Abstract
Fragile X syndrome, the most common form of inherited mental retardation, is caused by the absence of the RNA-binding protein fragile X mental retardation protein (FMRP). FMRP regulates local protein synthesis in dendritic spines. Dopamine (DA) is involved in the modulation of synaptic plasticity. Activation of DA receptors can regulate higher brain functions in a protein synthesis-dependent manner. Our recent study has shown that FMRP acts as a key messenger for DA modulation in forebrain neurons. Here, we demonstrate that FMRP is critical for DA D1 receptor-mediated synthesis of synapse-associated protein 90/PSD-95-associated protein 3 (SAPAP3) in the prefrontal cortex (PFC). DA D1 receptor stimulation induced dynamic changes of FMRP phosphorylation. The changes in FMRP phosphorylation temporally correspond with the expression of SAPAP3 after D1 receptor stimulation. Protein phosphatase 2A, ribosomal protein S6 kinase, and mammalian target of rapamycin are the key signaling molecules for FMRP linking DA D1 receptors to SAPAP3. Knockdown of SAPAP3 did not affect surface expression of α-amino-3-hydroxyl-5-methyl-4-isoxazole-4-propionate (AMPA) GluR1 receptors induced by D1 receptor activation but impaired their subsequent internalization in cultured PFC neurons; the subsequent internalization of GluR1 was also impaired in Fmr1 knock-out PFC neurons, suggesting that FMRP may be involved in subsequent internalization of GluR1 through regulating the abundance of SAPAP3 after DA D1 receptor stimulation. Our study thus provides further insights into FMRP involvement in DA modulation and may help to reveal the molecular mechanisms underlying impaired learning and memory in fragile X syndrome.
Keywords: G Protein-coupled Receptors (GPCR); Glutamate Receptors Ionotropic (AMPA, NMDA); Neurobiology; Neurological Diseases; Protein Synthesis; AMPA Receptor; FMRP; Prefrontal Cortex; SAPAP3; Dopamine
Introduction
Fragile X syndrome, the most common form of inherited mental retardation, is caused by the absence of the RNA-binding protein fragile X mental retardation protein (FMRP),3 because of silencing of the FMR1 gene (1–6). FMRP interacts with its mRNA targets in brain, the functional consequence of which is presently known for few messages (1, 7, 8). FMRP associates with neuronal polysomes and mRNPs and regulates local protein synthesis, especially in dendritic spines (1, 2, 7, 9–13). This is supported by the effects of group I metabotropic glutamate receptor stimulation on FMRP synthesis and transport (2, 8, 11, 14, 15).
The phosphorylation status of FMRP has been identified as a regulator for FMRP function (16, 17). Previous studies showed that phosphorylated FMRP may associate with stalled ribosomes (18). FMRP phosphorylation might be a key regulatory step in activity-dependent protein synthesis (16, 17, 19). The group I mGluR stimulation induces rapid changes in FMRP phosphorylation (dephosphorylation and rephosphorylation) in hippocampus. The changes in FMRP phosphorylation correlate with the expression of synapse-associated protein 90/PSD-95-associated protein 3 (SAPAP3) (1, 16, 17), a postsynaptic scaffolding protein whose mRNA has been identified as an FMRP target (20–22). It further supports the roles of FMRP in synaptic stimulation-induced protein synthesis.
Dopamine (DA), a well known neurotransmitter involved in the synaptic modulation, is important for cognitive functions (23–29). DA transmission is mediated by five G protein-coupled receptors classified as either D1 (D1 and D5 subtypes) or D2 (D2–D4 subtypes) receptors (25, 26, 28, 30–32). DA D1 receptors are positively coupled to protein kinase A through Gs proteins and modulate AMPA GluR1 receptor (33–35). The activation of DA receptors also regulates long term memory formation in a protein synthesis-dependent manner (36–39). Activation of DA D1 receptor stimulates local protein synthesis in the neuronal dendrites (39). Our recent study has identified FMRP as a key messenger for DA modulation in forebrain (40, 41). However, it is still unknown whether FMRP could be involved in protein synthesis induced by DA receptor stimulation.
In the present study, we demonstrate that DA D1 receptor stimulation could induce expression of SAPAP3 and dynamic changes of FMRP phosphorylation in the prefrontal cortex (PFC). DA D1 receptor-induced SAPAP3 expression is protein synthesis-dependent and requires FMRP. Protein phosphatase 2A (PP2A), mammalian target of rapamycin (mTOR), and ribosomal protein S6 kinase (S6K1) are the key signaling molecules in regulation of FMRP phosphorylation and SAPAP3 expression by DA D1 receptors. Knockdown of SAPAP3 did not affect surface expression of AMPA GluR1 receptors induced by D1 receptor stimulation but impaired their subsequent internalization in cultured PFC neurons. Similarly, the subsequent internalization of surface GluR1 was also impaired in Fmr1 knock-out (KO) PFC neurons. Our study thus provides evidence for DA receptor-mediated synapse-associated protein synthesis and reveals one possible molecular mechanism by which FMRP contributes to DA modulation in forebrain.
EXPERIMENTAL PROCEDURES
Animals
Adult male C57Bl/6 mice were used in most of the experiments. Fmr1 wild type (WT) and KO mice of the FVB.129P2-Fmr1tm1Cgr strain were generously provided by Dr. W. T. Greenough (University of Illinois). The mice were generated and maintained as reported before (42–44). All of the mice were housed under a 12:12 light cycle with food and water provided ad libitum. All of the mouse protocols were in accordance with National Institutes of Health guidelines and approved by the Animal Care and Use Committee of the University of Toronto.
Drugs and Antibodies
SKF81997, dihydrexidine, bromocriptine, SCH23390, okadaic acid, and rapamycin were purchased from Tocris Bioscience (Ellisville, MO). Anisomycin, protease inhibitor mixture, and phosphatase inhibitor mixtures 1 and 2 were purchased from Sigma. The anti-FMRP (1C3) and anti-FMRP (7G1) (developed by Dr. Stephen T. Warren) antibodies were from Millipore Corporation (Temecula, CA) and the Developmental Studies Hybridoma Bank (University of Iowa), respectively; anti-PP1 or PP2A antibodies, horseradish peroxidase-linked goat anti-mouse IgG, and goat anti-rabbit IgG antibodies for Western blot were purchased from Millipore. Anti-phospho-mTOR (Ser2448), anti-mTOR, anti-phospho-S6K1 (Thr389), anti-S6K1 antibody, anti-phosphoserine, or anti-phosphotyrosine antibodies were purchased from Cell Signaling Technology (Danvers, MA), and anti-SAPAP3 antibody was a gift from Dr. Guoping Feng (Duke University) or purchased from Prosci Incorporation (Poway, CA). The anti-actin antibody was from Sigma. The small interference RNA (siRNA) was from Thermo Scientific Dharmacon (Lafayette, CO). Transfection reagent for siRNA was from Invitrogen.
Brain Slice Preparation and Drug Treatment
Adult male mice were anesthetized with 1–2% halothane. Transverse slices containing PFC (300 μm) will be prepared using standard methods (40, 42). The slices were slowly brought to a final temperature of 30 °C in ACSF gassed with 95% O2, 5% CO2 and incubated for at least 1 h before experiments. The mouse brain slices were treated with DA receptor agonists for 5 min in ACSF. After treatment, the slices were briefly washed with fresh ACSF and incubated in ACSF without agonists until the time points indicated in the experiments.
Immunoprecipitation
Immunoprecipitation was carried out as reported previously (45). For detection of FMRP phosphorylation, the solubilized protein samples were prepared with lysis buffer and precipitated with 50 μl of protein G-agarose precoupled with anti-FMRP (7G1) antibody for at least 4 h at 4 °C. The reaction mixtures were then washed three times, eluted by boiling in loading buffer, and subjected to Western blot using anti-FMRP (1C3), anti-phosphoserine, or anti-phosphotyrosine antibodies.
Western Blot
Western blot analysis was done as described previously (40, 46, 47). The cultured cortical neurons or brain tissues were harvested and homogenized in lysis buffer containing protease inhibitor mixture and phosphatase inhibitor mixture. Electrophoresis of equal amounts of total protein was done on SDS-polyacrylamide gels. Separated proteins were transferred to polyvinylidene fluoride membranes overnight at 4 °C for Western blot analysis. The membranes were probed with primary antibodies overnight at 4 °C or 4–6 h at room temperature. The membranes were then incubated in the appropriate horseradish peroxidase-coupled secondary antibody for 2 h at room temperature followed by ECL detection of the proteins with Western Lightning Plus ECL substrate (PerkinElmer Life Sciences) according to the manufacturer's instructions. The density of immunoblots was measured using National Institutes of Health ImageJ software.
RT-PCR
The total RNA from the ACC was isolated using the RNAspin mini kit (GE Healthcare). RT-PCR was carried out using procedures reported before (42, 45). The exponential phase of the PCR was determined by amplifying equivalent amounts of input over different number of PCR cycles and by amplifying dilutions of input RNA over the same number of PCR cycles. 25 PCR cycles were used. The primers for Sapap3 used in this experiment were as follows: sense, 5′-ACTATTTGCAGGTGCCGCAAG-3′; and antisense, 5′-GGGCTACCATCTGAGTCTCC-3′. Glyceraldehyde-3-phosphate dehydrogenase was amplified as an internal control by using the primer sets: sense, 5-AACGACCCCTTCATTGAC-3′; and antisense, 5′-TCCACGACATACTCAGCAC-3′. RT-PCR products were electrophoresed on 1.5% agarose gels and visualized under UV light by ethidium bromide staining. The relative density of bands was analyzed by the National Institutes of Health ImageJ program.
PP1 and PP2A Enzyme Activity Assay
PP1 and PP2A activity were measured as reported before (16, 48) using a serine-threonine phosphatase assay kit (Millipore) and following the manufacturer's directions. To immunoprecipitate PP1 or PP2A, rabbit anti-PP1 or mouse anti-PP2A antibodies were added to tissue lysate and gently mixed at 4 °C overnight, followed by 50% protein A-agarose bead slurry and incubation for 2 h at 4 °C. The beads were washed three times with PBS, followed by the enzyme assay according to the manufacturer's manual.
Primary Culture of Prefrontal Cortical Neurons
Prefrontal cortical neurons were prepared from postnatal day 0 mice using methods described previously (40, 46). The prefrontal cortices were dissected, minced, and trypsinized for 15 min using 0.125% trypsin (Invitrogen). The cultures were grown in Neurobasal A medium supplemented with B27 and 2 mm GlutaMax (Invitrogen) and incubated at 37 °C in 95% air, 5% CO2 with 95% humidity. The cultures were used for experiments between days in vitro 14 and 18.
Surface Biotinylation Assay
Surface AMPA receptor subunits were detected by a biotinylation assay, followed by Western blot analysis (40). Briefly, ice-cold PBS (with calcium and magnesium, pH 7.4; Invitrogen) were added to the cultures or slices to prevent receptor internalization. The cultures or slices were incubated in sulfo-NHS-LC-biotin (0.3 mg/ml in cold PBS; Pierce) for 30 min. The surface biotinylation were stopped by removal of the above solution and incubation in 10 mm ice-cold glycine in PBS for 20 min. Then the samples were washed three times with cold PBS and lysed by radioimmune precipitation assay buffer. Biotinylated proteins were precipitated with 100 μl of ImmunoPure immobilized streptavidin (Pierce). Biotinylated proteins were separated on 4–12% SDS-PAGE gel and detected by Western blot using specific antibodies for AMPA receptor subunits.
Data Analysis
The data were presented as the mean values ± S.E. Statistical comparisons were made using the paired or unpaired t test. In all cases, p < 0.05 was considered significant.
RESULTS
Synapse-associated Protein SAPAP3 Expression and FMRP Phosphorylation Mediated by DA Receptors in PFC
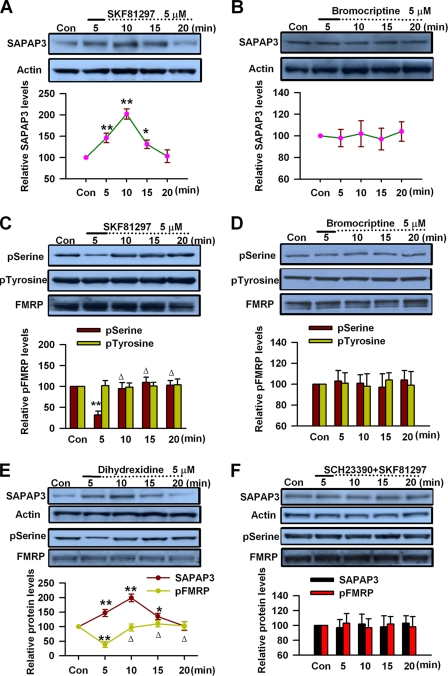
Dopaminergic afferent from the ventral tegmental area plays important roles in PFC cognitive functions, including working memory, reward, and attention (49–53). Activation of DA receptors regulates higher brain functions in a protein synthesis-dependent manner (25, 37, 38, 54). A previous study has shown that DA D1 receptor stimulation induces local protein synthesis in neuronal dendrites (39). Our recent study has shown that FMRP is critical for the DA modulation in forebrain (40, 41). The expression of SAPAP3, a synapse-associated protein that is critical for synaptic function and related to anxiety, obsessive-compulsive disorder, and other psychiatric disorders (21), can be induced by group I mGluR stimulation in hippocampus (1, 16, 17). In this study, to further elucidate the molecular mechanisms for FMRP involvement in DA modulation, we investigated whether DA receptor stimulation could induce synapse-associated protein expression in adult PFC slices. By Western blot, we found that application of DA D1 receptor agonist SKF81297 (5 μm, 5 min) could increase SAPAP3 expression in a time-dependent manner in PFC slices, the increase could be observed immediately after 5 min treatment, and the highest level was reached at the 10-min time point (n = 6; Fig. 1A). Then the SAPAP3 expression started to decrease and reached the control basal levels at the 20-min time point (Fig. 1A). By contrast, application of DA D2 receptor agonist bromocriptine (5 μm, 5 min) did not affect SAPAP3 expression in PFC slices (n = 5; Fig. 1B). These data indicate that DA D1 receptor stimulation can regulate SAPAP3 expression in PFC neurons.
FIGURE 1.
The effect of DA receptor activation on SAPAP3 and FMRP phosphorylation in prefrontal cortical slices. A, DA D1 receptor agonist SKF81297 (5 μm, 5 min) caused a time-dependent increase of SAPAP3 expression in PFC slices. B, DA D2 receptor agonist bromocriptine (5 μm, 5 min) did not affect SAPAP3 expression. C, SKF81297 (5 μm, 5 min) caused time-dependent changes of FMRP phosphorylation at serine residues, whereas the phosphorylation at tyrosine residues was not affected. D, bromocriptine (5 μm, 5 min) did not affect FMRP phosphorylation at serine or tyrosine residues. E, D1 receptor agonist dihydrexidine (5 μm, 5 min) treatment caused time-dependent increase of SAPAP3 expression and changes of FMRP phosphorylation at serine residues in PFC slices. F, D1 receptor agonist SKF81297 (5 μm, 5 min) did not affect SAPAP3 expression or FMRP phosphorylation at serine residues in the presence of D1 receptor antagonist SCH23390 (5 μm); SCH23390 was applied 5 min prior to and during SKF81297 treatment. The slices were treated with DA D1 or D2 receptor agonists for the first 5 min (solid lines) and incubated to the time points as indicated by the dotted lines after the drugs were washed out. Representative Western blots (top panels) and quantification data (bottom panels) are shown. n = 6 mice in A and C; n = 5 mice in B and D–F; *, p < 0.05, and **, p < 0.01, compared with control; Δ, p < 0.01, compared with 5-min time point. The data were calculated as ratios to loading controls and then normalized by the values of control conditions. Con, control.
It has been shown that group I mGluR stimulation induces the changes of FMRP phosphorylation, which correspond to SAPAP3 expression in hippocampus (1, 16, 17). We next investigated whether DA receptor stimulation could affect the phosphorylation status of FMRP in PFC. By immunoprecipitation and Western blot, we found that application of DA D1 receptor agonist SKF81297 (5 μm, 5 min) could cause rapid changes of FMRP phosphorylation at serine residues in PFC slices, the dramatic decrease of FMRP phosphorylation could be observed immediately after 5 min of treatment (n = 6; Fig. 1C), and then FMRP phosphorylation levels rapidly went up to the control levels after the 10-min time point (p < 0.01, compared with the values at the 5-min time point, n = 6; Fig. 1C), closely paralleling the temporal profile of SAPAP3 expression seen in Fig. 1A. However, the phosphorylation levels of FMRP at tyrosine residues were not affected by SKF81297 (5 μm, 5 min) treatment (n = 5; Fig. 1C). By contrast, application of DA D2 receptor agonist bromocriptine (5 μm, 5 min) did not affect FMRP phosphorylation at serine or tyrosine residues in PFC slices (n = 5; Fig. 1D). These results indicate that DA D1 receptor stimulation induces dynamic changes of FMRP phosphorylation, including transient dephosphorylation and subsequent rapid rephosphorylation after stimulation.
To further confirm the roles of DA D1 receptors in SAPAP3 expression and FMRP phosphorylation, we then tested the effect of another DA D1 receptor agonist dihydrexidine in PFC slices. We found that application of dihydrexidine (5 μm, 5 min) could cause the changes in both SAPAP3 expression and FMRP phosphorylation at serine residues in PFC slices, similar to the effects of SKF81297; the dynamic changes of SAPAP3 expression corresponded with the temporal pattern of FMRP phosphorylation after D1 receptor stimulation (Fig. 1E). In the presence of DA D1 receptor antagonist SCH23390 (5 μm, 5 min prior to and during SKF81297 treatment), DA D1 receptor agonist SKF81297 (5 μm, 5 min) did not cause any change in SAPAP3 expression or FMRP phosphorylation at serine residues in PFC slices (n = 5; Fig. 1F). These data further support that DA D1 receptors can regulate SAPAP3 expression and FMRP phosphorylation in PFC neurons. The correspondence between temporal patterns of SAPAP3 expression and FMRP phosphorylation may suggest roles for FMRP in DA-mediated synapse-associated protein expression.
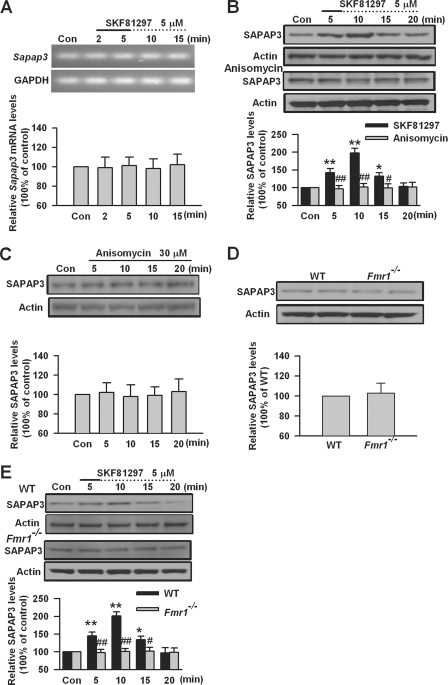
FMRP Is Required for Protein Synthesis-dependent SAPAP3 Expression Mediated by DA D1 Receptors
DA D1 receptors regulate protein synthesis-dependent long term recognition memory and contribute to induction of late phase of LTP in a protein synthesis-dependent manner (25, 38, 39, 54). To investigate whether DA D1 receptor-mediated SAPAP3 expression is protein synthesis-dependent, we investigated the Sapap3 mRNA levels after DA D1 receptor agonist SKF81297 treatment. By RT-PCR, we found that SKF81297 (5 μm, 2 and 5 min) treatment did not affect the Sapap3 mRNA levels in PFC slices (n = 4; Fig. 2A). This suggests that DA D1 receptor stimulation may not regulate SAPAP3 expression at the transcriptional level. We then tested the effect of protein synthesis inhibitor anisomycin on D1 receptor agonist SKF81297-induced SAPAP3 expression in PFC slices. Anisomycin (30 μm) was applied 5 min prior to and during SKF81297 (5 μm, 5 min) treatment. We found that anisomycin could completely block the changes of SAPAP3 expression induced by SKF81297 in PFC slices (p < 0.01, compared with SKF81297 treatment, n = 6; Fig. 2B). However, application of anisomycin (30 μm) did not affect basal expression levels of SAPAP3 in PFC slices (n = 4; Fig. 2C). These results indicate that DA D1 receptor-induced SAPAP3 expression is protein synthesis-dependent.
FIGURE 2.
DA D1 receptor-mediated SAPAP3 expression was protein synthesis-dependent and abolished in PFC of Fmr1−/− mice. A, DA D1 receptor agonist SKF81297 (5 μm, 2 and 5 min) did not affect the levels of Sapap3 mRNA in PFC slices, as shown by RT-PCR. The sizes of PCR products are 142 and 191 bp for Sapap3 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), respectively. Representative gels (top panel) and quantification data (bottom panel) of Sapap3 mRNA are shown. B, the protein synthesis inhibitor anisomycin (30 μm) could block SKF81297-induced (5 μm, 5 min) SAPAP3 expression in PFC slices. Anisomycin was applied 5 min prior to and during SKF81297 treatment. C, anisomycin (30 μm) did not affect SAPAP3 expression in PFC slices. D, the basal SAPAP3 expression was not altered in PFC of Fmr1−/− mice. E, SKF81297 (5 μm) did not affect SAPAP3 expression in PFC of Fmr1−/− mice. The slices were treated with DA D1 receptor agonist for the first 5 min (solid line) and incubated to the time points as indicated by the dotted lines after the drugs were washed out. Representative Western blots (top panel) and quantification data (bottom panel) of protein levels are shown in B–E. *, p < 0.05, and **, p < 0.01, compared with control in B and E; #, p < 0.05, and ##, p < 0.01, compared with SKF81297 treatment in B, compared with WT in E. The data were calculated as ratios to loading controls and then normalized by the values of control conditions in A–C and E and by WT values in D; n = 6 mice for each group in B and D, n = 4 mice in A and C, and n = 5 mice in E. Con, control.
FMRP binds to its mRNA targets in brain and regulates local protein synthesis (1, 2, 13, 55). It is necessary for neurotransmitter-induced protein translation at synapses (1, 56). SAPAP3 has been identified as a FMRP ligand (17, 20). Can FMRP be involved in SAPAP3 synthesis mediated by DA D1 receptors? To test this, we investigated the effect of DA D1 receptor stimulation on SAPAP3 expression in PFC slices of Fmr1 KO mice. We found that the basal levels of SAPAP3 was not changed in PFC of Fmr1 KO mice compared with WT mice (n = 6; Fig. 2D). Application of DA D1 receptor agonist SKF81297 (5 μm, 5 min) could increase SAPAP3 expression in PFC slices of WT mice but did not affect SAPAP3 expression in PFC slices of Fmr1 KO mice (p < 0.01, compared with WT, n = 5; Fig. 2E). These data indicate that a lack of FMRP can block D1 receptor agonist-induced SAPAP3 expression, suggesting that FMRP is critical for DA D1 receptor-mediated SAPAP3 synthesis in PFC neurons.
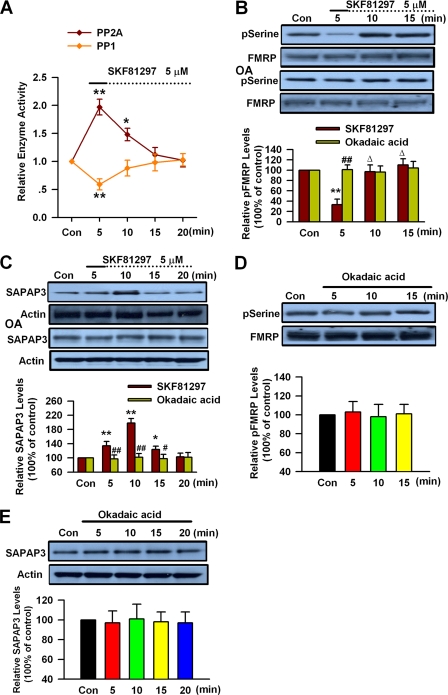
PP2A and Regulation of FMRP Phosphorylation and SAPAP3 Expression by DA D1 Receptor
Protein phosphatases have been shown to play key roles in mGluR-mediated SAPAP3 expression through modulation of FMRP phosphorylation (16, 17). The PP2A has been identified as a major FMRP phosphatase in primary neurons (16). DA D1 receptor activation can regulate PP1 and PP2A activity via protein kinase A and dopamine and cAMP-regulated phosphoprotein (DARPP-32) (57–60). To investigate the roles of protein phosphatases in modulation of FMRP phosphorylation and SAPAP3 expression by DA D1 receptors, we performed PP1/2A enzyme assay in PFC slices after D1 receptor agonist treatment. We found that application of DA D1 receptor agonist SKF81297 (5 μm, 5 min) could transiently activate PP2A, whereas PP1 activity was slightly inhibited in PFC slices (n = 5; Fig. 3A). These data indicate that DA D1 receptor stimulation can activate PP2A in PFC neurons.
FIGURE 3.
PP2A is involved in DA-mediated changes of FMRP phosphorylation and SAPAP3 expression. A, measurement of PP2A and PP1 activity. D1 agonist SKF81297 (5 μm, 5 min) caused transient activation of PP2A and slight inhibition of PP1 in PFC slices. B and C, the PP2A inhibitor okadaic acid (OA, 1 nm) abolished the changes of FMRP phosphorylation (B) and SAPAP3 expression (C) induced by SKF81297 (5 μm, 5 min). Okadaic acid was applied 5 min prior to and during SKF81297 treatment. D and E, okadaic acid (1 nm) did not affect the basal FMRP phosphorylation (D) and SAPAP3 expression (E) in PFC slices. The slices were treated with DA D1 receptor agonist for the first 5 min (solid lines) and incubated to the time points as indicated by the dotted lines after the drugs were washed out. Representative Western blots (top panel) and quantification data (bottom panel) are shown in B–E. *, p < 0.05, and **, p < 0.01, compared with control in A–C; #, p < 0.05, and ##, p < 0.01, compared with SKF81297 treatment in B and C; Δ, p < 0.01, compared with 5-min time point in B. The data were normalized by control values in A. For Western blot, the data were calculated as ratios to loading controls and then normalized by the values of control conditions in B–E. n = 5 mice for each group in A–C, and n = 4 mice in D and E. Con, control.
To further characterize the roles of PP2A in DA D1 receptor stimulation-induced changes of FMRP phosphorylation and SAPAP3 expression, we then tested the effect of protein phosphatase inhibitor okadaic acid, which is relatively specific for PP2A at a concentration of 0.2–1 nm (16, 61). We found that application of okadaic acid (1 nm) almost completely blocked the changes of FMRP phosphorylation and SAPAP3 expression induced by D1 receptor agonist SKF81297 (5 μm, 5 min) treatment (n = 5; Fig. 3, B and C). However, application of okadaic acid (1 nm) itself did not affect the levels of basal FMRP phosphorylation or SAPAP3 expression in PFC slices (n = 4; Fig. 3, D and E). These data indicate that PP2A activation is critically involved in DA D1 receptor-regulated FMRP phosphorylation and SAPAP3 expression in PFC neurons.
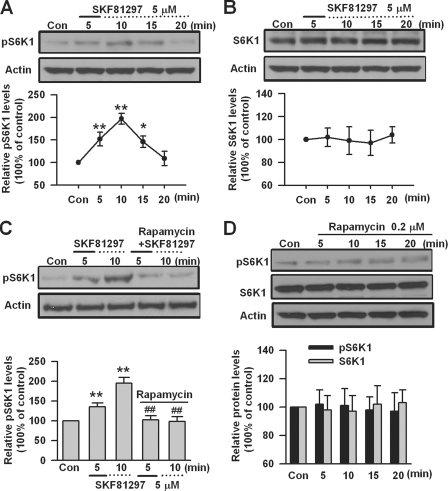
Activation of S6K1 by DA D1 Receptors
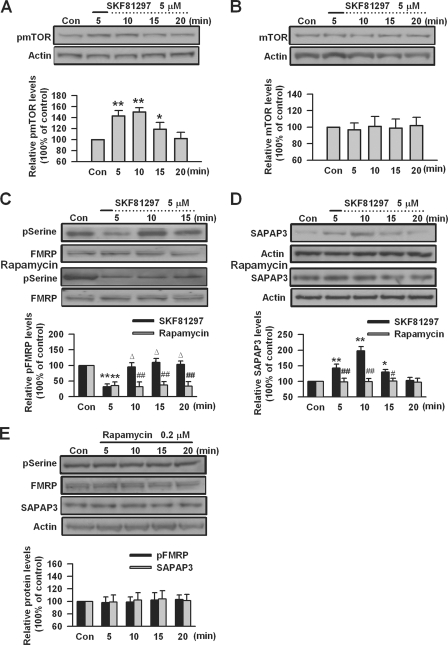
S6K1, which is downstream of mTOR in mTOR signaling pathway (62–65), has been identified as the key kinase for FMRP during group I mGluR stimulation (1, 17). S6K1 can be activated through its phosphorylation by mTOR (17, 65, 66). To investigate whether S6K1 is involved in DA D1 receptor-mediated effects, we measured the phosphorylation of S6K1 (Thr389) in PFC slices by Western blot. We found that DA D1 receptor agonist SKF81297 (5 μm, 5 min) caused a rapid increase in the phosphorylation of S6K1; the highest phosphorylation level was seen at the 10-min time point (n = 5; Fig. 4A). However, application of SKF81297 (5 μm, 5 min) did not affect the total protein levels of S6K1 in PFC slices (n = 5; Fig. 4B). These data indicate that DA D1 receptor stimulation activates S6K1 in PFC neurons.
FIGURE 4.
The activation of S6K1 by DA D1 receptors. A, DA D1 agonist SKF81297 (5 μm, 5 min) increased the phosphorylation of S6K1 (Thr389) in a time-dependent manner. B, SKF81297 (5 μm, 5 min) did not affect the basal levels of S6K1 in PFC slices. C, the mTOR inhibitor rapamycin (0.2 μm) could block SKF81297-induced S6K1 phosphorylation (Thr389) in PFC slices. Rapamycin was applied 5 min prior to and during SKF81297 (5 μm, 5 min) treatment. D, application of rapamycin (0.2 μm) did not affect the phosphorylation or basal expression of S6K1 in PFC slices. The slices were treated with DA D1 receptor agonist for the first 5 min (solid lines) and incubated to the time points as indicated by the dotted lines after the drugs were washed out. Representative Western blots (top) and quantification data (bottom) are shown in A–D. *, p < 0.05, and **, p < 0.01, compared with control in A and C; #, p < 0.05, and ##, p < 0.01, compared with SKF81297 treatment in C. The data were calculated as ratios to loading controls and then normalized by the values of control conditions. n = 5 mice for each group in A–C, and n = 4 mice for each group in D. Con, control.
To determine the roles of mTOR in S6K1 activation by DA D1 receptors, we tested the effect of mTOR inhibitor rapamycin (16, 65, 67, 68) in PFC slices. We found that application of rapamycin (0.2 μm) 5 min prior to and during D1 receptor agonist SKF81297 (5 μm, 5 min) treatment, could block SKF81297-induced S6K1 phosphorylation in PFC slices (n = 5; Fig. 4C). However, application of rapamycin (0.2 μm) itself did not affect the phosphorylation or basal expression of S6K1 in PFC slices (n = 4; Fig. 4D). These data indicate that activation of S6K1 by DA D1 receptors is mTOR-dependent.
S6K1 and DA D1 Receptor-mediated Effects on FMRP and SAPAP3
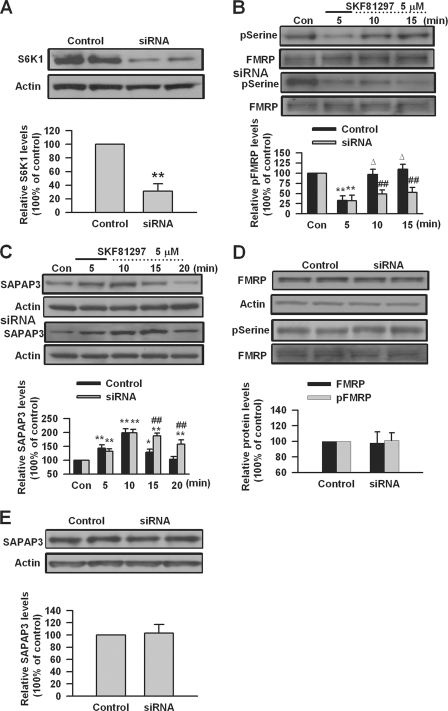
S6K1 has been shown to directly phosphorylate FMRP during group I mGluR stimulation (1, 17). To investigate whether S6K1 could be involved in FMRP phosphorylation by DA D1 receptor stimulation, we tested the effect of knockdown of S6K1 in cultured PFC neurons. We found that transfection of S6K1 siRNA could knock down the expression of S6K1 in cultured PFC neurons (31 ± 10% of control levels, p < 0.01, compared with control, n = 4; Fig. 5A). Knockdown of S6K1 by siRNA did not affect dephosphorylation of FMRP but blocked the FMRP rephosphorylation after DA D1 receptor agonist SKF81297 (5 μm, 5 min) treatment in PFC neurons (n = 5; Fig. 5B). We also found that knockdown of S6K1 did not affect the expression or phosphorylation status of FMRP in cultured PFC neurons at the basal condition (n = 4; Fig. 5D). These data indicate that S6K1 is required for FMRP phosphorylation after DA D1 receptor stimulation and suggest that S6K1 might be a key kinase for modulation of FMRP phosphorylation by DA D1 receptors in PFC neurons.
FIGURE 5.
The effect of S6K1 knockdown on FMRP phosphorylation and SAPAP3 expression mediated by DA D1 receptors. A, S6K1 siRNA could reduce S6K1 expression in cultured PFC neurons. B, transfection of S6K1 siRNA did not affect FMRP dephosphorylation at serine residues but blocked the subsequent rephosphorylation after DA D1 receptor agonist SKF81297 (5 μm, 5 min) treatment. C, S6K1 siRNA did not affect the up-regulation of SAPAP3 but could impair the decrease of up-regulated SAPAP3 after SKF81297 (5 μm, 5 min) treatment in cultured PFC neurons. D and E, S6K1 siRNA did not affect basal expression, phosphorylation of FMRP (D), and SAPAP3 expression (E). siRNA was transfected into cultured PFC neurons (day in vitro 16) 40 h before experiments. PFC neurons were treated with DA D1 receptor agonist for the first 5 min (solid lines) and incubated to the time points as indicated by the dotted lines after the drugs were washed out. Representative Western blots (top panels) and quantification data (bottom panels) are shown in A–E. *, p < 0.05, and **, p < 0.01, compared with control siRNA in A–C; ##, p < 0.01, compared with control siRNA in B and C; Δ p < 0.01, compared with 5-min time point in B. The data were calculated as ratios to loading controls and then normalized by the values of control conditions. n = 4 dishes for each group in A, D, and E, and n = 5 dishes in B and C. Con, control.
We next investigated the roles of S6K1 in DA D1 receptor-mediated SAPAP3 expression in PFC neurons. We found that knockdown of S6K1 by siRNA did not affect the up-regulation of SAPAP3 but blocked the subsequent decrease of up-regulated SAPAP3 after DA D1 receptor agonist SKF81297 (5 μm, 5 min) treatment in cultured PFC neurons (n = 5; Fig. 5C). Knockdown of S6K1 did not affect the basal expression of SAPAP3 in cultured PFC neurons (n = 4; Fig. 5E). These data indicate that S6K1 is involved in the dynamic regulation of SAPAP3 and suggest that S6K1 may act as a SAPAP3 translational repressor through FMRP phosphorylation after DA D1 receptor stimulation.
mTOR and SAPAP3 Expression Mediated by DA D1 Receptors
The mTOR signaling pathway has been shown to regulate protein synthesis-dependent changes in synaptic strength (67–70). Previous studies suggest that DA D1 receptors are linked to the mTOR signaling pathway (38, 71). The activity of mTOR is related to its phosphorylation status (16, 67). To investigate the role of mTOR in DA D1 receptor-mediated SAPAP3 expression, we measured the phosphorylation of mTOR (Ser2448) after DA D1 receptor agonist stimulation in PFC slices. We found that DA D1 receptor agonist SKF81297 (5 μm, 5 min) could cause dynamic changes in the phosphorylation of mTOR, the mTOR phosphorylation was increased immediately after SKF81297 treatment, the highest levels were seen at 10 min, and then the phosphorylation decreased and reached basal levels at the 20-min time point (n = 5; Fig. 6A). However, the basal expression levels of mTOR were not affected by SKF81297 (5 μm, 5 min) treatment (n = 4; Fig. 6B). These data indicate that DA D1 receptor agonist stimulation can activate mTOR in PFC neurons.
FIGURE 6.
mTOR is involved in SAPAP3 expression mediated by DA D1 receptors. A, DA D1 agonist SKF81297 (5 μm, 5 min) increased the phosphorylation of mTOR (Ser2448) in a time-dependent manner. B, SKF81297 (5 μm, 5 min) did not affect the basal levels of mTOR in PFC slices. C, the mTOR inhibitor rapamycin (0.2 μm) did not affect FMRP dephosphorylation at serine residues but blocked the subsequent rephosphorylation after SKF81297 (5 μm, 5 min) treatment. D, rapamycin (0.2 μm) could block SKF81297-induced SAPAP3 expression in PFC slices. Rapamycin was applied 5 min prior to and during SKF81297 treatment. E, rapamycin (0.2 μm) did not affect the basal levels of FMRP phosphorylation and SAPAP3 expression in PFC slices. The slices were treated with DA D1 receptor agonist for the first 5 min (solid lines) and incubated to the time points as indicated by the dotted lines after the drugs were washed out. Representative Western blots (top panels) and quantification data (bottom panels) are shown in A–E. *, p < 0.05, and **, p < 0.01, compared with control in A, C, and D; #, p < 0.05, and ##, p < 0.01, compared with SKF81297 treatment in C and D; Δ, p < 0.01, compared with the 5-min time point in C. The data were calculated as ratios to loading controls and then normalized by the values of control conditions. n = 5 mice in A, C, and D, and n = 4 mice in B and E. Con, control.
To further elucidate the roles of mTOR in DA D1 receptor-mediated changes of FMRP phosphorylation and SAPAP3 expression, we tested the effect of mTOR inhibitor rapamycin in PFC slices. We found that application of rapamycin (0.2 μm) did not affect the dephosphorylation of FMRP but blocked FMRP rephosphorylation after DA D1 receptor agonist SKF81297 (5 μm, 5 min) treatment in PFC slices (n = 5; Fig. 6C), similar to what was observed when S6K1 was knocked down in cultured PFC neurons. In the presence of rapamycin (0.2 μm), D1 receptor agonist SKF81297 (5 μm, 5 min) did not cause any change of SAPAP3 expression (p < 0.01 or p < 0.05, compared with SKF81297 treatment, n = 5; Fig. 6D). In addition, application of rapamycin (0.2 μm) did not affect FMRP phosphorylation and SAPAP3 expression in PFC slices at the basal condition (p > 0.05, compared with control, n = 4; Fig. 6E). The data indicate that inhibition of mTOR can block SAPAP3 expression caused by DA D1 receptor stimulation, whereas its effect on FMRP phosphorylation is similar to that of S6k1 knockdown. These findings suggest that in addition to its function through S6K1, mTOR is critically involved in SAPAP3 synthesis that is caused by PP2A-catalyzed dephosphorylation of FMRP.
Inhibition of mTOR in S6K1 Knockdown PFC Neurons
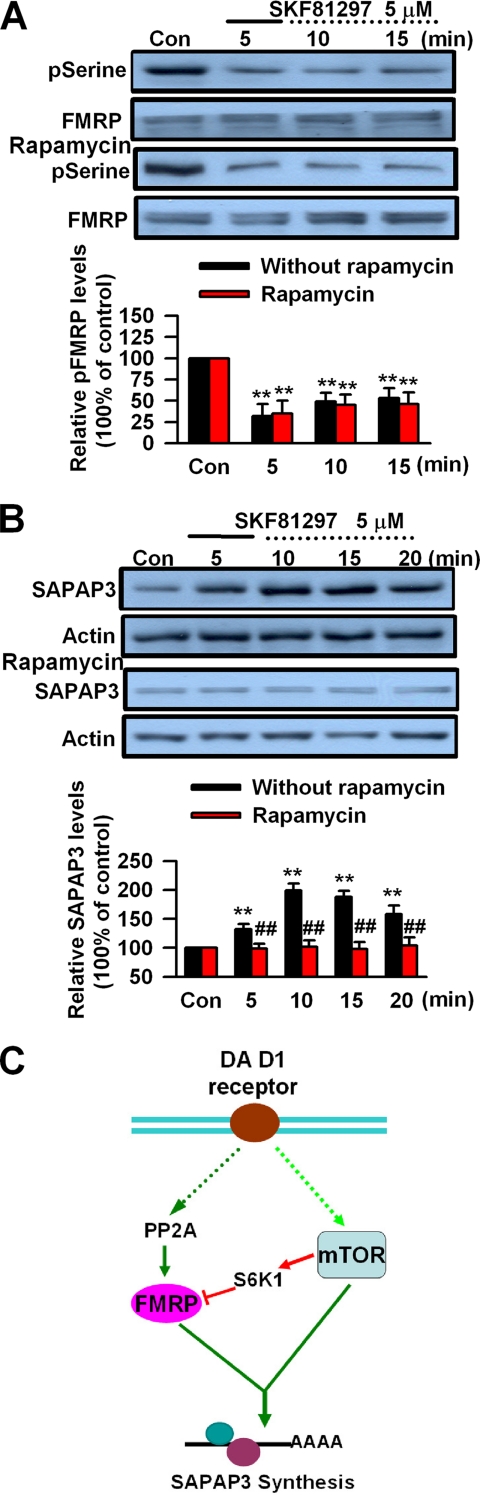
To further characterize the roles of mTOR in SAPAP3 synthesis, we applied the mTOR inhibitor rapamycin to S6K1 knockdown PFC neurons. We found that application of rapamycin did not affect the phosphorylation state of FMRP in S6K1 knockdown PFC neurons (n = 5 dishes; Fig. 7A). However, rapamycin could block the initial increase of SAPAP3 expression caused by DA D1 receptor stimulation, which actually was not affected by S6K1 knockdown in cultured PFC neurons (n = 5 dishes; Fig. 7B). These results further support that mTOR and S6K1 contribute differently to SAPAP3 synthesis, although inhibition of mTOR and knockdown of S6K1 have shown similar effects on phosphorylation of FMRP after dopamine D1 receptor stimulation. Taken together, we have identified PP2A, mTOR, and S6K1 as the key signaling molecules for DA D1 receptor modulation of FMRP phosphorylation and SAPAP3 expression (Fig. 7C).
FIGURE 7.
Pharmacological inhibition of mTOR in S6K1 knockdown PFC neurons. A, the mTOR inhibitor rapamycin (0.2 μm) did not affect FMRP phosphorylation at serine residues in S6K1 knockdown-cultured PFC neurons treated with SKF81297 (5 μm, 5 min). B, rapamycin (0.2 μm) blocked SKF81297-induced (5 μm, 5 min) SAPAP3 expression in S6K1 knockdown-cultured PFC neurons. C, a model of signaling pathways for DA D1 receptor-mediated SAPAP3 expression. Both FMRP and mTOR are required for initiation of SAPAP3 synthesis. PP2A dephosphorylates FMRP, and dephosphorylated FMRP is involved in the initiation of SAPAP3 synthesis. The mTOR also activates S6K1, which phosphorylates FMRP, and phosphorylated FMRP acts as a repressor for SAPAP3 synthesis. PFC neurons were treated with DA D1 receptor agonist for the first 5 min (solid lines) and incubated to the time points as indicated by the dotted lines after the drugs were washed out. Representative Western blots (top panels) and quantification data (bottom panels) are shown in A and B. **, p < 0.01, compared with control in A and B; ##, p < 0.01, compared with S6K1 siRNA only in B. The data were calculated as ratios to loading controls and then normalized by the values of control conditions. n = 5 dishes for each group in A and B. Con, control.
SAPAP3 and Surface AMPA GluR1 Receptors after DA D1 Receptor Activation
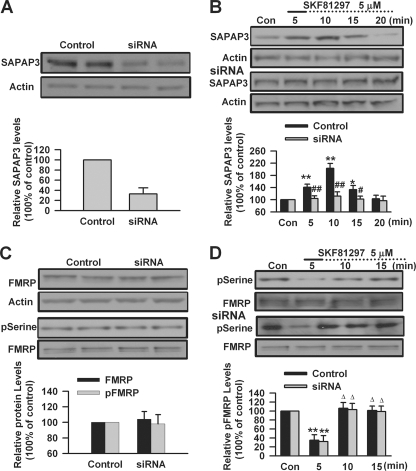
DA D1 receptors are positively coupled to protein kinase A signaling pathway through Gs proteins and regulate the phosphorylation and trafficking of AMPA GluR1 receptor (33–35, 72, 73). Our recent study has shown that DA D1 receptor stimulation can increase surface expression of GluR1 in cultured PFC neurons; the surface expression of GluR1 triggered by D1 receptor agonist is impaired in Fmr1 KO PFC neurons (40). SAPAP3, a postsynaptic scaffolding protein at excitatory synapses, plays key roles in regulating synaptic function and plasticity (21, 22). Can SAPAP3 be involved in surface expression of GluR1 triggered by DA D1 receptor stimulation? To test this, we used SAPAP3 siRNA to knock down the expression of SAPAP3 in cultured PFC neurons. We found that transfection of SAPAP3 siRNA could knock down the expression of SAPAP3 in cultured PFC neurons (33 ± 12% of control levels, p < 0.01, compared with control, n = 4; Fig. 8A). SAPAP3 siRNA could block DA D1 receptor agonist SKF81297-induced (5 μm, 5 min) changes of SAPAP3 expression that were observed in PFC neurons transfected with control siRNA (compared with control siRNA, n = 5; Fig. 8B). However, it did not affect the basal levels of FMRP expression and phosphorylation in cultured PFC neurons (n = 4; Fig. 8C). In addition, the changes of FMRP phosphorylation caused by SKF81297 (5 μm, 5 min) treatment were not affected by knockdown of SAPAP3 (n = 5; Fig. 8D). These data suggest that FMRP could be an upstream regulator for DA D1 receptor-mediated SAPAP3 expression.
FIGURE 8.
Knockdown of SAPAP3 in cultured PFC neurons. A, SAPAP3 siRNA could knock down SAPAP3 expression in cultured PFC neurons. B, SAPAP3 siRNA could block DA D1 receptor agonist SKF81297-induced (5 μm, 5 min) changes of SAPAP3 expression that were observed in cultured PFC neurons transfected with control siRNA. C, transfection of SAPAP3 siRNA did not affect the basal expression and phosphorylation (serine residues) levels of FMRP. D, SKF81297 (5 μm, 5 min) could induce the changes of FMRP phosphorylation in cultured PFC neurons; Transfection of SAPAP3 siRNA did not affect the changes of FMRP phosphorylation caused by SKF81297 treatment. siRNA was transfected into cultured PFC neurons (day in vitro 16) 40 h before experiments. PFC neurons were treated with DA D1 receptor agonist for the first 5 min (solid lines) and incubated to the time points as indicated by the dotted lines after the drugs were washed out. Representative Western blots (top) and quantification data (bottom) are shown in A–D. *, p < 0.05, and **, p < 0.01, compared with control in A, B, and D; #, p < 0.05, and ##, p < 0.01, compared with control siRNA in B; Δ, p < 0.01, compared with 5-min time point in D. The data were calculated as ratios to loading controls and then normalized by the values of control conditions. n = 4 dishes in A and C, and n = 5 dishes for each group in B and D. Con, control.
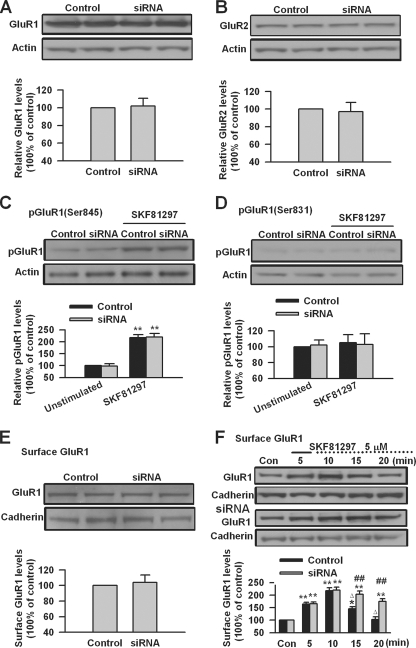
We next investigated the effect of SAPAP3 knockdown on AMPA receptors in cultured PFC neurons. We found that SAPAP3 knockdown did not affect the total levels of both GluR1 and GluR2 nor the phosphorylation and surface expression of GluR1 in cultured PFC neurons at the basal condition (n = 4; Fig. 9, A–E). We then compared the surface expression of GluR1 after DA D1 receptor stimulation. We found that DA D1 receptor agonist SKF81297 (5 μm, 5 min) treatment can cause a similar increase of surface GluR1 in control and SAPAP3 siRNA transfected PFC neurons; the highest levels of surface GluR1 could be observed at the 10-min time point (n = 6; Fig. 9F). After 10 min, the surface GluR1 started to decrease and reached basal levels at the 20-min time point in control siRNA transfected PFC neurons (p < 0.01, compared with the 10-min time point, n = 6; Fig. 9F). However, the decrease of surface GluR1 was impaired in SAPAP3 siRNA transfected PFC neurons (p < 0.01, compared with control siRNA, n = 6; Fig. 9F). These results indicate that SAPAP3 may not be required for the surface delivery of GluR1 expression but could be involved in subsequent internalization of surface GluR1 after DA D1 receptor stimulation.
FIGURE 9.
The effect of SAPAP3 knockdown on AMPA receptors after D1 receptor activation in PFC neurons. A and B, transfection of SAPAP3 siRNA did not affect the basal levels of AMPA receptor GluR1 (A) or GluR2 (B) subunits. C and D, SAPAP3 siRNA did not affect the basal levels of GluR1 phosphorylation at residues Ser845 (C) or Ser831 (D) in cultured PFC neurons. E, SAPAP3 siRNA did not affect basal GluR1 surface expression in cultured PFC neurons as measured by biotinylation assay. F, SAPAP3 siRNA did not affect the increase of surface GluR1 but impaired the decrease of the increased surface GluR1 after SKF81297 (5 μm, 5 min) treatment in cultured PFC neurons. siRNA was transfected into cultured PFC neurons (day in vitro 16) 40 h before experiments. PFC neurons were treated with DA D1 receptor agonist for the first 5 min (solid lines) and incubated to the time points as indicated by the dotted lines after the drugs were washed out. Representative Western blots (top) and quantification data (bottom) are shown in A–F. *, p < 0.05, and **, p < 0.01, compared with control in C and F; ##, p < 0.01, compared with control siRNA in F; Δ, p < 0.01, compared with the 10-min time point in F. The data were calculated as ratios to loading controls and then normalized by the values of control conditions. n = 4 dishes in A–E, and n = 6 dishes for each group in F. Con, control.
Requirement of FMRP for Subsequent Internalization of Surface GluR1
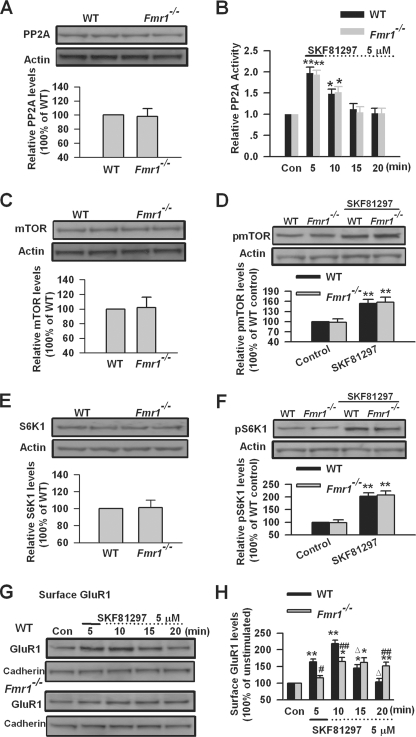
DA D1 receptor-mediated SAPAP3 expression requires FMRP; PP2A, mTOR, and S6K1 are the key signaling molecules for FMRP involvement in this process. To further characterize the roles of FMRP in these signaling pathways, we investigated the expression and activity of PP2A, mTOR, and S6K1 in PFC slices from Fmr1 KO mice. By Western blot, we found that the total levels of PP2A, mTOR, and S6K1 were not altered in PFC slices from Fmr1 KO mice compared with that of WT mice (n = 6; Fig. 10, A, C, and E). DA D1 receptor agonist SKF81297 (5 μm, 5 min) treatment can cause a similar increase of PP2A activity in WT and Fmr1 KO PFC slices n = 5; Fig. 10B). In addition, no difference was found in the phosphorylation of mTOR or S6K1 at the basal condition or after SKF81297 (5 μm, 5 min) treatment between WT and Fmr1 KO PFC slices (n = 6; Fig. 10, D and F). The data indicate that the lack of FMRP does not affect PP2A, mTOR, and S6K1 in DA D1 receptor-mediated SAPAP3 synthesis in PFC neurons. It also suggests that activation of pp2A and mTOR signaling are not sufficient to induce SAPAP3 synthesis when FMRP is absent, although both pp2A and mTOR are required for SAPAP3 synthesis caused by DA D1 receptor stimulation.
FIGURE 10.
AMPA receptor GluR1 surface expression after DA D1 receptor activation in Fmr1 KO PFC neurons. A and B, the basal PP2A levels (A) and PP2A activation (B) after SKF81297 (5 μm, 5 min) treatment were not altered in a PFC slice of Fmr1 KO mice. C and D, the basal mTOR levels (C) and mTOR phosphorylation (D) after SKF8129 treatment were not altered in a PFC slice of Fmr1 KO mice. E and F, the basal S6K1 levels (E) and S6K1 phosphorylation (Thr389) (F) after SKF81297 treatment were not altered in a PFC slice of Fmr1 KO mice. G and H, the increase of surface GluR1was partially blocked, and the decreases of the increased surface GluR1 levels were impaired after SKF81297 (5 μm, 5 min) treatment in PFC slices of Fmr1 KO mice compared with WT mice, as measured by biotinylation assay. The slices were treated with DA D1 receptor agonist for the first 5 min (solid lines) and incubated to the time points as indicated by the dotted lines after the drugs were washed out. *, p < 0.05, and **, p < 0.01, compared with control in B, D, F, and H; ##, p < 0.01, compared with WT in H; Δ, p < 0.01, compared with 10-min time point in H. The data were calculated as ratios to loading controls and then normalized by the values of control conditions. n = 6 mice in A and C–F, and n = 5 mice for each group in B, G, and H. Con, control.
Because DA D1 receptor-mediated SAPAP3 expression, which requires FMRP, could be involved in subsequent surface GluR1 internalization after DA D1 receptor stimulation, we next determined surface expression of GluR1 in PFC slices of Fmr1 KO mice. As we have reported before (40), we found that the increase of surface GluR1 triggered by DA D1 receptor agonist SKF81297 (5 μm, 5 min) was impaired in Fmr1 KO PFC slices (n = 5; Fig. 10, G and H). The surface GluR1 started to decrease after 10 min and reached basal levels at the 20-min time point in WT PFC neurons (p < 0.01, compared with 10-min time point, n = 5; Fig. 10, G and H). However, the decrease of surface GluR1 was impaired in Fmr1 KO PFC slices (n = 5; Fig. 10, G and H). These results indicate that the lack of FMRP not only affects the initial surface GluR1 expression but also impairs subsequent internalization of surface GluR1 after DA D1 receptor stimulation.
DISCUSSION
Our previous studies suggest that FMRP is required for the physiological function of cingulate cortex (42, 43). Recently, we have identified FMRP as a key messenger for DA modulation in forebrain neurons (40). Here, we provided evidence that FMRP is involved in DA D1 receptor-mediated synthesis of SAPAP3, a synapse-associated protein. DA D1 receptor stimulation may control the abundance of SAPAP3 through dynamic modulation of FMRP phosphorylation in PFC neurons. FMRP is not only required for the surface expression of AMPA GluR1 receptors but also necessary for their subsequent internalization after surface delivery by D1 receptor stimulation. Regulation of SAPAP3 abundance could be the key mechanism for FMRP involvement in GluR1 receptor internalization after D1 receptor stimulation.
FMRP- and DA-mediated SAPAP3 Expression
The DA system can modulate synaptic plasticity and higher brain functions in a protein synthesis-dependent manner (26, 36–38, 54). Dopaminergic stimulation induces protein synthesis in neuronal dendrites (39). In this study, we found that DA D1 receptor stimulation can induce SAPAP3 synthesis in PFC neurons. These data provide direct evidence that DA receptors can regulate the synthesis of synapse-associated protein.
FMRP regulates local protein synthesis in dendritic spines (2, 11, 74). Previous studies suggested that FMRP phosphorylation might be a key regulator for activity-dependent protein synthesis (16–18). We found that DA D1 receptor stimulation could cause dynamic changes of FMRP phosphorylation at serine residues in PFC slices. The changes of FMRP phosphorylation correspond to the temporal pattern of SAPAP3 after D1 receptor stimulation. This suggests that FMRP could be involved in DA mediated synapse-associated protein expression. Our recent study has shown that FMRP plays key roles for DA modulation in forebrain neurons (40). In this study, we found that DA D1 receptor-mediated SAPAP3 expression was abolished in Fmr1 KO PFC neurons. It indicates the requirement of FMRP for DA D1 receptor-induced synapse-associated protein synthesis.
Key Signaling Molecules for FMRP Linking DA Receptors to SAPAP3
Dynamic changes of FMRP phosphorylation have been found during group I mGluR-mediated SAPAP3 expression (16). PP2A, a major FMRP phosphatase, is critical for FMRP dephosphorylation after group I mGluR stimulation (16, 17). DA D1 receptor activation is believed to regulate PP1 and PP2A activity via protein kinase A and DARPP-32 (57, 59, 60). Here, we found that application of D1 receptor agonist SKF81297 could transiently activate PP2A, whereas PP1 activity was slightly inhibited in PFC slices. Pharmacological inhibition of PP2A almost completely blocked the changes of FMRP phosphorylation and SAPAP3 expression induced by D1 receptor agonist treatment. The data indicate that DA D1 receptor stimulation can activate PP2A in PFC neurons; PP2A is the key protein phosphatase for FMRP dephosphorylation after DA D1 receptor stimulation. It also suggests that PP2A-mediated FMRP dephosphorylation might be a key step for induction of SAPAP3 synthesis by D1 receptor stimulation in PFC neurons.
S6K1, which is downstream of mTOR, can be activated by mTOR during group I mGluR activation (17, 62, 63, 66, 75). FMRP can be directly phosphorylated by S6K1 (17). S6K1 has been identified as a major FMRP kinase during group I mGluR stimulation (1, 17). In this study, we have shown that S6K1 can be activated by DA D1 receptor stimulation, and activation of S6K1 by DA D1 receptors is mTOR-dependent. Knockdown of S6K1 did not affect the dephosphorylation of FMRP after DA D1 receptor stimulation but blocked the phosphorylation of FMRP after its dephosphorylation in cultured PFC neurons. Interestingly, the up-regulation of SAPAP3 after DA D1 receptor stimulation was not affected, whereas the decrease of up-regulated SAPAP3 was impaired by knockdown of S6K1 in cultured PFC neurons. The data indicate that S6K1 is the key kinase for FMRP phosphorylation triggered by DA D1 receptor activation. It also suggests that S6K1 via phosphorylating FMRP plays a critical role in the regulation of SAPAP3 abundance by DA D1 receptors.
DA D1 receptors are linked to mTOR (38, 71), which regulates protein synthesis-dependent changes in synaptic plasticity (67, 69, 70). The mTOR has been shown to be involved in FMRP phosphorylation and SAPAP3 expression triggered by group I mGluRs (16, 17). Because DA D1 receptor stimulation could induce changes of FMRP phosphorylation and mediate SAPAP3 synthesis, we next investigated the roles of mTOR in this process. We found that DA D1 receptor stimulation could activate mTOR, as shown by the increased phosphorylation of mTOR in PFC slices. Pharmacological inhibition of mTOR could block the phosphorylation of FMRP after its dephosphorylation in PFC slices, which is similar to what was observed in cultured PFC neurons when S6K1 was knocked down. Pharmacological inhibition of mTOR also completely blocked DA D1 receptor agonist-induced up-regulation of SAPAP3 expression, which was actually not affected by knockdown of S6K1 in cultured PFC neurons. Theses results indicate that mTOR and S6K1 contribute differently to SAPAP3 synthesis induced by DA D1 receptor stimulation.
It is well known that mTOR plays critical roles in protein synthesis (1, 70). The mTOR, as a kinase, phosphorylates eukaryotic initiation factor 4E-binding protein 1; phosphorylation of eukaryotic initiation factor 4E-binding protein 1 causes it to dissociate from eukaryotic initiation factor 4E and activates cap-dependent translation initiation (1, 70, 76). Inhibition of mTOR with rapamycin has been shown to block activity-dependent or brain-derived neurotrophic factor (BDNF)-induced dendritic protein synthesis (77–79). This is consistent with our finding that rapamycin blocked SAPAP3 expression triggered by D1 receptor activation. Although we cannot rule out the possibility that S6K1 is also involved in SAPAP3 synthesis triggered by DA D1 receptor stimulation, our data do show that knockdown of S6K1 by siRNA blocked the subsequent decrease of up-regulated SAPAP3 but did not affect the initial increase of SAPAP3 expression in this process. It could be possible that the function of S6K1 in SAPAP3 synthesis might be compensated by other kinases after S6K1 is knocked down. This was also supported by the study in S6K1 KO mice, which actually showed that SAPAP3 was slightly elevated in the hippocampi of S6K1 KO mice (17), suggesting that S6K1 may not be necessary for initial SAPAP3 synthesis.
Although mTOR plays a critical role in SAPAP3 synthesis, our data actually indicate that FMRP and mTOR are both required for SAPAP3 synthesis triggered by DA D1 receptor stimulation, instead of a direct route from mTOR to SAPAP3 synthesis that does not go through FMRP (see Fig. 7C for model). Our results from Fmr1 KO mice suggest that FMRP is definitely needed for SAPAP3 expression triggered by DA D1 receptor stimulation; activation of mTOR is not sufficient to induce SAPAP3 expression without FMRP, because the mTOR and its activation by D1 receptor stimulation are actually not altered in PFC neurons of Fmr1 KO mice. Meanwhile, activation of mTOR is also necessary for SAPAP3 synthesis, because dephosphorylation of FMRP is unable to cause SAPAP3 expression when DA D1 receptors were stimulated in the presence of mTOR inhibitor rapamycin. The dissociation between the FMRP phosphorylation state and SAPAP3 synthesis in the presence of rapamycin reflects the critical roles of mTOR in SAPAP3 synthesis induced by DA D1 receptor stimulation. To further elucidate the detailed mechanisms by which FMRP and mTOR are involved in SAPAP3 synthesis induced by DA D1 receptor stimulation will be interesting for future studies.
FMRP, SAPAP3, and DA Modulation of AMPA Receptors
DA D1 receptor stimulation induces surface expression of AMPA GluR1 receptors in cultured PFC neurons (33, 40). Because SAPAP3 synthesis can be induced by DA D1 receptor stimulation, we investigated its role in surface expression of AMPA GluR1 receptors. We found that knockdown of SAPAP3 did not affect DA D1 receptor agonist-induced increase of surface GluR1 cultured PFC neurons. However, the decrease of surface GluR1 was impaired in SAPAP3 siRNA transfected PFC neurons compared with control neurons. These results indicate that SAPAP3 may not be required for the surface delivery of GluR1 expression but could be involved in subsequent internalization of surface GluR1 after DA D1 receptor stimulation. Previous study has shown that PSD-95 is necessary for GluR1 internalization induced by neurotransmitter stimulation (80). It is possible that SAPAP3, a PSD-95-associated protein, may be involved in subsequent GluR1 internalization after DA D1 receptor stimulation through its interaction with PSD-95.
Our recent study has shown that DA D1 receptor stimulation-induced GluR1 surface expression is impaired in Fmr1 KO PFC neurons (40). Here, we further demonstrated that the decrease of surface GluR1 was also impaired in Fmr1 KO PFC slices compared with WT ones. It indicates that FMRP is not only required for GluR1 surface expression but also necessary for its subsequent internalization after DA D1 receptor stimulation. Because the lack of FMRP and the knockdown of SAPAP3 have similar effects on subsequent internalization of GluR1 after DA D1 receptor stimulation in PFC neurons, it suggests that FMRP may be involved in internalization of GluR1 through regulating the abundance of SAPAP3.
In summary, we have demonstrated that FMRP is critical for DA D1 receptor-mediated SAPAP3 synthesis in PFC neurons. DA D1 receptor stimulation controls the abundance of SAPAP3 through dynamic regulation of FMRP phosphorylation. FMRP is not only involved in surface expression of AMPA GluR1 receptors but also required for their subsequent internalization after surface delivery by D1 receptor stimulation. FMRP may be involved in GluR1 receptor internalization after D1 receptor stimulation through regulation of SAPAP3 abundance in PFC neurons. Our study thus provides further insights into FMRP involvement in DA receptor-mediated responses in PFC. It may help to elucidate the molecular mechanisms underlying impaired learning and memory in fragile X syndrome.
Acknowledgments
We thank Dr. William T. Greenough for the generous gift of Fmr1 WT and KO breeding pairs and Dr. Guoping Feng for providing part of the SAPAP3 antibody used in this study.
This work was supported by grants from the EJLB Canadian Institutes of Health Research Michael Smith Chair in Neurosciences and Mental Health, Canada Research Chair, and NeuroCanada and by Canadian Institutes of Health Research Operating Grants CIHR66975 and CIHR81086 (to M. Z.).
- FMRP
- fragile X mental retardation protein
- DA
- dopamine
- SAPAP3
- synapse-associated protein 90/PSD-95-associated protein 3
- PFC
- prefrontal cortex
- PP2A
- protein phosphatase 2A
- mTOR
- mammalian target of rapamycin
- S6K1
- ribosomal protein S6 kinase
- KO
- knock-out
- GPCR
- G protein-coupled receptor
- mGluR
- metabotropic glutamate receptor
- AMPA
- α-amino-3-hydroxyl-5-methyl-4-isoxazole-4-propionate
- WT
- wild type
- siRNA
- small interference RNA
- RT
- reverse transcription
- PBS
- phosphate-buffered saline
- ACSF
- artificial cerebrospinal fluid.
REFERENCES
- 1.Bassell G. J., Warren S. T. (2008) Neuron 60, 201–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bear M. F., Huber K. M., Warren S. T. (2004) Trends Neurosci. 27, 370–377 [DOI] [PubMed] [Google Scholar]
- 3.Belmonte M. K., Bourgeron T. (2006) Nat. Neurosci. 9, 1221–1225 [DOI] [PubMed] [Google Scholar]
- 4.Feng Y., Zhang F., Lokey L. K., Chastain J. L., Lakkis L., Eberhart D., Warren S. T. (1995) Science 268, 731–734 [DOI] [PubMed] [Google Scholar]
- 5.Terracciano A., Chiurazzi P., Neri G. (2005) Am. J. Med. Genet. C Semin. Med. Genet. 137c, 32–37 [DOI] [PubMed] [Google Scholar]
- 6.O'Donnell W. T., Warren S. T. (2002) Annu. Rev. Neurosci. 25, 315–338 [DOI] [PubMed] [Google Scholar]
- 7.Garber K., Smith K. T., Reines D., Warren S. T. (2006) Curr. Opin. Genet. Dev. 16, 270–275 [DOI] [PubMed] [Google Scholar]
- 8.Hou L., Antion M. D., Hu D., Spencer C. M., Paylor R., Klann E. (2006) Neuron 51, 441–454 [DOI] [PubMed] [Google Scholar]
- 9.Bagni C., Greenough W. T. (2005) Nat. Rev. Neurosci. 6, 376–387 [DOI] [PubMed] [Google Scholar]
- 10.Banko J. L., Hou L., Poulin F., Sonenberg N., Klann E. (2006) J. Neurosci. 26, 2167–2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grossman A. W., Aldridge G. M., Weiler I. J., Greenough W. T. (2006) J. Neurosci. 26, 7151–7155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huber K. M., Gallagher S. M., Warren S. T., Bear M. F. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 7746–7750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin P., Warren S. T. (2003) Trends Biochem. Sci. 28, 152–158 [DOI] [PubMed] [Google Scholar]
- 14.Ronesi J. A., Huber K. M. (2008) Sci. Signal. 1, pe6. [DOI] [PubMed] [Google Scholar]
- 15.Weiler I. J., Irwin S. A., Klintsova A. Y., Spencer C. M., Brazelton A. D., Miyashiro K., Comery T. A., Patel B., Eberwine J., Greenough W. T. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 5395–5400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Narayanan U., Nalavadi V., Nakamoto M., Pallas D. C., Ceman S., Bassell G. J., Warren S. T. (2007) J. Neurosci. 27, 14349–14357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Narayanan U., Nalavadi V., Nakamoto M., Thomas G., Ceman S., Bassell G. J., Warren S. T. (2008) J. Biol. Chem. 283, 18478–18482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ceman S., O'Donnell W. T., Reed M., Patton S., Pohl J., Warren S. T. (2003) Hum. Mol. Genet. 12, 3295–3305 [DOI] [PubMed] [Google Scholar]
- 19.Siomi M. C., Higashijima K., Ishizuka A., Siomi H. (2002) Mol. Cell. Biol. 22, 8438–8447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown V., Jin P., Ceman S., Darnell J. C., O'Donnell W. T., Tenenbaum S. A., Jin X., Feng Y., Wilkinson K. D., Keene J. D., Darnell R. B., Warren S. T. (2001) Cell 107, 477–487 [DOI] [PubMed] [Google Scholar]
- 21.Welch J. M., Lu J., Rodriguiz R. M., Trotta N. C., Peca J., Ding J. D., Feliciano C., Chen M., Adams J. P., Luo J., Dudek S. M., Weinberg R. J., Calakos N., Wetsel W. C., Feng G. (2007) Nature 448, 894–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Welch J. M., Wang D., Feng G. (2004) J. Comp. Neurol. 472, 24–39 [DOI] [PubMed] [Google Scholar]
- 23.Chen L., Bohanick J. D., Nishihara M., Seamans J. K., Yang C. R. (2007) J. Neurophysiol. 97, 2448–2464 [DOI] [PubMed] [Google Scholar]
- 24.Gurden H., Takita M., Jay T. M. (2000) J. Neurosci. 20, RC106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang Y. Y., Kandel E. R. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 2446–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lemon N., Manahan-Vaughan D. (2006) J. Neurosci. 26, 7723–7729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paspalas C. D., Goldman-Rakic P. S. (2005) J. Neurosci. 25, 1260–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan Z., Feng J., Fienberg A. A., Greengard P. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 11607–11612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang C. R., Seamans J. K. (1996) J. Neurosci. 16, 1922–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao W. J., Krimer L. S., Goldman-Rakic P. S. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 295–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Snyder S. H. (2006) Neuron 49, 484–485 [DOI] [PubMed] [Google Scholar]
- 32.Chen G., Greengard P., Yan Z. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 2596–2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun X., Zhao Y., Wolf M. E. (2005) J. Neurosci. 25, 7342–7351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishi A., Kuroiwa M., Miller D. B., O'Callaghan J. P., Bateup H. S., Shuto T., Sotogaku N., Fukuda T., Heintz N., Greengard P., Snyder G. L. (2008) J. Neurosci. 28, 10460–10471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Price C. J., Kim P., Raymond L. A. (1999) J. Neurochem. 73, 2441–2446 [DOI] [PubMed] [Google Scholar]
- 36.Huang Y. Y., Simpson E., Kellendonk C., Kandel E. R. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 3236–3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nagai T., Takuma K., Kamei H., Ito Y., Nakamichi N., Ibi D., Nakanishi Y., Murai M., Mizoguchi H., Nabeshima T., Yamada K. (2007) Learn. Mem. 14, 117–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schicknick H., Schott B. H., Budinger E., Smalla K. H., Riedel A., Seidenbecher C. I., Scheich H., Gundelfinger E. D., Tischmeyer W. (2008) Cereb. Cortex 18, 2646–2658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith W. B., Starck S. R., Roberts R. W., Schuman E. M. (2005) Neuron 45, 765–779 [DOI] [PubMed] [Google Scholar]
- 40.Wang H., Wu L. J., Kim S. S., Lee F. J., Gong B., Toyoda H., Ren M., Shang Y. Z., Xu H., Liu F., Zhao M. G., Zhuo M. (2008) Neuron 59, 634–647 [DOI] [PubMed] [Google Scholar]
- 41.Weinshenker D., Warren S. T. (2008) Nature 455, 607–608 [DOI] [PubMed] [Google Scholar]
- 42.Wang H., Wu L. J., Zhang F., Zhuo M. (2008) J. Neurosci. 28, 4385–4397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao M. G., Toyoda H., Ko S. W., Ding H. K., Wu L. J., Zhuo M. (2005) J. Neurosci. 25, 7385–7392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shang Y., Wang H., Mercaldo V., Li X., Chen T., Zhuo M. (2009) J. Neurochem. 111, 635–646 [DOI] [PubMed] [Google Scholar]
- 45.Wang H., Fukushima H., Kida S., Zhuo M. (2009) J. Biol. Chem. 284, 18953–18962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang H., Gong B., Vadakkan K. I., Toyoda H., Kaang B. K., Zhuo M. (2007) J. Biol. Chem. 282, 1507–1517 [DOI] [PubMed] [Google Scholar]
- 47.Gong B., Wang H., Gu S., Heximer S. P., Zhuo M. (2007) Eur. J. Neurosci. 26, 275–288 [DOI] [PubMed] [Google Scholar]
- 48.Mao L., Yang L., Arora A., Choe E. S., Zhang G., Liu Z., Fibuch E. E., Wang J. Q. (2005) J. Biol. Chem. 280, 12602–12610 [DOI] [PubMed] [Google Scholar]
- 49.Phillips P. E., Stuber G. D., Heien M. L., Wightman R. M., Carelli R. M. (2003) Nature 422, 614–618 [DOI] [PubMed] [Google Scholar]
- 50.Schultz W. (2002) Neuron 36, 241–263 [DOI] [PubMed] [Google Scholar]
- 51.Williams G. V., Goldman-Rakic P. S. (1995) Nature 376, 572–575 [DOI] [PubMed] [Google Scholar]
- 52.Margolis E. B., Mitchell J. M., Ishikawa J., Hjelmstad G. O., Fields H. L. (2008) J. Neurosci. 28, 8908–8913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Matsuda Y., Marzo A., Otani S. (2006) J. Neurosci. 26, 4803–4810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang Y. Y., Kandel E. R. (2007) Learn. Mem. 14, 497–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Greenough W. T., Klintsova A. Y., Irwin S. A., Galvez R., Bates K. E., Weiler I. J. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 7101–7106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weiler I. J., Spangler C. C., Klintsova A. Y., Grossman A. W., Kim S. H., Bertaina-Anglade V., Khaliq H., de Vries F. E., Lambers F. A., Hatia F., Base C. K., Greenough W. T. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 17504–17509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ahn J. H., McAvoy T., Rakhilin S. V., Nishi A., Greengard P., Nairn A. C. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 2979–2984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Beaulieu J. M., Sotnikova T. D., Marion S., Lefkowitz R. J., Gainetdinov R. R., Caron M. G. (2005) Cell 122, 261–273 [DOI] [PubMed] [Google Scholar]
- 59.Calabresi P., Gubellini P., Centonze D., Picconi B., Bernardi G., Chergui K., Svenningsson P., Fienberg A. A., Greengard P. (2000) J. Neurosci. 20, 8443–8451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Slobodyansky E., Aoki Y., Gaznabi A. K., Aviles D. H., Fildes R. D., Jose P. A. (1995) Am. J. Physiol. 268, F279–F284 [DOI] [PubMed] [Google Scholar]
- 61.Dounay A. B., Forsyth C. J. (2002) Curr. Med. Chem. 9, 1939–1980 [DOI] [PubMed] [Google Scholar]
- 62.Antion M. D., Hou L., Wong H., Hoeffer C. A., Klann E. (2008) Mol. Cell. Biol. 28, 2996–3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Antion M. D., Merhav M., Hoeffer C. A., Reis G., Kozma S. C., Thomas G., Schuman E. M., Rosenblum K., Klann E. (2008) Learn. Mem. 15, 29–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Holz M. K., Ballif B. A., Gygi S. P., Blenis J. (2005) Cell 123, 569–580 [DOI] [PubMed] [Google Scholar]
- 65.Polakiewicz R. D., Schieferl S. M., Gingras A. C., Sonenberg N., Comb M. J. (1998) J. Biol. Chem. 273, 23534–23541 [DOI] [PubMed] [Google Scholar]
- 66.Saitoh M., Pullen N., Brennan P., Cantrell D., Dennis P. B., Thomas G. (2002) J. Biol. Chem. 277, 20104–20112 [DOI] [PubMed] [Google Scholar]
- 67.Hou L., Klann E. (2004) J. Neurosci. 24, 6352–6361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tang S. J., Reis G., Kang H., Gingras A. C., Sonenberg N., Schuman E. M. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 467–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kelleher R. J., 3rd, Govindarajan A., Tonegawa S. (2004) Neuron 44, 59–73 [DOI] [PubMed] [Google Scholar]
- 70.Costa-Mattioli M., Sossin W. S., Klann E., Sonenberg N. (2009) Neuron 61, 10–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Santini E., Heiman M., Greengard P., Valjent E., Fisone G. (2009) Sci. Signal. 2, ra36. [DOI] [PubMed] [Google Scholar]
- 72.Wolf M. E., Mangiavacchi S., Sun X. (2003) Ann. N.Y. Acad. Sci. 1003, 241–249 [DOI] [PubMed] [Google Scholar]
- 73.Swayze R. D., Lisé M. F., Levinson J. N., Phillips A., El-Husseini A. (2004) Neuropharmacology 47, 764–778 [DOI] [PubMed] [Google Scholar]
- 74.Garber K. B., Visootsak J., Warren S. T. (2008) Eur. J. Hum. Genet. 16, 666–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cota D., Matter E. K., Woods S. C., Seeley R. J. (2008) J. Neurosci. 28, 7202–7208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sharma A., Hoeffer C. A., Takayasu Y., Miyawaki T., McBride S. M., Klann E., Zukin R. S. (2010) J. Neurosci. 30, 694–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schratt G. M., Nigh E. A., Chen W. G., Hu L., Greenberg M. E. (2004) J. Neurosci. 24, 7366–7377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Takei N., Inamura N., Kawamura M., Namba H., Hara K., Yonezawa K., Nawa H. (2004) J. Neurosci. 24, 9760–9769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gong R., Park C. S., Abbassi N. R., Tang S. J. (2006) J. Biol. Chem. 281, 18802–18815 [DOI] [PubMed] [Google Scholar]
- 80.Bingol B., Schuman E. M. (2004) Neuropharmacology 47, 755–763 [DOI] [PubMed] [Google Scholar]