Abstract
Exocytosis from synaptic vesicles is driven by stepwise formation of a tight α-helical complex between the fusing membranes. The complex is composed of the three SNAREs: synaptobrevin 2, SNAP-25, and syntaxin 1a. An important step in complex formation is fast binding of vesicular synaptobrevin to the preformed syntaxin 1·SNAP-25 dimer. Exactly how this step relates to neurotransmitter release is not well understood. Here, we combined different approaches to gain insights into this reaction. Using computational methods, we identified a stretch in synaptobrevin 2 that may function as a coiled coil “trigger site.” This site is also present in many synaptobrevin homologs functioning in other trafficking steps. Point mutations in this stretch inhibited binding to the syntaxin 1·SNAP-25 dimer and slowed fusion of liposomes. Moreover, the point mutations severely inhibited secretion from chromaffin cells. Altogether, this demonstrates that the trigger site in synaptobrevin is crucial for productive SNARE zippering.
Keywords: Intracellular Trafficking, Membrane Fusion, Neurotransmitters, Synapses, Vesicles, SNARE Protein, Coiled Coil, Isothermal Titration Calorimetry
Introduction
Upon Ca2+ influx into the synaptic terminal, neurotransmitters are rapidly released from synaptic vesicles that fuse with the plasma membrane. Electron microscopy studies have revealed that neurotransmitter-laden vesicles are docked to the plasma membrane. Genetic approaches that target the key proteins involved in the process have demonstrated that the release machinery must be rendered fusion-ready (“primed”) to facilitate rapid exocytosis after morphological docking of the vesicles. Fusing vesicles can be detected by electrophysiological recordings or advanced microscopic techniques. However, it is not possible to monitor the molecular machinery that drives docking, priming, and fusion of vesicles directly. For insights into the molecular events during secretion, we are therefore forced to rely on indirect biochemical and structural methods (for an overview, see Refs. 1–7).
The core of the release machinery that drives the fusion of synaptic vesicles with the plasma membrane consists of the three SNARE4 proteins: syntaxin 1a, SNAP-25 (synaptosomal associated protein of 25 kDa), and synaptobrevin 2. Synaptobrevin 2 resides on synaptic vesicles, whereas SNAP-25 and syntaxin 1a reside in the plasma membrane. According to the “zipper” model, their assembly into a tight four-helix bundle (8, 9) between the vesicle and the plasma membrane, starting from their membrane-distal N termini, pulls the membranes together and initiates membrane fusion (10).
To shed light on this assembly process, we have analyzed the soluble portions of the proteins in previous studies. Our kinetic and thermodynamic investigations have culminated in a model that suggests that the two plasma membrane SNAREs, syntaxin 1 and SNAP-25, first assemble into a transient 1:1 heterodimer, which then provides a high affinity binding site for the vesicular synaptobrevin (11, 12). Subsequently, we were able to demonstrate that the same path is followed when the SNAREs are inserted into liposome membranes (13, 14). Ultimately, however, this chain of events needs to be tested by correlative in vitro and in vivo studies. Notably, formation of the syntaxin·SNAP-25 heterodimer is rather slow in vitro (11), indicating that this step needs to be supported by additional factors. By contrast, synaptobrevin binding is rapid in vitro (12, 13), suggesting that this protein is able to act rapidly when a binding site is available. It is challenging, however, to relate the capacity of synaptobrevin to the status of the SNARE machinery in vivo. Not surprisingly, it is debated whether or not the SNARE complex is already partially or even completely formed between the fusing membranes before the influx of Ca2+ (7, 15–19).
To understand how synaptobrevin is able to react quickly and how this is controlled in vivo, we first of all need to gain deeper insights into the binding mechanism biochemically. As a means to study binding directly, we developed a syntaxin·SNAP-25 heterodimer that is artificially stabilized by a short C-terminal fragment of synaptobrevin. This so-called “ΔN complex” offers an accessible binding site for the N-terminal region of synaptobrevin. In fact, our previous investigations have demonstrated that the N-terminal portion of the coiled coil domain of synaptobrevin is critical for rapid binding (12, 13).
Here, we have employed computational methods to explore how the rapid reactivity of synaptobrevin is encoded in its amino acid sequence. We found a well conserved sequence stretch between the coiled coil layers −4 and −2 that is suited to function as a so-called coiled coil “trigger site.” Guided by this finding, we then tested the role of this region by point mutations. Remarkably, biochemical and electrophysiological investigations strongly support the idea that this stretch is crucial for productive SNARE complex assembly.
EXPERIMENTAL PROCEDURES
Protein Constructs
All of the recombinant proteins were derived from cDNAs from Rattus norvegicus, with the exception of Sec22b, for which cDNA from Mus musculus was used. All of the expression constructs were subcloned into the pET28a expression vector (Novagen). As basic expression constructs for the neuronal SNARE, we used the SNARE domain of syntaxin 1a (soluble portion, aa 180–262, SyxH3; including the transmembrane region, aa 183–288, SyxH3TMR), a cysteine-free variant of SNAP-25A (aa 1–206), and synaptobrevin 2 (soluble portion, aa 1–96, Syb1–96; including TMR, aa 1–116, SybTMR). In addition, we used a soluble synaptobrevin with a single cysteine at position 28 (Syb1–96Cys-28), a synaptobrevin fragment without (aa 49–96, Syb49–96) or with a single cysteine residue at position 79 (Syb49–96Cys-79), the soluble portion of endobrevin (Eb1–74), and the SNARE domains of VAMP4 (aa 47–117) and of tomosyn (aa 1031–1116); these constructs have been described before (11, 13, 20–22). Expression constructs of the SNARE domain of Ykt6 (aa 132–198) and Sec22b (aa 125–195) were cloned into the pET28a vector (Novagen) via NdeI and XhoI sites. Furthermore, various synaptobrevin mutants were generated: Syb1–96I45A, SybTMRI45A, Syb1–96M46A, SybTMRM46A, Syb1–96I45A,M46A, SybTMRI45A,M46A, and Syb1–96N49A,V50A. We also generated a mutation of endobrevin, Eb1–74I25A,M26A. Finally, two constructs of the SNARE domain of tomosyn were generated, within which the putative trigger sequences of synaptobrevin (aa 44–49, DIMRVN; TomSybChim) and of endobrevin (aa 24–29, NIMTQN; TomEbChim), respectively, were introduced into the corresponding region in tomosyn, aa 1064–1069, GELARA. Constructs containing a mouse synaptobrevin 2 open reading frame followed by an internal ribosome entry site and enhanced green fluorescent protein were made in the pSFV1 plasmid. The mutations were introduced into the pSFV1-based plasmid by PCR mutagenesis.
Protein Purification
All of the proteins were expressed in Escherichia coli strain BL21 (DE3) and purified by Ni2+-nitrilotriacetic acid chromatography followed by ion exchange chromatography on an Äkta system (GE Healthcare) essentially as described (20). His6 tags were generally removed using thrombin. The proteins with a transmembrane region, syntaxin 1A (183–288) and synaptobrevin 2 (1–116), were purified by ion exchange chromatography in the presence of 15 mm CHAPS essentially as described (13, 23). All of the SNARE complexes were purified using a Mono Q column (GE Healthcare) after overnight assembly of the purified monomers at 4 °C. The following ternary complexes were employed: Syb1–96·SyxH3·SNAP-25, Syb1–96I45A,M46A·SyxH3·SNAP-25, Syb1-96N49A,V50A·SyxH3·SNAP-25, and Syb49–96·SyxH3·SNAP-25 (ΔN complex). The ΔN complex for liposome fusion experiments (i.e. Syb49–96·SyxH3TMR·SNAP-25) was purified by ion exchange chromatography in the presence of 15 mm CHAPS (13, 23). Protein concentrations were determined by absorption at 280 nm in 6 m guanidine HCl and/or using the Bradford assay.
Isothermal Titration Calorimetry (ITC)
ITC was performed on a VP-ITC instrument (GE Healthcare) at 25 °C essentially as described (12, 24). The samples were dialyzed twice against a degassed phosphate buffer (20 mm sodium phosphate, pH 7.4, 150 mm NaCl, 1 mm dithiothreitol). Typically, an initial 5-μl injection was followed by several 15-μl injections. The heat change/injection was integrated to yield the molar enthalpy for each injection. Blank titrations, which were carried out by injecting a ligand into the buffer, were subtracted from each data set. All of the ITC experiments were carried out at least twice. The resulting binding isotherms were analyzed using the ITC software package to obtain the binding enthalpy (ΔH), the stoichiometry (n), and the association constant (Ka). We used a one-site binding model that assumes that one or more ligands can bind independently. The dissociation constant (Kd) and the free binding energy (ΔG) were calculated using the basic thermodynamic relationships Kd = Ka−1, ΔG = −RT lnKa, and ΔG = ΔH − TΔS.
Fluorescence Spectroscopy
All of the measurements were carried out in a Fluorolog 3 spectrometer in a T configuration equipped for polarization (model FL322; Horiba Jobin Yvon). Single cysteine variants were labeled with Alexa 488 C5 maleimide according to the manufacturer's instructions (Invitrogen). All of the experiments were performed at 25 °C in 1-cm quartz cuvettes (Hellma) in a phosphate buffer (20 mm sodium phosphate, pH 7.4, 150 mm NaCl, 1 mm dithiothreitol). Fluorescence anisotropy, which is used to indicate the local flexibility of the labeled residue and which increases upon complex formation and decreases upon dissociation, were measured essentially as described (11–13). The G factor was calculated according to G = IHV/IHH, where I is the fluorescence intensity, the first subscript letter indicates the direction of the exciting light, and the second subscript letter indicates the direction of emitted light. We also measured the intensity of the vertically (V) and horizontally (H) polarized emission light after excitation by vertically polarized light. The anisotropy (r) was determined according to r = (IVV − G IVH)/(IVV + 2 G IVH).
CD Spectroscopy
CD measurements were performed essentially as described (12, 13, 18) using a Chirascan instrument (Applied Photophysics). Generally, Hellma quartz cuvettes with path lengths of 0.1 cm were used. For thermal denaturation experiments, the purified complexes were dialyzed against a phosphate buffer. The ellipticity at 222 nm was recorded between 25 and 95 °C at temperature increments of 30 °C/h.
Electrophysiological Measurements of Secretion
After transfection of the chromaffin cells with Semliki Forest Virus, expression of the protein was allowed for 4 h essentially as described previously (18). Control and mutant constructs were expressed in cells from the same preparations of synaptobrevin 2/cellubrevin double knock-out mice to cancel variability between preparations. Whole cell patch clamp, membrane capacitance measurements, amperometry, ratiometric intracellular calcium [Ca2+]i measurements, and flash photolysis of caged Ca2+ were performed as described previously (18). The displayed calcium concentrations, capacitance traces, and amperometric currents are the averages over all of the cells recorded. For statistical analysis, we distinguished between the membrane capacitance of the cell prior to stimulation (cell size), the first second of capacitance increase following stimulation (burst), and the capacitance increase occurring during the subsequent 4 s (sustained component). Electrophysiological data are presented as the means ± S.E., and the nonparametric Mann-Whitney test was used for significance testing.
Immunocytochemistry
Chromaffin cells were cultured on poly-l-lysine-coated coverslips, fixed in 3.7% paraformaldehyde in phosphate-buffered saline for 20 min, washed, and permeabilized in 0.2% Triton X-100. The remaining paraformaldehyde was neutralized in 50 mm NH4Cl. The cells were blocked in 2% bovine albumin serum (Sigma) and incubated with primary antibodies (mouse anti-synaptobrevin 2 (69.1) at a dilution of 1:500; and rabbit anti-synaptotagmin 1 (R33) at a dilution of 1:200) for 2 h. The cells were washed four times, incubated with secondary antibodies (Alexa 546-conjugated goat anti-mouse or Alexa 647-conjugated goat anti-rabbit, dilution 1:200; Invitrogen), washed, and imaged. Fluorescence quantification was performed on a Zeiss Axiovert 200 microscope, fluorescence excitation was achieved by monochromatic light (Polychrome V, TILL Photonics), and the images were analyzed using TILLvisION v4.0.1 (TILL Photonics). Fluorescence levels were quantified as the integrated intensity of a square region of the image containing the cell minus the intensity of a background region of the same size. The data were averaged over cells and are represented as the means and S.E.
Liposome Fusion Assay
Liposomes were prepared as previously described (23). Lipids (Avanti, Alabaster, AL) were mixed in chloroform to yield (molar ratios): phosphatidylcholine (5 m), phosphatidylethanolamine (2 m), phosphatidylserine (1 m), phosphatidylinositol (1 m), and cholesterol (1 m). After drying, they were resuspended in 20 mm HEPES/KOH, pH 7.4, 100 mm KCl, 5 mm dithiothreitol, 5% (w/v) sodium cholate at a total lipid concentration of 13.5 mm. SNARE proteins in 20 mm sodium cholate were added (lipid to protein ratio of 200:1 n/n), followed by size exclusion chromatography on a PC 3.2/10 fast desalting column (GE Healthcare) equilibrated in 20 mm HEPES/KOH, pH 7.4, 150 mm KCl, 1 mm dithiothreitol. For the preparation of N-(7-nitro-2-1,3-benzoxadiazol-4-yl)/rhodamine-labeled liposomes 1.5% (n/n) 1,2-dioleyl-sn-glycero-3-phosphoethanol-amine-N-lissamine rhodamine B sulfonyl, 1.5% (n/n) 1,2-dioleyl-sn-glycero-3-phosphoethanol-amine-N-(7-nitro-2–1,3-benzoxadiazol-4-yl), and 17% (n/n) phosphatidylethanolamine were used instead of 20% (n/n) phosphatidylethanolamine. Fusion reactions were performed at 30 °C (13, 23). For reactions, 15 μl of labeled and unlabeled liposomes were mixed in a total volume of 1.2 ml, resulting in final protein concentrations of ∼200 nm for both liposome populations. Fluorescence dequenching, indicating lipid mixing between both liposome populations, was measured using 460 nm for excitation and 538 nm for emission. The fluorescence intensities were normalized to the initial fluorescence intensity.
Computational Biology
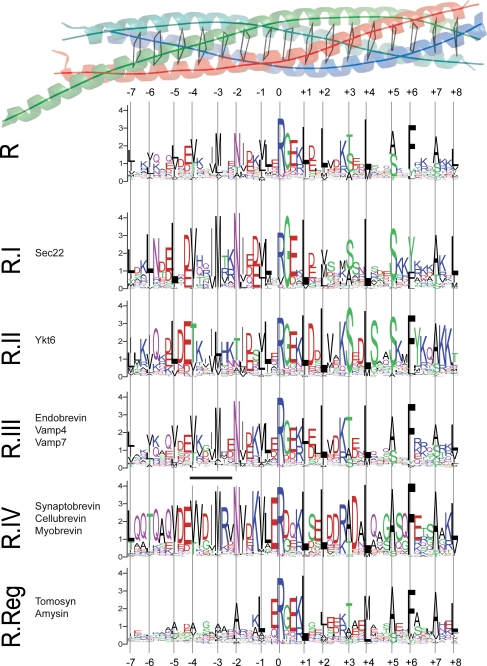
To explore the conserved features of R-SNARE sequences, 1359 R-SNARE sequences were downloaded from our SNARE database and grouped according to our previously established classifications (25): R.I (229 sequences), R.II (236 sequences), R.III (463 sequences), R.IV (278 sequences), and R.Reg (153 sequences). We then aligned the SNARE motif and used the WebLogo software (26) to generate the sequence logos. To shed further light on important sites within the different subtypes, we employed the computational method of Hannenhalli and Russell (27).
RESULTS
Sequence Analysis Indicates an Important Role for a Conserved Region between the Coiled Coil Layers −4 and −2 of Synaptobrevin
To explore the characteristics of the synaptobrevin amino acid sequence, we took advantage of our large collection of classified SNARE sequences (25). Our earlier investigation had shown that SNARE protein sequences generally split into four major phylogenetic classes, the Qa-, Qb-, Qc-, and R-SNAREs, which represent the four different helix positions in the SNARE bundle. The four basic types, in turn, split into 20 basic subtypes that operate in different vesicular trafficking steps within the cell (25). Synaptobrevin belongs to the R-SNARE class that can be separated into five different conserved subtypes (I–V); each of these subtypes most likely was present in the eukaryotic ancestor. Notably, the collection of sequences can also be arranged according to phylogenetic relationships, which allows discrimination between a common feature and a special adaptation of only a certain lineage (28, 29). Overall, we based our analysis on an alignment of 1359 R-SNAREs from more than 200 eukaryotic species.
It is thought that most coiled coil proteins contain short sequences that encode for autonomous helical folding units (30–33). Such trigger sites are thought to speed up the formation of coiled coils. However, whether trigger sequences occur in SNARE coils is unknown so far. We thus searched our collection for occurrences of the two regular expressions (I/V/L)X(D/E)IX(R/K)X and (I/V/L)(D/E)XIX(R/K)X that have been described to represent coiled coil trigger sequences (30). In fact, we found that the region between layer −4 and −2 in a large variety of R-SNAREs corresponds to these expressions. For example, the first regular expression (corresponding to residues 42–48, VVDIMRV, in synaptobrevin 2) is present in almost all of the animal synaptobrevins (subgroup IV) we inspected, suggesting that the region between layer −4 and −2 may represent a functionally important segment, possibly a coiled coil trigger site.
To independently inspect the sequence of synaptobrevin for functionally important sites, we employed the computational method of Hannenhalli and Russell (27) next. The method is able to identify sites in sequence alignments that are conserved within each subtype but differ among them. Interestingly, the two residues that the method identified to be most important for R-SNARE subtype specificity were between the layer position −3 and the site following layer −2. However, none of the sites in the motif exceed the restrictive significance cut-off given by the authors of the method (27).
To further investigate the specificity of the −4 to −2 region in R-SNAREs, we decided to study the entropies and conservation of each of the sites in the R-SNARE motif. We generated a graphical representation of the conservation patterns using the software WebLogo (26) (Fig. 1). Confirming earlier investigations, our analysis reveals that, in general, the primarily apolar residues that constitute the coiled coil interface are highly conserved. When we inspected the WebLogos of the other subgroups, it became evident that a stretch, located roughly between layers −4 and −2, is generally highly conserved. Interestingly, this region is also conserved within the R.II group (Ykt6) but exhibits a striking difference in its amino acid sequence, whereas it is not well conserved in the R.Reg group (tomosyn; Fig. 1). Note that the low conservation of the N-terminal part of the R.Reg group sequence might explain the somewhat reduced significance of the results of the Hannenhalli and Russell method mentioned above.
FIGURE 1.
Conservation of the R-SNARE motif. Multiple sequence alignments of the central coiled coil region of synaptobrevin homologs from more than 200 different species are represented using the software WebLogo (26). This program provides a precise illustration of sequence similarities in a sequence alignment. In detail, the height of a column in a sequence logo reflects its conservation, and the height of a letter within a column indicates its relative frequency. WebLogos of all available R-SNARE motifs (R) in our database and of the five different conserved subgroups (R.I, R.II, R.III, R.IV, and R.Reg) established previously (25) are shown. At the top, the layer structure of the parallel four-helix bundle of the SNARE complex (9, 47) is shown as a ribbon diagram (blue, red, and green for synaptobrevin 2, syntaxin 1a, and SNAP-25a, respectively). The layers in the core of the bundle (−7 to +8) are indicated by virtual bonds between the corresponding Cα positions. Below, the six different logos are drawn according to the layer structure. The region corresponding to the regular expression for trigger sequences defined in (30) is indicated as a black line on top of the logo for the R.IV subtype. Interestingly, although this region is highly conserved in all R-SNAREs of type IV, exact matches of the trigger sequences were not discovered in R.IV-SNAREs from nematodes, for example. This appears to be in line with recent evidence that such sequences can show considerable diversity, suggesting that no general consensus sequence exists and that, rather, stabilizing effects along a protein sequence determine the ability to fold into a coiled coil. For example, in the endosomal SNARE complex, the methionine in layer −3 forms a weak hydrogen bond with asparagine in the neighboring layer −2 (42). It is thus possible that this intramolecular interaction increases the α-helical propensity of this segment before being part of the four-helix bundle. Note that the logos represent R-SNARE sequences of species belonging to a variety of different eukaryotic kingdoms. The conservation patterns of fungal and metazoan R-SNARE subtypes are given in supplemental Fig. S5.
Mutations in the Conserved Sequence Region Interfere with Binding of Synaptobrevin in Vitro
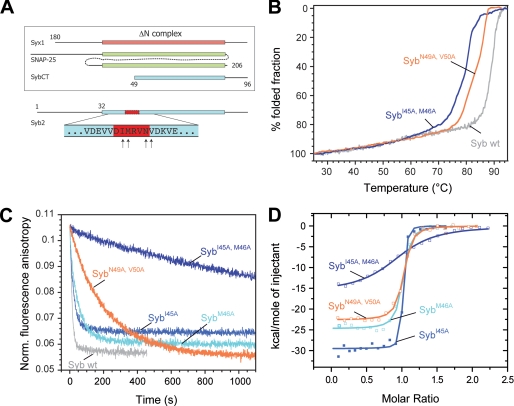
Although our sequence analyses suggest an important role for the region between layer −4 and layer −2 of R-SNAREs, ultimately, its function needs to be brought to light biochemically. To probe the role of the conserved residues identified between the coiled coil layers −4 and −2 of synaptobrevin 2, we initially generated two double mutations (Fig. 2A), in which the layer residue −3 (Met-46) or −2 (Asn-49) and an adjacent conserved residue were mutated to alanines: I45A and M46A (SybI45A,M46A) and N49A and V50A (SybN49A,V50A). Alanine, with its small apolar side chain, was used to avoid drastic effects on the overall stability of the assembled ternary SNARE complex (18). To investigate the effect of the double mutations on the overall structural integrity of the SNARE complex, we monitored thermal unfolding of the purified complex by circular dichroism spectroscopy. Compared with the SNARE complex containing wild-type synaptobrevin (≈88 °C; Fig. 2B), the double mutants only reduced the stability of the complex slightly (≈78 °C and ≈82 °C, respectively).
FIGURE 2.
Mutations in the putative coiled coil trigger site strongly inhibit binding of synaptobrevin 2 to the ΔN complex. A, depiction of the constructs used for binding experiments between the ΔN complex and synaptobrevin. The purified ΔN complex was composed of SyxH3 (aa 180–262), full-length SNAP-25a (aa 1–206), and Syb49–96. The arrows at the bottom of the diagram indicate the residues mutated in synaptobrevin (aa 1–96). B, thermal unfolding of SNARE complexes was monitored by CD spectroscopy at 222 nm. Purified SNARE complexes containing the double mutants SybI45A,M46A and SybN49A,V50A unfolded at slightly lower temperatures than the complex containing wild-type (wt) synaptobrevin (18). C, upon binding, the Alexa 488-labeled synaptobrevin fragment Syb49–96 is displaced from the ΔN complex (SyxH3·SNAP-25·Syb49–96C79Alexa488). As demonstrated before (12, 13), displacement is visible by a decrease in fluorescence anisotropy. Approximately 100 nm of the ΔN complex was incubated with different synaptobrevin mutants (500 nm). In agreement with our previous findings, the labeled fragment was quickly displaced upon addition of synaptobrevin (12, 13). Displacement was significantly slower when the double mutants SybI45A,M46A and SybN49A,V50A were used. D, isothermal titrations of different synaptobrevin mutants into the ΔN complex. Titration of SybI45A (18 μm), SybM46A (20 μm), and SybN49A,V50A (22 μm) into 2.5 μm purified ΔN complex (SyxH3·SNAP-25·Syb49–96); 38 μm SybI45A,M46A was titrated in 4 μm ΔN complex.
To monitor binding of synaptobrevin, we used a stabilized syntaxin·SNAP-25 heterodimer, the so-called ΔN complex. As demonstrated previously (12, 13), this experimental strategy allows us to isolate the binding step by avoiding the 2:1 off-pathway reaction. In an earlier study, we found that N-terminally truncated fragments of synaptobrevin were unable to bind to the ΔN complex, whereas C-terminally truncated fragments were able to bind quickly (12, 13), supporting the notion that binding commences N-terminally. At this point, it needs to be pointed out that the ΔN complex contains a synaptobrevin fragment encompassing residues 49–96. Noticeably, the accessible binding site of the ΔN complex only partly includes the layer positions −3 and −2 (Fig. 2A), which were highlighted by our bioinformatic investigations. Interestingly, however, when we used a synaptobrevin peptide comprising only the open binding site of the ΔN complex, i.e. residues 16–44, no binding to the ΔN complex was observable. Likewise, this peptide was not able to compete with wild-type synaptobrevin for binding to the ΔN complex (data not shown). This suggests that the region C-terminal to residue 44 must play a crucial, although unexpected, role for binding as well.
For our analysis, we first used a ΔN complex that contained a peptide that was labeled at position Cys-79 with the fluorophore Alexa 488 (Syb49–96C79Alexa). Upon binding of synaptobrevin, the peptide was readily displaced, which was visible by a decrease in fluorescence anisotropy (Fig. 2C). Noticeably, compared with wild-type synaptobrevin, binding of SybN49A,V50A was clearly retarded. An even more pronounced defect was found for SybI45A,M46A. Intriguingly, alanine mutations in the coiled coil layers upstream of this region (i.e. Syb1–96L32A,T35A and Syb1–96V39A,V42A, corresponding to layers −7 and −6 and layers −5 and −4, respectively) had a much less severe effect on the ability to displace the peptide (supplemental Fig. S1) (19).
To compare the binding affinities of the different mutants, we next employed isothermal titration calorimetry (ITC). Using this technique, we recently showed that wild-type synaptobrevin binds with an apparent affinity of ∼2 nm to the ΔN complex (12). In agreement with the kinetic measurements, we observed a slightly reduced affinity of ≈9 nm for the binding of SybN49A,V50A to the ΔN complex (Fig. 2D), whereas the binding affinity of the second mutant, SybI45A,M46A, was greatly reduced (≈350 nm; Table 1). Again, N-terminal alanine mutations in the upstream coiled coil layers had a much milder effect on the binding affinity to the ΔN complex (Table 1).
TABLE 1.
Thermodynamic parameters of the interaction of synaptobrevin 2 mutants and different R-SNARE homologs with the neuronal ΔN complex measured by ITC at 25 °C
The experimental ITC data are shown in Figs. 2D and 5B. No heat changes were detected when the SNARE motifs of tomosyn or of Ykt6 were mixed with the ΔN complex (SyxH3·SNAP-25·Syb49–96). For comparison, the thermodynamic parameters for wild-type synaptobrevin 2 (12) and of three additional alanine layer mutations (19) are given as well.
| Syringe | Kd | ΔH | −TΔS | ΔG | N |
|---|---|---|---|---|---|
| nm | kcal mol−1 | kcal mol−1 | kcal mol−1 | ||
| Syb1–96a | 2.1 ± 0.6 | −29.9 ± 0.3 | 18.1 | −11.8 | 1.05 |
| Syb1–96I45A | 0.9 ± 0.4 | −29.5 ± 0.3 | 17.2 | −12.3 | 0.98 |
| Syb1–96M46A | 7.2 ± 1.4 | −24.7 ± 0.2 | 13.6 | −11.1 | 0.98 |
| Syb1–96I45A,M46A | 344.8 ± 44.7 | −15.7 ± 0.4 | 6.9 | −8.8 | 0.97 |
| Syb1–96N49A,V50A | 8.8 ± 1.6 | −22.6 ± 0.2 | 11.6 | −11.0 | 0.98 |
| Endobrevin | 0.8 ± 0.8 | −21.3 ± 0.4 | 8.9 | −12.4 | 0.99 |
| VAMP4 | 1.1 ± 0.6 | −15.3 ± 0.2 | 3.1 | −12.2 | 1.04 |
| Sec22 | 21.2 ± 8.2 | −7.2 ± 0.2 | −3.3 | −10.5 | 1.05 |
| Syb1–96F77Ab | 0.8 ± 0.4 | −18.8 ± 0.2 | 6.4 | −12.4 | 0.99 |
| Syb1–96L32A,T35Ab | 44.1 ± 12.3 | −16.6 ± 0.4 | 6.5 | −10.1 | 1.01 |
| Syb1–96V39A,V42Ab | 8.4 ± 2.8 | −23.4 ± 0.4 | 12.3 | −11.1 | 1.02 |
a Thermodynamic parameters for wild-type synaptobrevin 2.
b Thermodynamic parameters for three additional alanine layer mutations.
We therefore decided to concentrate in this work on the more drastic effect of SybI45A,M46A in the binding reaction. To measure the slowdown of the binding reaction caused by the point mutations in SybI45A,M46A directly, we introduced a cysteine at position 28 for labeling purposes. In a previous study, this label position had been instrumental for measuring the on-rate of wild-type synaptobrevin to the ΔN complex as ≈250,000 m−1 s−1 (12, 13). When we used the same strategy for SybI45A,M46A, we determined a severely reduced on-rate of ≈860 m−1 s−1 (supplemental Fig. S2).
To find out which of the two point mutations was mostly responsible for the severe effect, we generated constructs carrying only one or the other of the two mutations SybI45A and SybM46A. Interestingly, the two synaptobrevin variants carrying single point mutations were only slightly slower than wild-type synaptobrevin in displacing the labeled fragment from the ΔN complex (Fig. 2C). Their milder effect was confirmed when we investigated the binding of the two variants to the ΔN complex by ITC. Although the binding of SybI45A was nearly indistinguishable from wild-type synaptobrevin, SybM46A bound with only slightly reduced affinity (≈20 nm; Table 1). This suggests that the two neighboring residues act together to enable synaptobrevin to bind quickly.
SybI45A,M46A Reduces Liposome Fusion Efficiency
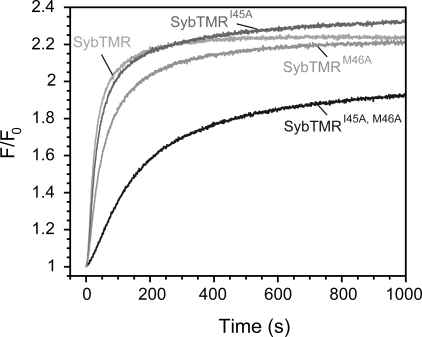
In the above experiments, we have demonstrated that point mutations in the conserved region between layers −4 and −2 severely affect the binding ability of the soluble domain of synaptobrevin. To investigate whether these findings also hold true for membrane-inserted synaptobrevin, we generated versions of the double mutant and of the corresponding single point mutations carrying a transmembrane region (SybTMR). The synaptobrevin mutants were reconstituted into one population of liposomes. Fusion with liposomes carrying the ΔN complex was then measured by lipid dequenching (13). In agreement with our results on the soluble domain of synaptobrevin, the liposome fusion rate was greatly reduced compared with wild-type synaptobrevin when SybTMRI45A,M46A was used (Fig. 3). The fusion rate in the presence of SybTMRI45A was comparable with the reaction in the presence of wild-type synaptobrevin, whereas only a moderate slowdown was observable for the other single point mutation, SybTMRM46A (Fig. 3). In line with previous investigations (13, 14, 23, 34–36), this again demonstrates that the presence of membranes alone does not alter the molecular properties of synaptobrevin significantly.
FIGURE 3.
Mutations in the trigger site greatly decrease the efficiency of synaptobrevin in mediating liposome fusion. Previously, we showed that SNARE-driven liposome fusion can be greatly accelerated when a purified ΔN complex (SyxH3TMR·SNAP-25·Syb49–96) is used in one population of liposomes. This complex serves as an available acceptor site for liposomes containing synaptobrevin (13, 14, 23, 34–36). Fusion was monitored by lipid dequenching. At a final protein concentration of ∼200 nm for both liposome populations, the liposomes containing wild-type synaptobrevin fused very rapidly, whereas fusion in the presence of the double mutant SybTMRI45A,M46A was much slower.
SybI45A,M46A Severely Inhibits Secretion from Chromaffin Cells
This result encouraged us to investigate the consequence of the synaptobrevin point mutations during Ca2+-dependent exocytosis of neurotransmitters. For this, we made use of adrenal chromaffin cells, a powerful model system for studying neurotransmitter release (37). In these cells, secretion can be activated by releasing calcium from a photolysable cage. A strong flash of UV light leads to an abrupt, step-like increase in the calcium concentration. This approach allows us to distinguish between vesicles that can undergo rapid release upon sudden elevation of the intracellular Ca2+ concentration (the exocytotic burst) and vesicles that are released at a much slower pace (the sustained phase of release). Vesicles released during the burst phase are thought to be already docked to the plasma membrane and primed for release, whereas vesicles that are released during the sustained phase, according to this model, need to prime first.
We used chromaffin cells from mice lacking both synaptobrevin 2 (38) and cellubrevin (also referred to as VAMP3) (39). In these cells, Ca2+-dependent neurotransmitter secretion is abolished, but it can be restored by viral overexpression of wild-type synaptobrevin 2 (15, 17). After overexpression, secretion was monitored by membrane capacitance measurements and carbon fiber amperometry. All of the constructs studied here (including wild-type synaptobrevin 2) led to ≈>10-fold overexpression over endogeneous levels, as revealed by quantitative immunostaining (supplemental Fig. S3). It was further shown that overexpressed synaptobrevin variants were sorted correctly to secretory vesicles by immunostaining. This is in contrast to an earlier investigation in which the region of amino acids 39–53 was suspected to constitute a signal for synaptic vesicle targeting (40).
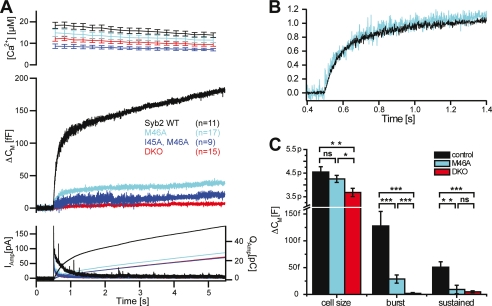
Remarkably, overexpression of synaptobrevin carrying the double mutation SybTMRI45A,M46A in chromaffin cells was not able to restore secretion (Fig. 4A). In fact, the secretion defect of the double mutant was too severe to allow us to interpret its nature. Therefore, we next overexpressed a synaptobrevin carrying only the M46A mutation, SybTMRM46A. For this mutant, exocytosis was restored to roughly 20% of the control values (Fig. 4A). The reduction in secretion was found both in the burst component (0–1 s after the flash photorelease of calcium) and in the sustained component (1–5 s after the flash) (Fig. 4C). A similar reduction was seen by concurrent amperometric measurements (Fig. 4A, bottom panel).
FIGURE 4.
Mutations in the trigger site greatly decrease the ability of synaptobrevin to support neurotransmitter release. Chromaffin cells of synaptobrevin 2/cellubrevin double null mice were isolated, and mutant or wild-type (WT) synaptobrevin was overexpressed. Catecholamine release was measured electrochemically and by membrane capacitance increase, which is proportional to changes in surface area. A, overexpression of synaptobrevin carrying the point mutations I45A and M46A or M46A shows a drastically reduced secretory response compared with wild-type synaptobrevin. Top panel, means ± S.E. of intracellular Ca2+ concentration after UV-induced Ca2+ uncaging at time = 0.5 s. Middle panel, mean capacitance increase. Bottom panel, mean amperometric current (thick traces, left ordinate) and amperometric charge (thin traces, right ordinate). Note that because the double mutant showed only very little response, it was not analyzed in more detail. B, a detailed view of normalized capacitance traces of wild-type synaptobrevin and the M46A mutant (A) shows that the kinetics of the remaining release is unaffected. C, burst and sustained release is reduced in the M46A mutant (wild-type rescue: n = 11; M46A: n = 17; double knock-out (DKO): n = 15. *, p < 0.05; **, p < 0.01; ***, p < 0.001 in nonparametric Mann-Whitney U test). The basal cell size is smaller in the double knock-out cells, probably because of impaired exocytosis before the stimulus.
To analyze the changes in the kinetics of neurotransmitter release better, we scaled the capacitance traces to the same amplitude at 0.5 s after the flash (Fig. 4B). This analysis showed that the release kinetics in the presence of SybTMRM46A are similar to those in the presence of wild-type synaptobrevin 2. This indicates that SybTMRM46A does not significantly change the release properties but severely affects the pool size of release-ready vesicles. This phenotype is consistent with compromised vesicle priming, the reaction during which vesicles are matured to a fusogenic state (41). Probably, the sequence region between layers −4 and −2 is important for the binding of synaptobrevin to the syntaxin·SNAP-25 acceptor site during priming of the vesicle. In this regard, it needs to be mentioned that in another study we encountered phenotypes that were comparable, although less severe, when we mutated two consecutive coiled coil layers at the very N terminus of the synaptobrevin 2 SNARE motif (i.e. SybTMRL32A,T35A and SybTMRV39A,V42A) (19).
As outlined above, the very N-terminal coiled coil layers were shown to be involved in binding (12, 13). Thus the question arose as to why the downstream layers −3 and −2 are important for binding as well. One possible explanation, supported by our sequence analysis described above, is that this region in synaptobrevin constitutes a coiled coil trigger site that renders the entire N-terminal region able to engage readily.
The Putative Trigger Sequence Is Conserved in Many Synaptobrevin Homologs
The sequence between layers −4 and −2 is highly conserved in several (but not all) subtypes of R-SNAREs (Fig. 1). Therefore, we next tested whether the degree of conservation is reflected in the ability of other R-SNARE homologs to bind to the ΔN complex. For these experiments, we chose examples of each of the other conserved R-SNARE subtypes from rat: Sec22 (R.I), Ykt6 (R.II), endobrevin/VAMP8 and VAMP4 (both R.III), and tomosyn (R.Reg). Note that, like synaptobrevin 2, endobrevin, VAMP4, and tomosyn have been shown to form very similar four-helix bundle SNARE complexes (21, 22, 42). For each R-SNARE type, we focused on the SNARE domain region as depicted in supplemental Fig. S4.
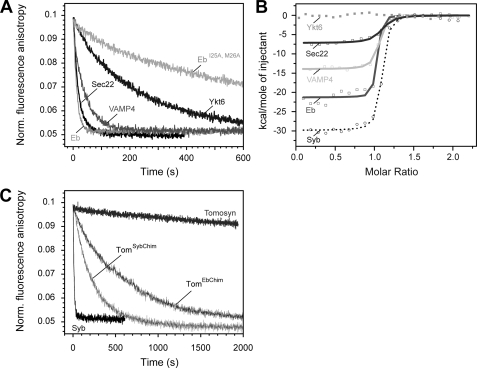
When we measured the displacement of the labeled synaptobrevin peptide from the ΔN complex, we found that Sec22 and endobrevin exhibited rates that were comparable with synaptobrevin 2 (Fig. 5A), whereas displacement by VAMP4 was slightly slower. By contrast, the displacement rate of Ykt6 was clearly delayed (Fig. 5A), and remarkably, tomosyn was not able to displace the synaptobrevin fragment at all (Fig. 5C). In fact, when we used ITC, we found that tomosyn did not bind to the ΔN complex (data not shown). Similarly, no binding was detected for Ykt6, possibly because the binding process is too slow to be detectable by ITC (Fig. 5B). By contrast, the other R-SNAREs were able to bind with high affinity, although different enthalpies of binding were recorded that are probably caused by the overall varying amino acid compositions of the different R-SNAREs (Fig. 5B and Table 1). In general, the observed binding pattern for the different R-SNAREs reflects the conservation of the putative trigger site between layers −4 and −2. Whereas this region is conserved in Sec22, endobrevin, and VAMP4, its sequence pattern clearly deviates in Ykt6, and it is not conserved in tomosyn (Fig. 1).
FIGURE 5.
Different abilities of synaptobrevin homologs to bind to the ΔN complex. A, the rate of displacement of Syb49–96 from the ΔN complex monitored by fluorescence anisotropy. As in Fig. 2C, ∼100 nm labeled ΔN complex (SyxH3·SNAP-25·Syb49–96C79Alexa488) was incubated with the SNARE domain of different synaptobrevin homologs (500 nm) (12, 13). A depiction of the different constructs used is shown in supplemental Fig. S3. Endobrevin and Sec22 were able to quickly displace Syb49–96, whereas VAMP4 was slightly slower. Possibly the somewhat reduced binding efficiency of VAMP4 compared with synaptobrevin 2 is due to an exchange of the conserved isoleucine to a valine in the site preceding layer −3 in this R.III-SNARE subtype, which is specific to animals. Only slow displacement was observed for Ykt6, whereas tomosyn was unable to displace the fragment (C). B, interaction of the SNARE domain of different synaptobrevin homologs and the ΔN complex measured by ITC. Titrations of VAMP4 and Eb (15 μm) into a purified ΔN complex (1.7 μm). Sec22 (40 μm) was titrated into 4.25 μm ΔN complex. Upon titration of Ykt6 or tomosyn into the ΔN complex, no binding was detected. For comparison, the titration of Syb1–96 into the ΔN complex is shown again (12). C, chimeras of tomosyn containing a functional trigger site are able to displace the Syb49–96 from the ΔN complex. As in A, displacement was monitored by fluorescence anisotropy of 100 nm labeled ΔN complex (SyxH3·SNAP-25·Syb49–96C79Alexa488) after the addition of 500 nm of the tomosyn chimeras. For comparison, the reaction is shown in the presence of Syb1–96 and tomosyn.
To test the assumption that several other R-SNAREs, e.g. endobrevin (Eb), also contain a coiled coil trigger site, we introduced point mutations into this protein (EbI25A,M26A) that correspond to the positions mutated in synaptobrevin (SybI45A,M46A). Indeed, we discovered that EbI25A,M26A could only slowly displace the labeled fragment from the ΔN complex (Fig. 5A), corroborating that this region is very important for binding.
As shown above, the SNARE domain of tomosyn is not able to bind to the ΔN complex quickly. According to our idea, tomosyn should be rendered capable of binding, if it contained a trigger site. We therefore generated tomosyn constructs carrying the putative trigger site of synaptobrevin 2 (aa 1064–1069 of tomosyn, GELARA, replaced by aa 44–49 of Syb2, DIMRVN; TomSybChim) or endobrevin (aa 1064–1069 of tomosyn, GELARA, replaced by 24–29 of endobrevin, NIMTQN; TomEbChim). Remarkably, these two tomosyn chimeras were able to bind to the ΔN complex, supporting the existence of a conserved trigger site that is crucial for binding (Fig. 5C). It should be noted, however, that the two chimeras still bound more slowly than synaptobrevin 2 or endobrevin, indicating that other residues in the N-terminal portion contribute to binding as well.
DISCUSSION
In this study, we combined computational, biochemical, and in vivo methods to gain deeper insights into the synaptobrevin binding step during neurotransmitter release. Our previous in vitro studies have shown that binding of synaptobrevin starts with the N-terminal region of the SNARE domain (12, 13). Our computational analysis now points to a short conserved stretch within the N-terminal region of synaptobrevin that probably acts as a coiled coil trigger site. Intriguingly, the same region is most important for subtype specificity. For biochemical analysis, we took advantage of the fact that, nowadays, we are able to discriminate between different steps of SNARE complex formation. Our earlier in vitro studies have revealed that the association of the two plasma membrane SNAREs syntaxin and SNAP-25 occurs at a limited rate, whereas binding of synaptobrevin to the preformed syntaxin·SNAP-25 heterodimer is rapid (11–13).
Remarkably, point mutations in the putative trigger site had drastic effects on the ability of synaptobrevin to engage with a stabilized syntaxin·SNAP-25 heterodimer, the ΔN complex. A comparable defect emerged when liposome fusion experiments were carried out. In addition, electrophysiological experiments in chromaffin cells revealed that the synaptobrevin mutations severely inhibited neurotransmitter release. Together, these findings strongly suggest that the defect in secretion is caused by the reduced ability of the synaptobrevin mutants to engage with the acceptor complex during priming, or, more commonly speaking, vesicles are less likely to be rendered fusion-ready when the trigger site in synaptobrevin is mutated.
Kinetic analysis indicates that the few vesicles carrying the mutated synaptobrevin can mature into a fusion-ready state and are able to release their contents at a rate comparable with that of the wild-type synaptobrevin. It is therefore likely that in the fusion-ready state, synaptobrevin must be assembled, probably partly, with syntaxin and SNAP-25 in a trans-SNARE complex between the fusing membranes (18, 19). In this configuration, the complex could serve as a binding platform for the Ca2+ sensor synaptotagmin and the late-acting factor complexin. Biochemically, the point mutations diminished the reactivity of synaptobrevin. Because point mutations in more N-terminal layers had much milder effects, it seems likely that the conserved region between layers −4 and −2 indeed constitutes a trigger site for coiled coil formation. This assessment is corroborated by the finding that the trigger sequence could be transferred onto the “inactive” R-SNARE domain of tomosyn, increasing its binding efficiency.
Intriguingly, a recent NMR study on full-length synaptobrevin (Syb1–116) has uncovered that not only the transmembrane domain and surrounding regions adopt α-helical confirmation (amino acids 77–88) but also the N-terminal region between amino acids 35 and 53. Notably, the center of the helix in the N-terminal segment of the SNARE motif coincides with the trigger site identified by us. The NMR experiments were carried out in the presence of the detergent dodecylphophocholine (43), which is widely used for studying membrane proteins because it forms membrane-mimicking micelles. The detergent was found to induce α-helical structure in the N-terminal region of the soluble portion of synaptobrevin (Syb1–96) as well. This indicates that the N-terminal helix is not induced by the insertion of the protein into a membrane. Rather, it seems that the membrane distal N-terminal stretch has an independent, inherent tendency to adopt an α-helical conformation, corroborating the notion that this segment might serve as a trigger site for synaptobrevin binding. Very likely, the point mutations tested reduce the propensity of this stretch to form a helix. Nevertheless, whether this segment is already (partially) folded in vivo before binding or whether it “zips up” upon contact with the cognate acceptor site needs to be tested in the future.
The reduced binding activity of synaptobrevin mutants appears to translate directly into a smaller primed vesicle pool. This evokes a scenario in which synaptobrevin can readily bind when the acceptor complex is made available. In general, we observed a good correlation between the effect of the synaptobrevin mutations in vivo and in vitro. For example, we noted that, biochemically, the double mutation of the two neighboring residues, i.e. I45A and M46A, caused a very strong binding defect. In vivo, the double mutant led to only very little response. The single point mutation M46A was less drastic biochemically and also caused a less drastic secretory phenotype (≈ 80% reduction of the exocytotic burst). Double mutations in the more N-terminal layers, L32A,T35A and V39A,V42A, had an even milder effect biochemically (supplemental Fig. S1) and caused a less reduced exocytotic burst (≈ 50%) (19), yet the degree of the in vitro and in vivo effects seems to be slightly different for some mutations. We came across similar subtle differences in earlier studies (18, 19). Although it is possible that our biochemical tools are yet not suited to detecting small nuances in the reactivity of synaptobrevin, the more severe defect in secretion might also suggest that additional factors such as Munc-18 and Munc-13 are involved in establishing the trans-SNARE complex during priming.
Notably, only the R-SNARE types with deviating sequences in the putative trigger sequence, Ykt6 and tomosyn, exhibited a markedly reduced ability to bind to the ΔN complex, whereas even Sec22, for example, an R-SNARE involved in endoplasmic reticulum-Golgi trafficking, was able to bind with high affinity. With regard to the fact that the ΔN complex does not represent the cognate binding site for most of the other R-SNAREs, the low binding specificity revealed is particularly remarkable. Moreover, the ability of endobrevin, VAMP4, and Sec22 to bind rapidly to the ΔN complex might even suggest that these R-SNAREs, which function in other trafficking steps within the cell, are also generally localized to transport vesicles and bind to a homologous acceptor complex consisting of three different Q-SNARE helices, a “Qabc complex,” sitting in the target membrane.
By contrast, our findings show that the binding mechanism of Ykt6 and tomosyn, two R-SNAREs that, interestingly enough, do not possess transmembrane anchors, differs. This is particularly interesting, because tomosyn, which is unable to bind to the ΔN complex, is thought to bind to the same syntaxin 1a·SNAP-25 acceptor complex as synaptobrevin 2 (44, 45). Furthermore, the structures of both of these core complexes are virtually congruent (9, 21).
So why is the binding of tomosyn so different from that of the other proteins? Notably, tomosyn contains a large N-terminal domain consisting of two consecutive seven-bladed β-propellers (45, 46). Our computational analysis reveals that the tomosyn SNARE domain is mostly conserved in the central and C-terminal region, suggesting that this segment comprises the functionally important site (Fig. 1). It is thus conceivable that only the C-terminal half of this SNARE domain is tailored to form a tight SNARE bundle. The two different regions of its SNARE motif might allow tomosyn to act as a regulatory SNARE protein that serves as a placeholder for synaptobrevin.
Supplementary Material
Acknowledgments
We thank W. Berning-Koch and U. Ries for expert technical assistance and S. Munro for critical reading of the manuscript.
This work was supported by Deutsche Forschungsgemeinschaft Grants SFB523-B11 (to D. F.) and SFB523-B16 (to J. B. S.) and by a Lundbeck Foundation Junior Group Leader Fellowship (to J. B. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- SNARE
- soluble N-ethylmaleimide-sensitive factor attachment protein receptors
- ITC
- isothermal titration calorimetry
- VAMP
- vesicle-associated membrane protein
- Syx
- syntaxin
- Syb
- synaptobrevin
- aa
- amino acid(s)
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid
- Eb
- endobrevin.
REFERENCES
- 1.Jahn R., Scheller R. H. (2006) Nat. Rev. Mol. Cell Biol. 7, 631–643 [DOI] [PubMed] [Google Scholar]
- 2.Martens S., McMahon H. T. (2008) Nat. Rev. Mol. Cell Biol. 9, 543–556 [DOI] [PubMed] [Google Scholar]
- 3.Rizo J., Rosenmund C. (2008) Nat. Struct. Mol. Biol. 15, 665–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Südhof T. C., Rothman J. E. (2009) Science 323, 474–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Verhage M., Sørensen J. B. (2008) Traffic 9, 1414–1424 [DOI] [PubMed] [Google Scholar]
- 6.Wojcik S. M., Brose N. (2007) Neuron 55, 11–24 [DOI] [PubMed] [Google Scholar]
- 7.Sørensen J. B. (2009) Annu. Rev. Cell Dev. Biol. 25, 513–537 [DOI] [PubMed] [Google Scholar]
- 8.Stein A., Weber G., Wahl M. C., Jahn R. (2009) Nature 460, 525–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sutton R. B., Fasshauer D., Jahn R., Brunger A. T. (1998) Nature 395, 347–353 [DOI] [PubMed] [Google Scholar]
- 10.Hanson P. I., Heuser J. E., Jahn R. (1997) Curr. Opin. Neurobiol. 7, 310–315 [DOI] [PubMed] [Google Scholar]
- 11.Fasshauer D., Margittai M. (2004) J. Biol. Chem. 279, 7613–7621 [DOI] [PubMed] [Google Scholar]
- 12.Wiederhold K., Fasshauer D. (2009) J. Biol. Chem. 284, 13143–13152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pobbati A. V., Stein A., Fasshauer D. (2006) Science 313, 673–676 [DOI] [PubMed] [Google Scholar]
- 14.Siddiqui T. J., Vites O., Stein A., Heintzmann R., Jahn R., Fasshauer D. (2007) Mol. Biol. Cell 18, 2037–2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borisovska M., Zhao Y., Tsytsyura Y., Glyvuk N., Takamori S., Matti U., Rettig J., Südhof T., Bruns D. (2005) EMBO J. 24, 2114–2126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerber S. H., Rah J. C., Min S. W., Liu X., de Wit H., Dulubova I., Meyer A. C., Rizo J., Arancillo M., Hammer R. E., Verhage M., Rosenmund C., Südhof T. C. (2008) Science 321, 1507–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kesavan J., Borisovska M., Bruns D. (2007) Cell 131, 351–363 [DOI] [PubMed] [Google Scholar]
- 18.Sørensen J. B., Wiederhold K., Müller E. M., Milosevic I., Nagy G., de Groot B. L., Grubmüller H., Fasshauer D. (2006) EMBO J. 25, 955–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walter A. M., Wiederhold K., Bruns D., Fasshauer D., Sørensen J. B. (2010) J. Cell Biol. 188, 401–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fasshauer D., Antonin W., Margittai M., Pabst S., Jahn R. (1999) J. Biol. Chem. 274, 15440–15446 [DOI] [PubMed] [Google Scholar]
- 21.Pobbati A. V., Razeto A., Böddener M., Becker S., Fasshauer D. (2004) J. Biol. Chem. 279, 47192–47200 [DOI] [PubMed] [Google Scholar]
- 22.Zwilling D., Cypionka A., Pohl W. H., Fasshauer D., Walla P. J., Wahl M. C., Jahn R. (2007) EMBO J. 26, 9–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stein A., Radhakrishnan A., Riedel D., Fasshauer D., Jahn R. (2007) Nat. Struct. Mol. Biol. 14, 904–911 [DOI] [PubMed] [Google Scholar]
- 24.Burkhardt P., Hattendorf D. A., Weis W. I., Fasshauer D. (2008) EMBO J. 27, 923–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kloepper T. H., Kienle C. N., Fasshauer D. (2007) Mol. Biol. Cell 18, 3463–3471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. (2004) Genome Res. 14, 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hannenhalli S. S., Russell R. B. (2000) J. Mol. Biol. 303, 61–76 [DOI] [PubMed] [Google Scholar]
- 28.Kienle N., Kloepper T. H., Fasshauer D. (2009) BMC Evol. Biol. 9, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kloepper T. H., Kienle C. N., Fasshauer D. (2008) Mol. Biol. Evol. 25, 2055–2068 [DOI] [PubMed] [Google Scholar]
- 30.Frank S., Lustig A., Schulthess T., Engel J., Kammerer R. A. (2000) J. Biol. Chem. 275, 11672–11677 [DOI] [PubMed] [Google Scholar]
- 31.Lee D. L., Lavigne P., Hodges R. S. (2001) J. Mol. Biol. 306, 539–553 [DOI] [PubMed] [Google Scholar]
- 32.Mason J. M., Arndt K. M. (2004) Chembiochem. 5, 170–176 [DOI] [PubMed] [Google Scholar]
- 33.Steinmetz M. O., Jelesarov I., Matousek W. M., Honnappa S., Jahnke W., Missimer J. H., Frank S., Alexandrescu A. T., Kammerer R. A. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7062–7067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holt M., Riedel D., Stein A., Schuette C., Jahn R. (2008) Curr. Biol. 18, 715–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Domanska M. K., Kiessling V., Stein A., Fasshauer D., Tamm L. K. (2009) J. Biol. Chem. 284, 32158–32166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van den Bogaart G., Holt M. G., Bunt G., Riedel D., Wouters F. S., Jahn R. (2010) Nat. Struct. Mol. Biol. 17, 358–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neher E. (2006) Pflugers Arch. Eur. J. Physiol. 453, 261–268 [DOI] [PubMed] [Google Scholar]
- 38.Schoch S., Deák F., Königstorfer A., Mozhayeva M., Sara Y., Südhof T. C., Kavalali E. T. (2001) Science 294, 1117–1122 [DOI] [PubMed] [Google Scholar]
- 39.Yang C., Mora S., Ryder J. W., Coker K. J., Hansen P., Allen L. A., Pessin J. E. (2001) Mol. Cell. Biol. 21, 1573–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grote E., Hao J. C., Bennett M. K., Kelly R. B. (1995) Cell 81, 581–589 [DOI] [PubMed] [Google Scholar]
- 41.Sørensen J. B. (2004) Pflugers Arch. Eur. J. Physiol. 448, 347–36214997396 [Google Scholar]
- 42.Antonin W., Fasshauer D., Becker S., Jahn R., Schneider T. R. (2002) Nat. Struct. Biol. 9, 107–111 [DOI] [PubMed] [Google Scholar]
- 43.Ellena J. F., Liang B., Wiktor M., Stein A., Cafiso D. S., Jahn R., Tamm L. K. (2009) Proc. Natl. Acad. Sci. U. S. A. 106, 20306–20311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ashery U., Bielopolski N., Barak B., Yizhar O. (2009) Trends Neurosci. 32, 275–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fasshauer D., Jahn R. (2007) Nat. Struct. Mol. Biol. 14, 360–362 [DOI] [PubMed] [Google Scholar]
- 46.Hattendorf D. A., Andreeva A., Gangar A., Brennwald P. J., Weis W. I. (2007) Nature 446, 567–571 [DOI] [PubMed] [Google Scholar]
- 47.Fasshauer D., Sutton R. B., Brunger A. T., Jahn R. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 15781–15786 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.