Abstract
Kar2p, an essential Hsp70 chaperone in the endoplasmic reticulum of Saccharomyces cerevisiae, facilitates the transport and folding of nascent polypeptides within the endoplasmic reticulum lumen. The chaperone activity of Kar2p is regulated by its intrinsic ATPase activity that can be stimulated by two different nucleotide exchange factors, namely Sil1p and Lhs1p. Here, we demonstrate that the binding requirements for Lhs1p are complex, requiring both the nucleotide binding domain plus the linker domain of Kar2p. In contrast, the IIB domain of Kar2p is sufficient for binding of Sil1p, and point mutations within IIB specifically blocked Sil1p-dependent activation while remaining competent for activation by Lhs1p. Taken together, these results demonstrate that the interactions between Kar2p and its two nucleotide exchange factors can be functionally resolved and are thus mechanistically distinct.
Keywords: ATPases, Endoplasmic Reticulum (ER), Heat Shock Protein, Protein Folding, Protein Secretion, Yeast, Grp170, Hsp110
Introduction
The Hsp70 Kar2p is an essential molecular chaperone of the yeast endoplasmic reticulum (ER)2 that is structurally and functionally homologous to mammalian BiP (1). Kar2p is essential for cellular homeostasis and participates in the transport of nascent polypeptides into the ER lumen, polypeptide folding, and the selection of misfolded proteins for degradation (2–4). Hsp70 proteins are highly conserved throughout eukaryotes and have a characteristic domain structure consisting of an N-terminal nucleotide binding domain (NBD) connected by a flexible 12-amino acid “linker domain” to the C-terminal substrate binding domain (SBD) (5). The affinity of the SBD for substrates, usually exposed hydrophobic peptides, is regulated by the nucleotide-bound status of the NBD, and this is communicated to the SBD via the linker domain (5–7). High affinity substrate association is triggered by ATP hydrolysis, and substrate binding is maintained until bound ADP is released (8, 9). The ATPase cycle of Kar2p, like other typical Hsp70 proteins, is driven by its interactions with two classes of cofactors. Hsp40/DnaJ-like proteins stimulate ATP hydrolysis, whereas nucleotide exchange factors (NEFs) facilitate ADP release thus promoting the next cycle of ATP binding (10, 11). These cofactors allow Kar2p to cycle through multiple stages of substrate binding and release thus facilitating protein translocation and folding. Two NEFs have been identified for Kar2p in the ER lumen of Saccharomyces cerevisiae, namely Lhs1p and Sil1p (12, 13). Individual deletions of LHS1 (Δlhs1) and SIL1 (Δsil1) in yeast cells are viable (13, 14), but the double deletion (Δlhs1Δsil1) is lethal suggesting that nucleotide exchange is essential for Kar2p activity and cell viability (13). In addition, the expression of the mammalian homologue of Lhs1p, Grp170 (also known as ORP150), has been closely correlated with cell survival in response to hypoxic/ischemic stress (15–17), and loss of function mutations in SIL1 are associated with Marinesco Sjörgren syndrome (18, 19) highlighting the importance of both NEFs in cell homeostasis.
Lhs1p and Grp170 are members of an Hsp70 subfamily with ∼30% sequence similarity to Kar2p (14, 20). A further subgroup of Hsp70s called the Hsp110s are found in the cytoplasm of eukaryotes and are also thought to function as NEFs for their cytoplasmic Hsp70 partners (21–23). Lhs1p and the Hsp110s appear structurally similar to Hsp70s but have an additional loop domain that separates a series of β-sheets from the α-helical domain in the C-terminal SBD (22). However, the additional loop region appears more extended in Lhs1p than in the Hsp110s, although the significance of this difference, if any, is unknown (22). Despite the structural similarities between their NBD regions, the canonical Hsp70s, the Hsp110s, and Lhs1p/Grp170 have quite different nucleotide binding and hydrolysis characteristics (12, 23, 24). In contrast to Kar2p, Lhs1p binds ATP to form a remarkably stable complex, and it is only this nucleotide-bound form of Lhs1p that interacts with Kar2p provided the latter is itself not in the ATP-bound form (25). However, although nucleotide binding by Lhs1p is essential for its NEF activity, the intrinsic holdase activity of Lhs1p is independent of nucleotide in a manner that is similar to the Hsp110s (25–27). The second Kar2p NEF Sil1p is unrelated at sequence level to either Kar2p or Lhs1p but shows very limited sequence similarity to a defined subset of cytosolic NEFs, including yeast Fes1p and mammalian HspBP1 (14, 28). Both Fes1p and HspBP1 are thought to stimulate nucleotide exchange by destabilizing the NBD and triggering opening of the nucleotide binding cleft to allow ADP release (28, 29), yet the mechanism by which Sil1p stimulates nucleotide exchange has not been examined. Indirect evidence suggests that Sil1p and Lhs1p bind in a mutually exclusive manner to promote the release of nucleotide from Kar2p (12), yet little has been done to characterize the interactions further.
In this study, we have created a structural model of Kar2p to facilitate our analysis of Sil1p and Lhs1p binding. The model allowed us to predict the subdomain structure of Kar2p and define several recombinant fragments of Kar2p for binding analysis in vitro. These studies identified a role for the IIB domain of Kar2p in Sil1p binding and Sil1p-stimulated nucleotide exchange. We demonstrate that mutation of highly conserved residues within the IIB domain inhibits the ability of Sil1p to stimulate the ATPase activity of Kar2p, but it fails to perturb Lhs1p binding and Lhs1p-specific nucleotide exchange. Furthermore, we have identified a role for the linker domain of Kar2p in the interaction between Lhs1p and the NBD of Kar2p, suggesting that the NEFs bind very differently to Kar2p and that the mechanisms of nucleotide exchange are distinct.
EXPERIMENTAL PROCEDURES
Plasmids
GST-tagged Kar2p and His-Lhs1p were expressed in Escherichia coli from plasmids pDF1 and pETLhs1 described previously (12). pSM11 encodes a 10-histidine-tagged version of SIL1 minus its signal sequence and -DEL retention motif (residues 20–407), created by PCR and inserted into the T7 expression vector pET16-b (Novagen) using the restriction enzyme sites NdeI and BamHI. The fragments of Kar2p (NBD (residues 45–425), NBD-linker (residues 45–437), lobe I (residues 45–234), lobe II (residues 235–432), and IIB (residues 273–352)) were constructed by PCR and cloned into pGEX4T-3 (N-terminal GST tag; GE Healthcare) using the restriction sites BamHI and SalI. KAR2 mutants (E311A and R317A) were made by site-directed mutagenesis using the protocol from the QuikChange kit (Stratagene) and introduced into full-length Kar2p in the pDF1 plasmid for expression as GST-tagged fusions.
Protein Purification
GST- and His-tagged proteins were expressed in DH5α and BL21 E. coli cells, respectively. Cleared cell extracts were made as described previously (13) and purified by binding to equilibrated 1 ml of chelating Hi-Trap or GST-Trap columns (GE Healthcare) as described (12).
GST Pulldowns
The GST-pulldown assays were performed as described previously (13). Briefly, 10 μg of purified GST-tagged protein was incubated with glutathione-agarose (Sigma) for 1 h at 4 °C. After washes with GST-binding buffer (20 mm HEPES, pH 7.4, 100 mm KCl, 5 mm MgCl2, 1 mm EDTA, 1 mm dithiothreitol, 0.1% Nonidet P-40, 2% glycerol + protease inhibitor mixture (Sigma)), the beads were incubated with 10 μg of His-tagged protein (4 °C, 1 h). After washing, protein was eluted from the glutathione-agarose beads in SDS sample buffer at 95 °C and analyzed by SDS-PAGE followed by Coomassie Brilliant Blue staining or Western blotting. The antibodies used for Western blotting were specific to the His or GST tag sequences (Sigma and Santa Cruz Biotechnology, respectively). To test the nucleotide dependence of the observed interactions, the pulldown was repeated in the presence of 2 mm ADP or ATP.
Structural Modeling of Kar2p
A structural model of Kar2p was made using comparative modeling techniques. Bovine Hsc70 chaperone (Protein Data Bank code 1YUW (5)) was used as the template. Kar2p was aligned to Hsc70 using ClustalW (30). We were able to model residues 47–611 of Kar2p (residues 1–46 correspond to the signal sequence of Kar2p), and sequence identity over the region modeled was 66%. The Kar2p model was produced with Modeler (31). 15 models were produced, and the one with lowest pseudo-energy was selected. The final model has a root mean square deviation of 0.65 Å from the template structure.
Steady State ATPase Assays
The ATPase activity of Kar2p and the Kar2p mutants was analyzed by a colorimetric assay as described previously (12, 32). Briefly, 1 μm Kar2p (K) or the Kar2p mutants (311K and 317K) were incubated alone or in the presence of 2 μm Sec63 soluble J domain (J), 0.8 μm Lhs1p (L), or 0.8 μm Sil1p (S) in 30 μl of ATPase buffer (50 mm Tris, pH 7.4, 50 mm KCl, 5 mm MgCl2) at 25 °C for 1 h. The ratio of K, J, S, and L used in the assay is within the optimum range identified for Hsp70 activity based on previous experiments with DnaK and Kar2p (see supplemental Fig. S1) (10, 12). The amount of free phosphate was calculated following the addition of malachite green solution by monitoring the absorbance at A640 nm. The amount of phosphate released was calculated compared with KH2PO4 standard curve, and values were corrected for breakdown of ATP in the absence of added protein. Mean hydrolysis rates were determined for each condition from a minimum of three experiments.
Protease Protection Assay
The ability of wild-type and mutant protein to undergo a nucleotide-dependent conformational change was assessed using proteinase K digestion as described previously (33). 5 μg of protein was incubated in the presence/absence of 1 mm nucleotide (ADP or ATP) for 30 min at 20 °C in assay buffer (20 mm HEPES, pH 7.4, 25 mm KCl, 2 mm MgCl2, 0.5 mm dithiothreitol, 0.1 mm EDTA). 30 μg/ml proteinase K was added for 5 min at 20 °C, and the reaction was quenched with ice-cold trichloroacetic acid to a final concentration of 10%. Trichloroacetic acid-precipitated proteins were analyzed by SDS-PAGE and Coomassie Brilliant Blue staining.
RESULTS
Interaction of Kar2p with Its Nucleotide Exchange Factors
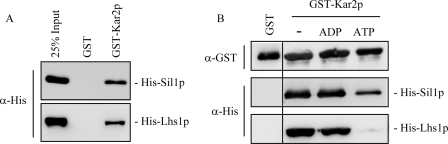
Kar2p has previously been shown to interact with both Sil1p and ATP-bound Lhs1p (13, 25). To analyze these interactions further, we established a GST pulldown assay in vitro using tagged recombinant proteins expressed and purified from E. coli. Previous data have shown that GST-Kar2p is able to bind His-Lhs1p (12, 25), and here we show that His-Sil1p is also able to bind to GST-Kar2p (Fig. 1A), allowing the binding of both NEFs to be analyzed in parallel. As shown previously, the interaction of Lhs1p (purified as a stable ATP-bound complex (25)) with Kar2p was sensitive to the presence of 2 mm ATP but not ADP confirming that Lhs1p binds most efficiently to the apo- and ADP-bound forms of Kar2p (Fig. 1B) (25). Similarly, we show that the binding of Sil1p to Kar2p was also reduced in the presence of ATP but not ADP (Fig. 1B). Interestingly, the binding of Sil1p appears less sensitive to the presence of ATP than the Kar2p-Lhs1p interaction.
FIGURE 1.
Sil1p and Lhs1p bind to Kar2p in a nucleotide-sensitive manner. A, purified GST or GST-Kar2p was immobilized onto glutathione-agarose followed by incubation with His-Sil1p or His-Lhs1p (expressed in E. coli from pSM11 and pETLhs1, respectively). Binding was analyzed by SDS-PAGE and Western blotting with an antibody specific for the His sequence. The input in this figure and subsequent figures corresponds to 25% of the total His-tagged protein added to the assay. B, to assess the sensitivity of the Kar2p-NEF interactions to adenosine nucleotides, the binding assay was repeated in the presence of 2 mm ADP or 2 mm ATP, and the amount of NEF binding was analyzed by Western blotting with antibodies specific to the His and GST sequences.
It has been suggested that Sil1p and Lhs1p bind to Kar2p in a mutually exclusive manner to stimulate nucleotide exchange (12). To analyze the binding of both Sil1p and Lhs1p to Kar2p, we incubated GST-Kar2p with His-Sil1p alone or in the presence of an excess of His-Lhs1p. The binding of Sil1p to Kar2p was greatly reduced in the presence of an excess of Lhs1p suggesting that the NEFs are unable to bind to Kar2p simultaneously (supplemental Fig. S2).
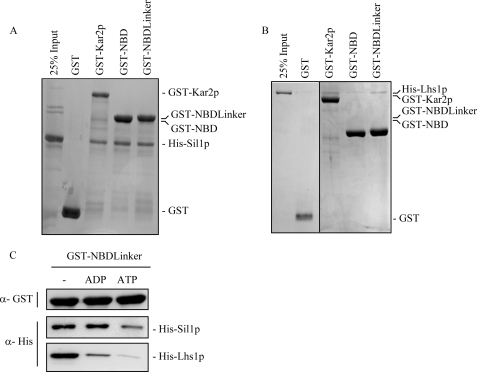
It has previously been shown that Sil1p binds to the Kar2p NBD (13), and this is confirmed here using a GST-NBD construct to pull down His-tagged Sil1p (Fig. 2A). We next examined whether Lhs1p might also bind to the Kar2p NBD but, to our surprise, found very little coprecipitation of His-Lhs1p with GST-NBD in this assay. However, when the GST fusion was extended to include both the NBD and linker domains of Kar2p, we then found coprecipitation of Lhs1p at a level similar to that observed for the full-length GST-Kar2p control (Fig. 2B). Sil1p also bound to GST-NBD-linker, so we used this construct to next examine the effects of nucleotide on these NEF interactions. We observed that the binding of both Sil1p and Lhs1p to NBDlinker was reduced in the presence of ATP in a similar manner to that observed for the full-length proteins (compare Fig. 1B and Fig. 2C). However, the interaction of the NBDlinker with Lhs1p appeared to be more sensitive to ADP than the interaction with full-length Kar2p (Fig. 2C), suggesting that the Kar2p SBD also contributes to the recognition of the ADP-bound form of Kar2p by Lhs1p. Notably, Sil1p was able to bind to the ADP bound NBDlinker implying that Sil1p binding is independent of the presence of the SBD.
FIGURE 2.
Lhs1p, but not Sil1p, requires the linker domain of Kar2p to bind to the nucleotide binding domain. A and B, ability of Sil1p and Lhs1p to bind to the N terminus of Kar2p (NBD + linker), and the NBD alone was assayed. Immobilized GST, GST-Kar2p, GST-NBDlinker (residues 45–438), and GST-NBD (residues 45–425) were incubated with His-Sil1p and His-Lhs1p for 1 h followed by analysis by SDS-PAGE and Coomassie staining. C, binding of both His-Sil1p and His-Lhs1p to GST-NBDlinker was assayed in the presence of 2 mm ADP or 2 mm ATP, and the amount of His-tagged protein bound was analyzed by Western blotting.
Sil1p Binds to the IIB Domain of Kar2p to Stimulate Nucleotide Exchange
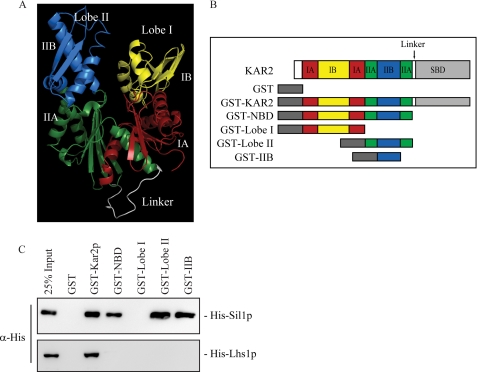
Despite the obvious role for the linker domain of Kar2p in Lhs1p binding, it appears that Sil1p is able to bind to the NBD of Kar2p independently of the linker domain. To facilitate our investigation into the binding of Sil1p to Kar2p, we built a comparative model of Kar2p using the structure of bovine Hsc70 (Protein Data Bank code 1YUW (5)). We were able to model the mature Kar2p protein minus its signal sequence and extreme C terminus allowing us to predict the domain boundaries of Kar2p. Sequence identity between Kar2p and Hsc70 is 66% over the region modeled, suggesting that the domain boundary assignment is likely to be accurate.
Using the structural model, we assigned domain boundaries to the regions of the Kar2p NBD. The NBD of an Hsp70 has two distinct lobes (I and II) that are subdivided into two domains (A and B). We have assigned these domains to Kar2p as follows: Kar2p NBD encompasses residues 45–425 and the linker domain residues 426–438. Lobe I of the NBD includes residues 45–234 (IA, residues 45–85 and 162–230; IB, residues 86–161) and lobe II includes residues 235–425 (IIA, 235–273 and 352–425; IIB, 274–351) (Fig. 3A). To determine the location of the Sil1p-binding site within the NBD of Kar2p, we constructed further GST-tagged fragments of Kar2p encompassing lobe I and lobe II of the NBD (Fig. 3B). Analysis of Sil1p binding to these fragments showed that Sil1p interacted specifically with lobe II of the Kar2p NBD and showed no binding to lobe I (Fig. 3C). In an attempt to isolate the binding site for Sil1p further, binding of Sil1p to a fragment corresponding to the IIB domain of lobe II was analyzed. Loss of the IIA domain from this construct appeared to have no effect on the Sil1p-binding site as Sil1p was able to bind to the IIB domain alone (Fig. 3C), and from these data we propose that the IIB domain of Kar2p is sufficient for Sil1p binding.
FIGURE 3.
Sil1p binds to the IIB domain of Kar2p. A, structural model of the Kar2p NBDlinker created using sequence alignments and modeling software. The NBD consists of two lobes, lobe I (right) and lobe II (left). The subdomains of the lobes are colored as follows: IA, red; IB, yellow; IIA, green; IIB, blue; and the flexible linker domain connecting the NBD to the SBD is colored white. B, domain organization of Kar2p. The recombinant fragments designed for use in this study are represented below, namely GST, GST-Kar2p, GST-NBD, GST-Lobe I, GST-Lobe II, and GST-IIB. C, binding of His-Sil1p and His-Lhs1p to the recombinant fragments of Kar2p followed by analysis by Western blotting.
In contrast, Lhs1p was unable to bind to any of the Kar2p NBD fragments expressed individually (Fig. 3C). The lack of Lhs1p binding implies that if the domains of the NBD contain Lhs1p-binding sites then they are not oriented correctly to bind to Lhs1p in the absence of the intact NBD and the linker domain.
Mutations in the IIB Domain of Kar2p Disrupt Sil1p Binding
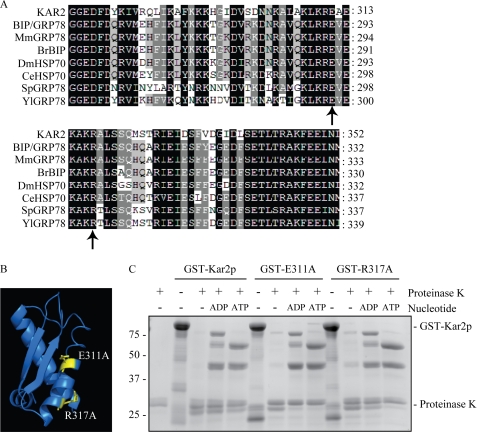
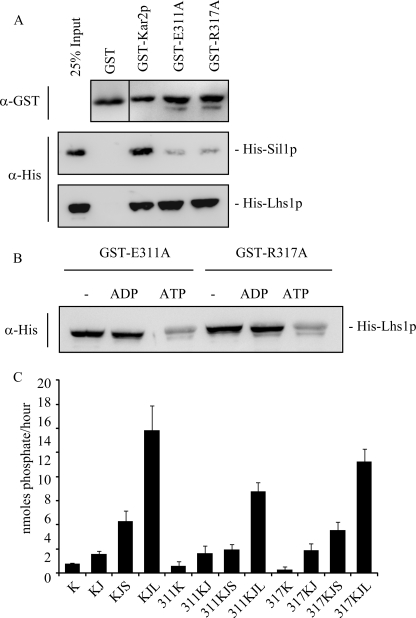
To confirm the role of the IIB domain in Sil1p binding, amino acid substitutions were made to residues within the IIB domain of Kar2p by site-directed mutagenesis. Residues Glu-311 and Arg-317 were chosen for mutagenesis as they are highly conserved within the IIB domain of Kar2p homologues (Fig. 4A) and are in the vicinity of residues reported to be involved in the binding of the NEF HspBP1 to its Hsp70 partner (34). In addition, Glu-311 and Arg-317 are predicted to be exposed on a surface of the IIB domain of Kar2p in our structural model (Fig. 4B). These residues were mutated to alanine (E311A and R317A) in the GST-Kar2p sequence in an attempt to disrupt any potential Sil1p interactions at these sites.
FIGURE 4.
Mutations in the IIB domain of Kar2p do not perturb the Kar2p nucleotide-dependent conformational change. A, sequence alignment of the IIB domain of Kar2p with other Kar2p homologues using ClustalW software. Sequences aligned are as follows: Mm, Mus musculus; Br, Brachydanio rerio; Dm, Drosophila melanogaster; Ce, Caenorhabditis elegans; Sp, Schizosaccharomyces pombe, and Yl, Yarrowia lipolytica, and the residues numbers are annotated. Residues chosen for mutagenesis are indicated with a black arrow. Black shading represents 100% sequence identity; dark gray shading represents 80% sequence identity, and light gray shading represents 60% sequence identity. B, location of the residues chosen for mutagenesis in the IIB domain of the structural model of Kar2p. C, protease protection assay of wild-type and mutant Kar2p. 5 μg of protein was incubated with nucleotide for 30 min followed by 5 min of digestion with proteinase K. Digestion products were analyzed by Coomassie staining.
Data suggest that the conformational changes triggered by nucleotide binding to canonical Hsp70s can be detected by altered protease sensitivity of the protein in the presence of ADP and ATP (33, 35, 36). To confirm that the introduction of point mutations into the IIB domain of Kar2p has not perturbed the ability of Kar2p to alter its conformation in response to nucleotide, purified GST-Kar2p, GST-Kar2p E311A, and GST-Kar2p R317A were preincubated with ADP or ATP followed by digestion for 5 min with proteinase K. In the apo state, the majority of Kar2p, Kar2p E311A, and Kar2p R317A was digested by the protease to fragments <25 kDa (Fig. 4C), whereas digestion of the ADP- or ATP-bound states of both Kar2p and the mutant forms of Kar2p produced several protease protected fragments that differed in size depending on the nucleotide bound (Fig. 4C). The fragments appeared to be of similar abundance and size for both wild-type and mutant Kar2p suggesting that the addition of the point mutations had little or no effect on the ability of Kar2p to bind and alter its conformation in response to nucleotide. Next, we tested the effects of these point mutations on NEF interactions. We found that both E311A and R317A exhibited dramatically reduced efficiency of interaction with Sil1p when compared with wild-type Kar2p as a control. In contrast, both mutants remained competent to bind Lhs1p (Fig. 5A) in a manner whose nucleotide sensitivity was identical to the pattern observed for wild-type Kar2p (Fig. 5B). Thus, the E311A and R317A mutations in the IIB domain of Kar2p specifically interfere with Sil1p binding but not with the binding of Lhs1p.
FIGURE 5.
Mutation of residues Glu-311 and Arg-317 affects Sil1p binding and function. A, ability of Kar2p E311A and R317A to bind to His-Sil1p and His-Lhs1p was assayed in a GST pulldown alongside wild-type Kar2p. Binding of the His-tagged proteins was visualized by Western blotting. B, effect of nucleotide on the interaction between His-Lhs1p and the Kar2p mutants was assayed by repeating the binding experiments in the presence of 2 mm ADP or ATP. C, steady state ATPase activity of Kar2p was measured by a colorimetric assay at A640 nm, and the amount of phosphate released was calculated using a standard curve. Kar2p (K) or the Kar2p mutants (311K or 317K) were incubated in the presence of 0.8 μm GST-J domain of Sec63 (J) and with His-Sil1p (S) or His-Lhs1p (L) (as indicated) for 1 h at 25 °C in the presence of 2 mm ATP.
The ability of Sil1p to stimulate the activity of Kar2p E311A and Kar2p R317A was investigated using a steady state ATPase assay. As seen previously, the ATPase activity of wild-type Kar2p was stimulated by addition of the soluble lumenal loop of Sec63p (GST-63J), which contains a J domain (Fig. 5C) (12). The activity of Kar2p was then stimulated further by the addition of Sil1p and more so by the addition of Lhs1p (Fig. 5C) (12). However, no additive effect was observed by adding both Sil1p and Lhs1p to the reaction (supplemental Fig. S3) (12). The stimulation of Kar2p activity by GST-63J and the NEFs represents the activation of ATP hydrolysis and nucleotide exchange, respectively. The ATPase activity of Kar2p E311A and Kar2p R317A was stimulated by the addition of GST-63J to a similar extent as wild-type Kar2p (Fig. 5C), suggesting that the mutant proteins are still active in the ATP hydrolysis phase of their ATPase cycle, yet no increase in simulation was observed following addition of Sil1p to Kar2p E311A, with only a small increase in activity following the addition of Sil1p to Kar2p R317A (Fig. 5C). These data suggest that the point mutations also affect the ability of Sil1p to trigger the release of nucleotide from Kar2p. Nevertheless, the activity of Kar2p E311A and R317A was stimulated by the addition of Lhs1p (Fig. 5C), although to a slightly lesser extent than wild-type Kar2p, demonstrating that the IIB mutants can still be stimulated to release bound nucleotide. These data confirm a role for IIB in Sil1p binding and stimulation of nucleotide exchange and provide evidence that the regulation of nucleotide exchange by Lhs1p is distinct to the mechanism employed by Sil1p.
DISCUSSION
The ability of Kar2p to bind and release substrate is regulated by its ATPase cycle. Sil1p and Lhs1p act as NEFs for Kar2p to stimulate the release of bound nucleotide allowing Kar2p to cycle through multiple rounds of substrate binding and release. In this study, we observed the interaction of Kar2p with its NEFs Sil1p and Lhs1p in vitro. The binding of both Sil1p and Lhs1p to Kar2p was sensitive to the presence of nucleotide, with both Sil1p and Lhs1p binding preferentially to the ADP-bound or apo-forms of Kar2p. These findings agree with data from studies on other canonical Hsp70s that suggest that NEFs have a higher affinity for their Hsp70 partners when they are apo- or ADP-bound (11, 25). Interestingly, although the observed interaction between Kar2p and Sil1p was reduced by ∼50% in the presence of ATP, the interaction between Kar2p and Lhs1p was essentially abolished under these conditions suggesting some mechanistic differences in the binding of the two NEFs.
With the aid of a structural model of Kar2p, we have identified a role for the IIB domain of Kar2p in Sil1p binding and have shown that IIB alone is sufficient for Sil1p binding in vitro. Mutations of residues Glu-311 and Arg-317 within the Kar2p IIB domain disrupt Sil1p binding to Kar2p and so abolish Sil1p-dependent stimulation of the Kar2p ATPase activity. These results indicate that the IIB region of Kar2p plays an essential role in the stimulation of nucleotide exchange by Sil1p. The yeast cytoplasmic NEF Fes1p and its mammalian homologue HspBP1 only share 15.9 and 8.7% sequence identity with Sil1p, respectively, yet are also thought to stimulate nucleotide exchange by interacting with the IIB domain of their Hsp70 partners (21, 28). Structural analysis of the core domain of human HspBP1 (BP1c residues 84–359) in complex with lobe II of Hsp70 revealed multiple contacts between BP1c and the IIB domain of Hsp70, with no contacts observed to the IIA domain (34). Indeed, the major interaction site between the two proteins is formed between BP1c and the β-hairpin of the IIB domain of Hsp70 (34). The residues mutated in our study lie within the β-hairpin structure in the IIB domain of Kar2p in our structural model suggesting that, despite the low sequence similarity, Sil1p and HspBP1 may bind to their respective Hsp70s in very similar ways.
In striking contrast, the amino acid substitutions in the IIB domain of Kar2p had no effect on the ability of Lhs1p to bind to Kar2p; we also did not observe any interaction between Lhs1p and the IIB domain alone in this assay. Recent crystal structures of an Hsp70 in complex with an Hsp110 indicate multiple contacts between the NBDs of these proteins (37, 38). As the Hsp110s are homologous to Lhs1p, we might predict that Kar2p and Lhs1p would form a similar complex. However, although Hsp110 can bind to the NBD of its respective cytosolic Hsp70 (26, 39), our data demonstrate that Lhs1p requires the additional presence of the linker domain (NBDlinker) to stably interact, thereby suggesting a novel role for the linker domain in the interaction of these lumenal chaperones. The linker region of Hsp70s is thought to be intimately involved in transmitting the nucleotide-bound status of the NBD to the SBD to regulate substrate binding (5–7). Upon ATP binding, the linker domain is thought to bind to a hydrophobic patch at the base of the NBD bringing the NBD and SBD into close proximity. Subsequent substrate binding and ATP hydrolysis reverse this movement causing the linker domain to become exposed and the NBD and SBD to dissociate (5–7). It is this movement, and domain association, that is thought to regulate substrate binding affinity. Lhs1p binding to the NBD of Kar2p is sensitive to the presence of the linker domain, and Lhs1p appears to favor the apo- or ADP-bound states of Kar2p in which the linker domain is predicted to be exposed. Interestingly, unlike the binding of Sil1p, Lhs1p binding to Kar2p is completely abolished in the presence of ATP, suggesting that the linker “in” conformation is not favorable for Lhs1p binding. This strict regulation of Lhs1p binding to Kar2p suggests that the role of these Hsp70s homologues may be tightly coordinated ensuring that Lhs1p would be preferentially recruited to the ADP-bound form of Kar2p representing the form that most stably binds substrate. Such strict regulation of Lhs1p recruitment, combined with the ability of Lhs1p to bind to unfolded substrate (25), would be consistent with a model in which substrate could be passed efficiently from Kar2p to Lhs1p following Lhs1p-dependent nucleotide exchange. The coordinated transfer of substrate between Kar2p and Lhs1p might be predicted to facilitate processes such as polypeptide translocation into the ER. In contrast, Sil1p, which stimulates Kar2p nucleotide exchange independently of Lhs1p, is not thought to bind to substrate and may activate the chaperone function of Kar2p for other cellular processes such as protein folding. However, these hypotheses remain speculative and will require further study. In summary, our data demonstrate that the binding sites within Kar2p for Sil1p and Lhs1p are distinct and can be functionally resolved. The complex nature of the Lhs1p interaction suggests an intimate interaction with Kar2p, which may have important implications for our understanding of this essential chaperone network.
Supplementary Material
Acknowledgment
We thank Dr. M. R. Pool for comments on the manuscript.
This work was supported by Wellcome Trust.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- ER
- endoplasmic reticulum
- NBD
- nucleotide binding domain
- SBD
- substrate binding domain
- NEF
- nucleotide exchange factor
- GST
- glutathione S-transferase.
REFERENCES
- 1.Rose M. D., Misra L. M., Vogel J. P. (1989) Cell 57, 1211–1221 [DOI] [PubMed] [Google Scholar]
- 2.Brodsky J. L., Werner E. D., Dubas M. E., Goeckeler J. L., Kruse K. B., McCracken A. A. (1999) J. Biol. Chem. 274, 3453–3460 [DOI] [PubMed] [Google Scholar]
- 3.Corsi A. K., Schekman R. (1997) J. Cell Biol. 137, 1483–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simons J. F., Ferro-Novick S., Rose M. D., Helenius A. (1995) J. Cell Biol. 130, 41–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang J., Prasad K., Lafer E. M., Sousa R. (2005) Mol. Cell 20, 513–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swain J. F., Dinler G., Sivendran R., Montgomery D. L., Stotz M., Gierasch L. M. (2007) Mol. Cell 26, 27–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vogel M., Mayer M. P., Bukau B. (2006) J. Biol. Chem. 281, 38705–38711 [DOI] [PubMed] [Google Scholar]
- 8.Mayer M. P., Bukau B. (2005) Cell. Mol. Life Sci. 62, 670–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morano K. A. (2007) Ann. N. Y. Acad. Sci. 1113, 1–14 [DOI] [PubMed] [Google Scholar]
- 10.Liberek K., Marszalek J., Ang D., Georgopoulos C., Zylicz M. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 2874–2878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szabo A., Langer T., Schröder H., Flanagan J., Bukau B., Hartl F. U. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 10345–10349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steel G. J., Fullerton D. M., Tyson J. R., Stirling C. J. (2004) Science 303, 98–101 [DOI] [PubMed] [Google Scholar]
- 13.Tyson J. R., Stirling C. J. (2000) EMBO J. 19, 6440–6452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Craven R. A., Egerton M., Stirling C. J. (1996) EMBO J. 15, 2640–2650 [PMC free article] [PubMed] [Google Scholar]
- 15.Kuwabara K., Matsumoto M., Ikeda J., Hori O., Ogawa S., Maeda Y., Kitagawa K., Imuta N., Kinoshita T., Stern D. M., Yanagi H., Kamada T. (1996) J. Biol. Chem. 271, 5025–5032 [DOI] [PubMed] [Google Scholar]
- 16.Ozawa K., Kuwabara K., Tamatani M., Takatsuji K., Tsukamoto Y., Kaneda S., Yanagi H., Stern D. M., Eguchi Y., Tsujimoto Y., Ogawa S., Tohyama M. (1999) J. Biol. Chem. 274, 6397–6404 [DOI] [PubMed] [Google Scholar]
- 17.Tamatani M., Matsuyama T., Yamaguchi A., Mitsuda N., Tsukamoto Y., Taniguchi M., Che Y. H., Ozawa K., Hori O., Nishimura H., Yamashita A., Okabe M., Yanagi H., Stern D. M., Ogawa S., Tohyama M. (2001) Nat. Med. 7, 317–323 [DOI] [PubMed] [Google Scholar]
- 18.Anttonen A. K., Mahjneh I., Hämäläinen R. H., Lagier-Tourenne C., Kopra O., Waris L., Anttonen M., Joensuu T., Kalimo H., Paetau A., Tranebjaerg L., Chaigne D., Koenig M., Eeg-Olofsson O., Udd B., Somer M., Somer H., Lehesjoki A. E. (2005) Nat. Genet. 37, 1309–1311 [DOI] [PubMed] [Google Scholar]
- 19.Senderek J., Krieger M., Stendel C., Bergmann C., Moser M., Breitbach-Faller N., Rudnik-Schöneborn S., Blaschek A., Wolf N. I., Harting I., North K., Smith J., Muntoni F., Brockington M., Quijano-Roy S., Renault F., Herrmann R., Hendershot L. M., Schröder J. M., Lochmüller H., Topaloglu H., Voit T., Weis J., Ebinger F., Zerres K. (2005) Nat. Genet. 37, 1312–1314 [DOI] [PubMed] [Google Scholar]
- 20.Park J., Easton D. P., Chen X., MacDonald I. J., Wang X. Y., Subjeck J. R. (2003) Biochemistry 42, 14893–14902 [DOI] [PubMed] [Google Scholar]
- 21.Dragovic Z., Broadley S. A., Shomura Y., Bracher A., Hartl F. U. (2006) EMBO J. 25, 2519–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Easton D. P., Kaneko Y., Subjeck J. R. (2000) Cell Stress Chaperones 5, 276–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raviol H., Bukau B., Mayer M. P. (2006) FEBS Lett. 580, 168–174 [DOI] [PubMed] [Google Scholar]
- 24.Huang P., Gautschi M., Walter W., Rospert S., Craig E. A. (2005) Nat. Struct. Mol. Biol. 12, 497–504 [DOI] [PubMed] [Google Scholar]
- 25.de Keyzer J., Steel G. J., Hale S. J., Humphries D., Stirling C. J. (2009) J. Biol. Chem. 284, 31564–31571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andréasson C., Fiaux J., Rampelt H., Mayer M. P., Bukau B. (2008) J. Biol. Chem. 283, 8877–8884 [DOI] [PubMed] [Google Scholar]
- 27.Shaner L., Trott A., Goeckeler J. L., Brodsky J. L., Morano K. A. (2004) J. Biol. Chem. 279, 21992–22001 [DOI] [PubMed] [Google Scholar]
- 28.Kabani M., McLellan C., Raynes D. A., Guerriero V., Brodsky J. L. (2002) FEBS Lett. 531, 339–342 [DOI] [PubMed] [Google Scholar]
- 29.McLellan C. A., Raynes D. A., Guerriero V. (2003) J. Biol. Chem. 278, 19017–19022 [DOI] [PubMed] [Google Scholar]
- 30.Thompson J. D., Higgins D. G., Gibson T. J. (1994) Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sali A., Blundell T. L. (1993) J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 32.Lanzetta P. A., Alvarez L. J., Reinach P. S., Candia O. A. (1979) Anal. Biochem. 100, 95–97 [DOI] [PubMed] [Google Scholar]
- 33.McClellan A. J., Endres J. B., Vogel J. P., Palazzi D., Rose M. D., Brodsky J. L. (1998) Mol. Biol. Cell 9, 3533–3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shomura Y., Dragovic Z., Chang H. C., Tzvetkov N., Young J. C., Brodsky J. L., Guerriero V., Hartl F. U., Bracher A. (2005) Mol. Cell 17, 367–379 [DOI] [PubMed] [Google Scholar]
- 35.Kamath-Loeb A. S., Lu C. Z., Suh W. C., Lonetto M. A., Gross C. A. (1995) J. Biol. Chem. 270, 30051–30059 [DOI] [PubMed] [Google Scholar]
- 36.Kassenbrock C. K., Kelly R. B. (1989) EMBO J. 8, 1461–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Polier S., Dragovic Z., Hartl F. U., Bracher A. (2008) Cell 133, 1068–1079 [DOI] [PubMed] [Google Scholar]
- 38.Schuermann J. P., Jiang J., Cuellar J., Llorca O., Wang L., Gimenez L. E., Jin S., Taylor A. B., Demeler B., Morano K. A., Hart P. J., Valpuesta J. M., Lafer E. M., Sousa R. (2008) Mol. Cell 31, 232–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andréasson C., Fiaux J., Rampelt H., Druffel-Augustin S., Bukau B. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 16519–16524 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.