Abstract
Focal adhesion kinase (FAK) associates with both integrins and growth factor receptors in the control of cell motility and survival. Loss of FAK during mouse development results in lethality at embryonic day 8.5 (E8.5) and a block in cell proliferation. Because FAK serves as both a scaffold and signaling protein, gene knock-outs do not provide mechanistic insights in distinguishing between these modes of FAK function. To determine the role of FAK activity during development, a knock-in point mutation (lysine 454 to arginine (R454)) within the catalytic domain was introduced by homologous recombination. Homozygous FAKR454/R454 mutation was lethal at E9.5 with defects in blood vessel formation as determined by lack of yolk sac primary capillary plexus formation and disorganized endothelial cell patterning in FAKR454/R454 embryos. In contrast to the inability of embryonic FAK−/− cells to proliferate ex vivo, primary FAKR454/R454 mouse embryo fibroblasts (MEFs) were established from E8.5 embryos. R454 MEFs exhibited no difference in cell growth compared with normal MEFs, and R454 FAK localized to focal adhesions but was not phosphorylated at Tyr-397. In E8.5 embryos and primary MEFs, FAK R454 mutation resulted in decreased c-Src Tyr-416 phosphorylation. R454 MEFs exhibited enhanced focal adhesion formation, decreased migration, and defects in cell polarity. Within immortalized MEFs, FAK activity was required for fibronectin-stimulated FAK-p190RhoGAP association and p190RhoGAP tyrosine phosphorylation linked to decreased RhoA GTPase activity, focal adhesion turnover, and directional motility. Our results establish that intrinsic FAK activity is essential for developmental processes controlling blood vessel formation and cell motility-polarity but not cell proliferation. This work supports the use of FAK inhibitors to disrupt neovascularization.
Keywords: Cell Migration, Embryo, Fibroblast, Fibronectin, Src, Tyrosine-Protein Kinase (Tyrosine Kinase), Gene Knock-in, Focal Adhesion Kinase (FAK), p190RhoGAP, Vasculogenesis
Introduction
Vascular and endothelial cell (EC)4 development requires combined signals from extracellular matrix proteins and growth factor receptors (1). A functional circulatory system is the first organ system to develop and is essential for survival and growth of the embryo. ECs differentiate from hemangioblasts in yolk sac blood islands at embryonic day 7 (E7.0) in the mouse. By E.8.0, ECs form a vascular plexus in a process termed “vasculogenesis” that encompasses the growth of a primitive blood vessel network (2). Further expansion, remodeling, and EC tube formation within this primitive vasculature into a primary capillary plexus occurs by both sprouting and non-sprouting angiogenesis, leading to a functional circulatory system (3). Integrin binding to extracellular matrix facilitates vascular development by promoting cell adhesion, controlling cell motility, and regulating cell proliferation-survival (4). However, because integrin receptors do not possess intrinsic catalytic activity, signals generated by integrins are transduced by associated proteins (5, 6).
Focal adhesion kinase (FAK) is a cytoplasmic tyrosine kinase that associates with and is activated by integrins in cells at substratum contact sites termed focal adhesions (FAs) (7, 8). FAK activation is characterized in part by autophosphorylation at Tyr-397, leading to the formation of a multiprotein signaling complex (9). At FAs, FAK functions as both a cytoskeleton-associated scaffolding protein and a signaling kinase promoting integrin-stimulated tyrosine phosphorylation events (10). Studies have identified FA-associated proteins, such as paxillin (11–13), p130Cas (14), c-Src (15), PKL (16, 17), α-actinin (18), Grb7 (19), N-WASP (20), PDZ-RhoGEF (21), p190RhoGAP (22), and p190RhoGEF (23) as substrates for FAK that are involved in modulating FA dynamics during cell motility (24, 25).
Recent studies have established that FAK expression and activity promote the recruitment of p190A (p190RhoGAP-A) to leading edge FAs and enhance p190A tyrosine phosphorylation and the establishment of cell polarity in fibroblasts, ECs, and colon carcinoma cells in connection with integrin signaling (26). Tyrosine-phosphorylated p190A is active and associated with RhoA GTPase inhibition upon cell spreading on fibronectin (27, 28). p190A also forms a complex with p120RasGAP in the control of directed cell movement (29, 30). Interestingly, the development of small molecule ATP-competitive inhibitors to FAK limit FA turnover dynamics and prevent cell movement and polarity but do not readily block cell growth in culture (26, 31–33).
Developmental mouse studies have revealed that total or EC-specific FAK gene deletion result in early embryonic lethality with defects in blood vessel formation (34–36). FAK−/− ECs and fibroblasts exhibit elevated p53 tumor suppressor levels, motility defects, and an inability to proliferate in culture (34). FAK and the related Pyk2 (proline-rich kinase 2) protein-tyrosine kinase can inhibit p53 in a kinase-independent manner by promoting increased p53 ubiquitination-degradation via N-terminal FAK/Pyk2 FERM (band 4.1, ezrin, radixin, moesin homology) domain nuclear translocation, p53 binding, and enhancement of p53 ubiquitination-turnover (37, 38). Thus, although FAK loss implicates FAK signaling in controlling cell cycle progression, this control point may operate independently of FAK kinase activity (39).
To gain insights into the developmental role for FAK activity, we generated a kinase-inactive point mutation (FAK lysine 454 to arginine, R454) in fak exon 21 and created a knock-in mouse by homologous recombination. We report that FAKR454/R454 embryos are viable until E9.5, 1 day later than FAK−/− lethality at E8.5 (40), yet more than 5 days earlier than mice containing a deletion of fak exon 15 encompassing 19 residues (semipenetrant lethality after E14.5) spanning the major autophosphorylation site at FAK Tyr-397 (41). E9.5 FAKR454/R454 embryos exhibited defective EC patterning and tubule formation within the yolk sac and disorganized EC staining within embryos. Surprisingly, development can proceed further with an active FAK protein that is missing the Tyr-397 site (deletion of FAK exon 15) compared with embryos expressing kinase-inactive R454 FAK. Remarkably, primary mouse embryo fibroblasts (MEFs) were established from FAKR454/R454 embryos and exhibited no proliferation defects. Instead, R454 FAK MEFs showed increased FA formation, deregulated membrane ruffling, and decreased motility associated with defects in polarity and directional persistence. We find that FAK activity controls p190A tyrosine phosphorylation linked to decreased RhoA GTPase activity initiated by integrin binding to fibronectin. Our results from the first kinase-dead FAK knock-in mouse support the conclusion that FAK activity is essential in promoting cell motility-polarity but not required for adherent cell growth-proliferation.
EXPERIMENTAL PROCEDURES
Mice
FAK R454 knock-in mutation was generated by homologous recombination (InGenious Targeting Laboratory, Stony Brook, NY) with the cloning strategy and methods shown in supplemental Fig. 1. Heterozygous wild type (WT) and R454 knock-in (FAKWT/R454Neo) mice were maintained on a mixed C57BL/6 × 129/SvEv background. Transgenic mice expressing the Saccharomyces cerevisiae FLP1 recombinase gene were obtained from The Jackson Laboratory (catalog no. 003800) and were crossed with FAKWT/R454Neo mice to inactivate the neomycin cassette. Mice were housed and bred according to Association for Assessment and Accreditation of Laboratory Animal Care International-approved institutional guidelines.
Cells
Primary FAKR454/R454 and FAKWT/WT MEFs were isolated from E8.5 embryo explant culture within a drop of Matrigel (BD Biosciences) as described (37). After expansion and limited passage, primary MEFs were immortalized via retrovirus-mediated expression of human telomerase reverse transcriptase (hTERT) or large T-antigen (T-Ag) obtained from Addgene (Cambridge, MA), followed by puromycin selection. Embryo explants and MEFs were maintained on dishes precoated with 0.1% gelatin in Dulbecco's modified Eagle's medium with 10% fetal bovine serum, non-essential amino acids for minimum Eagle's medium, sodium pyruvate (1 mm), penicillin (50 units/ml), streptomycin (50 μg/ml), and ciprofloxacin (20 μg/ml). For analysis of primary MEF proliferation, 25,000 cells were plated onto 0.1% gelatin-coated 6-well plates, harvested every 24 h in triplicate, stained with trypan blue for viability, and counted (ViCell XR, Beckman Coulter).
Reagents and Antibodies
Antibodies to CD31 (PECAM-1, clone MEC13.3), Pyk2 (clone 11), paxillin (clone M107), p190RhoGAP (clone 30), FAK (610087), and p130Cas (clone 21) were from BD Biosciences. Antibodies to β-actin (AC-17) and purified bovine fibronectin were from Sigma. Antibodies to Src (Src-2) and RhoA (sc-179) were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Antibodies to FAK (4.47), p120RasGAP (clone B4F8), phosphotyrosine (4G10), and Rac1 (clone 102) were from Millipore. Anti-mouse p53 (CM5) was from Novocastra. Alexa 350 phalloidin, anti-Tyr(P)-402 Pyk2 (44-618G), and anti-paxillin Tyr(P)-31 (44-720G) were from Invitrogen. Antibodies to activated c-Src Tyr(P)-416 (2101) and phosphorylated p130Cas Tyr(P)-410 (4011) were from Cell Signaling Technology. Anti-β-Cop was from Calbiochem-EMD, and anti-HEF1/NEDD9 (2G9) was from Novus Biologicals. Fluorescein isothiocyanate-conjugated anti-rat secondary was from eBiosciences.
Yolk Sac and Whole Mount Embryo Anti-CD31 Staining
Brightfield images of yolk sacs and dissected embryos were obtained using a Zeiss M2-Bio stereomicroscope equipped with a Luminera color CCD camera. Freshly isolated yolk sacs were mounted on poly-l-lysine-coated coverslips, fixed in 3.7% paraformaldehyde, permeabilized with 0.1% Triton X-100 for 4 h, and then blocked with phosphate-buffered saline containing 1% bovine serum albumin and 1% goat normal serum for 2 h. Vascular and primary capillary plexus structures were visualized by rat anti-mouse CD31 staining (1:100 for 4 h) and detected by incubation with fluorescein-conjugated goat anti-rat IgG. Images were collected using an IX81 Olympus confocal microscope with a Hamamatsu ORCA-ER monochrome camera. Whole embryo CD31 staining was visualized by Olympus OV100 and IX81 spinning disk confocal imaging. Images were cropped using Adobe Photoshop CS3 software.
Cell Migration
Millicell serum chemotaxis assays were performed as described (23), and data points represent enumerations of three migration chambers from at least two independent experiments. For scratch wound closure motility assays, cells were seeded in 6-well plates and starved for 24 h. Cells were wounded by scratching with pipette tip, washed with phosphate-buffered saline, photographed (0 h point), and placed into growth media. After 16 h, matched pair wound regions were photographed, and the wound edge distance was measured using Image J. For phase-contrast time lapse video microscopy, cells were plated onto fibronectin (FN)-coated (10 μg/ml) MatTek glass bottom microwell dishes in the presence of 10% fetal bovine serum at 37 °C with humidity and CO2 regulation. After 1 h, images were collected at 2-min intervals over 5 h with a ×20 lens on an automated stage (Olympus IX81). Cell trajectories were measured by tracking nucleus position over time using Image J.
Biochemical Analyses
Cell lysis, immunoprecipitation, and immunoblotting analyses were performed as described (37). Integrin-stimulated signaling assays were performed by serum starvation (0.5% fetal bovine serum) of cells overnight, limited trypsin treatment (0.06% with 2 mm EDTA) to collect cells, inactivation of trypsin with soybean trypsin inhibitor (0.25 mg/ml), and resuspension of cells in migration medium (Dulbecco's modified Eagle's medium with 0.5% bovine serum albumin). Cells were held in suspension for 30 min prior to replating on fibronectin-coated (10 μg/ml) plates for the indicated times. Rho activity was measured by pull-down assays using glutathione S-transferase (GST)-rhotekin Rho binding domain as described (42). Rac activity was measured by binding to the GST-PAK residues 67–150 purified from bacterial lysates as described (43). Relative Rho or Rac activity was calculated from two independent experiments by analyzing densitometry of blots using Image J and normalized to total RhoA or Rac1 levels.
Immunofluorescence Staining
Cells were serum-starved, removed by trypsin treatment, and replated onto 10 μg/ml FN-coated glass coverslips in the absence of serum for the indicated times. Cells were fixed in 3.7% paraformaldehyde for 15 min and permeabilized with 0.1% Triton X-100 for 10 min. Anti-paxillin (1:100) and anti-FAK (1:25; BD Biosciences) were incubated overnight at 4 °C. FA number was determined by paxillin staining by integrated morphometry analysis using Image J on thresholded images to select classified objects of a size range of 0.1–20 pixels as FAs. The “analyze particles” command was used to determine both FA numbers.
Golgi Reorientation-Polarity Assay
Golgi reorientation analyses were performed in a scratch wound assay as described (26). To quantify Golgi reorientation of cells at the wounded edge, a square was drawn over the nucleus and divided into four quadrants. Quadrant A was assigned to the area of the cell between the nucleus and the leading edge. A properly reoriented Golgi was indicated by β-Cop staining entirely within quadrant A. Analyses were performed on 100 cells/experiment to determine the percentage of cells with reoriented Golgi.
RESULTS
Generation of Kinase-inactive FAK (R454) Knock-in Mice
To investigate the role of FAK kinase activity during development, a kinase-inactive (R454) knock-in mutation was generated by homologous recombination (supplemental Fig. 1A). Mice contained a neomycin-selectable marker, and crosses of heterozygous FAKWT/R454Neo mice did not yield homozygous FAKR454Neo/R454Neo mice at birth (data not shown). The numbers of FAKWT/WT and FAKWT/R454Neo pups (1:2) were consistent with Mendelian ratios, indicating that the R454 mutation did not create a trans-dominant lethal phenotype. Both male and female FAKWT/R454Neo mice were small in size at postnatal day 6, but within 3–4 weeks, FAKWT/WT and FAKWT/R454Neo mice were indistinguishable (data not shown). Because the neomycin marker transcribes mRNA in the opposite orientation of FAK mRNA transcription, the neomycin cassette was removed by crossing FAKWT/R454Neo mice with FLP1 recombinase transgenic mice (supplemental Fig. 1B). Crosses of FAKWT/R454 mice did not yield homozygous FAKR454/R454 mice at birth (Table 1), and heterozygous FAKWT/R454 mice were visually indistinguishable from FAKWT/WT mice. Timed pregnancies, ultrasound analyses (supplemental Fig. 2), and embryo extraction revealed that FAKR454/R454 mouse development stopped at E9.5 (Table 1). Genotyping of embryos from FAKWT/R454 crosses revealed normal Mendelian ratios at E7.5, E8.5, and E9.5, but no FAKR454/R454 embryos were found at E10.5 or beyond. Analyses of multiple E8.5 litters showed no morphological differences between FAKWT/WT, FAKWT/R454, and FAKR454/R454 embryos (data not shown). Thus, FAKR454/R454 embryos survive 1 day longer than FAK−/− embryos (44).
TABLE 1.
Genotypes of embryos obtained from crosses between FAKWT/R454 mice
Indicated is the total number of embryos obtained at various embryonic ages and at birth.
| E7.5 | E8.5 | E9.5 | E10.5 | Born | |
|---|---|---|---|---|---|
| FAKWT/WT | 5 | 28 | 74 | 31 | 378 |
| FAKWT/R454 | 7 | 56 | 140 | 59 | 768 |
| FAKR454/R454 | 3 | 26 | 67 | 0 | 0 |
| Total | 15 | 110 | 281 | 90 | 1146 |
FAKR454/R454 Embryos Exhibit Mesodermal and Vessel Morphogenesis Defects
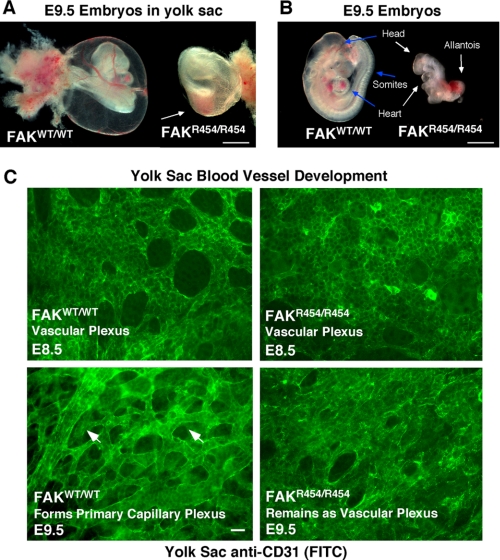
Light microscopic analysis of more than 50 E9.5 embryos did not reveal blood filled vessels in FAKR454/R454 embryo yolk sacs compared with FAKWT/WT littermates (Fig. 1A). Upon dissection, FAKR454/R454 embryos were abnormal but with recognizable anterior head and heart structures (Fig. 1B). Hearts were beating in FAKWT/WT embryos at this stage, but this was not observed in FAKR454/R454 embryos. No organized somite formation was detected in E9.5 FAKR454/R454 embryos (Fig. 1B), consistent with mesodermal defects present in FAK−/− embryos (40). FAKR454/R454 embryos consistently exhibited an enlarged and unfused allantois structure (Fig. 1B). In normal embryos, the allantois fuses with the chorion membrane, and this connects the fetal and maternal circulatory systems between E8.5 and E9.5.
FIGURE 1.
FAKR454/R454 kinase-inactive knock-in mutation is embryonic lethal with vascular defects. A, bright field images of excised yolk sacs with embryos reveals a lack of blood-filled vessels on FAKR454/R454 compared with FAKWT/WT embryos at E9.5. The red color within the FAKR454/R454 yolk sac (arrow) is associated with internal embryo hemorrhage. Bar, 100 μm. B, at E9.5, FAKWT/WT embryos have undergone turning and have a defined head, beating heart, and multiple segmental somites (arrows). FAKR454/R454 embryos at E9.5 have undergone turning, are small, and possess head and heart structures that are malformed (arrows). No somites or beating hearts were detectable in FAKR454/R454 embryos. The allantois is enlarged, does not undergo chorion fusion, and is frequently a site of hemorrhage within FAKR454/R454 embryos. Bar, 100 μm. C, whole mount anti-CD31 staining of yolk sacs at E8.5 reveals equivalent vascular plexus formation in FAKR454/R454 and FAKWT/WT embryos. At E9.5, ECs within yolk sacs of FAKWT/WT embryos form a primary capillary plexus characterized by tubules and branched structures (arrowheads). The primary capillary plexus is not formed in FAKR454/R454 yolk sacs at E9.5 because this remains a vascular plexus structure. Bar, 20 μm.
It is known that FAK expression becomes elevated at E8.5, a period of extensive morphogenic cell movement during development (45). Anti-CD31 staining (a marker of ECs) of E8.5 yolk sacs revealed equivalent EC-associated vascular plexus structures formed in FAKWT/WT and FAKR454/R454 embryos (Fig. 1C). This is consistent with FAK not being essential for angioblast to EC differentiation (34). Between E8.5 and E9.0, ECs within the yolk sac undergo morphogenic movements to form a primary capillary plexus characterized by EC tubules and branched structures. By E9.5, FAKWT/WT embryo yolk sac ECs formed a capillary plexus (Fig. 1C, arrowheads), but this was not observed in FAKR454/R454 embryo yolk sacs. Defective capillary plexus and EC branch formation was not accompanied by increased cell apoptosis in E9.5 FAKR454/R454 embryo yolk sacs (supplemental Fig. 3). This FAKR454/R454 embryo defect is similar to that observed in conditional β1 integrin or FAK knock-outs where yolk sacs exhibit disordered vascular beds (36, 46).
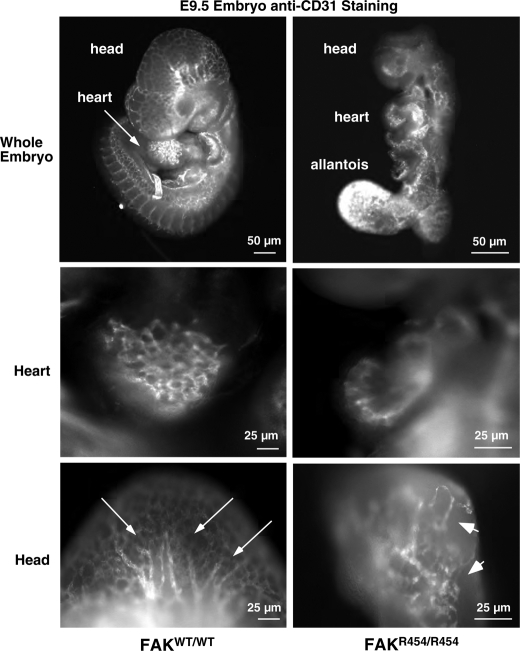
This defect in FAKR454/R454 vascular patterning was also apparent upon analyzing whole embryos by anti-CD31 staining for ECs (Fig. 2). In E9.5 FAKWT/WT embryos, a network of CD31 staining was evident and concentrated in head, heart, and somite structures. In head regions, tubule-like structures were apparent (Fig. 2, arrows). In FAKR454/R454 embryos, intense CD31 staining was detected in the extended allantois (Fig. 2), and this was also a common site of hemorrhage (Fig. 1B). However, the general pattern throughout FAKR454/R454 embryos was that of limited patches of connected CD31-positive cells (Fig. 2, arrowheads). These results support the conclusion that FAK activity is important in the processes of embryonic vascular network formation.
FIGURE 2.
Defective EC patterning within FAKR454/R454 embryos. Whole embryos at E9.5 were stained with anti-CD31 to visualize ECs. Comparisons are between FAKWT/WT and FAKR454/R454 littermates. Defined EC branchlike and tubule structures (arrows) in the head and heart of FAKWT/WT embryos are shown. FAKR454/R454 embryos exhibit intense EC staining in the unfused allantois. Clusters (arrowheads) of ECs are present in FAKR454/R454 head and heart structures, but these were not organized within a defined network.
Loss of FAK Activity Affects Tyrosine Phosphorylation in Vivo
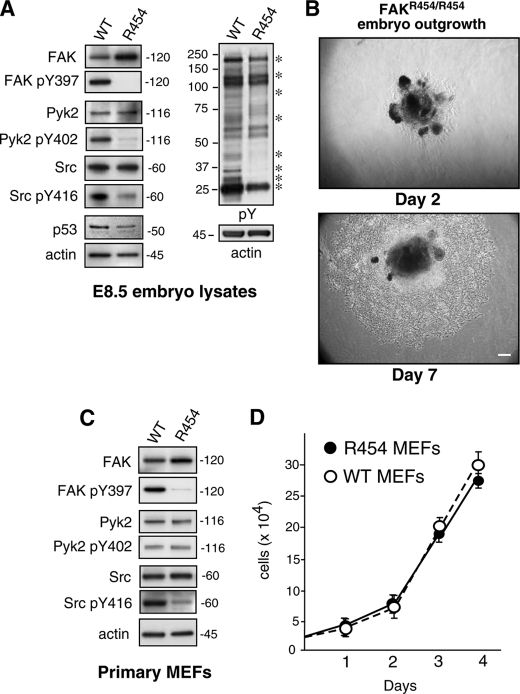
To verify that the observed developmental defects are due to the lack of FAK signaling, whole E8.5 embryos were extracted and solubilized for protein immunoblotting analyses (Fig. 3A). When normalized to the level of β-actin expression, R454 FAK levels were ∼2-fold higher than WT FAK. Additionally, whereas WT FAK was strongly phosphorylated at Tyr-397 (pY397), R454 FAK was not detectably phosphorylated at this site within E8.5 embryo lysates (Fig. 3A). This result verifies the loss of FAK activity in vivo. Moreover, these results are consistent with FAK Tyr-397 autophosphorylation and that FAK activity is linked to increased FAK polyubiquitination and turnover (47). When embryo lysates were analyzed by anti-phosphotyrosine (pY) immunoblotting, protein bands at 25, 32, 35, 42, 65–70, 85, 130, and 190 kDa (indicated by asterisks) were of lower intensity or absent in FAKR454/R454 compared with FAKWT/WT embryos (Fig. 3A). Known proteins, such as Pyk2 and c-Src, were equally expressed but decreased in activation-associated tyrosine phosphorylation events at Pyk2 Tyr-402 (pY402), and c-Src Tyr-416 (pY416), respectively. This result supports the notion that FAK may promote Pyk2 and c-Src activity in vivo during development.
FIGURE 3.
R454 FAK expression and FAK phosphorylation target identification within E8.5 embryo lysates and establishment of primary FAKR454/R454 MEFs. A and C, protein lysates from FAKWT/WT (WT) and FAKR454/R454 (R454) embryos harvested at E8.5 or lysates from primary MEFs were analyzed by immunoblotting for total FAK, Tyr(P)-397 (pY397) FAK, total Pyk2, Pyk2 Tyr(P)-402 (pY402), total c-Src, and Src Tyr(P)-416 (pY416). Anti-phosphotyrosine (pY) immunoblotting revealed decreased target protein tyrosine phosphorylation (asterisks) in lysates from E8.5 R454 embryos. Anti-actin immunoblotting was used as a loading control. Decreased c-Src activation (as detected by Src Tyr(P)-416 blotting) was detected in R454 embryos and MEFs. A, p53 tumor suppressor expression was lower in R454 E8.5 embryos as determined by immunoblotting. B, phase-contrast images of E8.5 FAKR454/R454 embryo outgrowth in culture after 2 and 7 days. Shown is Matrigel-embedded embryo (dark center) and surrounding cell outgrowth. Bar, 100 μm. D, primary WT and R454 MEFs proliferate equally in culture. Results are the mean cell number (n = 3 independent points ± S.D. (error bars)).
FAKR454/R454 MEFs Proliferate ex Vivo
Previous analyses of E8.5 FAK+/+, FAK+/−, and FAK−/− embryo lysates revealed elevated p53 tumor suppressor levels as a function of FAK inactivation (37). Increased p53 activation blocked FAK−/− cell outgrowth from embryos ex vivo and prevented the establishment of primary FAK−/− (p53+/+) MEFs. FAK regulates p53 via its FERM domain, and R454 FAK was shown to inhibit p53 equally as WT FAK (37). Interestingly, p53 levels were slightly lower in FAKR454/R454 compared with FAKWT/WT E8.5 embryo lysates (Fig. 3A). To determine whether cells from FAKR454/R454 embryos exhibit proliferation defects, E8.5 embryos were embedded in Matrigel and grown ex vivo (Fig. 3B). Within 2 days, embryonic cells became adherent, and within 7 days, an expanding ring of proliferating cells surrounded both FAKR454/R454 (Fig. 3B) and FAKWT/WT embryos (data not shown). The growth of FAKR454/R454 MEFs in culture is consistent with the kinase-independent regulation of p53 by FAK and establishes that FAK activity is not essential for adherent cell proliferation.
These embryonic cells were further expanded as primary MEFs and revealed slightly elevated R454 FAK protein expression compared with WT MEFs (Fig. 3C). In contrast to WT FAK, R454 FAK was not phosphorylated at Tyr-397 (pY397). Pyk2 expression and activation (pY402) were equivalent between WT and R454 MEFs whereas c-Src activation (pY416) was lower in R454 MEFs (Fig. 3C) and consistent with the E8.5 embryo immunoblotting results. This result is consistent with FAK-mediated phosphorylation of c-Src at Tyr-416 mediating direct c-Src activation (15). Analyses of growth rates in culture revealed no significant differences between WT and R454 MEFs (Fig. 3D), confirming the fact that FAK activity is not essential for fibroblast proliferation in vitro.
Loss of FAK Activity Affects Adhesion and Protrusion Formation
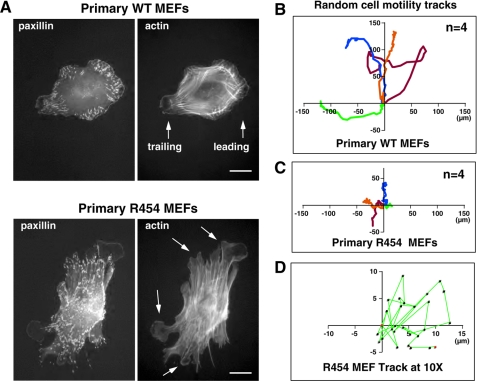
Cell movement is a coordinated process involving adhesion, spreading, FA formation, arrangement of a protruding leading edge, and the coordinated retraction of a trailing edge involving FA disassembly. When plating the primary MEFs onto FN, R454 MEFs bound and spread equally compared with WT MEFs (supplemental Videos 1 and 2). Analysis of actin and paxillin distribution showed that after 1 h on FN in the absence of serum, WT MEFs formed a characteristic pattern of FAs around the cell periphery with the generation of leading and trailing edge cell projections (Fig. 4A). In contrast, R454 MEFs formed an exaggerated number of FAs connected to elevated numbers of actin stress fibers after 1 h on FN (Fig. 4A). Additionally, R454 MEFs exhibited multiple lamellipodial projections around the cell periphery without a defined front-back distribution (Fig. 4A, arrows). These results add to earlier findings using FAK−/− MEFs (48, 49) that integrin-stimulated FAK activation is important in promoting both leading edge formation and the coordination of FA turnover upon FN matrix engagement.
FIGURE 4.
Primary R454 MEFs exhibit increased FA formation and lack directional persistence of cell movement. A, WT and R454 MEFs were plated onto FN-coated glass slides for 1 h and co-stained for paxillin and actin. WT MEFs formed defined leading and trailing edge cell protrusions, whereas R454 MEFs formed protrusions randomly around the cell periphery (arrows). Increased numbers of paxillin-stained FAs formed in R454 MEFs. Bar, 10 μm. B, analysis of WT MEF migration paths by time lapse imaging of MEFs plated on FN in the presence of serum. Cell tracks were determined by cell nuclei position, and migration origin was superimposed at 0. The scale is in μm, n = 4 cells. C, analysis of R454 MEF migration paths as in B. The scale is μm, n = 4 cells. D, ×10 magnified migration track from C shows the lack of directionality of R454 MEF movement.
Migration and Polarity Defects of R454 MEFs
Excessive FA and protrusion formation exhibited by R454 MEFs suggest that these cells may exhibit motility defects. WT MEFs plated on FN in the presence of serum possess defined leading and trailing edge structures and exhibit directional persistence of movement in time lapse imaging studies (supplemental Video 3). In contrast, R454 MEFs exhibited limited cell movement upon serum addition with enhanced spreading and membrane ruffling occurring at multiple projections (supplemental Video 4). The average migration speed of WT and R454 MEFs was 0.37 ± 0.06 and 0.17 ± 0.04 μm/min (±S.D.), respectively. This is consistent with 2-fold differences in random motility measured for FAK−/− and FAK-reconstituted MEFs (50). Notably, the enlargement of an R454 MEF time lapse migration track reveals a random motility pattern, whereas WT MEFs exhibit directional persistence of movement (Fig. 4, B–D). These results support the notion that FAK activity is important in the establishment of cell polarity.
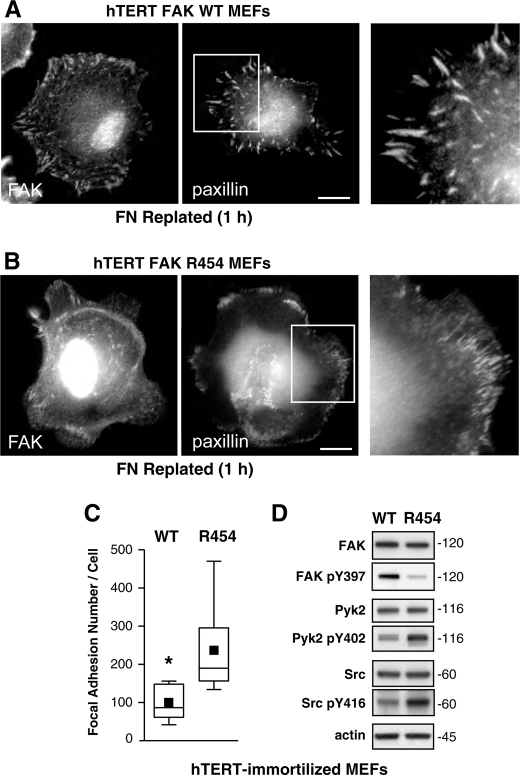
Because both primary WT and R454 MEFs were established from early embryos and possess a finite number of cell doublings, immortalized populations of WT and R454 FAK MEFs were established by retroviral transduction with either hTERT (Fig. 5) or T-Ag (supplemental Fig. 4). Importantly, both WT and R454 FAK were recruited to FAs upon plating of hTERT (Fig. 5, A and B) and T-Ag (supplemental Fig. 4, A and B) MEFs onto FN for 1 h as visualized by indirect immunofluorescent cell staining. This result shows that the R454 mutation does not disrupt FAK localization to FAs. Notably, both hTERT R454 (Fig. 5, B and C) and T-Ag R454 (supplemental Fig. 4, B and C) MEFs form significantly greater numbers of FAs as determined by paxillin staining. Immunoblotting analyses showed equivalent FAK, Pyk2, and c-Src expression in hTERT R454 and WT MEFs (Fig. 5D). FAK Tyr(P)-397 phosphorylation was low in R454 MEFs, whereas both Pyk2 Tyr(P)-402 and Src Tyr(P)-416 levels were higher in R454 compared with WT hTERT MEFs (Fig. 5D). Similar up-regulation of Pyk2 and c-Src activity occurred in R454 T-Ag-immortalized MEFs (supplemental Fig. 4D) compared with immunoblotting results with primary MEFs (Fig. 3). Thus, immortalization or increased cell passage yielded changes in Pyk2 and Src activation within an R454 FAK background. However, this did not affect the phenotype of increased FA formation observed in both primary and immortalized R454 MEFs.
FIGURE 5.
Immortalized FAK R454 MEFs exhibit increased FA formation. Primary WT and R454 MEFs were immortalized by retroviral hTERT expression, and pooled populations of cells were characterized. A and B, MEFs were plated on FN for 1 h in the absence of serum and stained with antibodies to FAK and paxillin. Bar, 10 μm. Inset (right), enlargement of paxillin-stained FAs. C, increased FA formation in R454 MEFs. Box-and-whisker plots of paxillin-positive stained points were enumerated within WT and R454 MEFs plated on FN for 1 h (n = 15 cells/point). Box-and-whisker diagrams show the distribution of the data: mean (square), 25th percentile (bottom line), median (middle line), 75th percentile (top line), and 5th or 95th percentiles (whiskers). Significance was determined using a two-tailed Student's t test (*, p < 0.0001). D, lysates from WT and R454 MEFs were by analyzed by total FAK, Tyr(P)-397 (pY397) FAK, total Pyk2, Pyk2 Tyr(P)-402 (pY402), total c-Src, Src Tyr(P)-416 (pY416), and actin immunoblotting. R454 FAK is expressed but only weakly phosphorylated at Tyr-397. Increased levels of Pyk2 and c-Src activation were detected in hTERT-immortalized R454 compared with WT MEFs.
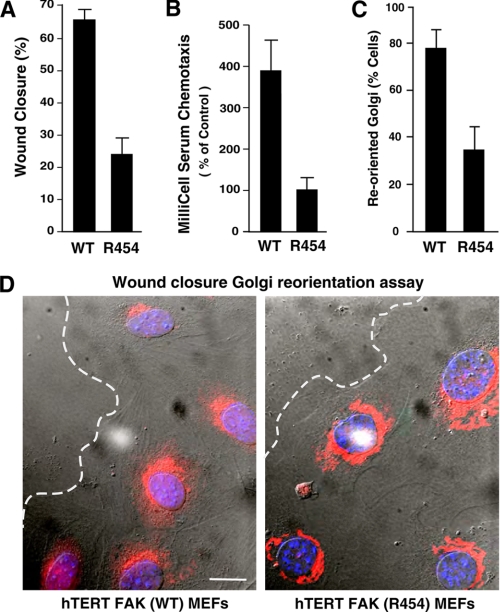
Despite increased Pyk2 and c-Src activation as a function of hTERT R454 MEF immortalization, R454 MEFs exhibited 2.5-fold reduced wound closure (Fig. 6A) and ∼4-fold reduced chemotaxis motility (Fig. 6B) compared with hTERT WT MEFs. Similar results were obtained by comparisons of R454 and WT T-Ag-immortalized MEFs (data not shown). Because primary R454 MEFs exhibited a random motility phenotype (Fig. 4B), Golgi reorientation was evaluated in wound healing assays to determine the effects of R454 FAK inactivation on cell polarity (Fig. 6, C and D). In unwounded confluent cell populations, Golgi orientation is randomly distributed. Upon creating a wound edge and allowing for initial cell movement within the denuded space (4 h), cells will establish an axis of polarity, with microtubules and Golgi markers facing the wounded edge (51). Whereas ∼80% of WT MEFs at the wound edge exhibit Golgi reorientation, less than 40% of R454 MEFs exhibit this response (Fig. 6, C and D). These results show that FAK activity is essential for efficient fibroblast motility and polarity.
FIGURE 6.
Immortalized FAK R454 MEFs exhibit motility and polarity defects. A, wound closure motility of WT and R454 MEFs was calculated by measuring the distance change between 0 and 16 h in time lapse experiments. Values are means ± S.D. from 10 independent analyses. B, chamber motility was measured over 4 h with WT and R454 MEFs with FN and serum. Values are means ± S.D. from three experiments. C, FAK activity is required for Golgi reorientation and cell polarization. 100 cells were analyzed, and values represent percentage of cells ± S.D. D, representative images from Golgi reorientation assay. WT and R454 MEFs were grown to confluence, wounded with a pipette tip, and allowed to migrate in the presence of serum for 4 h. Cells were fixed, imaged in phase, and stained for Golgi (β-Cop, red) and nuclear (Hoechst, blue) markers. Position of the leading lamella (dashed line) is indicated. Bar, 10 μm.
FAK Activity Serves as a Master Regulator of p190A-p120RasGAP Complex Formation and RhoA Regulation Downstream of Integrins
The phenotype of increased FA formation in primary and immortalized R454 MEFs is consistent with elevated RhoA GTPase activity (28). Rho GTPases are held in balance by the opposing action of guanine nucleotide exchange factors that facilitate GTP loading and GTPase-activating proteins (GAPs) that promote Rho inactivation by stimulating intrinsic GTP hydrolysis (52). The p190RhoGAP proteins termed p190A and p190B inhibit RhoA activity in cells (53). p190A tyrosine phosphorylation is associated with p190A activation, leading to Rho inhibition (27) and the formation of a complex with p120RasGAP (a GTPase-activating protein for Ras) (54).
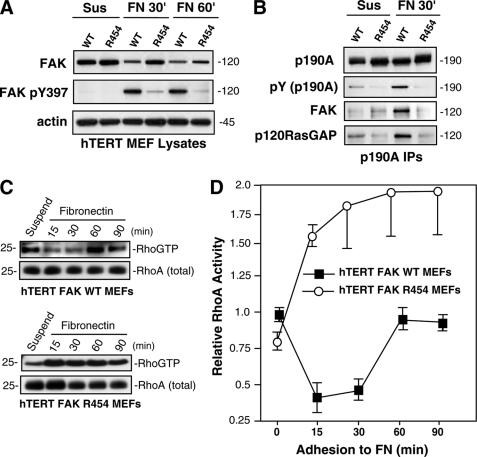
To determine if a linkage exists between FAK activity and p190A tyrosine phosphorylation, signaling was analyzed in hTERT-immortalized MEFs by replating onto FN to stimulate integrin signaling (Fig. 7). In WT MEFs, FAK was activated within 30 min and remained active at 60 min, as analyzed by Tyr(P)-397 immunoblotting and compared with cells held in suspension (Fig. 7A). R454 FAK was minimally phosphorylated at Tyr-397 at 30 or 60 min upon FN plating, confirming that R454 FAK is not active (Fig. 7A). Co-immunoprecipitation analyses using antibodies to p190A revealed increased p190A tyrosine phosphorylation and p190A association with FAK and p120RasGAP upon FN adhesion within 30 min compared with suspended cells (Fig. 7B). Notably, p190A was not detectably tyrosine-phosphorylated or associated with R454 FAK or p120RasGAP in R454 MEFs upon FN adhesion (Fig. 7B). Thus, despite enhanced c-Src activity in R454 FAK MEFs (Fig. 5C), FAK activity is essential for p190A tyrosine phosphorylation and the formation of a p190A-p120RasGAP-FAK complex upon FN-initiated cell stimulation.
FIGURE 7.
FAK promotes p190RhoGAP phosphorylation after FN stimulation of MEFs; connection to RhoA regulation. A, R454 FAK is not activated upon MEF adhesion to FN. Lysates from suspended and FN-replated (30 and 60 min) hTERT WT and R454 MEFs were analyzed by total FAK, Tyr(P)-397 (pY397) FAK, and actin blotting. B, p190A forms a complex with FAK and p120RasGAP upon WT but not R454 MEF adhesion to FN. p190A immunoprecipitations were made from lysates of hTERT WT or R454 MEFs held in suspension or FN-replated (30 min) and sequentially analyzed by anti-Tyr(P) (pY) and -p190A blotting. The 120 kDa region of the p190A immunoprecipitations (IPs) was sequentially analyzed by anti-FAK and -120RasGAP immunoblotting. C, transient decrease in RhoA GTP binding upon WT but not R454 MEF adhesion to FN. GTP-bound RhoA was determined by GST-rhotekin Rho-binding domain affinity pull-down assays from lysates of suspended and FN-replated cells, followed by blotting for total RhoA levels. D, elevated RhoA GTP binding was observed in R454 FAK MEFs upon FN adhesion. Quantitation of RhoA-GTP binding from two independent experiments ± S.D. Values are normalized to total RhoA levels are relative to RhoA-GTP levels in suspended WT FAK MEFs.
Previous studies have established that a transient decrease in RhoA-GTP levels occurs during the initial phases of cell spreading on FN, followed by a period of RhoA activation (55). Affinity binding and pull-down assays were performed to determine the levels of GTP-bound RhoA in WT and R454 MEFs (Fig. 7, C and D). In WT MEFs, a transient decrease in RhoA GTP levels coincided with FAK activation and p190A tyrosine phosphorylation at 15–30 min on FN (Fig. 7, C and D). This pattern of RhoA GTP regulation is consistent with published studies (42). In contrast, RhoA GTP levels were not regulated by FN adhesion and were significantly elevated in R454 MEFs upon FN adhesion (Fig. 7, C and D). Similar results were obtained with T-Ag-immortalized WT and R454 MEFs (supplemental Fig. 4). Thus, increased p190A tyrosine phosphorylation upon WT but not R454 MEFs is consistent with the regulation of RhoA GTPase activity in WT but not R454 MEFs. Loss of p190A tyrosine phosphorylation in R454 MEFs may prevent full p190A GAP activation and result in high RhoA GTP levels. Taken together, because increased RhoA activation is tightly connected to increased FA formation (28) and previous studies have established that FAK re-expression in FAK−/− MEFs suppresses RhoA activity to promote FA turnover (42), our results establish that FAK-mediated p190A tyrosine phosphorylation may be a control point for FN-stimulated RhoA regulation and FA dynamics.
Alterations in R454 MEFs Facilitate Elevated Rac Activation after FN Plating
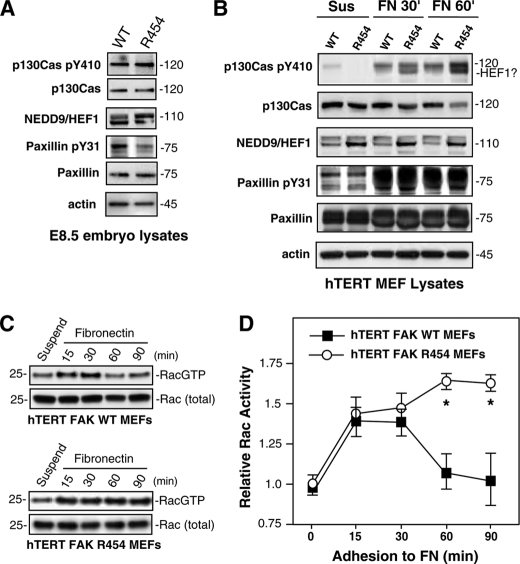
Constitutively active RhoA expression in MEFs increases FA formation and decreases cell spreading on FN (42). This is the phenotype of FAK−/− MEFs, and treatment with Rho-associated kinase inhibitor enhances FAK−/− spreading on FN (56). R454 MEFs exhibit elevated RhoA activity and FA formation but also exhibit normal initial spreading (supplemental Video 2) and enhanced membrane ruffling after 1 h on FN (Fig. 4A and supplemental Video 4). Because these latter phenotypes are consistent with Rac GTPase activation, FAK targets, such as paxillin, p130Cas, and HEF1/NEDD9, known to facilitate Rac activation (11, 57) were evaluated in WT and R454 E8.5 embryo lysates, and hTERT MEFs were replated onto FN (Fig. 8). p130Cas expression and substrate domain tyrosine phosphorylation (pY410 p130Cas) were equivalent in E8.5 embryos (Fig. 8A). Slightly elevated HEF1/NEDD9 expression (as two bands) was detected in R454 embryos, whereas equal paxillin expression but reduced paxillin Tyr(P)-31 tyrosine phosphorylation were detected in R454 compared with WT embryos (Fig. 8A).
FIGURE 8.
Elevated Rac GTP levels in FAK R454 MEFs upon FN replating are associated with increased HEF1 expression and p130Cas tyrosine phosphorylation. Protein lysates from FAKWT/WT (WT) and FAKR454/R454 (R454) embryos harvested at E8.5 (A) or protein lysates from hTERT-immortalized WT or R454 MEFs held in suspension for 30 min (Sus) or replated onto fibronectin (FN) for 30 or 60 min in the absence of serum (B) were evaluated by immunoblotting for total p130Cas, Tyr(P)-410 (pY410) p130Cas, NEDD9/HEF1, total paxillin, Tyr(P)-31 (pY31) paxillin, and actin. C, elevated GTP-bound Rac1 in R454 MEFs upon FN adhesion. GTP-bound Rac1 was determined by affinity pull-down assays (GST-PAK residues 67–150) from lysates of suspended and fibronectin-replated WT and R454 FAK hTERT-immortalized MEFs followed by blotting for total Rac1 levels. D, quantitation of Rac1-GTP binding and values normalized to total Rac1 levels and plotting relative to Rac1-GTP levels in suspended WT FAK MEFs. Data are from two independent experiments ± S.D. Significance was determined by two-way analysis of variance (*, p < 0.01).
To determine if these FAK targets were tyrosine-phosphorylated after FN replating of WT or R454 MEFs, immunoblotting analyses were performed and compared with lysates of cells held in suspension (Fig. 8B). FN replating equally stimulated p130Cas and paxillin tyrosine phosphorylation in WT and R454 MEFs. Interestingly, the p130Cas Tyr(P)-410 antibody detected two bands in R454 MEF lysates, the lower of which co-migrated with HEF1/NEDD9 that also showed increased expression in R454 MEFs (Fig. 8B). Additionally, p130Cas Tyr(P)-410 signal intensity was reproducibly stronger at 60 min after FN plating in R454 compared with WT MEFs. Correspondingly, Rac1 activation, as determined by affinity binding and pull-down assays, was elevated at 15–30 min upon FN replating of both WT and R454 MEFs (Fig. 8, C and D). However, Rac1 GTP-binding remained significantly elevated in R454 MEFs at 60 and 90 min after FN replating compared with WT MEFs (Fig. 8D). This elevated level of Rac1 activation in R454 MEFs may be associated with the enhanced spreading and membrane ruffling phenotypes of these cells. Future studies will be aimed at determining the molecular linkages regulating Rac1 activity in cells lacking FAK activity and whether up-regulated NEDD9/HEF1 expression is a common compensatory event associated with FAK inhibition.
DISCUSSION
During development, there is a complex interplay between the receptors for angiogenic growth factors, the extracellular matrix, and integrins in the processes of vasculogenesis and angiogenesis (1, 4, 58). FAK is activated by both growth factors and integrins and functions as a nexus between these signaling pathways. FAK knock-out is permissive for the formation of ECs and blood islands within yolk sacs but interferes with later steps of functional blood vessel formation (34, 40). Through the analysis of the first knock-in mutation generating a kinase-inactive FAK mouse (R454), we show that loss of FAK activity but not expression is essential for the proper formation of various mesoderm-associated structures (somites, heart, and EC-associated vascular network) during early embryonic development (Fig. 1). Similar vasculogenesis-associated defects occur in the yolk sacs of FAK−/− and FAKR454/R454 embryos due to the inability of ECs to become organized within vascular networks. Lethality of FAK−/− embryos occurs at E8.5 and 1 day later at E9.5 for FAKR454/R454 embryos. Although FAK−/− embryos did not possess ECs, FAKR454/R454 embryos exhibited a pattern of disorganized EC staining lacking the defined tubule structures that were detected in FAKWT/WT littermates at E9.5 (Fig. 2). We speculate that this defect in primitive vessel formation may be associated with observed blood hemorrhage in the head, heart, and allantois regions of FAKR454/R454 embryos.
The ability of ECs to form tubule structures during development is related to the processes of angiogenesis that involve alterations in cell movement-organization (59). Although it remains undefined whether the EC-associated morphogenic defects in FAKR454/R454 embryos is associated with EC-intrinsic or extrinsic loss of FAK activity in other vessel-associated cells, inactivation of FAK expression within ECs disrupts vascular network stability and prevents angiogenesis (35, 36). Because transgenic overexpression of FAK can promote angiogenesis (60) and small molecule FAK inhibitors possess antiangiogenic activity (32, 39), phenotypes associated with FAKR454/R454 knock-in mice support an essential role for FAK activity in facilitating angiogenesis. Although our inability to recover sufficient numbers of ECs from E8.5 embryos prevented further analyses, our genetic studies support the use of FAK inhibitors to disrupt neovascularization. Further, because enhanced FAK signaling is associated with the anchorage-independent survival pathway in tumor cells (33), we hypothesize that pharmacological inhibition of FAK activity may block tumor progression via selected effects on both the tumor and surrounding stroma (61).
In addition to pharmacological inhibition of FAK activity, inhibitors can block FAK Tyr-397 autophosphorylation via interaction at this site (62). Canonical FAK signaling models have placed emphasis on the role of FAK Tyr-397 phosphorylation, which creates a binding site for Src family kinases and leads to the formation of an active multiprotein FAK-Src signaling complex (8). This model is built upon the premise that FAK serves as an activatable scaffold via autophosphorylation with less emphasis on the role for FAK activity in trans-phosphorylating other substrate proteins that may be bound to FAK. Surprisingly, developmental studies characterizing mice with a deletion of FAK exon 15 encompassing the Tyr-397 site found that lethality did not occur until after E14.5 (41). This mutation creates a truncated FAK protein (Δ15 FAK) that is expressed and active at normal levels. Remarkably, development can proceed further with an active FAK protein that is missing the Tyr-397 site (Δ15 FAK) compared with embryos expressing kinase-inactive R454 FAK. One interpretation from this result is that FAK functions to directly phosphorylate targets to affect signaling pathways during development that occur independently of FAK autophosphorylation at Tyr-397. We speculate that Δ15 FAK facilitates development further than R454 FAK because Δ15 FAK may be able to phosphorylate specific targets at E9.5 that are dependent upon intrinsic FAK activity.
One of the targets we identified as being dependent on intrinsic FAK activity within embryos and primary MEFs was c-Src (Figs. 3 and 4). Because R454 FAK was not detectably phosphorylated at Tyr-397, our results are consistent with c-Src binding to WT FAK at Tyr-397 and FAK-mediated activation of c-Src via direct phosphorylation at c-Src Tyr-416 (15). Interestingly, many of the identified substrates for FAK, such as paxillin (11–13), p130Cas (14), and p190RhoGAP (22), were first identified as substrates for Src family kinases (63). MEFs expressing Δ15 FAK exhibit decreased p130Cas and paxillin tyrosine phosphorylation upon FN plating (41). This was interpreted as FAK Tyr-397 autophosphorylation being required for the recruitment of Src family kinases into a signaling complex needed for p130Cas and paxillin phosphorylation.
However, our analyses of E8.5 embryos and immortalized MEFs revealed normal to elevated paxillin and p130Cas phosphorylation upon FN plating (Fig. 8). In combination with increased NEDD9/HEF1 expression in R454 MEFs, this could contribute to the elevated level of Rac1 activation observed upon FN stimulation of R454 MEFs. Moreover, deregulated Rac1 activity could also contribute to the observed enhanced spreading and membrane ruffling phenotypes of R454 MEFs (Fig. 4). It remains unclear why paxillin and p130Cas would be normally tyrosine-phosphorylated in R454 but not Δ15 FAK MEFs. One explanation is that enhanced and potentially compensatory changes in Pyk2 and c-Src activation were detected in immortalized R454 MEFs (Fig. 5). Because both p130Cas and paxillin are substrates of Pyk2 in fibroblasts (64), we speculate that Pyk2-Src signaling may partially compensate for the loss of FAK activity, as first documented in FAK−/− MEFs (65). Importantly, the fact that p130Cas and paxillin tyrosine phosphorylation occur normally in R454 MEFs yet these cells exhibit increased defects in FA formation and motility suggests that p130Cas or paxillin are not linked to the observed R454-associated phenotypes. There must be other selective targets of FAK activity that are not being phosphorylated and correctly regulated within R454 MEFs.
Notably, our past studies with FAK−/− MEFs (26) and our current results with R454 MEFs (Fig. 7) support the notion that p190A tyrosine phosphorylation after FN plating is a specific target of FAK activity. Notably, FAK−/−, Δ15 FAK, and R454 FAK MEFs all exhibit common phenotypes of increased FA formation, abnormal lamellipodial projections, and decreased motility (Figs. 4–6) (41, 49, 65). Although p190A tyrosine phosphorylation was not evaluated in Δ15 FAK MEFs (41), loss of p190A tyrosine phosphorylation was also linked to increased RhoA-associated cell tension in FAK-null keratinocytes (17). Because treatment of FAK−/− (56) and R454 MEFs5 with a Rho-kinase inhibitor (Y27632) promotes actin reorganization and FA turnover, deregulated and elevated RhoA activity is a common phenotype observed in the absence of FAK expression or activity.
Tyrosine phosphorylation of p190A is associated with p190A activation leading to Rho inhibition (27) and the formation of a complex with p120RasGAP (54). The lack of p190A tyrosine phosphorylation in R454 MEFs would prevent full GAP activation and allow for elevated RhoA activity. Additionally, R454 FAK does not form a complex with p190A and p120RasGAP, as does WT FAK (Fig. 7). Notably, we previously showed through reconstitution studies in FAK−/− MEFs that FAK activation, the formation of a complex with p190A-p120RasGAP, and the recruitment of this complex to leading edge FAs upon FN plating promote cell polarity by facilitating RhoA inactivation at leading edge protrusions (26). We found that FAK phosphorylation at Tyr-397 and the binding of the p120RasGAP SH2-SH3-SH2 region to FAK formed a bridge between FAK and p190A. Thus, although R454 FAK is correctly localized to FAs (Fig. 5), it does not function to recruit the p120RasGAP-p190A complex, and this may contribute to the cell polarity defects observed for R454 FAK MEFs. Thus, the lack of RhoA regulation in R454 MEFs may prevent directional persistence of cell movement by affecting both FA formation and cell polarity. Notably, inactivating mutations of RASA1 (encoding p120RasGAP) have been shown to cause vascular defects in mice and humans (66, 67). Although it is unknown whether R454 FAK vascular defects are due to a lack of a functional FAK-p120-p190A complex, future studies may be able to address this issue.
Last, comparison of FAK−/−, R454 FAK, and Δ15 FAK homozygous developmental phenotypes reveals another surprising result. A small percentage of FAK Δ15 homozygous mice survive to adulthood and are fertile (41). After analyzing over 1000 embryos or pups, no homozygous FAK R454 mice were born or detected beyond E9.5 (Table 1). Thus, the absence of FAK autophosphorylation in Δ15 FAK embryos but not the absence of FAK activity in R454 FAK embryos can be compensated in vivo. The phenotype of FAK−/− embryos is more severe and is associated with a p53-dependent block in cell proliferation during development (37). FAK inhibition of p53 occurs via FAK FERM domain nuclear translocation, p53 binding, and the promotion of enhanced p53 degradation in a FAK kinase-independent manner. Importantly, p53 levels were low in E8.5 R454 FAK embryo lysates, and primary R454 MEFs exhibited no proliferation defects in culture (Fig. 3). These results are consistent with R454 FAK acting to inhibit p53 and enable cell proliferation. Alternatively, it is also possible that Pyk2 expression in R454 embryos and MEFs could act to inhibit p53 in a similar kinase-independent manner (38). Nevertheless, the proliferation of R454 MEFs in culture reinforces findings from the use of FAK-specific pharmacological inhibitors (31) that FAK activity is not essential for normal adherent cell proliferation. Although elevated FAK expression and signaling may enhance tumor growth (68, 69), our results show that FAK activity is not essential for cell cycle progression. In summary, our studies using a novel kinase-inactive FAK mouse highlight the essential role of intrinsic FAK activity in promoting blood vessel morphogenesis and cell motility-polarity but not cell proliferation.
Supplementary Material
Acknowledgments
We thank Sara Weis and David Cheresh for assistance in the early phase of mouse studies, Dusko Ilic for expertise in embryo development, Jacqueline Corbeil and Yuko Kono for ultrasound analyses, and Dwayne Stupack for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants HL093156 and GM087400. This work was also supported by American Heart Association Established Investigator Award 0540115N (to D. D. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–5 and Videos 1–4.
A. Tomar, unpublished results.
- EC
- endothelial cell
- En
- embryonic day n
- FA
- focal adhesion
- FAK
- focal adhesion kinase
- FERM domain
- band 4.1, ezrin, radixin, moesin domain
- FN
- fibronectin
- GAP
- GTPase-activating protein
- hTERT
- human telomerase reverse transcriptase
- MEF
- mouse embryonic fibroblast
- T-Ag
- large T-antigen
- WT
- wild type
- R454
- K454R mutation.
REFERENCES
- 1.Serini G., Valdembri D., Bussolino F. (2006) Exp. Cell Res. 312, 651–658 [DOI] [PubMed] [Google Scholar]
- 2.Jain R. K. (2003) Nat. Med. 9, 685–693 [DOI] [PubMed] [Google Scholar]
- 3.Risau W. (1997) Nature 386, 671–674 [DOI] [PubMed] [Google Scholar]
- 4.Hynes R. O. (2007) J. Thromb. Haemost. 5, Suppl. 1, 32–40 [DOI] [PubMed] [Google Scholar]
- 5.Giancotti F. G., Ruoslahti E. (1999) Science 285, 1028–1032 [DOI] [PubMed] [Google Scholar]
- 6.Schwartz M. A. (2001) Trends Cell Biol. 11, 466–470 [DOI] [PubMed] [Google Scholar]
- 7.Parsons J. T. (2003) J. Cell Sci. 116, 1409–1416 [DOI] [PubMed] [Google Scholar]
- 8.Mitra S. K., Schlaepfer D. D. (2006) Curr. Opin. Cell Biol. 18, 516–523 [DOI] [PubMed] [Google Scholar]
- 9.Schlaepfer D. D., Hauck C. R., Sieg D. J. (1999) Prog. Biophys. Mol. Biol. 71, 435–478 [DOI] [PubMed] [Google Scholar]
- 10.Mitra S. K., Hanson D. A., Schlaepfer D. D. (2005) Nat. Rev. Mol. Cell Biol. 6, 56–68 [DOI] [PubMed] [Google Scholar]
- 11.Deakin N. O., Turner C. E. (2008) J. Cell Sci. 121, 2435–2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zouq N. K., Keeble J. A., Lindsay J., Valentijn A. J., Zhang L., Mills D., Turner C. E., Streuli C. H., Gilmore A. P. (2009) J. Cell Sci. 122, 357–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pasapera A. M., Schneider I. C., Rericha E., Schlaepfer D. D., Waterman C. M. (2010) J. Cell Biol. 188, 877–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Defilippi P., Di Stefano P., Cabodi S. (2006) Trends Cell Biol. 16, 257–263 [DOI] [PubMed] [Google Scholar]
- 15.Wu L., Bernard-Trifilo J. A., Lim Y., Lim S. T., Mitra S. K., Uryu S., Chen M., Pallen C. J., Cheung N. K., Mikolon D., Mielgo A., Stupack D. G., Schlaepfer D. D. (2008) Oncogene 27, 1439–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown M. C., Cary L. A., Jamieson J. S., Cooper J. A., Turner C. E. (2005) Mol. Biol. Cell 16, 4316–4328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schober M., Raghavan S., Nikolova M., Polak L., Pasolli H. A., Beggs H. E., Reichardt L. F., Fuchs E. (2007) J. Cell Biol. 176, 667–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Izaguirre G., Aguirre L., Hu Y. P., Lee H. Y., Schlaepfer D. D., Aneskievich B. J., Haimovich B. (2001) J. Biol. Chem. 276, 28676–28685 [DOI] [PubMed] [Google Scholar]
- 19.Han D. C., Shen T. L., Guan J. L. (2000) J. Biol. Chem. 275, 28911–28917 [DOI] [PubMed] [Google Scholar]
- 20.Wu X., Suetsugu S., Cooper L. A., Takenawa T., Guan J. L. (2004) J. Biol. Chem. 279, 9565–9576 [DOI] [PubMed] [Google Scholar]
- 21.Chikumi H., Fukuhara S., Gutkind J. S. (2002) J. Biol. Chem. 277, 12463–12473 [DOI] [PubMed] [Google Scholar]
- 22.Holinstat M., Knezevic N., Broman M., Samarel A. M., Malik A. B., Mehta D. (2006) J. Biol. Chem. 281, 2296–2305 [DOI] [PubMed] [Google Scholar]
- 23.Lim Y., Lim S. T., Tomar A., Gardel M., Bernard-Trifilo J. A., Chen X. L., Uryu S. A., Canete-Soler R., Zhai J., Lin H., Schlaepfer W. W., Nalbant P., Bokoch G., Ilic D., Waterman-Storer C., Schlaepfer D. D. (2008) J. Cell Biol. 180, 187–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tomar A., Schlaepfer D. D. (2009) Curr. Opin. Cell Biol. 21, 676–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaller M. D. (2010) J. Cell Sci. 123, 1007–1013 [DOI] [PubMed] [Google Scholar]
- 26.Tomar A., Lim S. T., Lim Y., Schlaepfer D. D. (2009) J. Cell Sci. 122, 1852–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arthur W. T., Petch L. A., Burridge K. (2000) Curr. Biol. 10, 719–722 [DOI] [PubMed] [Google Scholar]
- 28.Hall A. (2005) Biochem. Soc. Trans. 33, 891–895 [DOI] [PubMed] [Google Scholar]
- 29.Arthur W. T., Burridge K. (2001) Mol. Biol. Cell 12, 2711–2720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kulkarni S. V., Gish G., van der Geer P., Henkemeyer M., Pawson T. (2000) J. Cell Biol. 149, 457–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Slack-Davis J. K., Martin K. H., Tilghman R. W., Iwanicki M., Ung E. J., Autry C., Luzzio M. J., Cooper B., Kath J. C., Roberts W. G., Parsons J. T. (2007) J. Biol. Chem. 282, 14845–14852 [DOI] [PubMed] [Google Scholar]
- 32.Roberts W. G., Ung E., Whalen P., Cooper B., Hulford C., Autry C., Richter D., Emerson E., Lin J., Kath J., Coleman K., Yao L., Martinez-Alsina L., Lorenzen M., Berliner M., Luzzio M., Patel N., Schmitt E., LaGreca S., Jani J., Wessel M., Marr E., Griffor M., Vajdos F. (2008) Cancer Res. 68, 1935–1944 [DOI] [PubMed] [Google Scholar]
- 33.Tanjoni I., Walsh C., Uryu S., Nam J. O., Mielgo A., Tomar A., Lim S. T., Liang C., Koenig M., Sun C., Kwok C., Patel N., McMahon G., Stupack D. G., Schlaepfer D. D. (2010) Cancer Biol. Ther. 9, 762–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ilic D., Kovacic B., McDonagh S., Jin F., Baumbusch C., Gardner D. G., Damsky C. H. (2003) Circ. Res. 92, 300–307 [DOI] [PubMed] [Google Scholar]
- 35.Shen T. L., Park A. Y., Alcaraz A., Peng X., Jang I., Koni P., Flavell R. A., Gu H., Guan J. L. (2005) J. Cell Biol. 169, 941–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Braren R., Hu H., Kim Y. H., Beggs H. E., Reichardt L. F., Wang R. (2006) J. Cell Biol. 172, 151–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lim S. T., Chen X. L., Lim Y., Hanson D. A., Vo T. T., Howerton K., Larocque N., Fisher S. J., Schlaepfer D. D., Ilic D. (2008) Mol. Cell 29, 9–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lim S. T., Miller N. L., Nam J. O., Chen X. L., Lim Y., Schlaepfer D. D. (2010) J. Biol. Chem. 285, 1743–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lim S. T., Mikolon D., Stupack D. G., Schlaepfer D. D. (2008) Cell Cycle 7, 2306–2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ilić D., Furuta Y., Kanazawa S., Takeda N., Sobue K., Nakatsuji N., Nomura S., Fujimoto J., Okada M., Yamamoto T., Aizawa S. (1995) Nature 377, 539–544 [DOI] [PubMed] [Google Scholar]
- 41.Corsi J. M., Houbron C., Billuart P., Brunet I., Bouvrée K., Eichmann A., Girault J. A., Enslen H. (2009) J. Biol. Chem. 284, 34769–34776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ren X. D., Kiosses W. B., Sieg D. J., Otey C. A., Schlaepfer D. D., Schwartz M. A. (2000) J. Cell Sci. 113, 3673–3678 [DOI] [PubMed] [Google Scholar]
- 43.Hsia D. A., Mitra S. K., Hauck C. R., Streblow D. N., Nelson J. A., Ilic D., Huang S., Li E., Nemerow G. R., Leng J., Spencer K. S., Cheresh D. A., Schlaepfer D. D. (2003) J. Cell Biol. 160, 753–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ilic D., Furuta Y., Suda T., Atsumi T., Fujimoto J., Ikawa Y., Yamamoto T., Aizawa S. (1995) Biochem. Biophys. Res. Commun. 209, 300–309 [DOI] [PubMed] [Google Scholar]
- 45.Furuta Y., Ilić D., Kanazawa S., Takeda N., Yamamoto T., Aizawa S. (1995) Oncogene 11, 1989–1995 [PubMed] [Google Scholar]
- 46.Carlson T. R., Hu H., Braren R., Kim Y. H., Wang R. A. (2008) Development 135, 2193–2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu E., Côté J. F., Vuori K. (2003) EMBO J. 22, 5036–5046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Webb D. J., Donais K., Whitmore L. A., Thomas S. M., Turner C. E., Parsons J. T., Horwitz A. F. (2004) Nat. Cell Biol. 6, 154–161 [DOI] [PubMed] [Google Scholar]
- 49.Tilghman R. W., Slack-Davis J. K., Sergina N., Martin K. H., Iwanicki M., Hershey E. D., Beggs H. E., Reichardt L. F., Parsons J. T. (2005) J. Cell Sci. 118, 2613–2623 [DOI] [PubMed] [Google Scholar]
- 50.Wang H. B., Dembo M., Hanks S. K., Wang Y. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 11295–11300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ridley A. J., Schwartz M. A., Burridge K., Firtel R. A., Ginsberg M. H., Borisy G., Parsons J. T., Horwitz A. R. (2003) Science 302, 1704–1709 [DOI] [PubMed] [Google Scholar]
- 52.Jaffe A. B., Hall A. (2005) Annu. Rev. Cell Dev. Biol. 21, 247–269 [DOI] [PubMed] [Google Scholar]
- 53.Bernards A., Settleman J. (2004) Trends Cell Biol. 14, 377–385 [DOI] [PubMed] [Google Scholar]
- 54.McGlade J., Brunkhorst B., Anderson D., Mbamalu G., Settleman J., Dedhar S., Rozakis-Adcock M., Chen L. B., Pawson T. (1993) EMBO J. 12, 3073–3081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ren X. D., Kiosses W. B., Schwartz M. A. (1999) EMBO J. 18, 578–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen B. H., Tzen J. T., Bresnick A. R., Chen H. C. (2002) J. Biol. Chem. 277, 33857–33863 [DOI] [PubMed] [Google Scholar]
- 57.Tikhmyanova N., Little J. L., Golemis E. A. (2010) Cell Mol. Life Sci. 67, 1025–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smyth S. S., Patterson C. (2002) J. Cell Biol. 158, 17–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carmeliet P. (2005) Nature 438, 932–936 [DOI] [PubMed] [Google Scholar]
- 60.Peng X., Ueda H., Zhou H., Stokol T., Shen T. L., Alcaraz A., Nagy T., Vassalli J. D., Guan J. L. (2004) Cardiovasc. Res. 64, 421–430 [DOI] [PubMed] [Google Scholar]
- 61.Walsh C., Tanjoni I., Uryu S., Nam J. O., Mielgo A., Tomar A., Luo H., Phillips A., Kwok C., Patel N., McMahon G., Stupack D. G., Schlaepfer D. D. (2010) Cancer Biol. Ther. 9, 776–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Golubovskaya V. M., Nyberg C., Zheng M., Kweh F., Magis A., Ostrov D., Cance W. G. (2008) J. Med. Chem. 51, 7405–7416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Frame M. C. (2004) J. Cell Sci. 117, 989–998 [DOI] [PubMed] [Google Scholar]
- 64.Bonnette P. C., Robinson B. S., Silva J. C., Stokes M. P., Brosius A. D., Baumann A., Buckbinder L. (2010) J. Proteomics 73, 1306–1320 [DOI] [PubMed] [Google Scholar]
- 65.Sieg D. J., Ilić D., Jones K. C., Damsky C. H., Hunter T., Schlaepfer D. D. (1998) EMBO J. 17, 5933–5947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Henkemeyer M., Rossi D. J., Holmyard D. P., Puri M. C., Mbamalu G., Harpal K., Shih T. S., Jacks T., Pawson T. (1995) Nature 377, 695–701 [DOI] [PubMed] [Google Scholar]
- 67.Eerola I., Boon L. M., Mulliken J. B., Burrows P. E., Dompmartin A., Watanabe S., Vanwijck R., Vikkula M. (2003) Am. J. Hum. Genet. 73, 1240–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ding Q., Grammer J. R., Nelson M. A., Guan J. L., Stewart J. E., Jr., Gladson C. L. (2005) J. Biol. Chem. 280, 6802–6815 [DOI] [PubMed] [Google Scholar]
- 69.Mitra S. K., Mikolon D., Molina J. E., Hsia D. A., Hanson D. A., Chi A., Lim S. T., Bernard-Trifilo J. A., Ilic D., Stupack D. G., Cheresh D. A., Schlaepfer D. D. (2006) Oncogene 25, 5969–5984 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.