Abstract
PEA-15/PED (phosphoprotein enriched in astrocytes 15 kDa/phosphoprotein enriched in diabetes) is a death effector domain-containing protein which is known to modulate apoptotic cell death. The mechanism by which PEA-15 inhibits caspase activation and increases ERK (extracellular-regulated kinase) activity is well characterized. Here, we demonstrate that PEA-15 is not only pivotal in the activation of the ERK pathway but also modulates JNK (c-Jun N-terminal kinase) signaling. Upon overexpression of PEA-15 in malignant glioma cells, JNK is potently activated. The PEA-15-induced JNK activation depends on the phosphorylation of PEA-15 at both phosphorylation sites (serine 104 and serine 116). The activation of JNK is substantially inhibited by siRNA-mediated down-regulation of endogenous PEA-15. Moreover, we demonstrate that glioma cells overexpressing PEA-15 show increased signs of autophagy in response to classical autophagic stimuli such as ionizing irradiation, serum deprivation, or rapamycin treatment. In contrast, the non-phosphorylatable mutants of PEA-15 are not capable of promoting autophagy. The inhibition of JNK abrogates the PEA-15-mediated increase in autophagy. In conclusion, our data show that PEA-15 promotes autophagy in glioma cells in a JNK-dependent manner. This might render glioma cells more resistant to adverse stimuli such as starvation or ionizing irradiation.
Keywords: Apoptosis, Autophagy, Brain, Cell Death, JNK, Brain Tumors, PEA-15, c-Jun N-terminal Kinase
Introduction
PEA-152/PED (phosphoprotein enriched in astrocytes 15 kDa/phosphoprotein enriched in diabetes) is a multifunctional protein involved in the regulation of proliferation, apoptosis, and glucose metabolism (1, 2). The PEA-15 protein consists of an N-terminal nuclear export sequence, a death effector domain, an ERK binding site, and two phosphorylation sites (Ser-104 and Ser-116) at the C terminus. Unphosphorylated PEA-15 binds ERK1/2 and prevents its translocation into the nucleus, thereby reducing the ERK1/2-mediated transcriptional activity (3). The phosphorylation of the seryl residues is mediated by PKC (4), calmodulin kinase II (5), and AKT/PKB (6). Phosphorylation of PEA-15 at serine 104 prevents ERK1/2-binding and phosphorylation at serine 116 enhances the binding to Fas-associated protein with death domain (FADD) and caspase 8, resulting in the inhibition of apoptosis (2).
Because of its functional role in ERK signaling and apoptosis, the PEA-15 protein is involved in the regulation of cell survival and proliferation, two key determinants of tumorigenesis (7). However, various studies reveal conflicting data on the functional role of PEA-15 in malignant tumors. PEA-15 mRNA is increased in metastatic squamous cell carcinoma compared with non-metastatic transformed cells (8). The overexpression of PEA-15 in a transgenic mouse model increases the susceptibility to chemically induced skin cancer (9). Moreover, we have shown in an earlier study that PEA-15 expression is increased in glioblastomas, and that the PEA-15 protein mediates resistance to glucose deprivation-induced apoptosis in glioma cells in an ERK-dependent manner (10), suggesting that PEA-15 promotes tumor cell survival in a poor microenvironment. Controversially, the adenovirus protein E1 decreases tumorigenicity and proliferation in ovarian cancer cells via increased PEA-15 expression (11). Moreover, it has recently been reported that PEA-15 induces cell death by autophagy in ovarian cancer cells (12).
Autophagy is a dynamic cellular process in which intracellular membrane structures, the so-called autophagosomes, sequester proteins, and organelles for degradation (13). It is an evolutionary highly conserved process which occurs in all eukaryotic cells from yeast to human. Autophagy provides energy to cells during periods of starvation. On the other hand, autophagy has recently been defined as a distinct type of cell death (so-called type-II programmed cell death) (14, 15). Autophagy starts by engulfing cytosol and/or organelles by double membrane-bound structures, known as autophagosomes. These autophagosomes finally fuse with lysosomes to form autolysosomes which mediate the subsequent degradation and recycling of the cellular components. Accumulating evidence argues for an important role of autophagy in the development of cancer (16). However, whether autophagy exerts a protective or destructive effect on cancer cells remains controversial. Breast cancer cell lines frequently contain a deletion of the autophagy gene beclin 1 (17), which is necessary to induce autophagy. An allelic deletion of chromosome 17q21 where beclin 1 is located is found in breast, ovarian, and prostate cancer (18–22). Moreover, mice with a heterozygous deletion of beclin 1 show increased incidence of lung cancer, hepatocellular carcinoma, and lymphoma (23). One major regulatory mechanism of autophagy is the PI3K/Akt/mTor pathway. In the presence of growth factors, phosphatidylinositol-3-phosphate kinase (PI3K) is activated at the plasma membrane. This activation subsequently leads to the activation of mTor (mammalian target of rapamycin) and to the inhibition of autophagy. Upon starvation of the cells, this pathway is blocked and autophagy is induced via class 3 PI3K and Beclin 1.
Several lines of evidence suggest that a cross-talk exists between the pathways leading to autophagy and apoptosis (14). Here, we describe that PEA-15, in addition to its ERK1/2-dependent anti-apoptotic function, is also involved in the regulation of autophagy in malignant glioma cells. We show that PEA-15 increases JNK activity and that JNK is required for the induction of autophagy. Promotion of autophagy might be another PEA-15-dependent protective mechanism which allows glioblastoma cells to survive under poor nutritional conditions.
EXPERIMENTAL PROCEDURES
Materials
The JNK inhibitor SP600125, 3-methyladenine, bafilomycin A1, acridine orange, and the LC3 antibody were purchased from Sigma. The rabbit polyclonal antibody specific for PEA-15 was described previously (24). The JNK1/3 antibody (c-17) was obtained from Santa Cruz Biotechnologies (Heidelberg, Germany). The pJNK was purchased from Cell Signaling (Frankfurt a.M., Germany). siRNA (short interfering RNA) was ordered from Ambion (Austin, TX).
Cell Lines and Transfections
The human glioma cell lines U87MG, U251MG, T98G, and LN18 were maintained in DMEM, high glucose (Invitrogen, Karlsruhe, Germany) containing 10% fetal calf serum (PAA Laboratories, Cölben, Germany), 1 mm glutamine (Invitrogen, Karlsruhe, Germany), and 1% penicillin/streptomycin (Invitrogen, Karlsruhe, Germany). The colony formation assay was carried out as described elsewhere (10). Cells stably expressing PEA-15 or PEA-15 variants were cultured in medium containing Geneticin® (G418) (Invitrogen, Karlsruhe, Germany). Cells were transfected by lipofection using Lipofectamin 2000 (Invitrogen) according to the manufacturer's protocol. Experiments using cells which transiently overexpress various plasmids were performed 18 h post-transfection if not indicated differently. The cells were transfected with the plasmids pcDNA3-neo (control), pcDNA3-FLAG-PEA-15, pcDNA3-FLAG-PEA- 15S104A, pcDNA3-FLAG-PEA-15S116A, pcDNA3-FLAG-PEA- 15S104/116A. Transient transfections with siRNA were carried out at a siRNA concentration of 50 nm. The following sequence was chosen to specifically target PEA-15 and Beclin 1, respectively: 5′-GCGAAAAGAGUGAGGAGAUtt-3′ (PEA- 15 sense); 5′-AUCUCCUCACUCUUUUCGCtt-3′ (PEA-15 antisense); 5′-CAGUUUGGCACAAUCAAUAtt-3′ (Beclin 1 sense); 5′-UAUUGAUUGUGCCAAACUGtt-3′ (Beclin 1 antisense). Cells transfected with scramble siRNA (scram) served as a control. The control siRNA had the following sequence: 5′-GAAGACGAAGAGUGAGGAUtt-3′ (sense) and 5′-AUCUCCACUCUCUGUUCUCtt-3′ (antisense). Cells were subjected to experiments 72 h post-transfection.
Site-directed Mutagenesis of PEA-15
The non-phosphorylatable variants of PEA-15 were generated as described previously (10). Briefly, to substitute the phosphorylation sites (Ser-104 and Ser-116) of PEA-15 by alanine, pcDNA3-PEA15 was used as a template using the QuikChange site-directed mutagenesis method (Stratagene, Amsterdam, Netherlands). To substitute Ser-104 by an Ala, the primer 5′-AACCCGTATTCCCGCTGCCAAGAAGTACA-3′ and its reverse complement were used. To exchange Ser-116 for Ala, pcDNA3- PEA-15served as template using the primer 5′-ATTCCGGCAGCCCGCTGAAGAAGAAATCA-3′ and its reverse complement. To generate the double phosphomutant pcDNA3- FLAG-PEA-15S104/116A the sequence of pcDNA3-FLAG- PEA-15S104A served as a template using the primer 5′-ATTCCGGCAGCCCGCTGAAGAAGAAATCA-3′ and its reverse complement. For each step 13 PCR cycles were performed. The presence of the desired mutations was confirmed by DNA sequencing (MWG, Martinsried, Germany).
FACS Analysis
The FACS analysis of glioma cells after acridine orange staining was performed using a FACS-calibur from Becton & Dickinson equipped with a 488 nm air-cooled argon laser. The green (510–530 nm) and red (>650 nm) fluorescence emission of 104 cells was measured. For plot analysis, the Cell Quest software (Becton & Dickinson, Franklin Lakes, NJ) was used. The cells were stained with 1 μg/ml acridine orange for 30 min in DMEM, high glucose lacking phenol red supplemented with 10% FCS. Post staining cells were washed in PBS and removed from the plate with trypsin-EDTA (Invitrogen). If the cells were treated with inhibitors the staining was performed in the presence of the corresponding inhibitor. FACS analysis was performed in DMEM, high glucose lacking phenol red.
Generation of Stably siRNA-infected Glioma Cell Lines
To target the PEA-15 protein for down-regulation by siRNA, the pSUPER-PEA-15 construct and the pSUPER-siRNA-scram (control) were generated as described previously (10). Briefly, siRNA sequence including the H1 promotor was subcloned from the pSuper construct into the lentiviral expression vector pG-Lenti (kindly provided by Carol Stocking-Harbers, Hamburg, Germany), which expresses green fluorescent protein (GFP) under the control of the SFFV promoter. Amphotropic retroviral supernatants were produced by transient transfection of 293T cells with the retroviral packaging vectors pMDLg/pRRE, pRSV-Rev, and pMD-G and the corresponding siRNA construct. U87MG and U251MG cells were infected at least two times with retroviral supernatant. Positive clones were selected by GFP expression and verified by Western blot analysis.
Cell Lysis and Immunoprecipitation
Cells were washed twice with ice-cold phosphate-buffered saline and lysed by the addition of lysis buffer. Cells which were subjected to Western blot analysis were lysed in SDS lysis buffer (62.5 mm Tris/HCl pH 6.8, 10% glycerol, 2% SDS). The cells were sonicated 2 times at 95% power for 30 s. For lysates subjected to co-immunoprecipitation experiment or in in vitro kinase assays, cells were lysed in lysis buffer (50 mm HEPES (pH 7.5), 150 mm NaCl, 1,5 mm MgCl2, 2 mm EGTA, 1% Triton X-100, 10% glycerol, 10 mm sodium fluoride, 1 mm Na4PPi, 100 μm β-glycerophosphate, 1 mm Na3VO4, 2 mm phenylmethylsulfonyl fluoride, and Complete (Roche, Mannheim, Germany) for 10 min on ice. The lysate was clarified by centrifugation for 20 min at 14,000 rpm in an Eppendorf centrifuge at 4 °C. The JNK C-17 antibody (Santa Cruz, Biotechnology, Heidelberg, Germany) directed against JNK1 and JNK3 was prebound to Protein G-Sepharose beads (50 ng/μl slurry) (Invitrogen) as described previously (25). The clarified lysate was incubated with 20 μl of antibody-bound protein G-Sepharose overnight at 4 °C. Immunoprecipitates were washed with HNTG buffer (20 mm HEPES (pH 7.5), 150 mm NaCl, 0.1% Triton X-100, 10% glycerol). Immunoprecipitates used for kinase assays were washed three times with HNTG buffer containing 1 m LiCl, three times with HNTG buffer, and twice with kinase assay buffer (50 mm Tris-HCl (pH 7.5), 100 mm NaCl, 1 mm MnCl2, 10 mm MgCl2, 0.1 mm Na3VO4).
Gel Electrophoresis and Western Blot Analysis
Lysates and immunoprecipitates were resolved by SDS-PAGE according to Laemmli (26). Proteins were transferred to nitrocellulose and immunoblotted using the antibodies indicated, followed by the appropriate horseradish peroxidase-conjugated secondary antibody (Bio-Rad). Western blots were developed by chemiluminescence. Multiple exposures of the Western blots were developed.
In Vitro Kinase Assays
Kinase assays were performed in 50 μl of kinase assay buffer containing 50 μm ATP and 5 μCi of [γ-32P]ATP (3000 Ci/mmol) (Perkin Elmer, Waltham, MA). For the JNK assays, 8 μg of GST-c-Jun was used as the substrate, and the reaction was carried out for 20 min at room temperature. Independent experiments showed that the reaction was linear within this time range. The reactions were terminated by the addition of an equal volume of 2× SDS sample buffer (100 mm Tris (pH 6.8), 4% SDS, 20% glycerol, 0,2% bromphenol blue, 100 mm dithiothreitol, 1% β-mercaptoethanol) containing 50 mm EDTA (pH 8,0). The pGEX-c-Jun-(1–115) vector was kindly provided by Dr. Kathleen A. Gallo (Michigan State University, East Lansing, MI). GST-c-Jun was expressed in Top10 Escherichia coli and purified by glutathione-Sepharose (Invitrogen) chromatography. Following the kinase assay, proteins were separated by SDS-PAGE. Gels were rinsed in phosphate-buffered saline, dried, and incorporation of radioactivity into GST-c-Jun was determined by autoradiography.
Acridine Orange Staining for Microscopy
To analyze autophagolysosome formation by fluorescence microscopy cells were stained in DMEM, high glucose supplemented with 10% fetal calf serum containing 1 μg/ml acridine orange. The color change from green to red was monitored under the microscope at different time points as indicated. Cells were analyzed with an Olympus CKX41 microscope, ×20 objective lens, and ColorView I FW camera. The exposure time was 200 ms. Cell imaging software was used for the acquisition of microscopic images.
Electron Microscopy
Electron microscopy was performed to analyze autophagosome formation. Cells were fixed in situ with 2,3% glutaraldehyde in 50 mm sodium cacodylate (pH, 7.2) for 30 min at 4 °C. Fixed cells were scraped from the plates, collected by low-speed centrifugation at 200 × g for 10 min at 4 °C, successively stained with 2% osmium tetroxide and 0.5% uranyl acetate. After dehydration and embedding in Epru the probes were processed for ultrathin section. Micrographs were taken with a Zeiss EM-10A electron microscope at 80 kV. The magnification indicator was routinely controlled by the use of a grating replica.
RESULTS
PEA-15 Increases JNK Activity in Glioma Cells
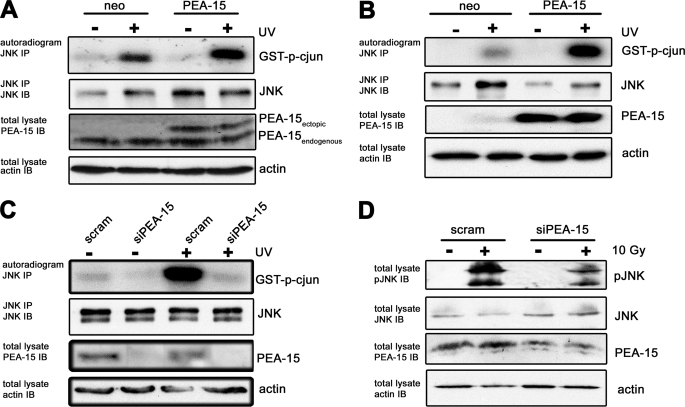
Although the pivotal role of PEA-15 in the ERK signaling pathway has been intensively studied, only little is known about the influence of PEA-15 on the stress-activated JNK pathway. We hypothesized that PEA-15 not only has the potential to regulate the physiological function of ERK1/2, but also impacts the JNK pathway in glioma cells. To investigate the effect of PEA-15 on JNK activity, PEA-15 was transiently and stably expressed in U251MG (Fig. 1A) and U87MG (Fig. 1B) cells, respectively. Endogenous JNK1 was immunoprecipitated from cellular lysates. The immunoprecipitates were subjected to an in vitro kinase assay using GST-c-Jun mimicking the endogenous substrate. These experiments showed that UV-induced JNK activity was substantially increased in PEA-15 overexpressing cells. Accordingly, siRNA-mediated down-regulation of endogenous PEA-15 strongly decreased JNK activity after UV (Fig. 1C) or γ-irradiation (Fig. 1D). A decrease in JNK activity in response to reduced PEA-15 levels was already observed in the absence of UV-induced JNK activation (Fig. 1C, lane 2). Taken together these results indicate that enhanced expression of PEA-15 increases JNK activity.
FIGURE 1.
PEA-15 increases JNK activity. A, endogenous JNK-1 was immunoprecipitated from cellular lysates of U251MG cells transiently expressing PEA-15. JNK activity was induced by UV irradiation (40 J/cm3). An in vitro immune complex assay for JNK activity was performed using GST-c-Jun as a substrate. The upper panel shows an autoradiogram with bands corresponding to GST-c-Jun. The second panel shows a JNK immunoblot (IB) of the same immunoprecipitated samples (IP) from the in vitro kinase assay. The third panel shows a PEA-15 immunoblot of cellular lysates. The lower panel shows an immunoblot for actin from cellular lysates confirming equal loading. B, endogenous JNK-1 was immunoprecipitated from cellular lysates of U87MG cells, stably expressing PEA-15. JNK activity was induced by UV irradiation (40 J/cm3). An in vitro immune complex assay for JNK activity was performed using GST-c-Jun as a substrate. C, endogenous JNK-1 was immunoprecipitated from cellular lysates of U251MG cells, in which PEA-15 was transiently down-regulated by small-interfering RNA. JNK activity was induced by UV irradiation (40 J/cm3). An in vitro immune complex assay for JNK activity was performed using GST-c-Jun as a substrate. D, lysates of U251MG cells in which PEA-15 is stably down-regulated by small-interfering RNA. Cells were lysed 24 h post γ-irradiation (10 Gy), and JNK activity was determined using a phosphospecific JNK antibody.
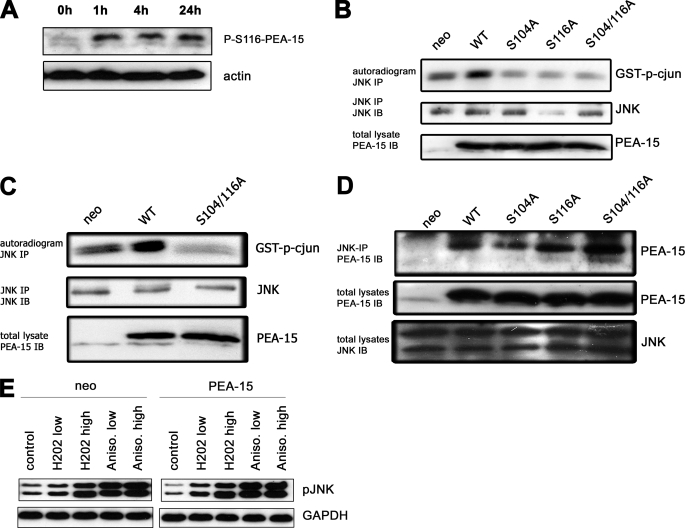
PEA-15 is a phosphoprotein that contains two C-terminal phosphorylation sites, the seryl residues 104 and 116. We wondered whether phosphorylation of PEA-15 is required for PEA-15-induced JNK activity. First, we investigated if γ-irradiation increases PEA-15 phosphorylation. Glioma cells were subjected to γ-irradiation and PEA-15 phosphorylation was analyzed using an antibody directed against the phosphorylated serine 116 (Fig. 2A). Phosphorylation of PEA-15 is still detected 24 h postirradiation. If phosphorylation of PEA-15 is required for JNK activity the inhibition of PEA-15 phosphorylation is supposed to decrease JNK activity. Therefore, we transiently (Fig. 2B) and stably (Fig. 2C) expressed various non-phosphorylatable PEA-15 mutants (PEA-15-S104A, PEA-15-S116A, PEA-15-S104/116A) in glioma cells. The experiments demonstrate that phosphorylation of both seryl residues is required for PEA-15-induced JNK activation. Because it was demonstrated that only non-phosphorylated PEA-15 is capable of binding ERK1/2 (3), we examined if the interaction between PEA-15 and JNK also depends on the phosphorylation status of PEA-15. Thus, endogenous JNK1 was immunoprecipitated from cellular lysates and subjected to an in vitro immunocomplex assay. Interestingly, not only wild-type PEA-15 but all of the tested phosphovariants of PEA-15 were capable to interact with JNK (Fig. 2D). In contrast to γ-irradiation, activation of JNK by H2O2 or anisomycin was independent of PEA-15 expression (Fig. 2E). Accordingly, H2O2 and anisomycin did not induce phosphorylation of PEA-15 (not shown).
FIGURE 2.
Phosphorylation of PEA-15 is required for JNK activity. A, U251MG cells were γ-irradiated with 10 Gy. Phosphorylation of PEA-15 at S116 was analyzed at the time points indicated. B, endogenous JNK-1 was immunoprecipitated from cellular lysates of LN18 glioma cells which transiently expressed various PEA-15 variants as indicated. JNK activity was induced by UV irradiation (40 J/cm3). An in vitro immune complex assay for JNK activity was performed using GST-c-Jun as a substrate. The upper panel shows an autoradiogram with bands corresponding to GST-c-Jun. The second panel shows a JNK immunoblot of the same immunoprecipitated samples from the in vitro kinase assay. The lower panel shows a PEA-15 immunoblot of cellular lysates. C, endogenous JNK-1 was immunoprecipitated from cellular lysates of U251MG glioma cells, which stably expressed a non-phosphorylatable variant of PEA-15 (S104A/S116A). JNK activity was induced by UV irradiation (40 J/cm3). An in vitro immune complex assay for JNK activity was performed using GST-c-Jun as a substrate. D, co-immunoprecipitation of PEA-15 phosphovariants and endogenous JNK1. Endogenous JNK-1 was immunoprecipitated from cellular lysates of U251MG glioma cells transiently overexpressing various PEA-15 variants as indicated. Co-immunoprecipitated PEA-15 was assessed by immunoblotting using a PEA-15 antibody. E, immunoblot analysis of phospho-JNK after treatment with H2O2 or anisomycin in U251MG glioma cells stably expressing PEA-15 or a neo control vector. Concentrations of H2O2 and anisomycin were as follows: H2O2 low, 1 mm; H2O2 high, 10 mm; anisomycin low, 0.1 μg/ml; anisomycin high, 0.5 μg/ml.
PEA-15 Promotes Autophagy in Glioma Cells
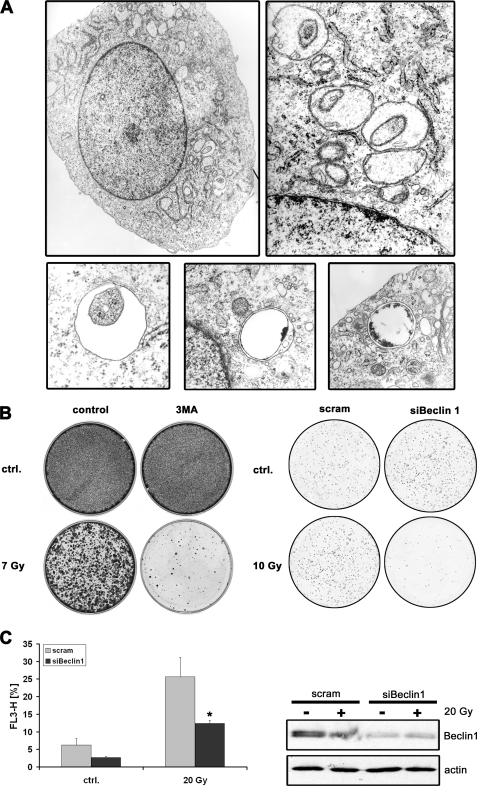
Growing evidence suggests that standard treatment of glioblastomas with temozolomide in combination with γ-irradiation induces autophagy (27–30). To set up an experimental system for the investigation of autophagy, we first confirmed that γ-irradiated glioblastoma cells show classical signs of autophagy. An electron microscopy analysis of several γ-irradiated glioblastoma cell lines clearly showed the massive appearance of double membrane vesicles, the so-called autophagosomes, which engulf cytoplasm and cellular organelles (Fig. 3A). Moreover, we were interested in the long term outcome of glioma cells undergoing autophagy after γ-irradiation. Thus, we performed clonogenicity assays after γ-irradiation in the presence or absence of 3-methyladenine, an inhibitor of autophagy, and after down-regulation of Beclin1, a protein important in the induction of autophagy. These experiments demonstrated that inhibition of autophagy either by a chemical autophagy inhibitor or by down-regulation of Beclin1 results in a substantial decrease in the number of colonies, suggesting that autophagy plays rather a cytoprotective than a cell death-promoting role (Fig. 3B). Because there is increasing evidence for Beclin1-independent autophagy (31) we investigated the role of Beclin-1 in γ-irradiation-induced autophagy of glioblastoma cells. To this end, we transiently down-regulated Beclin 1 in U251MG cells. Cells that were exposed to γ-irradiation showed a significant decrease in the number of autophagic cells when Beclin 1 was down-regulated (Fig. 3C), suggesting that γ-irradiation-induced autophagy is dependent on the expression of Beclin 1.
FIGURE 3.
Ionizing irradiation of glioma cells induces Beclin 1-dependent autophagy. A, electron microscopy analysis of U251MG cells 24 h after γ-irradiation (10 Gy). The electron micrographs show numerous double-membrane vesicles with the classical features of autophagosomes. No signs of apoptosis are present (e.g. nuclear condensation and membrane blebbing). B, colony formation assay of U251MG cells treated with γ-irradiation ± 3-methyladenine (3-MA, 500 μm) or treated with γ-irradiation after siRNA-mediated down-regulation of Beclin 1. After 7 days, cells were stained with crystal violet. C, U251MG Beclin 1-siRNA and control-siRNA cells were subjected to γ-irradiation (20 Gy). Induction of autophagy was determined by acridine orange staining (1 μg/ml) followed by flow cytometry (mean ± S.D., Student's t test; *, p < 0.05). Effective siRNA-mediated down-regulation of Beclin 1 was confirmed by immunoblot analysis. Actin served as a loading control.
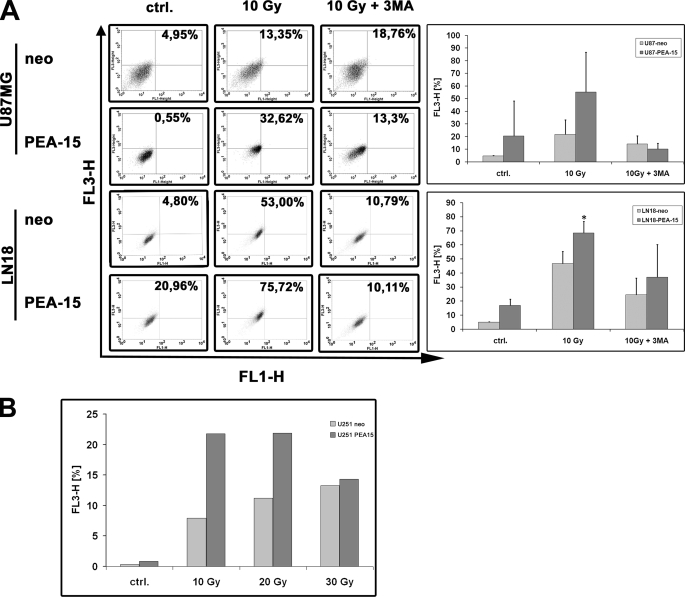
Given the known involvement of JNK in autophagy (32–34) and our finding of PEA-15-dependent JNK activation, we investigated whether the PEA-15 protein affects autophagy in glioma cells. To this end, various glioma cell lines were labeled with acridine orange and subjected to FACS after γ-irradiation. In U87 and LN18 glioma cell lines an increase in the number of autophagic cells after γ-irradiation was observed (Fig. 4A). The induction of autophagy was at least partially blocked by incubating the cells with 3-methyladenine. This result confirmed that the observed process was indeed autophagy. Importantly, overexpression of PEA-15 increased the autophagic activity in various glioma cell lines after γ-irradiation. The PEA-15-induced augmentation of autophagy was partly abolished in the presence of 3-methyladenine. In general, autophagy increased in a dose-dependent manner between an irradiation dose of 5 and 20 Gy. A further increase up to 30 Gy did not further increase autophagy (Fig. 4B).
FIGURE 4.
PEA-15 increases autophagy in glioma cells. A, U87 MG and LN 18 glioma cell lines were exposed to γ-irradiation (10 Gy) ± 3-methyladenine (MA, 500 μm). 24 h post irradiation the cells were stained with acridine orange (1 μg/ml) for 30 min ± 3-methyladenine (500 μm). Acridine orange staining was assayed by flow cytometry. FL1-H indicates intensity of green fluorescence, and FL3-H indicates intensity of red fluorescence. Bar graphs show the percentage of autophagic cells (mean ± S.D., Student's t test; *, p < 0.05, n = 3). B, U251MG cells were exposed to different doses of γ-irradiation as indicated. Bar graphs show percentage of autophagic cells as assessed by FACS analysis.
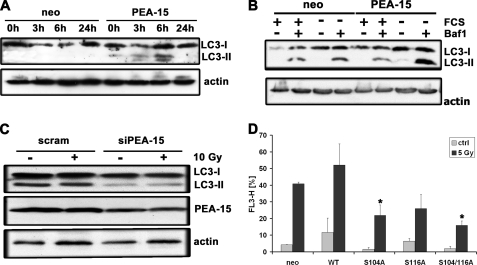
Next, the PEA-15-dependent promotion of autophagy was confirmed by examining the LC3 protein by immunoblot analysis. LC3 (microtubule-associated protein-1 light chain), the mammalian homolog of Atg8, is a reliable marker of autophagosomes. During autophagosome formation, LC3-I undergoes a conversion to LC3-II. The amount of LC3-II correlates well with the number of autophagosomes (35). First, the time course of LC3 conversion was examined in glioma cells upon γ-irradiation (Fig. 5A). U87MG glioma cells stably overexpressing PEA-15 showed a strong increase in LC3-II 6-h postirradiation. The absence of LC3-II 24-h postirradiation may be due to the fact that it is already degraded at this time point. Similarly, serum deprivation increased the amount of LC3-II in glioma cells (Fig. 5B). The increase of LC3-II was more sustained in the cells stably expressing PEA-15. To inhibit degradation of LC3 and to obtain an increased accumulation of the converted form after induction of autophagy bafilomycin A1 was used to stabilize LC3-II. In accordance with the previous experiments, the down-regulation of PEA-15 by small-interfering RNA resulted in a reduction of the LC3-II level (Fig. 5C). These results confirmed the data obtained by FACS analysis suggesting that PEA-15 increases autophagy in glioma cells.
FIGURE 5.
PEA-15-induced autophagy depends on phosphorylation of PEA-15. A, U87MG cells stably expressing PEA-15 were subjected to γ-irradiation (10 Gy) and analyzed for LC3-I to LC3-II conversion by immunoblot analysis at the time points indicated. Expression of actin was used as a loading control. B, U251MG cells stably expressing PEA-15 were serum-starved for 2 h in the presence or absence of bafilomycin A1 (10 nm). Bafilomycin A1 was used to stabilize LC3-II. The cells were lysed and conversion of LC3-I to LC3-II was analyzed by immunoblot analysis. C, LN18 glioma cells in which PEA-15 was transiently down-regulated were exposed to γ-irradiation (10 Gy) and analyzed for LC3-I and LC3-II by immunoblot analysis (upper panel). Expression of actin was used as a loading control. D, U251MG glioma cells stably expressing PEA-15 or various PEA-15 phosphomutants were exposed to γ-irradiation (5 Gy). 24 h postirradiation, the cells were stained with acridine orange (1 μg/ml) for 20 min. Acridine orange staining was assayed by flow cytometry (mean ± S.D., Student's t test; *, p < 0.05, compared with WT).
The Phosphorylation Status of PEA-15 Regulates Its Effect on Autophagy
Various reports emphasize the crucial role of the two phosphorylation sites Ser-104 and Ser-116 for the function of the PEA-15 protein (2). Thus, we investigated whether the phosphorylation status of PEA-15 is important for the PEA-15-induced autophagy. Glioma cells transiently expressing various non-phosphorylatable PEA-15 mutants were exposed to γ-irradiation and, 18-h post-treatment, to acridine orange staining followed by FACS analysis. The experiments showed that abrogation of PEA-15 phosphorylation strongly diminished the number of autophagic cells (Fig. 5D). The autophagic activity in cells expressing the non-phosphorylatable mutants was even lower than in the neo control-transfected cells. From these data, we conclude that the phosphorylation of PEA-15 is required for its autophagy-promoting function.
PEA-15-mediated Promotion of Autophagy Depends on JNK
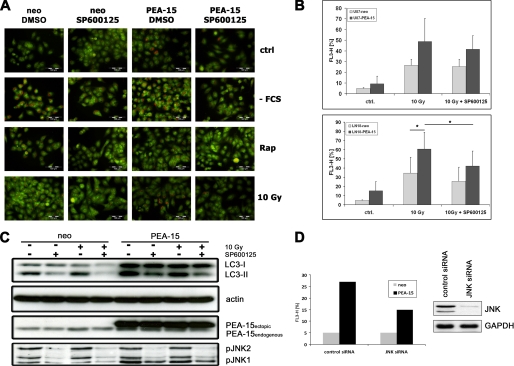
Although JNK is well known for its central role in the regulation of apoptosis there is increasing evidence that JNK is also a major regulator in the process of autophagy (32–34). Because PEA-15 increases JNK activity as well as the number of cells undergoing autophagy, we investigated whether the PEA-15-induced promotion of autophagy is mediated by the activation of JNK. To abrogate JNK activation we used the JNK inhibitor SP600125 (36, 37). Glioma cells stably expressing PEA-15 or the empty neo control vector were exposed to various autophagic stimuli including serum deprivation, γ-irradiation, or treatment with rapamycin, either in the presence or absence of JNK inhibitor. The cells were stained with acridine orange and then analyzed by fluorescence microscopy. In cells which undergo autophagy acridine orange will undergo a shift from a green/dim red to bright intense red which can be monitored by fluorescence microscopy (35). These experiments showed that in the presence of JNK inhibitor the number of autophagic cells strongly decreased (Fig. 6A). To confirm our observation by additional methods, the cells were subjected to FACS analysis (Fig. 6B) and immunoblot analysis of LC3 conversion (Fig. 6C). As expected, the JNK inhibitor substantially reduced the number of autophagic cells as assessed by FACS analysis. This result was further corroborated by immunoblot analysis of LC3 conversion, which showed a decrease in LC3-II in the presence of JNK inhibitor (Fig. 6C). Moreover, the PEA-15-dependent increase in autophagy was at least partly inhibited by siRNA-mediated down-regulation of JNK1/2 (Fig. 6D). Taken together, these data demonstrate that JNK activity contributes to autophagy. Moreover, we postulate that the PEA-15-mediated increase in autophagy in glioma cells is regulated in concert with JNK because inhibition of JNK strongly decreased PEA-15-induced autophagy.
FIGURE 6.
JNK is required for PEA-15-induced autophagy. A, U251MG glioma cells stably expressing PEA-15 were subjected to various autophagic stimuli as indicated. One hour prior to induction of autophagy, the indicated samples were treated with SP600125 (50 μm). Cells subjected to serum deprivation and rapamycin treatment were analyzed 4 h after autophagy induction. Cells subjected to 10 Gy γ-irradiation were analyzed after 24 h. The cells were stained for 1 h with acridine orange (1 μg/ml) and analyzed by fluorescence microscopy. B, U87MG, and LN18 glioma cell lines stably expressing PEA-15 were subjected to γ-irradiation (10 Gy). One hour prior to irradiation, the cells were treated with SP600125 (50 μm). 24 h postirradiation the cells were stained with acridine orange (1 μg/ml) for 1 h in the presence of SP600125. Acridine orange staining was assayed by flow cytometry. Bar graphs show the percentage of autophagic cells (mean ± S.D., Student's t test; *, p < 0.05, n = 3). C, U87MG glioma cells stably expressing PEA-15 were subjected to γ-irradiation (10 Gy). One hour prior to irradiation, cells were treated with SP600125 (50 μm). Six hours postirradiation the cells were lysed and subjected to immunoblot analysis. The upper panel indicates conversion from LC3-I to LC3-II. Actin served as loading control (second panel). The third panel shows PEA-15 expression. The lower panel indicates phosphorylation of JNK. D, autophagy was assessed by FACS analysis 24 h after siRNA-mediated down-regulation of JNK1/2 in U87MG cells stably expressing PEA-15 or a neo control vector. Immunoblot analysis demonstrates the successful down-regulation of JNK1/2.
DISCUSSION
Despite many reports describing the regulation of ERK1/2 by PEA-15, only very little is known about the role of PEA-15 in the JNK pathway. Here, we demonstrate that PEA-15 increases JNK activity in human glioblastoma cells. So far, two reports addressed the question whether overexpression of PEA-15 might influence JNK activity (38). However, these reports demonstrated either no effect of PEA-15 on JNK phosphorylation (39) or a PEA-15-dependent decrease in JNK activity in 293 cells (38). In contrast, we detect a consistent and substantial increase in JNK activity upon ectopic (stable and transient) expression of PEA-15 in various glioma cell lines. Accordingly, a decrease in JNK activity is observed after siRNA-mediated down-regulation of endogenous PEA-15 (Fig. 1). The cell type-specific features of PEA-15 and JNK might be one possible explanation for these different findings, because our experiments were performed in several glioblastoma cell lines, whereas Condorelli et al. (38) and Ramos et al. (39) analyzed 293 and CHO cells, respectively. In contrast to Formstecher et al. (3), we detect a physical interaction between JNK and PEA-15, which may be due to the different cell types used.
The functional role of JNK in cell survival and cell death is complex. JNK has a limited number of substrates but these mediate biological pleiotropic activities. For example, JNK activity is required to induce apoptosis upon nerve growth factor withdrawal (40). Transcription factors which are targets of JNK, such as c-Jun and c-Myc, belong to the group of oncogenes, whereas p53, another target of JNK (41), represents a tumor suppressor gene. A large number of publications state that JNK is important to promote apoptosis. However, there is also evidence that the activation of c-Jun is necessary for transformation by various oncogenes such as c-Ras, c-Fos, v-Src, v-Sis, and BCR-abl (42–47). Suppression of JNK in PC3 prostate cancer xenograft tumors diminished tumor growth and promoted tumor regression (48). Recent reports even demonstrate that JNK is required for tumorigenicity in human glioma cells (49, 50). Interestingly, the vast majority of glioblastomas (90.5%) was reported to exhibit constitutively activated JNK, whereas low grade gliomas show only little JNK activation (51), suggesting that JNK plays a role in the tumorgenesis and/or progression of malignant gliomas. Because PEA-15 is highly expressed in malignant gliomas (10), the high constitutive activity of JNK could be at least partially caused by PEA-15. The phosphorylation status of PEA-15 may play a central role in this process because non-phosphorylated PEA-15 lacks the capability to induce JNK phosphorylation.
JNK has been reported to be involved in the induction of autophagy (32–34). Given the JNK-activating property of PEA-15, we were interested to investigate whether PEA-15 regulates autophagy via JNK. Our experiments showed that the inhibition of JNK reduces autophagic activity, e.g. after γ-irradiation. Moreover, the PEA-15-dependent increase in autophagy is at least partly prevented by a JNK inhibitor. Therefore, we postulate that PEA-15 regulates autophagy in a JNK-dependent manner in glioblastoma cells. The process of PEA-15/JNK-induced autophagy might be mediated by Bcl-2. JNK leads to multisite phosphorylation of Bcl-2, thereby disrupting the Beclin 1/Bcl-2 complex, which results in the release of Beclin-1 and the subsequent induction of autophagy (34, 52).
Autophagy is a process which primarily aims at promoting cell survival. However, recent evidence suggests that autophagy is involved in cellular death, the so-called type II programmed cell death. Nevertheless, in the glioblastoma model examined here, colony formation assays showed that rather the inhibition than the induction of autophagy increases cellular death (Fig. 3B). Interestingly, a very recent report states that PEA-15 increases autophagy in ovarian cancer cell lines, confirming our results in glioblastoma cells (12). However, in ovarian cancer cells autophagy appears to induce cell death rather than survival. Thus, the final outcome of autophagy may be context- and cell type-dependent. In accordance with our data, Lomonaco et al. report that the induction of autophagy contributes to the radioresistance of glioma stem cells (53). Additionally, it has been reported that autophagy inhibitors increase the sensitivity of glioma cells to temozolomide (29). These data strongly point to a protective role of autophagy in glioma cells. Moreover, an increased expression of LC3 in colorectal cancer has been reported (54), again supporting the idea that autophagy may exert a protective role in certain tumors. Our recent report that glioma cells overexpressing PEA-15 are more resistant to apoptosis induced by glucose deprivation is in accordance with an overall cytoprotective effect of PEA-15 in glioblastomas. Moreover, 15–40% of primary glioblastomas harbor a PTEN mutation (55–57). As shown in a recent report, the loss of PTEN increases PEA-15 phosphorylation resulting in the inhibition of CD95-induced apoptosis (58). We hypothesize that this increase in PEA-15 phosphorylation could enhance autophagy, thereby promoting glioma cell survival. Further supporting evidence for the cellular protection by autophagy in glioblastomas comes from our observation that upon γ-irradiation no HMGB1 is released (data not shown), because it was recently suggested that the induction of cellular death accompanied by or executed by autophagy triggers the release of HMGB1 (59).
For cancer therapy it is essential to determine whether autophagy induces cellular death or promotes cellular survival. Therefore, it is of great importance to understand the molecular mechanism and the cellular context by which the cell decides to die. We show that PEA-15-induced JNK activation plays a pivotal role in the induction of autophagy. Because PEA-15, due to its variable phosphorylation status, acts as a molecular switch it may represent a crossroad between life and death, and also between apoptosis and autophagy. The outcome may be defined by the cell type and the cellular context. PEA-15 may play an important role in highly resistant cancer cells by orchestrating multiple processes, including the inhibition of apoptosis, the increase in proliferation by ERK activation, and the enhancement of autophagy by activation of JNK.
Acknowledgments
We thank Friedemann Kiefer for kindly providing the GST-c-Jun expression plasmid and Carol Stocking-Harbers for kindly providing the pG-Lenti expression plasmid. We are grateful to Annika Baude for critically reading the manuscript.
This work was supported by a grant from the Deutsche Krebshilfe (to W. R.) (German Cancer Aid, Max Eder Program).
- PEA
- phosphoprotein enriched in astrocytes
- JNK
- c-Jun N-terminal kinase
- DMEM
- Dulbecco's modified Eagle's medium
- FACS
- fluorescent-activated cell sorting
- GST
- glutathione S-transferase
- ERK
- extracellular signal-regulated kinase
- WT
- wild type.
REFERENCES
- 1.Renault F., Formstecher E., Callebaut I., Junier M. P., Chneiweiss H. (2003) Biochem. Pharmacol. 66, 1581–1588 [DOI] [PubMed] [Google Scholar]
- 2.Renganathan H., Vaidyanathan H., Knapinska A., Ramos J. W. (2005) Biochem. J. 390, 729–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Formstecher E., Ramos J. W., Fauquet M., Calderwood D. A., Hsieh J. C., Canton B., Nguyen X. T., Barnier J. V., Camonis J., Ginsberg M. H., Chneiweiss H. (2001) Dev. Cell 1, 239–250 [DOI] [PubMed] [Google Scholar]
- 4.Renault-Mihara F., Beuvon F., Iturrioz X., Canton B., De Bouard S., Léonard N., Mouhamad S., Sharif A., Ramos J. W., Junier M. P., Chneiweiss H. (2006) Mol. Biol. Cell 17, 5141–5152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kubes M., Cordier J., Glowinski J., Girault J. A., Chneiweiss H. (1998) J. Neurochem. 71, 1307–1314 [DOI] [PubMed] [Google Scholar]
- 6.Trencia A., Perfetti A., Cassese A., Vigliotta G., Miele C., Oriente F., Santopietro S., Giacco F., Condorelli G., Formisano P., Beguinot F. (2003) Mol. Cell. Biol. 23, 4511–4521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanahan D., Weinberg R. A. (2000) Cell 100, 57–70 [DOI] [PubMed] [Google Scholar]
- 8.Dong G., Loukinova E., Chen Z., Gangi L., Chanturita T. I., Liu E. T., Van Waes C. (2001) Cancer Res. 61, 4797–4808 [PubMed] [Google Scholar]
- 9.Formisano P., Perruolo G., Libertini S., Santopietro S., Troncone G., Raciti G. A., Oriente F., Portella G., Miele C., Beguinot F. (2005) Oncogene 24, 7012–7021 [DOI] [PubMed] [Google Scholar]
- 10.Eckert A., Böck B. C., Tagscherer K. E., Haas T. L., Grund K., Sykora J., Herold-Mende C., Ehemann V., Hollstein M., Chneiweiss H., Wiestler O. D., Walczak H., Roth W. (2008) Oncogene 27, 1155–1166 [DOI] [PubMed] [Google Scholar]
- 11.Bartholomeusz C., Itamochi H., Nitta M., Saya H., Ginsberg M. H., Ueno N. T. (2006) Oncogene 25, 79–90 [DOI] [PubMed] [Google Scholar]
- 12.Bartholomeusz C., Rosen D., Wei C., Kazansky A., Yamasaki F., Takahashi T., Itamochi H., Kondo S., Liu J., Ueno N. T. (2008) Cancer Res. 68, 9302–9310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levine B., Klionsky D. J. (2004) Dev. Cell 6, 463–477 [DOI] [PubMed] [Google Scholar]
- 14.Baehrecke E. H. (2005) Nat. Rev. Mol. Cell Biol. 6, 505–510 [DOI] [PubMed] [Google Scholar]
- 15.Gozuacik D., Kimchi A. (2007) Curr. Top Dev. Biol. 78, 217–245 [DOI] [PubMed] [Google Scholar]
- 16.Levine B. (2007) Nature 446, 745–747 [DOI] [PubMed] [Google Scholar]
- 17.Liang X. H., Jackson S., Seaman M., Brown K., Kempkes B., Hibshoosh H., Levine B. (1999) Nature 402, 672–676 [DOI] [PubMed] [Google Scholar]
- 18.Eccles D. M., Cranston G., Steel C. M., Nakamura Y., Leonard R. C. (1990) Oncogene 5, 1599–1601 [PubMed] [Google Scholar]
- 19.Futreal P. A., Söderkvist P., Marks J. R., Iglehart J. D., Cochran C., Barrett J. C., Wiseman R. W. (1992) Cancer Res. 52, 2624–2627 [PubMed] [Google Scholar]
- 20.Gao X., Zacharek A., Salkowski A., Grignon D. J., Sakr W., Porter A. T., Honn K. V. (1995) Cancer Res. 55, 1002–1005 [PubMed] [Google Scholar]
- 21.Russell S. E., Hickey G. I., Lowry W. S., White P., Atkinson R. J. (1990) Oncogene 5, 1581–1583 [PubMed] [Google Scholar]
- 22.Saito H., Inazawa J., Saito S., Kasumi F., Koi S., Sagae S., Kudo R., Saito J., Noda K., Nakamura Y. (1993) Cancer Res. 53, 3382–3385 [PubMed] [Google Scholar]
- 23.Qu X., Yu J., Bhagat G., Furuya N., Hibshoosh H., Troxel A., Rosen J., Eskelinen E. L., Mizushima N., Ohsumi Y., Cattoretti G., Levine B. (2003) J. Clin. Invest. 112, 1809–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharif A., Canton B., Junier M. P., Chneiweiss H. (2003) Ann. NY Acad. Sci. 1010, 43–50 [DOI] [PubMed] [Google Scholar]
- 25.Böck B. C., Vacratsis P. O., Qamirani E., Gallo K. A. (2000) J. Biol. Chem. 275, 14231–14241 [DOI] [PubMed] [Google Scholar]
- 26.Laemmli U. K., Beguin F., Gujer-Kellenberger G. (1970) J. Mol. Biol. 47, 69–85 [DOI] [PubMed] [Google Scholar]
- 27.Ito H., Daido S., Kanzawa T., Kondo S., Kondo Y. (2005) Int. J. Oncol. 26, 1401–1410 [PubMed] [Google Scholar]
- 28.Daido S., Yamamoto A., Fujiwara K., Sawaya R., Kondo S., Kondo Y. (2005) Cancer Res. 65, 4368–4375 [DOI] [PubMed] [Google Scholar]
- 29.Kanzawa T., Germano I. M., Komata T., Ito H., Kondo Y., Kondo S. (2004) Cell Death Differ. 11, 448–457 [DOI] [PubMed] [Google Scholar]
- 30.Kanzawa T., Bedwell J., Kondo Y., Kondo S., Germano I. M. (2003) J. Neurosurg. 99, 1047–1052 [DOI] [PubMed] [Google Scholar]
- 31.Scarlatti F., Maffei R., Beau I., Codogno P., Ghidoni R. (2008) Cell Death Differ. 15, 1318–1329 [DOI] [PubMed] [Google Scholar]
- 32.Cheng Y., Qiu F., Tashiro S., Onodera S., Ikejima T. (2008) Biochem. Biophys. Res. Commun. 376, 483–488 [DOI] [PubMed] [Google Scholar]
- 33.Cui Q., Tashiro S., Onodera S., Minami M., Ikejima T. (2007) J. Pharmacol. Sci. 105, 317–325 [DOI] [PubMed] [Google Scholar]
- 34.Wei Y., Pattingre S., Sinha S., Bassik M., Levine B. (2008) Mol. Cell 30, 678–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klionsky D. J., Cuervo A. M., Seglen P. O. (2007) Autophagy 3, 181–206 [DOI] [PubMed] [Google Scholar]
- 36.Bennett B. L., Sasaki D. T., Murray B. W., O'Leary E. C., Sakata S. T., Xu W., Leisten J. C., Motiwala A., Pierce S., Satoh Y., Bhagwat S. S., Manning A. M., Anderson D. W. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 13681–13686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heo Y. S., Kim S. K., Seo C. I., Kim Y. K., Sung B. J., Lee H. S., Lee J. I., Park S. Y., Kim J. H., Hwang K. Y., Hyun Y. L., Jeon Y. H., Ro S., Cho J. M., Lee T. G., Yang C. H. (2004) EMBO J. 23, 2185–2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Condorelli G., Trencia A., Vigliotta G., Perfetti A., Goglia U., Cassese A., Musti A. M., Miele C., Santopietro S., Formisano P., Beguinot F. (2002) J. Biol. Chem. 277, 11013–11018 [DOI] [PubMed] [Google Scholar]
- 39.Ramos J. W., Hughes P. E., Renshaw M. W., Schwartz M. A., Formstecher E., Chneiweiss H., Ginsberg M. H. (2000) Mol. Biol. Cell 11, 2863–2872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xia Z., Dickens M., Raingeaud J., Davis R. J., Greenberg M. E. (1995) Science 270, 1326–1331 [DOI] [PubMed] [Google Scholar]
- 41.Oleinik N. V., Krupenko N. I., Krupenko S. A. (2007) Oncogene 26, 7222–7230 [DOI] [PubMed] [Google Scholar]
- 42.Behrens A., Jochum W., Sibilia M., Wagner E. F. (2000) Oncogene 19, 2657–2663 [DOI] [PubMed] [Google Scholar]
- 43.Burgess G. S., Williamson E. A., Cripe L. D., Litz-Jackson S., Bhatt J. A., Stanley K., Stewart M. J., Kraft A. S., Nakshatri H., Boswell H. S. (1998) Blood 92, 2450–2460 [PubMed] [Google Scholar]
- 44.Clark G. J., Westwick J. K., Der C. J. (1997) J. Biol. Chem. 272, 1677–1681 [DOI] [PubMed] [Google Scholar]
- 45.Cripe L. D., Gelfanov V. M., Smith E. A., Spigel D. R., Phillips C. A., Gabig T. G., Jung S. H., Fyffe J., Hartman A. D., Kneebone P., Mercola D., Burgess G. S., Boswell H. S. (2002) Leukemia 16, 799–812 [DOI] [PubMed] [Google Scholar]
- 46.Franklin C. C., Sanchez V., Wagner F., Woodgett J. R., Kraft A. S. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 7247–7251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smeal T., Binetruy B., Mercola D., Grover-Bardwick A., Heidecker G., Rapp U. R., Karin M. (1992) Mol. Cell. Biol. 12, 3507–3513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang Y. M., Bost F., Charbono W., Dean N., McKay R., Rhim J. S., Depatie C., Mercola D. (2003) Clin. Cancer Res. 9, 391–401 [PubMed] [Google Scholar]
- 49.Cui J., Han S. Y., Wang C., Su W., Harshyne L., Holgado-Madruga M., Wong A. J. (2006) Cancer Res. 66, 10024–10031 [DOI] [PubMed] [Google Scholar]
- 50.Potapova O., Gorospe M., Bost F., Dean N. M., Gaarde W. A., Mercola D., Holbrook N. J. (2000) J. Biol. Chem. 275, 24767–24775 [DOI] [PubMed] [Google Scholar]
- 51.Li J. Y., Wang H., May S., Song X., Fueyo J., Fuller G. N., Wang H. (2008) J. Neuro-oncol. 88, 11–17 [DOI] [PubMed] [Google Scholar]
- 52.Pattingre S., Tassa A., Qu X., Garuti R., Liang X. H., Mizushima N., Packer M., Schneider M. D., Levine B. (2005) Cell 122, 927–939 [DOI] [PubMed] [Google Scholar]
- 53.Lomonaco S. L., Finniss S., Xiang C., Decarvalho A., Umansky F., Kalkanis S. N., Mikkelsen T., Brodie C. (2009) Int. J. Cancer 125, 717–722 [DOI] [PubMed] [Google Scholar]
- 54.Sato K., Tsuchihara K., Fujii S., Sugiyama M., Goya T., Atomi Y., Ueno T., Ochiai A., Esumi H. (2007) Cancer Res. 67, 9677–9684 [DOI] [PubMed] [Google Scholar]
- 55.Duerr E. M., Rollbrocker B., Hayashi Y., Peters N., Meyer-Puttlitz B., Louis D. N., Schramm J., Wiestler O. D., Parsons R., Eng C., von Deimling A. (1998) Oncogene 16, 2259–2264 [DOI] [PubMed] [Google Scholar]
- 56.Knobbe C. B., Merlo A., Reifenberger G. (2002) Neuro-oncology 4, 196–211 [PMC free article] [PubMed] [Google Scholar]
- 57.Tohma Y., Gratas C., Biernat W., Peraud A., Fukuda M., Yonekawa Y., Kleihues P., Ohgaki H. (1998) J. Neuropathol. Exp. Neurol. 57, 684–689 [DOI] [PubMed] [Google Scholar]
- 58.Peacock J. W., Palmer J., Fink D., Ip S., Pietras E. M., Mui A. L., Chung S. W., Gleave M. E., Cox M. E., Parsons R., Peter M. E., Ong C. J. (2009) Mol. Cell. Biol. 29, 1222–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thorburn J., Frankel A. E., Thorburn A. (2009) Autophagy 5, 247–249 [DOI] [PMC free article] [PubMed] [Google Scholar]