected (Fig. 3A). Immunoprecipitation with anti-FLAG antibody and Western blot analysis with anti-FLAG antibody or lectins revealed broad bands between 440 kDa and 220 kDa, apparently reflecting the heterogeneity of Muc21 glycosylation. Results of lectin blotting showed abundant binding of PNA and VVA-B4 to immunoprecipitated N-terminal FLAG-tagged Muc21 (Fig. 3A). The binding intensity of PNA and VVA-B4 increased with the expression of Muc21 (Fig. 3A). These results indicate that the Muc21 cDNA was translated to a mucin molecule having a large molecular mass with a high degree of O-glycosylation, displayed on the cell surface. The majority of carbohydrate moieties were suggested to be Galβ1–3GalNAc, recognized by PNA, and GalNAc, recognized by VVA-B4. The presence of sialic acids in the carbohydrate chains of expressed Muc21 was suggested by flow cytometry with SSA, which increased with Venus expression in 293T cells transfected with Muc21 with 84 TR (Fig. 3B), although binding of lectins specific for sialylated lectins was not clear in the lectin blotting analysis.
FIGURE 3.
Expression of the Muc21 resulted in large highly glycosylated glycoproteins with o-glycans. A, a mock (indicated as M) vector and a Muc21 expression vector having 84 TR and N-terminal FLAG tag (indicated as T) were transfected into 293T cells, and the cell lysates were precipitated with anti-FLAG antibody. The immunoprecipitates were separated on 4% polyacrylamide gels in the presence of 0.1% SDS, blotted onto polyvinylidene difluoride membranes, and detected with the same antibody or lectins. B, 293T cells transiently transfected with the Muc21-84TR-IRES2-Venus vector were stained with biotinylated PNA, VVA-B4, or SSA and analyzed by flow cytometry using biotinylated allophycocyanin-conjugated streptavidin as a fluorescence group at an excitation wavelength 638 nm and emission wavelength 660/20 nm.
Morphologic Changes Induced by Muc21 Expression
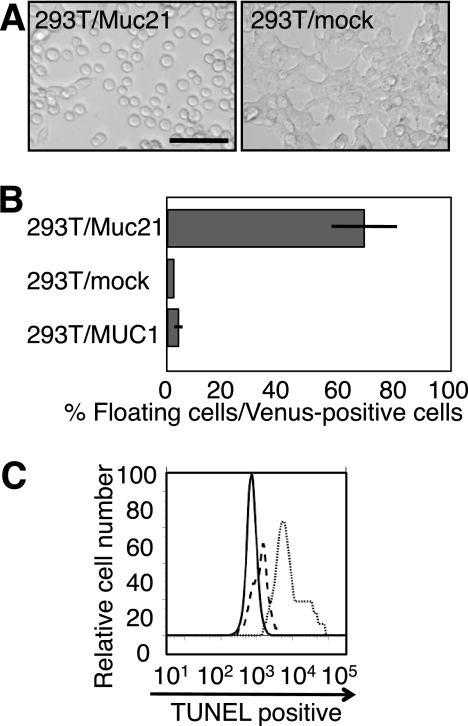
Upon transient expression of Muc21 in 293T cells, Venus-positive cells lost their extensions, became round, and remained floating (Fig. 4A). Nonadherent cells were collected from the medium supernatants, counted, and their percentage among Venus-positive cells was calculated. The results showed that more than half of the Venus-positive cells became nonadherent in 293T/Muc21 transfectants, whereas 293T/mock transfectant medium contained few floating cells (Fig. 4B). When instead of Muc21, the MUC1 gene was transfected into 293T cells using the same vector, 293T/MUC1 transfectants did not show nonadherent phenotypes, suggesting that loss of adhesion was unique to Muc21 (Fig. 4B). Floating cells obtained from the culture medium of Muc21-transfected cells were TUNEL-negative, clearly demonstrating that the floating phenotypes were not due to apoptosis (Fig. 4C).
FIGURE 4.
Muc21 expression in 293T cells induced morphologic changes. A, pictures of cells transiently transfected with mock or Muc21-84TR-IRES2-Venus vector are shown. Scale bar indicates 100 μm. B, the number of cells from the culture medium after transient transfection of Muc21-84TR-IRES2-Venus was evaluated against cells expressing the vectors as Venus-positive cells. C, the floating cells after transient expression of the N-terminal FLAG-tagged Muc21 vector were collected from culture medium and were subjected to TUNEL assays. The solid line, dashed line, and dotted line indicate the floating cells, negative control, and positive control, respectively.
Examination of Domains Responsible for the Nonadherent Phenotypes
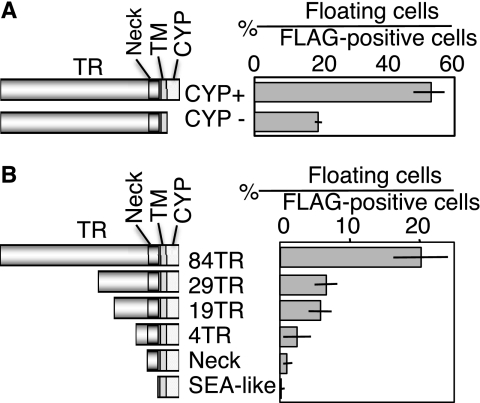
To test which domain of Muc21 was responsible for the morphological change, various Muc21 mutants were constructed. A cytoplasmic domain-deficient mutant having only 2 amino acids from the cytoplasmic domain of Muc21 but with the rest of the gene intact was constructed with an N-terminal FLAG tag in the pcDNA3.1(−) vector. When this vector was transfected into 293T cells, nonadherent phenotypes similar to those observed with intact cytoplasmic domain were still observed. The percentage of the floating cells among anti-FLAG-positive cells, however, was reduced (Fig. 5A).
FIGURE 5.
Long TR domain seemed to have a major effect on the antiadhesive property of Muc21. A, mutants lacking a cytoplasmic domain were constructed with an N-terminal FLAG tag and transfected into 293T cells. The number of floating cells in FLAG-positive cells was evaluated. B, extracellular domain mutants having various portions of Muc21 were constructed with an N-terminal FLAG tag and C-terminal c-myc tag and transfected into 293T cells. Note that the mutants in B were constructed using a different vector system from the mutants in A.
Because the cytoplasmic domain seemed not fully responsible for the nonadhesiveness, the contribution of the extracellular domain of Muc21 was examined. A 4 TR mutant that contained the atypical TR, a neck domain mutant, and an SEA-like sequence mutant were constructed with an N-terminal FLAG tag in the p3XFLAG-myc-CMV-25 expression vector. To investigate a correlation between the number of TR and the induction of antiadhesion, TR domain mutants having 84, 29, and 19 TR were constructed with N-terminal FLAG tag in the same vector. When these mutants were transfected into 293T cells, floating cells were observed with transfectants that had 84, 29, and 19 TR. The transfected cells with Muc21 with 4 TR did not float but displayed a round morphology (data not shown). Transfectants containing the neck domain mutant and the SEA-like sequence mutant were adherent. The percentage of the floating cells among the FLAG tag-positive cells clearly correlated with the number of TR, revealing that the large TR domain is responsible for the nonadherent phenotypes (Fig. 5B).
Homotypic Cell Aggregation and Adhesion to Extracellular Matrix
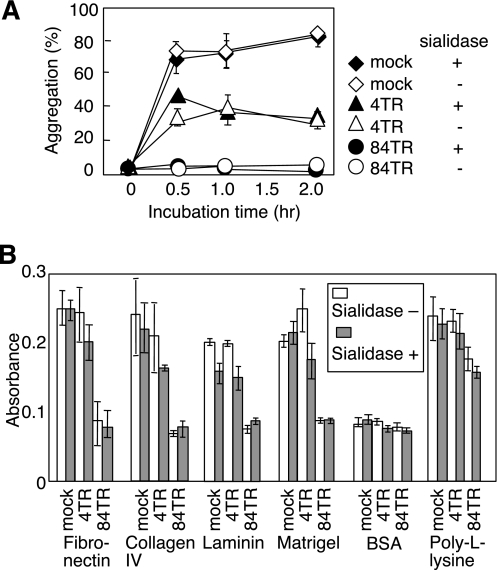
We further tested whether Muc21 interferes with homotypic cell-cell adhesion. After transient expression of the Muc21-IRES2-Venus vector, Venus-positive cells were collected by cell sorting to >90% purity. Sorted Venus-positive cells were subjected to homotypic aggregation assays. When the cells were allowed to aggregate by rotating at 37 °C, marked cell aggregates were observed in 293T/mock transfectants after 30 min. However, 293T cells transfected with Muc21 with 84 TR did not form aggregates at all, even after 2 h of incubation. The 293T cells transfected with Muc21 with 4 TR partially inhibited cell-cell interactions (Fig. 6A). These results indicate that Muc21 prevented homotypic cell-cell adhesion in a TR-dependent manner.
FIGURE 6.
Muc21 expression prevented homotypic cell-cell and cell-ECM interactions in a TR-dependent manner. A, mock-transfected cells (diamonds), cells transiently expressing Muc21 with 4 TR (triangles) or Muc21 with 84 TR (circles) were compared for homotypic aggregation. The cells were untreated (open symbols) or treated with sialidase (filled symbols) and sorted to collect Venus-positive populations. More than 90% Venus-positive cells were used. B, adhesion assays were performed with the same preparations of cells as in A. The six consecutive columns represent, from left to right, mock transfectants, sialidase-treated mock transfectants, cells transiently expressing Muc21 with 4 TR before and after sialidase treatment, and cells transiently expressing Muc21 with 84 TR before and after sialidase treatment. BSA, bovine serum albumin.
Adhesion of transfectants to extracellular matrix components was also tested. Venus-positive cells were seeded in wells coated with fibronectin, collagen IV, laminin, matrigel, bovine serum albumin, or poly-l-lysine. After 3 h of incubation, 293T cells transfected with Muc21 with 84 TR did not adhere to any of the surfaces except poly-l-lysine-coated plates (Fig. 6B), suggesting that Muc21 prevents integrin-mediated cell adhesion to extracellular matrix components (12). 293T cells transfected with Muc21 with 4 TR showed little difference in adhesion compared with 293T/mock transfectants. These results suggested that Muc21 hindered cell-ECM interaction through its large TR domain, likely by interfering with the function of integrins.
It was previously suggested that negative charges on the cell surface could induce antiadhesive effects by charge repulsion (13). Therefore, the contribution of sialic acid to the antiadhesive effect was evaluated by measuring antiadhesion before and after enzymatic removal of sialic acid. As shown in Fig. 6, A and B, sialic acids did not contribute to the antiadhesive effect of Muc21 because sialidase-treated and untreated cells showed the same levels of adhesion to cells and to ECM components.
Mechanism of the Antiadhesive Effect of Muc21
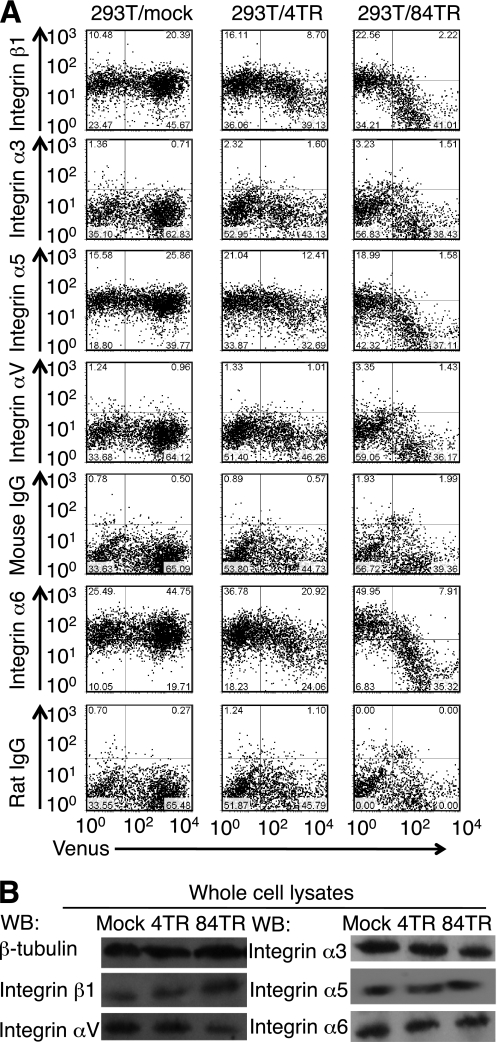
To investigate the possibility that integrin functions are modulated, accessibility of cell surface integrin α3, α5, α6, αV, and β1 by specific antibodies was evaluated by flow cytometry (Fig. 7A). Reduced binding of anti-α5, α6, and β1 integrin antibodies was observed with Venus-positive cells, judging from the decrease in the number of cells having allophycocyanin fluorescence above the control level shown by the dots. Antibody binding inversely correlated with the number of TR and Muc21 transfectants with the 84 TR showing the lowest antibody binding. The reduced binding of these antibodies to 293T/Muc21 transfectants was not due to the reduced expression of these integrin subunits because the protein expression levels were not reduced at the whole cell lysate levels as shown by the Western blot analysis (Fig. 7B). Therefore, the floating phenotypes of Muc21-transfectant cells could be explained at least in part by a reduced cell surface accessibility of integrins caused by the presence Muc21 TR possibly resulting in an impairment of integrin-mediated adhesion.
FIGURE 7.
Cell surface accessibilities of integrins α5, α6, and β1 were reduced by the expression of Muc21 with 84 TR. A, cells transiently transfected with mock, Muc21-4TR-IRES2-Venus, or Muc21-84TR-IRES2-Venus vectors were stained with anti-integrin α3, α5, α6, αV, or β1 antibodies, biotinylated second antibodies, and allophycocyanin-conjugated streptavidin and analyzed by flow cytometry. B, the adherent cells after transfection with mock or the Muc21-4TR-IRES2-Venus vector or the floating cells after transfection with the Muc21-84TR-IRES2-Venus vector were collected and lysed in lysis buffer. Cell lysates were separated electrophoretically on 7.5% polyacrylamide gels in the presence of 0.1% SDS and blotted onto polyvinylidene difluoride membranes. The blotted membranes were stained with anti-integrin α3, α5, α6, αV, or β1 antibodies or anti-β-tubulin antibodies. Although integrin subunits α3 and αV were below the control levels shown by the dots by flow cytometry, immunoprecipitates by the corresponding antibodies were observed by the Western blotting (WB).
DISCUSSION
TA3-Ha and TA3-St mouse mammary carcinoma variant cells show differential cellular characteristics, including the observation that TA3-Ha cells were less adhesive. Epiglycanin, displayed on TA3-Ha cells but not on TA3-St cells, was assumed to provide antiadhesive properties to the cells (9). However, evidence showing direct involvement of epiglycanin in the antiadhesive effect was missing until the epiglycanin gene was identified and the glycoform of the gene product was determined. In the present report, we clearly showed that Muc21, the molecular entity of epiglycanin, led to the nonadhesive phenotypes in 293T cells and prevented cell-cell and cell-ECM interactions. Furthermore, we showed that among the different domains of the molecule it was mainly the large TR domain that was responsible for induction of antiadhesion.
Muc21 is not yet fully cloned, but the gene structure and full-length TR were revealed by combining several sequence data results from 3′-RACE, 5′-RACE, partial sequencing, and NCBI database analysis. As expected from the gene structure, the expressed Muc21 protein having 84 TR showed a large molecular size and high glycosylation, reflecting the characteristics of epiglycanin. Transient transfection of the Muc21-IRES2-Venus vector induced a nonadhesive phenotype in Venus-positive cells. Nonadhesive phenotypes were observed not only with Muc21-transfected 293T cells, but also with B16-F1 melanoma cells and with NIH3T3 fibroblast cells (data not shown), strongly suggesting that the loss of adhesion mediated by Muc21 occurred independent of the recipient cells. Due to the highest transfection efficiency, we used 293T cells for further experiments. The floating 293T cells transfected with Muc21 with 84 TR survived and clonal populations were obtained. The growth rates of clones obtained from the transfectants with 84 TR and from mock transfectants were examined in regular tissue culture plates. All clones were shown to be growing, although growth rates were dependent on each clone (supplemental Fig. 1).
The mechanism of the loss of adhesion by mucins or glycoproteins having mucin domains was illustrated in various aspects, such as charge repulsion by sialic acids and steric hindrance by extended configurations. The contribution of sialic acids on the antiadhesive effects was previously shown for CD43 (14) and podocalyxin (12). However, the gene product of Muc21 in 293T cells seemed to contain mainly Galβ1–3GalNAc residues recognized by PNA and mAb 1A4-1. Furthermore, our results indicated that the antiadhesive effect of Muc21 with 84 TR was not significantly affected by sialidase treatment, indicating that contributions of sialic acids are negligible (Fig. 6, A and B). The direct role of the TR domain in the antiadhesive effect was proposed for MUC1 (15, 16) and sialomucin Muc4 (17), both by using mutants having different numbers of TR. Our present results clearly demonstrate the dominant role of the Muc21 TR domain in the induction of antiadhesion in Muc21-transfected cells (Figs. 5B and 6, A and B). However, Fig. 5A clearly showed a decrease in the antiadhesive effect of Muc21 lacking its cytoplasmic domain. There are several possible mechanisms by which the cytoplasmic domain could regulate cell-cell adhesion as discussed below.
Muc21 prevented homotypic cell-cell interactions and cell-ECM interactions. One possible mechanism may be a Muc21-mediated blocking of surface integrins resulting in an impairment of integrin function (Fig. 7A). Such concealing effects may not be specific for integrins. 293T cells transfected with Muc21 also showed reduced binding of antibody to human leukocyte antigen ABC and to several lectins (data not shown), thus there maybe a general masking effect due to the large size of the Muc21 molecule. The allotransplantability of TA3-Ha cells was previously explained by a similar masking effect by epiglycanin at the cell surface (18, 19). Epiglycanin was previously reported to be responsible for the lack of adhesiveness because capping of epiglycanin by its antibody led to increased adhesion to Kalinin and laminin (9).
Examination of the behavior of 293T cells transfected with Muc21 with 4 TR showed that cell-cell interactions were disrupted, but cell-ECM interactions were not. These results were consistent with the appearance of transfectants observed under the microscope, where a large population of Venus-positive cells after transfection by Muc21-4TR-IRES2-Venus vector showed a round phenotype but rarely became floating (data not shown). The length of integrins and cadherins from the cell surface is considered to be approximately 23 nm (17), and the length of Muc21 with 4 TR from the cell surface is predicted to be around 45 nm, as calculated by an extended polypeptide (10). Judging from this, loss of adhesion by a high expression of Muc21 with 4 TR is technically possible. The role of the cytoplasmic domain of Muc21 on the antiadhesiveness is currently unclear. The cytoplasmic domain of Muc21 had several potential functional motifs such as protein kinase C phosphorylation sites, a tyrosine kinase phosphorylation site, and a PDZ domain ligand site, which might induce morphologic changes to the cell. The incubation of 293T/Muc21 transfectants with a variety of protein kinase inhibitors, however, did not reverse the floating phenotypes induced by Muc21 expression (data not shown). The floating cells were observed in transfectants of mutant genes lacking the cytoplasmic domain. However, the appearance of the floating cells was delayed, as few floating cells were seen at 24 h after transfection (data not shown), and the percentage among vector-transfected cells was markedly reduced (Fig. 5A). Therefore, we could not exclude the possibility that the cytoplasmic domain actively participates in the antiadhesive effect, for instance, by stabilizing the cell surface display of Muc21. Although MUC1 is known for its antiadhesive property, 293T cells transfected with MUC1 with 22 TR did not show a phenotype similar to that seen with Muc21 under similar conditions. The reason for this strong antiadhesive effect of Muc21 might be due to its molecular structure, as discussed below. First, Muc21 is a large molecule, which could extend above the cell surface. Second, Muc21 has the highest number of potential O-glycosylation sites among all known mucin molecules. The percentage of serine and threonine residues among total amino acids of Muc21 is ∼60%, whereas other mucins remain at 20–40% (11). For example, MUC1 has 5 potential O-glycosylation sites in a TR consisting of 20 amino acids, whereas Muc21 has 10 potential O-glycosylation sites in a TR consisting of 15 amino acids. The combined effect of the large size and high glycosylation is likely the cause of the striking phenotypic changes when Muc21 is expressed.
Antiadhesiveness and steric hindrance potentially play important roles in various biological processes. To initiate metastasis, primary tumor cells need to detach from neighboring cells and from extracellular matrices. Cancer cells have a survival advantage against an attack by the host immune system. It was previously proposed that the steric hindrance effect of sialomucin Muc4 contributed to the immune evasion by cancer cells (20). Interestingly, several reports showed that mucins expressed on cancer cell surfaces limit the effectiveness of antitumor agents by preventing their cell accessibility (21, 22). Under normal physiological conditions, MUC1 and MUC16 prevented trophoblast adhesion to uterine endothelium (23, 24).
The consequences of the strong antiadhesive effect of Muc21 potentially affect many aspects. Whether Muc21 expression causes high metastatic potential should be examined using appropriate experimental models and taking the glycoforms into account. Importantly, the mRNA of Muc21 was detected in trachea, esophagus, and vagina, and protein expression was observed in the luminal surfaces of esophagus and vagina as detected by mAb 1A4-1.3 The manifested antiadhesive effects of Muc21 might provide an answer to the question of why Muc21 is uniquely expressed in those organs, which need an especially strong protection from harsh external environment and stimuli.
In summary, an antiadhesive property was observed in cells expressing a novel transmembrane mucin, Muc21. The underlying mechanism of this phenomenon can in part be explained by steric hindrance caused by the large highly glycosylated TR domain. Further investigations are necessary to show that antiadhesion caused by Muc21 is implicated in the behavior of cells in vivo.
Supplementary Material
Acknowledgments
We thank Kyoko Sakai and Miki Noji for assistance in the preparation of this manuscript; Miho Mori and Satomi Yoshinaga for technical assistance; Dr. Masaaki Yamazaki of Fujiya, Co., Ltd. for assistance in DNA sequencing; and Dr. Olivera Finn of the University of Pittsburgh for providing MUC1 cDNAs having 22 TR.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1 and Materials.
M. Hoshino, M. Kamata-Sakurai, K. Denda-Nagai, T. Itoh, K. Okada, K. Ishii-Schrade, Y. Yi, S. Toraya, and T. Irimura, unpublished data.
- TR
- tandem repeat
- RACE
- rapid amplification of cDNA ends
- BAC
- bacterial artificial chromosome
- IRES
- internal ribosome entry site
- SEA
- sea-urchin sperm protein, enterokinase, and agrin
- PNA
- peanut agglutinin
- VVA-B4
- Vicia villosa agglutinin-B4
- SSA
- Sambucus seiboldiana agglutinin
- TUNEL
- terminal deoxynucleotidyl transferase dUTP nick end labeling
- HCMF buffer
- HEPES buffer, Ca2+- and Mg2+-free
- ECM
- extracellular matrix.
REFERENCES
- 1.Codington J. F., Linsley K. B., Jeanloz R. W., Irimura T., Osawa T. (1975) Carbohydr. Res. 40, 171–182 [DOI] [PubMed] [Google Scholar]
- 2.Codington J. F., Sanford B. H., Jeanloz R. W. (1972) Biochemistry 11, 2559–2564 [DOI] [PubMed] [Google Scholar]
- 3.Hauschka T. S., Weiss L., Holdridge B. A., Cudney T. L., Zumpft M., Planinsek J. A. (1971) J. Natl. Cancer Inst. 47, 343–359 [PubMed] [Google Scholar]
- 4.Klein G. (1951) Exp. Cell Res. 2, 291–294 [Google Scholar]
- 5.Friberg S., Jr. (1972) J. Natl. Cancer Inst. 48, 1463–1476 [PubMed] [Google Scholar]
- 6.Codington J. F., Cooper A. G., Miller D. K., Slayter H. S., Brown M. C., Silber C., Jeanloz R. W. (1979) J. Natl. Cancer Inst. 63, 153–161 [PubMed] [Google Scholar]
- 7.Miller S. C., Hay E. D., Codington J. F. (1977) J. Cell Biol. 72, 511–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fung P. Y., Longenecker B. M. (1991) Cancer Res. 51, 1170–1176 [PubMed] [Google Scholar]
- 9.Kemperman H., Wijnands Y., Wesseling J., Niessen C. M., Sonnenberg A., Roos E. (1994) J. Cell Biol. 127, 2071–2080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Codington J. F., Haavik S. (1992) Glycobiology 2, 173–180 [DOI] [PubMed] [Google Scholar]
- 11.Itoh Y., Kamata-Sakurai M., Denda-Nagai K., Nagai S., Tsuiji M., Ishii-Schrade K., Okada K., Goto A., Fukayama M., Irimura T. (2008) Glycobiology 18, 74–83 [DOI] [PubMed] [Google Scholar]
- 12.Takeda T., Go W. Y., Orlando R. A., Farquhar M. G. (2000) Mol. Biol. Cell 11, 3219–3232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ligtenberg M. J., Buijs F., Vos H. L., Hilkens J. (1992) Cancer Res. 52, 2318–2324 [PubMed] [Google Scholar]
- 14.Ardman B., Sikorski M. A., Staunton D. E. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 5001–5005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wesseling J., van der Valk S. W., Hilkens J. (1996) Mol. Biol. Cell 7, 565–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wesseling J., van der Valk S. W., Vos H. L., Sonnenberg A., Hilkens J. (1995) J. Cell Biol. 129, 255–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Komatsu M., Carraway C. A., Fregien N. L., Carraway K. L. (1997) J. Biol. Chem. 272, 33245–33254 [DOI] [PubMed] [Google Scholar]
- 18.Cooper A. G., Codington J. F., Miller D. K., Brown M. C. (1979) J. Natl. Cancer Inst. 63, 163–169 [PubMed] [Google Scholar]
- 19.Miller S. C., Codington J. F., Klein G. (1982) J. Natl. Cancer Inst. 68, 981–988 [PubMed] [Google Scholar]
- 20.Komatsu M., Yee L., Carraway K. L. (1999) Cancer Res. 59, 2229–2236 [PubMed] [Google Scholar]
- 21.Nagy P., Friedländer E., Tanner M., Kapanen A. I., Carraway K. L., Isola J., Jovin T. M. (2005) Cancer Res. 65, 473–482 [PubMed] [Google Scholar]
- 22.Kalra A. V., Campbell R. B. (2009) Eur. J. Cancer 45, 164–173 [DOI] [PubMed] [Google Scholar]
- 23.Chervenak J. L., Illsley N. P. (2000) Biol. Reprod. 63, 294–300 [DOI] [PubMed] [Google Scholar]
- 24.Gipson I. K., Blalock T., Tisdale A., Spurr-Michaud S., Allcorn S., Stavreus-Evers A., Gemzell K. (2008) Biol. Reprod. 78, 134–142 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.