Abstract
Huntington disease is caused by expanded polyglutamine sequences in huntingtin, which procures its aggregation into intracellular inclusion bodies (IBs). Aggregate intermediates, such as soluble oligomers, are predicted to be toxic to cells, yet because of a lack of quantitative methods, the kinetics of aggregation in cells remains poorly understood. We used sedimentation velocity analysis to define and compare the heterogeneity and flux of purified huntingtin with huntingtin expressed in mammalian cells under non-denaturing conditions. Non-pathogenic huntingtin remained as hydrodynamically elongated monomers in vitro and in cells. Purified polyglutamine-expanded pathogenic huntingtin formed elongated monomers (2.4 S) that evolved into a heterogeneous aggregate population of increasing size over time (100–6,000 S). However, in cells, mutant huntingtin formed three major populations: monomers (2.3 S), oligomers (mode s20,w = 140 S) and IBs (mode s20,w = 320,000 S). Strikingly, the oligomers did not change in size heterogeneity or in their proportion of total huntingtin over 3 days despite continued monomer conversion to IBs, suggesting that oligomers are rate-limiting intermediates to IB formation. We also determined how a chaperone known to modulate huntingtin toxicity, Hsc70, influences in-cell huntingtin partitioning. Hsc70 decreased the pool of 140 S oligomers but increased the overall flux of monomers to IBs, suggesting that Hsc70 reduces toxicity by facilitating transfer of oligomers into IBs. Together, our data suggest that huntingtin aggregation is streamlined in cells and is consistent with the 140 S oligomers, which remain invariant over time, as a constant source of toxicity to cells irrespective of total load of insoluble aggregates.
Keywords: Amyloid, Biophysics, Huntington Disease, Protein Self-assembly, Ultracentrifugation, Analytical Ultracentrifugation, PolyQ, Polyglutamine
Introduction
The misfolding and aggregation of proteins is thought to underlie pathogenesis in a range of amyloid-related neurodegenerative diseases, including Alzheimer, Creutzfeldt-Jakob, and Parkinson diseases (1). Huntington disease (HD)2 is one example of a fatal neurodegenerative condition caused by mutations in the huntingtin gene that induce the huntingtin protein (Htt) to oligomerize into larger forms (2–5). The mutations lead to an expansion in an N-terminal polyglutamine (polyQ) sequence to beyond an HD threshold of 37 residues (6). Longer polyQ sequences correlate with faster aggregation rates and earlier ages of disease onset, which implicates the intrinsic aggregation potential as central to the mechanisms causing pathogenesis.
A key marker of pathology is the coalescence of mutant Htt into punctate intracellular reservoirs of aggregates known as inclusion bodies (IBs), yet despite intensive research over the last 20 years, it is still not well understood how this process, or how aggregation more generally, relates to cellular dysfunction. Although IB co-sequestration of other cellular components has been postulated as a mode of toxicity in HD (7, 8), more recent evidence suggests that IBs are not themselves toxic but are an active response by cells to sequester more toxic soluble forms (3).
Although it is now well established that SDS insolubility of mutant Htt loosely correlates to inclusions, there is a dearth of knowledge about the kinetics of aggregation of native Htt complexes expressed directly in the cellular environment (9). Knowledge of this process is fundamentally important to understanding the earliest molecular steps of pathogenesis and for determining how the cellular machinery engages with the mutant forms of Htt. Several studies examining mutant Htt in cellular contexts have unequivocally identified the presence of oligomers in the range of 5–80 nm (10–12). Other work has shown that SDS-insoluble oligomers accumulate in cell culture and transgenic mouse models of HD prior to visible inclusions (9). However, insight into the full heterogeneity, quantity, and flux of the soluble forms of Htt under non-denaturing conditions has been hampered by a lack of high resolution and unbiased approaches to directly characterize and quantify these forms (13).
We sought to investigate the aggregation kinetics of mutant huntingtin expressed in mammalian cells using fluorescence-adapted sedimentation velocity (SV) analysis in the analytical ultracentrifuge. SV analysis is a powerful approach for defining complex molecular size distributions and hydrodynamic properties of soluble proteins, including that of aggregating proteins (14, 15). However, only recently have advances in analytical ultracentrifugation hardware enabled green fluorescent protein (GFP)-tagged proteins to be uniquely tracked in complex solutions by virtue of the GFP fluorescence (16). In principle, this allows selective detection of the GFP-tagged protein from all other cellular components and extends the utility of SV analysis from purified proteins into more natural biological complexes, such as those in cell lysates. Recent studies have used this approach to measure the size and heterogeneity of fluorescently labeled purified proteins in the presence of other macromolecules, including serum, yet the approach has yet to be examined on endogenous proteins expressed directly in mammalian cells or on proteins that are known to aggregate intracellularly in neurodegenerative diseases, such as HD (17, 18).
This technique was applied to determine the kinetics of aggregation of the exon 1 fragment of huntingtin (Httex1), which has been commonly used to model HD, expressed directly in neuroblastoma cells (19–22). The proteins were examined in non-denaturing buffers and examined with no prepurification steps that may selectively remove molecules, such as large aggregates, from the sample. In comparison, we also investigated the aggregation kinetics of purified mutant Httex1 to investigate how the cellular machinery and environment mediates aggregate size and flux. We extended our analysis to probe how the cognate heat shock protein 70 chaperone, Hsc70, which has been implicated in mediating Httex1 toxicity and aggregation, binds to Httex1 and influences the heterogeneity of the intracellular pools of Httex1 when co-expressed with Httex1 in neuroblastoma cells (23).
EXPERIMENTAL PROCEDURES
The protein expression and purification, Western blotting, and confocal microscopy procedures are included in the supplemental material.
Cloning and Mutagenesis
Httex1-EGFP cDNA was amplified by PCR from the pGW1-Httex1-EGFP template (3) with primers NdeI-Htt forward (5′-aaaccccatatggcgaccctggaaaagctgatg) and XhoI-GFP end reverse (5′-gggtttctcgagttacttgtacagctcgtccatgcc). The PCR product was cloned into the NdeI/XhoI restriction sites of a modified pET28b vector (pet28-HMT) that encodes a His6-maltose-binding protein (MBP)-TEV site multiple cloning site (gift of Dan Minor and Rebecca Howard, University of California, San Francisco). The EGFP was converted to monomeric cyan fluorescent protein variant Cerulean by mutating residues Y66W, S72A, Y145A, N146I, M153T, V163A, H148D, and A206K (24) with the QuikChange multisite-directed mutagenesis kit (Stratagene).
To generate mammalian expression constructs, the cDNA sequences of Httex1-EGFP were amplified by PCR from the pET28-HMT constructs with primers Htt TOPO forward (5′-caccatggcgaccctggaaaagct) and EcoRI-GFP reverse (5′-aaaaagaattcttacttgtacagctcgtccatg). EGFP was converted to monomeric Emerald by mutations S72A, N149K, M153T, I167T, and A206K using the QuikChange mutagenesis kit (Stratagene) (25). The PCR product was inserted into pcDNA3.2/V5/ GW/D-TOPO via TOPO cloning (Invitrogen). The Httex1-EGFP construct was then transferred first to pDONR-221 vector and then to the pT-REx-Dest30 vector via gateway recombination (Invitrogen). To create an expression vector for Emerald alone, Emerald cDNA was amplified from pT-REx Httex1-Emerald with primers SacII GFP forward (5′-aggctccgcggccgcccccttcaccatgctggtgagcaagggcgaggagctgttc)and EcoRI-GFP reverse and religated back into the pT-REx Httex1-Emerald vector via the SacII/EcoRI sites. Vectors expressing the Httex146Q-Cherry and Httex125Q-Cherry were gifts of S. Finkbeiner and A. Tsvetkov (Gladstone Institutes, San Francisco, CA). A vector expressing β-galactosidase, pT-REx/GW-30/lacZ, was obtained from Invitrogen.
pCMV6-XL5-HSPA8 (Hsc70 gene; NM_006145) was purchased from Origene. The Hsc70 cDNA was amplified by PCR with the primers SacII-Hsc70 forward (5′-aggctccgcggccgcccccttcaccatgtccaagggacctgcagttgg) and BamHI link Hsc70 rev (5′-gctcaccagggatcccatggtggcgaccggtgggtcccgggctcgcggtaccgtatcaacctcttcaatggtgggccc) and cloned into the pGEM T-vector (Promega). The Hsc70 cDNA was cloned into the SacII/BamHI sites of pDONR Httex1-Emerald via the SacII/BamHI sites and then transferred to the pT-REx-Dest30 vector by gateway recombination (Invitrogen). This construct contains a linker region between Hsc70 and Emerald similar to that of a previously described construct that was validated to have normal chaperone activity (26).
The Y66L mutation was introduced into Emerald using the QuikChange site-directed mutagenesis kit. This mutation removes a critical residue involved in fluorophore formation but does not alter the overall structure of the protein (27). All vectors were sequenced and verified for accuracy within the cDNA regions. The resultant protein sequences expressed off these vectors are shown in supplemental Fig. S1.
Preparation of Cells and Lysates for Analytical Ultracentrifugation
2.5 × 106 Neuro2a cells were plated on a T25 culture flask and maintained in Opti-MEM (Invitrogen) containing 10% fetal calf serum, 1 mm glutamine, 100 μg/ml penicillin/streptomycin. The following day, the cells were transfected with 10 μg of total plasmid DNA and 25 μl of Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen). Medium was replaced daily. For the time course studies, cells were rinsed with Dulbecco's phosphate-buffered saline (PBS) and imaged on an AF6000 fluorescence microscope (Leica) with a HCX PL FLUOTAR 10.0 × 0.30 DRY objective. Data were collected with the GFP filter cube and recorded in 12-bit with a DFC360FX camera. Cells were harvested by rinsing in PBS before removal from the flask with a cell scraper in 4 ml of PBS. Cells were pelleted (1,000 × g; 2.5 min), resuspended in 1 ml PBS, transferred to a 2-ml microcentrifuge tube, and pelleted again (1,800 × g; 4 min). The pellet was snap frozen in liquid nitrogen and stored overnight at −80 °C. The pellet was resuspended in 1 ml of chilled lysis buffer (20 mm Tris, pH 8.0, 2 mm MgCl2, 1% (v/v) Triton X-100, EDTA-free protease inhibitor mixture tablets (Roche Applied Science; 1 tablet per 10 ml of solution), 20 units/ml Benzonase (Novagen)) and extruded through a 27-gauge syringe needle 25 times on ice. NaCl concentration was adjusted to 150 mm. Single-use aliquots were snap frozen in liquid nitrogen and stored at −80 °C. Total cellular protein concentrations were determined in triplicate by a BCA protein assay (Thermo Scientific) with bovine serum albumin as the standard.
Analytical Ultracentrifugation
For the recombinant proteins, proteins were prepared at 0.7 μm in diluent buffer (50 mm Tris, 150 mm NaCl, 12.5 mm imidazole, 1 mm tris(2-carboxyethyl)phosphine, 0.1% (w/v) sodium azide). For the lysates, samples were adjusted to 0.2 or 0.5 mg/ml total protein concentration and 0 or 2 m sucrose concentrations with lysis buffer (containing 150 mm NaCl) and freshly prepared 3 m sucrose, added by weight. Sucrose stock solutions were made by heating sucrose with water and stirring until dissolved and were calibrated for concentration and density by refractive index at 20 °C (28). Final sucrose concentrations were verified in the sample by refractive index.
Samples were loaded into two-channel sapphire window charcoal/Epon sedimentation velocity cells (Beckman-Coulter) with 50 μl of FC-43 heavy oil (Fluorinert). Cells were placed in a prechilled 8-hole AnTi rotor (Beckman-Coulter) and equilibrated to 7 °C in an XL-A analytical ultracentrifuge (Beckman-Coulter) fitted with a fluorescence detection system (Aviv) (16). Radial fluorescence scans were collected continuously at 50,000 or 3,000 rpm using a 488-nm laser for excitation and 520-nm cut-off emission filter and with the photomultiplier voltage kept constant in each experiment.
Data were analyzed using SEDFIT (29). High speed data sets were fitted to c(s) distributions (29) and van Holde-Weischet analysis (30). Low speed data sets were fitted to ls-g*(s) distributions (14). For c(s) and ls-g*(s) (except for the 2 m sucrose data), data were fitted with radial independent and, in some cases, time-independent noise and with confidence levels (F-ratio) of 0.95. For samples containing 2 m sucrose, data were fitted similarly except without radial or time-dependent noise, with the base line floated to account for the non-sedimenting material, and with a confidence level of 0.99. The meniscus and bottom positions were fixed. Sedimentation coefficients for samples lacking sucrose were corrected to s20,w using Sednterp (J. Philo). For samples containing 2 m sucrose, s20,w was calculated as described previously (32) using previously reported values for density and viscosity of concentrated sucrose solutions (33).
Statistics
Differences in IB frequency or Htt partitioning between cells expressing Httex146Q-Emerald/β-galactosidase with cells expressing Httex146Q-Emerald/Hsc70- Emerald(Y66L) over 3 days transient expression were evaluated using two-way analysis of variance (ANOVA) in Sigmaplot 11 software. Significance was evaluated using the Holm-Sidak method with p = 0.05.
RESULTS
Aggregation Kinetics of Recombinant Htt Exon 1
As a first step to understanding the aggregation kinetics, we examined recombinant Httex1 fused to the monomeric cyan fluorescent protein, Cerulean, which is a derivative of EGFP (24). Non-pathogenic Httex1-Cerulean (Httex125Q) and a pathogenic form (Httex146Q-Cerulean) were fused to MBP, which prevents aggregation until the MBP moiety is removed with TEV protease (supplemental Fig. S2A). Upon cleavage with TEV protease, Httex125Q-Cerulean did not aggregate, whereas the Httex146Q-Cerulean formed SDS-insoluble, fibrillar aggregates over 72 h (supplemental Fig. S2B).
Samples of recombinant protein were assessed using the analytical ultracentrifuge at different time points subsequent to TEV protease cleavage at a high angular velocity of 50,000 rpm, which is optimal for the detection of smaller mass species, such as monomers and low order oligomers (34). Freshly TEV protease-cleaved Httex125Q-Cerulean and material incubated for 72 h formed a highly monodisperse population with a sedimentation coefficient centered at 2.3 S (Fig. 1, A and B). Sedimentation coefficients are proportional to the size, shape, and density of the sedimenting species. Hence, 2.3 S best describes a monomer (37.4 kDa for Httex125Q-Cerulean) with a frictional ratio (f/f0) of 1.71, which suggests a reasonably elongated average conformation. This result is consistent with recent crystal structures of Httex1, suggesting a high level of flexibility and conformational heterogeneity (35). Httex146Q-Cerulean was also initially highly monodisperse as monomers upon TEV protease cleavage from MBP (s20,w = 2.4 S, which for a 40-kDa monomer mass gives an f/f0 of 1.72). After incubation at 37 °C for 72 h, Httex146Q-Cerulean had aggregated into much larger mass species that mostly pelleted before data could be acquired (Fig. 1A, inset). These data confirm that 25Q and 46Q counterparts of Httex1-Cerulean both initially exist mostly as elongated monomers; however, only the 46Q variant assembles into large aggregates over a 72-h incubation that sedimented too rapidly to be quantitatively analyzed under these experimental conditions.
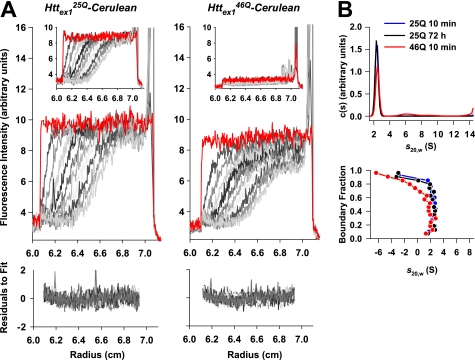
FIGURE 1.
Hydrodynamic properties of recombinant Httex1-Cerulean monomers. A, sedimentation velocity analysis of Httex125Q-Cerulean and Httex146Q-Cerulean at high speed (50,000 rpm). The initial data acquisition scans are shown in red and indicate all Httex1-Cerulean to be in solution. The remaining scans at subsequent time points are shown in incrementing grayscale (note the scans may be overlapping). The data were fitted to a c(s) continuous size distribution, which best describes the behavior of small, diffusing particles (residuals to the c(s) fit are shown in the lower panels). The inset shows the sedimentation behavior of samples incubated for 72 h at 37 °C prior to data analysis. B, sedimentation coefficient distributions based on the fits in A for the c(s) analysis (top panel); note the overlapping data for the Httex125Q-Cerulean samples. An alternate mode of analysis, the van Holde-Weischet analysis, which qualitatively describes the heterogeneity of sedimentation coefficients (lower panel) similarly indicates a population at 2–2.5 S.
We next determined whether we could define the molecular size distribution of the aggregated Httex146Q-Cerulean using SV analysis at lower centrifugal force. At 3,000 rpm, which we predicted would slow the rate of sedimentation of larger aggregates to within a detectable range, as has been previously described for other amyloid fibrils (15), we found that the Httex146Q-Cerulean produced a family of sedimenting boundaries upon centrifugation (Fig. 2A). The initial scan taken within 5 min of centrifugation (shown in red in Fig. 2A) revealed that no material had accumulated at the bottom of the cell prior to data acquisition, which indicated that the sedimenting boundaries encompass all of the huntingtin molecules present in the sample. Fitting the boundaries to a sedimentation coefficient size distribution model optimal for large, non-diffusing particles, ls-g*(s), indicated a heterogeneous population of species ranging from s20,w of 100 to 6000 S, slightly larger than the range of values observed with another amyloid-forming protein, apoC-II (15). These sedimentation values correspond to amyloid-like fibrillar structures detected by electron microscopy (supplemental Fig. S2B). Controls of freshly TEV protease-cleaved 25Q and 46Q Httex1-Cerulean and of the Httex125Q-Cerulean incubated for 72 h at 37 °C each showed no sedimentation, which confirmed an absence of aggregates and hence is consistent with these samples comprising just monomers (Fig. 2A).
FIGURE 2.
Kinetic assessment of aggregate size formed by recombinant Httex1-Cerulean aggregates by low speed centrifugation (3,000 rpm). A, under these conditions, only large aggregates form sedimenting boundaries, whereas monomers do not sediment (resulting in a plateau). The first scan is shown in red, with subsequent scans in incrementing grayscale. The data for the 72-h incubated Httex146Q-Cerulean were fitted to a ls-g*(s) continuous size distribution, which best describes the behavior of very large particles that have negligible diffusion (residuals to the fit shown in the inset). B, percentage of Httex146Q-Cerulean partitioned in the sedimenting boundary after different incubation times, reflecting a transfer of monomers to aggregates over time. C, time-dependent changes in aggregate size heterogeneity and sedimentation coefficients of aggregated Httex146Q-Cerulean were revealed by fits to ls-g*(s) size distributions at the different time points.
We next examined how the molecular size and heterogeneity changed for Httex146Q-Cerulean during the aggregation kinetics. Samples incubated at 37 °C for different times revealed a systemic increase in the fraction of the material that partitioned into the sedimenting boundary of aggregates, as anticipated for aggregation (Fig. 2B). The mode sedimentation coefficient and size heterogeneity of the sedimenting material also increased over 72 h, which is consistent with a systematic increase in fibril size and/or tangling (15) (Fig. 2C).
A notable feature of the recombinant protein is the apparent lack of species between monomers and ∼50 S oligomers, which roughly translates to a molecular mass of 3,500,000 Da or about 80 Httex146Q-Cerulean subunits. A similar phenomenon has been observed in the SV analysis of another amyloid-forming protein, apoC-II, and these data can be explained by the small oligomers being highly transient, dynamic, and/or infrequent under steady-state conditions (36).
Aggregation Kinetics of Httex1 in Cells
We next determined the molecular size of Httex1 expressed directly in the mouse neuroblastoma cell line Neuro2a. For the cell culture experiments, we replaced the Cerulean fluorescent protein moiety with Emerald, another monomeric EGFP variant that has a greater signal/noise ratio than Cerulean with the 488-nm laser used in SV experiments and is necessary to distinguish Httex1 sufficiently from nonspecific cellular autofluorescence. After 2 days of transient transfection, Httex146Q-Emerald formed SDS-insoluble material and was distributed into IBs in ∼35% of the cells as examined by confocal microscopy (Fig. 3, A and B). In contrast, the Httex125Q-Emerald did not form SDS-insoluble material and remained evenly distributed within the cytosol of all cells expressing Httex125Q-Emerald.
FIGURE 3.
Hydrodynamic assessment of Httex1-Emerald monomers in Neuro2a lysates by sedimentation velocity analysis. A, Western blot shows the expression levels over 2 days. Only Httex146Q-Emerald forms SDS-insoluble material over 2 days, indicative of aggregates. B, confocal microscopy shows Httex125Q-Emerald to remain distributed evenly through the cytosol of Neuro2a cells after 2 days. In contrast, the Httex146Q-Emerald forms punctate IBs in many cells. C, high speed sedimentation velocity analysis (50,000 rpm) of Httex1-Emerald in the lysate (0.5 mg/ml total protein) after 2 days of expression. The first scan is shown in red, with subsequent scans in incrementing grayscale. The first scan shows a fast pile up of material at the bottom for Httex146Q-Emerald for a fraction of the material. The lower panels show residuals to fits to a c(s) distribution. D, the sedimenting boundaries for Httex1-Emerald dictate a highly monodisperse population (elongated monomers) when assessed by c(s) analysis (upper panel) or van Holde-Weischet analysis (lower panel). Zoomed in rescaling of the c(s) distributions shows a minor fraction of material to sediment at 3–8 S (inset of upper panel).
Lysates were prepared in solutions containing mild detergents to minimize structural perturbations of Htt conformations or intermolecular interactions. At the high angular velocity of 50,000 rpm to examine more specifically the lower order oligomers, the Httex125Q-Cerulean formed a highly monodisperse distribution centered at s20,w = 2.2 S, which indicated a population of extended monomers, similar to that observed with the recombinant counterpart (Fig. 3, C and D). The slightly lower value obtained in the lysate relative to recombinant protein (2.3 versus 2.4 S) could reflect true differences in conformation or arise from differences in solution non-ideality from solvation, electrostatic interactions, or protein concentrations (16). Hence, as a control, we also performed SV analysis of Emerald (without Httex1) expressed in the Neuro2a cells. Emerald had a monodisperse population centered at s20,w = 2.3 S, which for its predicted mass of 26.8 kDa, gives an f/f0 of 1.37, indicating that in the same lysate conditions, Emerald has a more compact conformation than Httex1-Emerald (Fig. 3D). The relative “compactness” can be related to the known crystal structure of the Emerald parent protein, EGFP, which describes a reasonably compact β-barrel (37), hence confirming the elongated conformation of Httex1-Emerald monomers in the cell.
The Httex146Q-Emerald displayed two discernable populations when analyzed at the angular speed of 50,000 rpm. The first scan showed a rapidly moving boundary and a fraction of material already pelleted at the bottom of the column volume indicative of large molecular mass species (Fig. 3C). Subsequent scans revealed the remaining material to be highly monodisperse, with a mode sedimentation coefficient of s20,w = 2.3 S, indicative of an extended monomeric conformation (f/f0 = 1.79; Fig. 3D). For both the 25Q and 46Q counterparts, we noted a very minor fraction of the protein (less than 5%) to have a sedimentation coefficient at ∼3–8 S, which probably represents complexes of Htt with other cellular proteins or ligands (Fig. 3D, inset).
To assess the molecular size and heterogeneity of the larger aggregates of Httex146Q-Emerald, we performed SV analysis at the low angular velocity of 3,000 rpm. The first scan (red line in Fig. 4A, left) indicated that a fraction of the material had immediately pelleted prior to data acquisition. This suggests the presence of very large macromolecular aggregates, far larger than that observed with the recombinant protein, where all material could be visualized sedimenting at 3,000 rpm. In addition to the large material immediately pelleting, approximately one-third of the remaining Httex146Q-Emerald further evolved into a sedimenting boundary, which when fitted to an ls-g*(s) size distribution revealed a mode s20,w of 140 S. This population remained constant in size and heterogeneity between 2 and 3 days of expression (Fig. 4C, left). SV analysis of Httex125Q-Emerald showed a lack of sedimentation altogether at 3,000 rpm, consistent with it comprising more than 95% as monomers (Fig. 4B).
FIGURE 4.
Quantification of in-cell aggregate size heterogeneity with low speed and high viscosity (2 m sucrose) sedimentation velocity experiments (0.5 mg/ml total protein). A, sedimentation velocity analysis of the Httex146Q-Emerald at low speed (3,000 rpm). The first scan is shown in red and in the absence of sucrose indicates a proportion of material that has already piled up on the bottom. The remaining sedimenting boundary (incrementing grayscale) was fitted to an ls-g*(s) size distribution (residuals to fit shown in the lower panels). Sedimentation was slowed by increasing the viscosity with 2 m sucrose. B, sedimentation velocity analysis of Httex125Q-Emerald at low speed (3,000 rpm). Here, there is no sedimentation in the presence or absence of 2 m sucrose, indicative of a lack of aggregates. For comparison, lysates expressing β-galactosidase showed negligible signal intensity under the same photomultiplier voltage as Httex125Q-Emerald (purple scans), indicating negligible nonspecific fluorescence contributions to the samples. C, ls-g*(s) size distributions detected at low speed (3,000 rpm) for the oligomer boundary detected in the absence of sucrose (left) and IBs from the boundary detected in 2 m sucrose (right). Data shown are the mean ± S.D. of fits to three independent preparations of lysates.
In an attempt to characterize the largest aggregates present, we decreased the rates of sedimentation at 3,000 rpm by increasing the solution viscosity from 0.01 to 0.64 Poise with 2 m sucrose. Under these conditions, the largest material in the Httex146Q-Emerald sample sedimented as a well defined boundary, whereas no material sedimented for the Httex125Q-Emerald as expected (Fig. 4, A and B, right). A notable feature of the sedimenting material was its high apparent noise contribution relative to the other species present in solution, which could be attributed to particles having a sufficiently large size to move in and out of the laser beam path during data acquisition. Nevertheless, the data could be fitted to an ls-g*(s) size distribution, which gave a broad size distribution with a mode s20,w of 320,000 S over 3 days of expression (Fig. 4C, right). Although it is difficult to predict accurate masses from such large sedimentation coefficients, this value corresponds to a spherical proteinaceous particle of radius 0.62 μm assuming a partial specific volume typical of proteins (0.73 ml/g) and hence probably reflects the IBs. Although the IBs in our Neuro2a cells had an average radius of 3.1 μm ± 1.0 S.D; n = 280 IBs; supplemental Fig. S3), the estimate of the particle size from the sedimentation coefficient is probably a gross underestimate, considering that the IBs are probably far more asymmetrical and less dense than our modeling of a proteinaceous sphere. To determine whether the high sucrose concentration might induce artifactual aggregation, we also examined the sedimentation of recombinant Httex146Q fibrils and Httex146Q monomers in 2 m sucrose at 3,000 rpm. Under these conditions, we saw no sedimentation, further consistent with the conclusion that that the sedimentation boundaries obtained in the lysate truly reflect the inclusion population. Similar experiments were performed in 0.5 m sucrose, and fibrils gave a sedimentation coefficient distribution (s20,w) similar to those of samples without sucrose, indicating that fibrils are not noticeably dissociated or altered in size under the experimental conditions and time frame of the experiments.
In the context of the 320,000 S pool of Httex146Q-Emerald probably being the IBs, we hence concluded that the pool of Httex146Q-Emerald sedimenting at ∼140 S are oligomeric intermediates to IBs. 140 S corresponds to a spherical protein assembly of 7,700,000 Da, which is equivalent to ∼200 Httex146Q-Emerald molecules or particles with a radius of 13 nm. The oligomer pool, although vastly smaller in absolute size than the IB pool, still has a large tail after the 140 S mode, which indicates a high degree of complexity in the range of oligomer assembly states. For simplicity, we refer herein to this population as the “140 S” oligomers.
We next examined how the proportion of the three pools of Httex146Q changed over 3 days of transient expression in Neuro2a cell culture. Of the cells expressing Httex146Q-Emerald, the percentage containing IBs steadily increased over this time period (from 11 to 57%; p < 0.001, two-way analysis of variance (38)), consistent with a progressive shift in the cellular localization from a diffuse cytosolic pattern to IBs (Fig. 5A). Lysates were prepared and assessed using the combination of SV experiments described above to characterize the partitioning of the total population of Httex146Q-Emerald molecules in the cells. The mass proportion of Httex146Q-Emerald as monomers significantly decreased from 82% on day 1 to 28% on day 3 (Fig. 5B; p < 0.001; two-way ANOVA). Concurrently, the proportion in IBs significantly increased from 1 to 53% (p < 0.001; two-way ANOVA). Somewhat unexpectedly, the proportion of Httex146Q-Emerald in the 140 S pool (16–19%) did not significantly change over the 3-day time course (p = 0.546; two-way ANOVA).
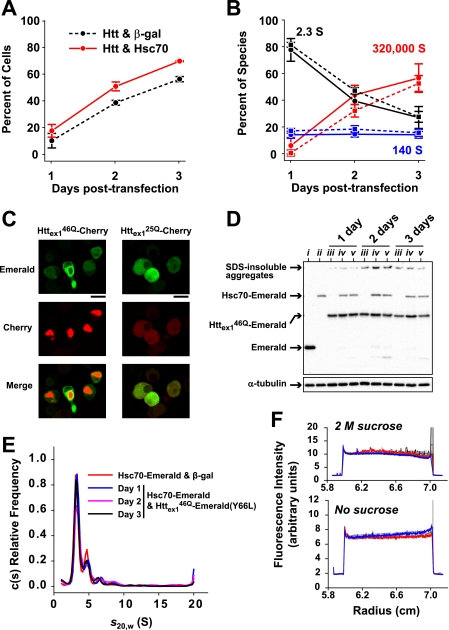
FIGURE 5.
The influence of Hsc70 on the evolution of three discrete species of Httex146Q-Emerald in Neuro2a cells over 3 days of transient expression. A, the percentage of cells expressing Httex146Q-Emerald containing IBs prior to lysis for assessment by SV analysis. Httex146Q-Emerald was co-expressed with Hsc70-Emerald(Y66L) or β-galactosidase. Values are mean ± S.D. (error bars) (n = 3 separate transfections). B, the relative population of 2.3 S monomers, 140 S oligomers, and 320,000 S IBs as assessed by SV analysis (dashed line, Httex146Q-Emerald and β-galactosidase; solid line, Httex146Q-Emerald and Hsc70-Emerald(Y66L)). Values are mean ± S.D. (n = 3 separate transfections). C, the localization of Hsc70-Emerald and Httex1-Cherry in Neuro2a cells after 3 days of transfection (imaged by confocal microscopy). Hsc70-Emerald is enriched at the periphery of the IBs in cells expressing Httex146Q-Cherry and remains diffuse in cells expressing Httex125Q-Cherry. Scale bar, 20 μm. D, Western blot probed with anti-GFP (top) and anti-α-tubulin (bottom) antibodies shows the expression levels and the presence of SDS-insoluble material over 3 days for the samples in A and B. Samples shown include Emerald and β-galactosidase (3-day expression; i), Hsc70-Emerald and β-galactosidase (3-day expression; ii), Httex146Q-Emerald and β-galactosidase (iii), Httex146Q-Emerald and Hsc70-Emerald(Y66L) (iv), and Httex146Q-Emerald(Y66L) and Hsc70-Emerald (v). For all combinations, equivalent masses of DNA were used in the transfections. E, c(s) size distributions of high speed (50,000 rpm) SV analysis showed no detectable interactions between Hsc70-Emerald and Httex146Q-Emerald(Y66L). F, ls-g*(s) size distributions of low speed (3,000 rpm) SV analysis showed no co-sedimentation of Hsc70-Emerald with Httex146Q-Emerald(Y66L) 140 S oligomers or 320,000 S IBs. The first scan is shown in red, the last scan is shown in blue, and intermediate scans are shown in grayscale (note that many are overlapping due to lack of sedimentation).
Chaperone-mediated Transport of Monomers into IBs
Using our SV experimental platform, we investigated how the cellular machinery might be involved in processing the Httex146Q-Emerald by co-expressing the cognate member of the Hsp70 family, Hsc70. Hsp70 proteins are components of the protein quality control network and prevent proteins from misfolding and aggregating by binding transiently to unfolded hydrophobic motifs in an ATP-dependent manner (39). Hsp70 members have been previously reported to co-localize with polyQ-related IBs and play a role in reducing toxicity of the polyQ-expanded proteins in animal and cell culture model systems (23, 40–45). Although some of these studies have reported Hsp70 members to suppress the formation of IBs, others have found no effect on IBs, which led us to speculate that mediation of toxicity might lie in Hsc70 altering the relative population of oligomers or monomers in the cell regardless of the total number of IBs formed.
To detect Hsc70 fluorometrically, we fused Emerald to the C terminus using the same paradigm as previously described, which has been validated to not interfere with its normal chaperone activity (26). Confocal microscopy revealed that Hsc70-Emerald was enriched at the periphery of IBs formed by Httex146Q fused to the red fluorescent protein Cherry, whereas it remained evenly diffuse through the cytosol when co-transfected with Httex125Q-Cherry (Fig. 5D). To determine the effect of Hsc70 on the rate of IB formation, we co-transfected Httex146Q-Emerald with Hsc70-Emerald(Y66L), whereby the Y66L mutation in Emerald renders Emerald non-fluorescent without altering its structure (27). When compared with cells co-expressing Httex146Q-Emerald with β-galactosidase as a negative control, we found a small but significant increase in the proportion of cells that contained IBs over 3 days (p < 0.001, two-way ANOVA; Fig. 5A). Expression levels of the Httex1 and Hsc70 moieties were equivalent in all the samples when examined by Western blot probed with an anti-GFP antibody, indicating that this difference was not due to variation in expression load (Fig. 5D).
To examine the effect of Hsc70-Emerald(Y66L) on Httex146Q-Emerald molecular partitioning in detail, we performed SV analysis of the samples over a 3-day co-transfection time course. In accordance with the number of cells containing IBs, we found Hsc70-Emerald(Y66L) to significantly increase the proportion of Htt molecules in IBs compared with β-galactosidase over 3 days (Fig. 5B; p = 0.044, two-way ANOVA). There was an apparent trend to Hsc70-Emerald(Y66L) decreasing the amount of Httex146Q-Emerald as monomers over 3 days, yet this was not statistically significant (Fig. 5B; p = 0.154, two-way ANOVA). However, the proportion of Httex146Q-Emerald in the 140 S pool was slightly but significantly lower in cells expressing Hsc70-Emerald(Y66L) compared with β-galactosidase over the 3 days (a decrease in average proportion from 17.8 to 15.0% ± 0.8% (S.E.), p = 0.030, two-way ANOVA). The differences in 140 S partitioning between days was not significant for cells expressing β-galactosidase or Hsc70-Emerald(Y66L) (p = 0.546; two-way ANOVA), indicating that although Hsc70 lowered the amount of Httex146Q-Emerald in oligomers, the oligomer population still remained static over time.
To further investigate the interaction of Hsc70 with Httex146Q, we measured the effect of Hsc70-Emerald(Y66L) on the molecular size and heterogeneity of Httex146Q-Emerald. Using a high velocity (50,000 rpm) SV experiment, we found no effect of Hsc70-Emerald(Y66L) on the hydrodynamic properties of Httex146Q-Emerald monomers, which suggested that either there was no direct binding between these species, or the interaction was too transient to detect (supplemental Fig. S4). Examination of lysates co-expressing Hsc70-Emerald with Httex146Q-Emerald(Y66L) also showed no evidence of Htt monomers, oligomers, or IBs altering the hydrodynamic properties of Hsc70-Emerald, further consistent with Hsc70-Emerald at best binding highly transiently to all forms of Httex146Q and at worst not binding at all (Fig. 5, E and F). In light of the confocal data showing an enrichment of Hsc70 with the periphery of Httex146Q IBs, these findings point to Hsc70 facilitating the transfer of Httex146Q monomers to IBs by binding in a highly dynamic fashion.
DISCUSSION
Using analytical ultracentrifugation for quantifying molecular size and heterogeneity under non-denaturing conditions, we identified several key findings. First, we found that purified mutant Htt self-associates from an expanded monomeric conformation into a heterogeneous population of aggregates that systematically increased in modal size over 3 days (50–6000 S). This is consistent with other studies showing that Httex1 forms amyloid-like fibrils and also that analytical ultracentrifugation can be used to measure fibrillization rates and fibrillar size heterogeneity (15, 34, 46). The finding that huntingtin monomers adopt an expanded conformation is also consistent with findings of others (35, 47). However, our work extends this knowledge into the cellular environment, whereby monomeric Httex1 remained in a remarkably similar expanded conformation and did not appear to bind appreciably to other intracellular ligands (less than 5% of the total “monomeric” pool of mutant and non-mutant Htt appeared in complex with other species). It is noteworthy that these results contrast with recent reports that polyQ fragments form mechanically collapsed disordered structures (48, 49), and the apparent discrepancy may be explained by the flanking residues of the polyQ sequence in Htt (notably the Pro-rich region C-terminal to the polyQ sequence) promoting more extended, non-compact conformations. Another possibility we cannot exclude is that the Emerald/Cerulean fusion tag alters the structural properties of the Htt exon 1 sequence. Alternatively, it remains possible that a subset of “collapsed” conformations remain undetected if these forms cause the Cerulean/Emerald moiety to not fold properly.
A striking and novel finding from our study was that oligomers formed by mutant Htt in neuroblastoma cells were static in the proportion of total Htt molecules and in their size heterogeneity despite continued conversion of monomers to IBs. Hence, the 140 S oligomers may present a rate-limiting step in the trafficking of small huntingtin aggregates to IBs through a series of highly compartmentalized intermediates (with a sharp mode of 140 S followed by an extended tail in the distribution extending up to ∼1600 S). These species may denote specific complexes with different cellular machineries or simply be larger oligomers and small fibrils that are constantly moved into the IBs via a rate limited mode of action. This result is supported by recent electron micrograph images of IBs in the human brain, showing oligomeric structures of a similar size to the oligomers in our study (∼30–60 nm) located at the periphery of an IB (50). Similar sized structures ∼30 nm in diameter have also been purified from Httex1-transfected cells or brain tissue of transgenic mouse models of HD (12, 51). Larger fibrillar structures have also been reported in IBs from ex vivo HD brain samples (52), which could reflect the larger species (>500 S) within the 140 S pool that more closely match the sedimentation coefficients of recombinant fibrils.
Genetic and biochemical evidence have shown that IB formation is a regulated process driven by dynein-mediated retrograde transport to the microtubule organizing center, consistent with our results suggesting that small oligomers are formed in a regulated fashion (38, 53, 54). It remains to be examined whether the different sized oligomers within the 140 S pool contain components of trafficking or quality control machinery or indeed have the same structural architecture relative to the other forms within the pool and with small fibrils formed by purified Htt. An alternative possibility is that the oligomers are off-track to IB formation and present a parallel partition that may be handled by distinct cellular machineries. For example, it remains an intriguing possibility that a subset of monomers indeed do adopt a collapsed coil conformation as proposed (48, 49) but that these forms are rapidly recruited into larger macromolecular complexes comprising other cellular machinery.
An important consequence of our results in light of studies showing IBs to correlate poorly with cellular toxicity is that oligomers are clearly uncorrelated to the presence of IBs or monomers (3). More importantly, if oligomers are indeed the most toxic molecular form, as previously postulated (4–5, 13, 55), then this implicates whole populations of cells under a steady and constant duress from mutant Httex1 toxicity. Hence, specifically mediating the pool of oligomers offers an attractive therapeutic target to modulate toxicity and provides more specificity than current approaches targeting total inclusion/SDS insoluble protein levels or monomer levels.
Our results also showed that Hsc70 facilitated the flux of Httex146Q monomers to IBs, a result consistent with other findings in that Hsp70 family proteins alone do not prevent the aggregation of Httex1 despite suppressing Htt toxicity (40–42). The binding appeared to be highly dynamic and transient, which is consistent with Hsp70 family proteins binding substrates by dynamic on-off cycling (56). Hsc70 has been suggested to bind polyQ-mediated IBs in a highly transient fashion (26, 57). Thus, Hsc70 seems to act as a carrier in the traffic of Httex1 to the IBs, yet we cannot determine from our studies whether this is through direct binding to monomers or is independent of the 140 S population. The efficacy of Hsc70 in the traffic and binding to Httex146Q may be limited by other components of the quality control network, such as Hsp40 co-chaperones, which have been shown to suppress IBs and to work in tandem with Hsp70 proteins to alter the morphology of Httex1 aggregates (23, 31).
In conclusion, we have used SV analysis to provide unprecedented insight into the molecular heterogeneity and hydrodynamic properties of all Htt molecules in the cellular milieu under non-denaturing conditions and without prepurification or prefractionation steps. Most noteworthy is the observation of a large pool of 140 S oligomers that are invariant in size and seem to be uncorrelated to the presence of monomers or IBs. This methodology will be useful for the study of parameters that mediate the aggregation pathway and for research in other neurodegenerative diseases that have been associated with oligomers, such as Alzheimer disease.
Supplementary Material
Acknowledgment
We thank Ken Goldie for performing the electron microscopy experiments.
This work was funded by a National Health and Medical Research Council (NHMRC) program grant (to A. F. H.) and NHMRC Project Grant 566640 (to D. M. H.). A. F. H. is an NHMRC CDA (Level 2) Fellow.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- HD
- Huntington disease
- polyQ
- polyglutamine
- IB
- inclusion body
- SV
- sedimentation velocity
- GFP
- green fluorescent protein
- EGFP
- enhanced green fluorescent protein
- Httex1
- exon 1 fragment of huntingtin
- MBP
- maltose-binding protein
- PBS
- Dulbecco's phosphate-buffered saline
- f/f0
- frictional ratio
- Hsp70
- heat shock protein 70
- ANOVA
- analysis of variance
- TEV
- tobacco etch virus.
REFERENCES
- 1.Chiti F., Dobson C. M. (2006) Annu. Rev. Biochem. 75, 333–366 [DOI] [PubMed] [Google Scholar]
- 2.Scherzinger E., Lurz R., Turmaine M., Mangiarini L., Hollenbach B., Hasenbank R., Bates G. P., Davies S. W., Lehrach H., Wanker E. E. (1997) Cell 90, 549–558 [DOI] [PubMed] [Google Scholar]
- 3.Arrasate M., Mitra S., Schweitzer E. S., Segal M. R., Finkbeiner S. (2004) Nature 431, 805–810 [DOI] [PubMed] [Google Scholar]
- 4.Takahashi Y., Okamoto Y., Popiel H. A., Fujikake N., Toda T., Kinjo M., Nagai Y. (2007) J. Biol. Chem. 282, 24039–24048 [DOI] [PubMed] [Google Scholar]
- 5.Takahashi T., Kikuchi S., Katada S., Nagai Y., Nishizawa M., Onodera O. (2007) Hum. Mol. Genet. 17, 345–346 [DOI] [PubMed] [Google Scholar]
- 6.MacDonald M. E., Ambrose C. M., Duyao M. P., Myers R. H., Lin C., Srinidhi L., Barnes G., Taylor S. A., James M., Groot N., MacFarlane H., Jenkins B., Anderson M. A., Wexler N. S., Gusella J. F., Bates G. P., Baxendale S., Hummerich H., Kirby S., North M., Youngman S., Mott R., Zehetner G., Sedlacek Z., Poustka A., Frischauf A.-M., Lehrach H., Buckler A. J., Church D., Doucette-Stamm L., O'Donovan M. C., Riba-Ramirez L., Shah M., Stanton V. P., Strobel S. A., Draths K. M., Wales J. L., Dervan P., Housman D. E., Altherr M., Shiang R., Thompson L., Fielder T., Wasmuth J. J., Tagle D., Valdes J., Elmer L., Allard M., Castilla L., Swaroop M., Blanchard K., Collins F. S., Snell R., Holloway T., Gillespie K., Datson N., Shaw D., Harper P. S. (1993) Cell 72, 971–9838458085 [Google Scholar]
- 7.Steffan J. S., Kazantsev A., Spasic-Boskovic O., Greenwald M., Zhu Y. Z., Gohler H., Wanker E. E., Bates G. P., Housman D. E., Thompson L. M. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 6763–6768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nucifora F. C., Jr., Sasaki M., Peters M. F., Huang H., Cooper J. K., Yamada M., Takahashi H., Tsuji S., Troncoso J., Dawson V. L., Dawson T. M., Ross C. A. (2001) Science 291, 2423–2428 [DOI] [PubMed] [Google Scholar]
- 9.Andreas W., Corinna K., Ben W., Kirupa S., Miriam B., Etienne R., Gillian P. B., Paolo P. (2008) J. Neurochem. 104, 846–85817986219 [Google Scholar]
- 10.Mukai H., Isagawa T., Goyama E., Tanaka S., Bence N. F., Tamura A., Ono Y., Kopito R. R. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 10887–10892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Behrends C., Langer C. A., Boteva R., Bottcher U. M., Stemp M. J., Schaffar G., Rao B. V., Giese A., Kretzschmar H., Siegers K., Hartl F. U. (2006) Mol. Cell 23, 887–897 [DOI] [PubMed] [Google Scholar]
- 12.Sathasivam K., Lane A., Legleiter J., Warley A., Woodman B., Finkbeiner S., Paganetti P., Muchowski P. J., Wilson S., Bates G. P. (2010) Hum. Mol. Genet. 19, 65–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hatters D. M. (2008) IUBMB Life 60, 724–728 [DOI] [PubMed] [Google Scholar]
- 14.Schuck P. (2000) Biophys. J. 78, 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.MacRaild C. A., Hatters D. M., Lawrence L. J., Howlett G. J. (2003) Biophys. J. 84, 2562–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.MacGregor I. K., Anderson A. L., Laue T. M. (2004) Biophys. Chem. 108, 165–185 [DOI] [PubMed] [Google Scholar]
- 17.Kingsbury J. S., Laue T. M., Klimtchuk E. S., Théberge R., Costello C. E., Connors L. H. (2008) J. Biol. Chem. 283, 11887–11896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ryan T. M., Howlett G. J., Bailey M. F. (2008) J. Biol. Chem. 283, 35118–35128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mangiarini L., Sathasivam K., Seller M., Cozens B., Harper A., Hetherington C., Lawton M., Trottier Y., Lehrach H., Davies S. W., Bates G. P. (1996) Cell 87, 493–506 [DOI] [PubMed] [Google Scholar]
- 20.Jackson G. R., Salecker I., Dong X., Yao X., Arnheim N., Faber P. W., MacDonald M. E., Zipursky S. L. (1998) Neuron 21, 633–642 [DOI] [PubMed] [Google Scholar]
- 21.Duennwald M. L., Jagadish S., Muchowski P. J., Lindquist S. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11045–11050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brignull H. R., Morley J. F., Garcia S. M., Morimoto R. I., Kheterpal I., Wetzel R. (2006) Methods Enzymol. 412, 256. [DOI] [PubMed] [Google Scholar]
- 23.Jana N. R., Tanaka M., Wang G.-h., Nukina N. (2000) Hum. Mol. Genet. 9, 2009–2018 [DOI] [PubMed] [Google Scholar]
- 24.Rizzo M. A., Springer G. H., Granada B., Piston D. W. (2004) Nat. Biotech. 22, 445–449 [DOI] [PubMed] [Google Scholar]
- 25.Tsien R. Y. (1998) Annu. Rev. Biochem. 67, 509–544 [DOI] [PubMed] [Google Scholar]
- 26.Kim S., Nollen E. A., Kitagawa K., Bindokas V. P., Morimoto R. I. (2002) Nat. Cell Biol. 4, 826–831 [DOI] [PubMed] [Google Scholar]
- 27.Rosenow M. A., Huffman H. A., Phail M. E., Wachter R. M. (2004) Biochemistry 43, 4464–4472 [DOI] [PubMed] [Google Scholar]
- 28.Lide D. R. (ed) (1996) CRC Handbook of Chemistry and Physics, pp. 8–76, CRC Press, Inc, Boca Raton, FL [Google Scholar]
- 29.Schuck P., Perugini M. A., Gonzales N. R., Howlett G. J., Schubert D. (2002) Biophys. J. 82, 1096–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Holde K. E., Weischet W. O. (1978) Biopolymers 17, 1387–1403 [Google Scholar]
- 31.Wacker J. L., Zareie M. H., Fong H., Sarikaya M., Muchowski P. J. (2004) Nat. Struct. Mol. Biol. 11, 1215–1222 [DOI] [PubMed] [Google Scholar]
- 32.Jacob L., Marc S. L., Peter S. (2002) Protein Sci. 11, 2067–207912192063 [Google Scholar]
- 33.Longinotti M. P., Horacio R. C. (2008) J. Phys. Chem. Ref. Data 37, 1503–1515 [Google Scholar]
- 34.Hatters D. M., MacPhee C. E., Lawrence L. J., Sawyer W. H., Howlett G. J. (2000) Biochemistry 39, 8276–8283 [DOI] [PubMed] [Google Scholar]
- 35.Kim M. W., Chelliah Y., Kim S. W., Otwinowski Z., Bezprozvanny I. (2009) Structure 17, 1205–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hatters D. M., Lindner R. A., Carver J. A., Howlett G. J. (2001) J. Biol. Chem. 276, 33755–33761 [DOI] [PubMed] [Google Scholar]
- 37.Ormo M., Cubitt A. B., Kallio K., Gross L. A., Tsien R. Y., Remington S. J. (1996) Science 273, 1392–1395 [DOI] [PubMed] [Google Scholar]
- 38.Wang Y., Meriin A. B., Zaarur N., Romanova N. V., Chernoff Y. O., Costello C. E., Sherman M. Y. (2009) FASEB J. 23, 451–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Minami Y., Hohfeld J., Ohtsuka K., Hartl F. U. (1996) J. Biol. Chem. 271, 19617–19624 [DOI] [PubMed] [Google Scholar]
- 40.Zhou H., Li S. H., Li X. J. (2001) J. Biol. Chem. 276, 48417–48424 [DOI] [PubMed] [Google Scholar]
- 41.Rujano M. A., Kampinga H. H., Salomons F. A. (2007) Exp. Cell Res. 313, 3568–3578 [DOI] [PubMed] [Google Scholar]
- 42.Wacker J. L., Huang S.-Y., Steele A. D., Aron R., Lotz G. P., Nguyen Q., Giorgini F., Roberson E. D., Lindquist S., Masliah E., Muchowski P. J. (2009) J. Neurosci. 29, 9104–9114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cummings C. J., Sun Y., Opal P., Antalffy B., Mestril R., Orr H. T., Dillmann W. H., Zoghbi H. Y. (2001) Hum. Mol. Genet. 10, 1511–1518 [DOI] [PubMed] [Google Scholar]
- 44.Muchowski P. J., Schaffar G., Sittler A., Wanker E. E., Hayer-Hartl M. K., Hartl F. U. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 7841–7846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Warrick J. M., Chan H. Y. E., Gray-Board G. L., Chai Y., Paulson H. L., Bonini N. M. (1999) Nat. Genet. 23, 425. [DOI] [PubMed] [Google Scholar]
- 46.Scherzinger E., Sittler A., Schweiger K., Heiser V., Lurz R., Hasenbank R., Bates G. P., Lehrach H., Wanker E. E. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 4604–4609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bennett M. J., Huey-Tubman K. E., Herr A. B., West A. P., Jr., Ross S. A., Bjorkman P. J. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 11634–11639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Crick S. L., Jayaraman M., Frieden C., Wetzel R., Pappu R. V. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 16764–16769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dougan L., Li J., Badilla C. L., Berne B. J., Fernandez J. M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 12605–12610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Legleiter J., Mitchell E., Lotz G. P., Sapp E., Ng C., DiFiglia M., Thompson L. M., Muchowski P. J. (2010) J. Biol. Chem. 285, 14777–14790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iwata A., Riley B. E., Johnston J. A., Kopito R. R. (2005) J. Biol. Chem. 280, 40282–40292 [DOI] [PubMed] [Google Scholar]
- 52.DiFiglia M., Sapp E., Chase K. O., Davies S. W., Bates G. P., Vonsattel J. P., Aronin N. (1997) Science 277, 1990–1993 [DOI] [PubMed] [Google Scholar]
- 53.Kopito R. R. (2000) Trends Cell Biol. 10, 524. [DOI] [PubMed] [Google Scholar]
- 54.Taylor J. P., Tanaka F., Robitschek J., Sandoval C. M., Taye A., Markovic-Plese S., Fischbeck K. H. (2003) Hum. Mol. Genet. 12, 749–757 [DOI] [PubMed] [Google Scholar]
- 55.Ross C. A., Poirier M. A. (2005) Nat. Rev. Mol. Cell Biol. 6, 891–898 [DOI] [PubMed] [Google Scholar]
- 56.Hartl F. U. (1996) Nature 381, 571–579 [DOI] [PubMed] [Google Scholar]
- 57.Matsumoto G., Kim S., Morimoto R. I. (2006) J. Biol. Chem. 281, 4477–4485 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.