Abstract
The INrf2 (Keap1)/Cul3-Rbx1 complex constantly degrades Nrf2 under normal conditions. When a cell encounters oxidative or electrophilic stress, Nrf2 dissociates from the INrf2/Cul3-Rbx1 complex and translocates into the nucleus. In the nucleus, Nrf2 activates a myriad of antioxidant and defensive genes that protect cells. Nrf2 is then exported out of the nucleus and degraded. INrf2 serves as a substrate adaptor to link Nrf2 to Cul3 and Rbx1. Cul3 and Rbx1 make up the ubiquitin ligase complex that is responsible for the ubiquitination and degradation of Nrf2. Previously we have shown a feedback autoregulatory loop between Nrf2 and INrf2 indicating that Nrf2 regulates INrf2 by controlling its transcription. Here we are extending this research by demonstrating the presence of another feedback autoregulatory loop between Cul3-Rbx1 and Nrf2. Experiments using Hepa-1 and HepG2 cells indicate that Nrf2 controls its own degradation by regulating expression and induction of Cul3-Rbx1 genes. Treatment with the antioxidant tert-Butylhydroquinone (t-BHQ) leads to induction of Cul3-Rbx1 genes. Mutagenesis and transfection experiments identified an antioxidant response element in the forward and reverse strands of the proximal Cul3 and Rbx1 promoters, respectively, that Nrf2 binds and regulates expression and antioxidant induction of the Cul3-Rbx1 genes. In addition, short interfering RNA inhibition and overexpression of Nrf2 led to a respective decrease and increase in Cul3-Rbx1 gene expression. The increase in Cul3-Rbx1 leads to ubiquitination and degradation of Nrf2. These data suggest that Nrf2 regulates Cul3-Rbx1 by controlling regulation of expression and induction of Cul3-Rbx1. The induction of Cul3-Rbx1 control Nrf2 by increasing degradation.
Keywords: Antioxidant, Protein Degradation, Transcription, Transcription Regulation, Ubiquitination, Autoregulatory Loop, Cul3-Rbx1, INrf2(Keap1), Nrf2
Introduction
The cellular defense system protects against oxidative stress caused by a vast range of xenobiotics, inflammation, and ionizing radiation (1–3). Disruption of these protective systems causes the accumulation of reactive oxygen species (ROS)2 and electrophiles that can contribute to diseases such as cancer, cardiovascular complications, acute and chronic inflammation, and neurodegenerative diseases (4). Therefore, it is obvious that cells must constantly labor to control levels of ROS, preventing them from accumulating. Cells have mechanisms to activate over two hundred defensive genes that protect against ROS and the diseases they contribute to (1, 5–7).
The antioxidant response element (ARE) was first identified as cis-element in the upstream regulatory region of the GSTA2 gene (8) and was found in the promoters of detoxifying enzyme genes such as glutathione S-transferases, NAD(P)H:quinone oxidoreductases, gastrointestinal glutathione peroxidase, and peroxiredoxin1 (9–13). The ARE is recognized by the family of Cap'n'Collar containing basic leucine zipper proteins including Nrf2. Among the family, Nrf2 is the most potent transcription factor in regulating the basal and inducible expression of antioxidant enzyme genes (14). Gene deletion studies also supported the important function of Nrf2 in cellular protection against oxidative stress and neoplasia (15).
At basal levels, Nrf2 resides within the cytoplasm of the cells by an interaction with an actin-bound cytosolic protein, INrf2 (inhibitor of Nrf2) or Keap1 (Kelch-like ECH-associated protein 1) (16–18). INrf2 functions as a substrate adaptor protein for a Cullin 3 (Cul3)-dependent ubiquitin-protein ligase complex to maintain the steady-state levels of Nrf2 (19). Covalent conjugation of proteins by ubiquitin usually involve three enzymatic activities for activating (E1), conjugating (E2), and ligating (E3) ubiquitin to a substrate (20). In this case, Nrf2 serves as the substrate, while Cul3 serves as a scaffold protein that forms the E3 ligase complex with Ring Box1 (Rbx1) that recruits a cognate E2 enzyme (6). INrf2, via its N-terminal BTB/POZ domain, binds to Cul3 (20, 22) and via its C-terminal Kelch domain binds to the substrate Nrf2, leading to the ubiquitination and subsequent degradation of Nrf2 through the 26 S proteasome (23–27).
Cellular exposure to oxidative stress leads to dissociation of Nrf2 from the INrf2/Cul3-Rbx1 complex (1, 5–7). Nrf2 escapes proteolysis and stabilizes, translocates into the nucleus, and causes activation of ARE-mediated genes leading to cytoprotection. Several reports suggest that persistent accumulation of Nrf2 in the nucleus is harmful. Nrf2 regulates the expression of several multidrug resistance-associated protein (MRP) efflux transporters in responses to oxidative stress (28) which could lead to chemotherapeutic drug resistance. INrf2-null mice demonstrated persistent accumulation of Nrf2 in the nucleus that led to postnatal death from malnutrition resulting from hyperkeratosis in the esophagus and forestomach (29). The capacity of INrf2 negative regulation of Nrf2 is also dependent upon Cul3 and Rbx1. The importance of Cul3 and Rbx1 is crucial because in their absence INrf2 may not be able to adapt ubiquitin ligases to degrade Nrf2. Taken together; unrestrained activation of Nrf2 in cells increases the risk of adverse effects, including tumorigenesis. On the other hand, stress-induced activation of the Nrf2 pathway in normal cells is tightly regulated and confers cytoprotection against oxidative and electrophilic stress and carcinogens. Therefore, it appears that cells contain mechanisms that autoregulate cellular abundance of Nrf2.
In previous studies we have demonstrated an autoregulatory feedback loop between Nrf2 and INrf2. In the current study, we extend this research to investigate if an autoregulatory loop exists between Nrf2 and Cul3-Rbx1. After Nrf2 activation by antioxidant, an increase in Cul3-Rbx1 expression was detected. Cul3-Rbx1 promoters and Nrf2 knockdown/overexpression studies show that Nrf2 induces promoter activity of Cul3-Rbx1 genes through Nrf2 binding to an ARE in the forward and reverse strands of the proximal promoters of Cul3-Rbx1 genes, respectively. The induced Cul3-Rbx1 proteins accelerate ubiquitination of Nrf2 for degradation. Therefore, Nrf2 controls its own degradation by regulating the levels of Cul3 and Rbx1 in cells.
MATERIALS AND METHODS
Cell Cultures
Human hepatoblastoma (HepG2) and mouse hepatoma (Hepa-1) cells were obtained from the American Type Culture Collection (Manassas, VA). HepG2 cells were grown in minimum essential α-medium and Hepa-1 in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, penicillin (40 units/ml), and streptomycin (40 μg/ml). The cells were grown in monolayer in an incubator at 37 °C in 95% air and 5% CO2.
Generation of Stable Flp-In T-REx HEK293 Cells Expressing Tetracycline-inducible Nrf2
Flp-In T-REx HEK293 cells purchased from Invitrogen were co-transfected with FLAG-Nrf2 cDNA in pcDNA5/FRT/TO and pOG44 plasmids (Invitrogen) by Effectene (Qiagen, Valencia, CA) according to manufacturer's instructions. Forty-eight hours after transfection, the cells were grown in medium containing 200 μg/ml hygromycin B (Invitrogen). The 293/FRT/FLAG-Nrf2 cells stably expressing tetracycline-inducible N-terminal FLAG-tagged Nrf2 were selected. The stably selected cells were grown and treated with 4 μg/ml of tetracycline (Sigma) for varying periods of time to follow the overexpression of FLAG-tagged Nrf2.
Plasmid Constructs
Genomic clones (BAC vector) containing mouse Rbx1 and Cul3 loci were purchased from BACPAC Resources Center, Children's Hospital Oakland Research Institute, Oakland, CA. A 2,724-bp fragment of Rbx1 promoter was isolated via PCR using the 5′- CTATGGTACCCTCCTGACAAGGACCTTTGTGGTTCAG-3′ and 5′-CATGCTCGAGCACGACAGACTGTGTGTTTCC-3′ primer pairs and high fidelity platinum TaqDNA polymerase (Invitrogen). The PCR-amplified promoter fragments were subcloned into pGL2-basic luciferase vector (Promega, Madison, WI) using Kpn1 and Xho1 restriction sites. The construct was designated as pGL2–2.7 kb. The sequence of the PCR forward primers for a series of deletion constructs is as follows: 2.0 kb forward, 5′-TACCGGTACCAAGCAGCGAGTGAATGCTCTTA-3′; 0.8 kb forward, 5′-TACCGGTACCATCAGAATGCCTCACCAGAACTCAA-3′; 0.4 kb forward 5′-TACCGGTACCGTTTCCAAAGACCAGCCCATG-3′. The same reverse primer, as used to construct 2.7-kb plasmid, was used. To locate and mutate the ARE of interest, new forward and reverse primers were used. The PCR promoter fragments were also subcloned into pGL2-basic luciferase vector using Kpn1 and Xho1 restriction sites. The constructs are designated as follows: 0.14 kb forward, 5′-TATTGGTACCCCTTTAAGGGGCGTGACC-3′ was used with both Wild type and mutant reverse primers. 0.14 kb Wild Type reverse 5′-ATATCTCGAGTCTGTCGTGTGACCACTGCG-3′; 0.14kb Mutant reverse 5′-ATATCTCGAGTCTGTCGTGCAGCCACTAAG-3′. To clarify which ARE has an essential role in Nrf2-induced Rbx1 promoter activity, oligonucleotides containing the ARE sequence were synthesized and cloned into the pGL2 promoter vector. The sequences of oligonucleotides of AREs are as follows: Rbx1 ARE Wild Type forward, 5′-TTACGGTACCAGGCGCAGTGGTCACACGCTCGAGTCGA-3′; Rbx1 ARE Wild Type reverse 5′-AATGCCATGGTCCGCGTCACCAGTGTGCGAGCTCAGCT-3′;Rbx1 ARE Mutant forward, 5′-TTACGGTACCAGGCATAGTGGCTGCACGCTCGAGTCGA-3′. Rbx1 ARE Mutant reverse, 5′-AATGCCATGGTCCGTATCACCGACGTGCGAGCTCAGCT-3′. A 2,606-bp fragment of Cul3 promoter was isolated via PCR using the 5′-TACCGGTACCGGGACTGTGGTTCCTAATTTTGTGATA-3′ and 5′-TATTCTCGAGTGTCACATTGAAGGCGGGAGGGCAGCC-3′ primer pairs and high fidelity platinum TaqDNA polymerase (Invitrogen). The PCR-amplified promoter fragments were subcloned into pGL2-basic luciferase vector (Promega, Madison, WI) using Kpn1 and Xho1 restriction sites. The construct was designated as pGL2–2.7 kb. The sequence of the PCR forward primers for a series of deletion constructs is as follows: 1.65 kb forward, 5′-TGTCGGTACCGCTTAACTCTCTAAGCTTAGTCATTAC-3′; 1.6 kb forward, 5′ TACCGGTACCGGGTCAATTCAACCATAAATAAACA-3′; 1.5 kb forward, 5′-TACCGGTACCTTCAAGACAGGGTTTCTCTGTGTA-3′; 0.6 kb forward, 5′-TATCGGTACCGTCTCCGACGCTCCTCTTT-3′; 0.5 kb Wild Type forward, 5′-TATCGGTACCGCTGACTACGCCCATTCCTT-3′; 0.5 kb Mutant forward, 5′-TACCGGTACCGCAACCTACTGCCATTCCTT-3′; 0.2kb forward, 5′-TACCGGTACCCTGCGCAGTGAGATGTTTGT-3′. The same reverse primer, as used to construct 2.7-kb plasmid, was used for all constructs. To clarify which ARE has an essential role in Nrf2-induced Cul3 promoter activity, oligonucleotides containing the ARE sequence were synthesized and cloned into the pGL2 promoter vector. The sequences of oligonucleotides of AREs are as follows: Cul3 ARE Wild Type forward, 5′-TTACGGTACCGCGCTGACTACGCCCATCTCGAGTCGA-3′;Cul3 ARE Wild Type reverse 5′-AATGCCATGGCGCGACTGATGCGGGTAGAGCTCAGCT-3′; Cul3 ARE Mutant forward, 5′-TTACGGTACCGCGCCAGCTACTACCATCTCGAGTCGA-3′. Cul3 ARE Mutant reverse, 5′-AATGCCATGGTCCGTATCACCGACGTGCGAGCTCAGCT-3′. The sequence accuracy of all constructs was confirmed by restriction enzyme digestion and sequencing by The Biopolymer Core Facility (Baltimore, MD). Plasmids pcDNA-Cul3 V5, and pcMX-Rbx1-Myc, pcMV-Nrf2-FLAG, pcDNA-INrf2-V5, and HA-Ub are also described previously (30).
Real Time PCR
Hepa-1 cells were seeded in 100-mm plates. Twenty-four hours later, cells were treated with either t-BHQ (Sigma) or 2 μg/ml Actinomycin D (Sigma) and harvested. RNA was extracted using RNeasy mini kit (Qiagen). RNA was converted to cDNA using High Capacity cDNA Reverse Transcription kit (Applied Biosystems) according to manufacturer's protocol. cDNA was used with Taqman Master Mix (Applied Biosystems) and Keap1 Primer and Probe amplicon Mm00497268_m1, or Cul3 Primer and Probe amplicon Mm00516747_m1 or Rbx1 Primer and Probe amplicon Mm01705487_s1 or NQO1 Primer and Probe amplicon Mm00500821_m1 or Nrf2 Primer and probe amplicon Mm00477784_m1 or GusB amplicon Mm00446953_m1 as an internal control (Applied Biosystems). Total mix was run on 7500 Real Time System (Applied Biosystems) using relative quantitation according to the manufacturer's protocols.
Western Blot Analysis
The cells were lysed in ice-cold RIPA-B buffer (20 mm Tris, pH 7.4, 150 mm NaCl, 1 mm EDTA, 1% Nonidet P-40, 0.5% deoxycholate, 1% Triton X-100), and protease inhibitor mixture (Roche Applied Science). Nuclear extracts were made using a nuclear extract kit from Active Motif (Carlsbad, CA) according to manufacturer's protocol. Fifty micrograms of proteins were separated by SDS-PAGE and transferred to nitrocellulose membranes. The membranes were incubated with anti-Cul3 (1:1000, Cell Signaling), anti-Rbx1 (1:1000, Bio Source) anti-INrf2 (1:1000, Santa Cruz Biotechnology), anti-Nrf2 (1:500, Santa Cruz Biotechnology), anti-FLAG (1:5000, Sigma), anti-V5 (1:5000, Invitrogen), anti-Myc (1:5000, Sigma), or anti-actin (1:5000, Sigma) antibodies, washed, and probed with electrochemiluminescence (Amersham Biosciences). To confirm the purity of nuclear-cytoplasmic fractionation, the membranes were reprobed with cytoplasm-specific, anti-lactate dehydrogenase (Chemicon) and nuclear specific, anti-lamin B antibodies (Santa Cruz Biotechnology). Protein expression was quantified by using NIH Image program (developed at the National Institutes of Health). In related experiments, the cells were treated with 100 μm t-BHQ in the absence or presence of 30 μg/ml cycloheximide for different time intervals. The cells were lysed and analyzed by Western blotting and probed with Cul3 and Rbx1 antibody. The blot was stripped and reprobed with anti-actin antibody.
Ubiquitination Assay
Hepa-1 cells were seeded in 100-mm plates and co-transfected with pCMV-FLAG-Nrf2 (1.0 μg), and pCMV-HA-Ub (0.5 μg). The transfected cells were treated with either DMSO or 100 μm t-BHQ for the indicated time. To check Nrf2 ubiquitination, 1 mg of protein lysate was used to immunoprecipitate with anti-FLAGM2 beads (Sigma). To analyze Nrf2 ubiquitination in cytoplasm and nuclear extracts, Hepa-1 cells in 100-mm plates were co-transfected with pcMX-FLAG-Nrf2 (1.0 μg) with or without plasmids encoding pcDNA-V5-INrf2 (0.5 μg), pcDNA-V5-Cul3 (0.5 μg), pCMX-myc-Rbx1, and HA-Ub (0.5 μg) in the absence or presence of 10 μm Mg-132. Nuclear and cytoplasmic extracts were prepared using the active motif kit, and 1 mg of extract was immunoprecipitated with anti-FLAG antibody. Immunoprecipitates were resolved on a 10% SDS-PAGE followed by immunoblotting with anti-HA antibody.
Transient Transfection and Luciferase Assay
Hepa-1, Flp-In T-REx HEK293, or 293/FRT/FLAG-Nrf2 cells were plated in 12-well plates at a density of 1 × 105 cells/well 24 h prior to transfection. The cells were transfected with 0.1 μg of the indicated luciferase plasmid using Effectene transfection reagent according to the manufacturer's instruction. The pRL-TK plasmid encoding Renilla luciferase (0.01 μg; Promega) was included as an internal control of transfection efficiency. After 32 h, transfected Hepa-1 cells were stimulated with DMSO or 100 μm t-BHQ for 16 h. Otherwise, transfected Flp-In T-REx HEK293 or 293/FRT/FLAG-Nrf2 cells were treated with 4.0 μg/ml of tetracycline for 8 or 16 h. The cells were harvested, lysed, and analyzed for luciferase activity using the dual-luciferase reporter assay system (Promega).
Chromatin Immunoprecipitation (ChIP) Assay
ChIP assay was performed using a kit from Active Motif as described previously (11). Briefly, 70% confluent Hepa-1 cells were treated with DMSO or 100 μm t-BHQ for 4 h and then fixed in 1% formaldehyde for 15 min. Cells were lysed and nuclei pelleted by centrifugation. Nuclei were resuspended and sheared using a sonicator (Misonix Inc., Farmingdale, NY) with five pulses of 20 s at 25% of maximum output. Sheared chromatin was immunoprecipitated with 2 μg of anti-Nrf2 or control IgG antibody. The cross-links reversed overnight at 65 °C and deproteinated with 20 μg/ml proteinase K. Nrf2-associated Cul3 and Rbx1-ARE was detected by PCR amplification with the primers as follows: Cul3 ARE, forward, 5′-GTCTCCGACGCTCCTCTTT-3′, and reverse, 5′-CTGCGCACTCACATGTTTGT-3′; Rbx1 ARE, forward, 5′-GCCTTTAAGGGGCGTGACC-3′, and reverse, 5′-ATATGGCTGGCAGGCCCGAG-3′. The PCR condition used for ChIP assay was 37 cycles of a denaturing step at 94 °C for 30 s, an annealing step at 65 °C for 30 s, and an extension step at 72 °C for 30 s. PCR products (387 bp with Cul3 ARE primers and 300bp with Rbx1 ARE primers) were separated on 2% agarose gel containing ethidium bromide and imaged using a Bio-Rad ChemiDoc XRS.
Nrf2 siRNA Interference Assay
Nrf2 siRNA was used to inhibit Nrf2 by a procedure described previously (11). Two types of Nrf2 siRNA (s70521 and s70523) and control siRNA were purchased from Applied Biosystems. Hepa-1 cells were transfected with 75 nm control siRNA or Nrf2 siRNA using Lipofectamine RNAiMAX reagent (Invitrogen) according to the manufacturer's instructions. Twenty-four hours later, cells were co-transfected with 0.1 μg of Cul3 and Rbx1 promoter-luciferase and 0.01 μg of pRL-TK Renilla plasmids. Thirty-two hours after the second transfection, cells were treated either with DMSO or with 100 μm t-BHQ for 24 h. The cells at the end of treatment were lysed and analyzed by measuring luciferase activity, Cul3, Rbx1, and Nrf2 RNA by Real-Time PCR, and Cul3, Rbx1, and Nrf2 protein by Western blot analysis and probing with Cul3, Rbx1, and Nrf2 antibodies.
Statistical Analyses
The data from luciferase and real time-PCR assays, and protein expression quantification were analyzed using a two-tailed Student's t test. Data are expressed as the mean ± S.D. (*, p < 0.05; **, p < 0.01; ***, p < 0.001) are shown within the figures.
RESULTS
Antioxidant t-BHQ Up-regulates Cul3 and Rbx1 Gene Expression
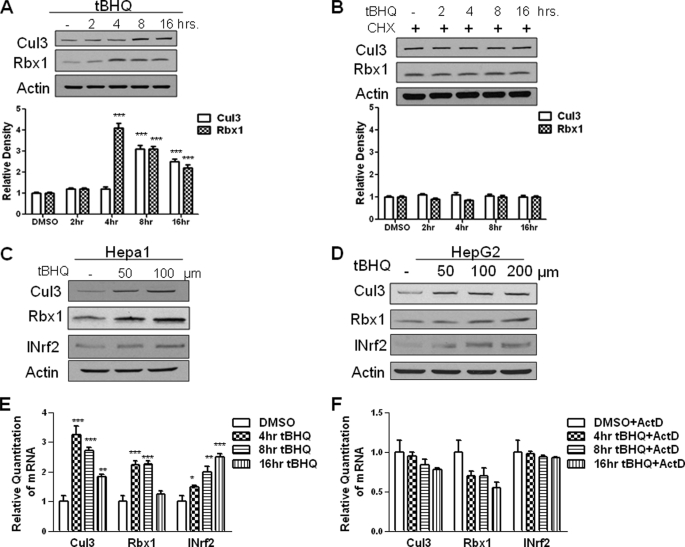
Western blot analysis shows a concentration-dependent and time-dependent increase in Cul3 and Rbx1 protein expression. The t-BHQ treatment of Hepa-1 cells showed a time-dependent increase in Cul3 and Rbx1 protein at 8 h and 4 h, respectively (Fig. 1A). The increase in Cul3 protein was reduced at 16 h. The increase in Rbx1 protein was reduced at 8 and 16 h after t-BHQ treatment. However, in the presence of the protein synthesis inhibitor cycloheximide, Cul3 and Rbx1 protein induction with t-BHQ was more or less blocked (Fig. 1B) confirming the increase in Cul3 and Rbx1 is because of new protein synthesis. Hepa-1 cells and HepG2 cells both demonstrated concentration dependent increase in Cul3 and Rbx1 protein in response to t-BHQ treatment (Fig. 1, C and D). Real-Time PCR analysis of mouse Hepa-1 cells treated with t-BHQ showed an increase in Cul3 and Rbx1 gene expression (Fig. 1E). Preincubation with the transcription inhibitor actinomycin D blocked the t-BHQ-mediated induced expression of Cul3 and Rbx1. INrf2 is shown as a positive control (Fig. 1F).
FIGURE 1.
Antioxidant t-BHQ induces INrf2 gene expression. A, Western analysis of Cul3 and Rbx1 expression in Hepa-1 cells. Hepa-1 cells were grown in a monolayer and treated with 100 μm t-BHQ for the indicated time intervals. Total cell lysate was analyzed with anti-Cul3 and Rbx1 antibodies. β-Actin was used as a loading control. Densitometry measurements of bands were quantitated and shown in graph blots below. B, Western analysis of Cul3 and Rbx1 protein in Hepa-1 cells. Total cell lysate was treated with 100 μm t-BHQ and 30 μg/ml cycloheximide (CHX) for the indicated time intervals and were analyzed for Cul3 and Rbx1 expression by Western blotting and probing with anti-Cul3 and Rbx1 antibody. β-Actin was used as a loading control. C and D, Hepa-1 and HepG2 cells were seeded in a monolayer and treated with 50, 100, and 200 μm t-BHQ for 16 h. Cells were then lysed and lysate was probed with anti-Cul3 and anti-Rbx1 antibodies. Anti-INrf2 was used as a positive control and β-actin was used as a loading control. E, Hepa-1 cells were seeded in a monolayer and treated with 100 μm t-BHQ for varying time points. Cells were lysed, and RNA was extracted and converted to cDNA. 50 ng of cDNA was mixed with 1× Taqman master mix and either Cul3, Rbx1, or INrf2 primers and probes and GusB primers and probes as control. Relative quantitation of mRNA was measured and plotted. F, in a similar experiment, Hepa-1 cells were seeded and were pretreated with 2 μg/ml actinomycin D (ActD) for 2 h followed by 100 μm t-BHQ + actinomycin D for the indicated time interval. RNA was extracted and converted to cDNA. 50 ng of cDNA was mixed with 1× Taqman master mix and either Cul3, Rbx1, or INrf2 primers and probes and GusB primers and probes as control. Relative quantitation of mRNA was measured and plotted. All experiments were repeated 3–5 times. The representative results are shown.
ARE in the Proximal Promoter on Forward and Reverse Strands Mediates Expression and t-BHQ Induction of Cul3 and Rbx1 Gene Expression
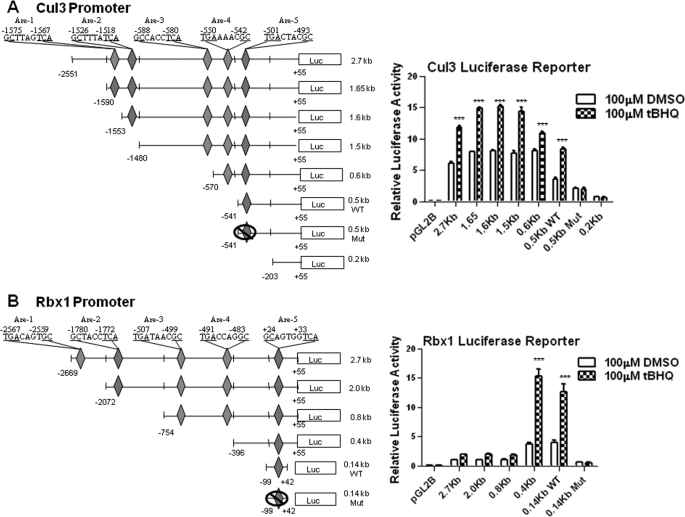
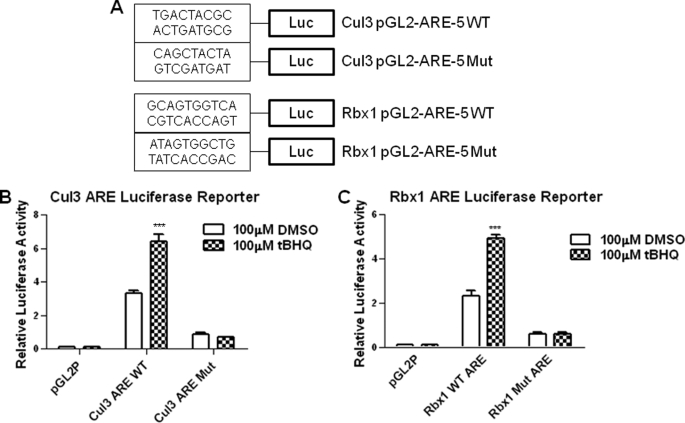
Deletion and mutagenesis followed by transfection assays investigated the cis-element(s) required for expression and t-BHQ induction of mouse Cul3 and Rbx1 genes (Fig. 2). A 2.7-kb Cul3 gene promoter and a 2.7 Rbx1 gene promoter attached to the luciferase gene upon transfection in Hepa-1 cells produced luciferase activity that was induced in response to t-BHQ treatment (Fig. 2, A and B, right panels). Mouse Cul3 and Rbx1 promoters were analyzed for AREs. Mouse-Cul3 and Rbx1 promoter analysis was done using Invitrogen's Vector NTI. Nucleotide sequence analyses of 2.7-kb Cul3 and Rbx1 promoters revealed the presence of five putative AREs (Fig. 2, A and B, left panels). In the Cul3 promoter, three of the elements were on the reverse strand at nucleotide positions −1575, −1526, −588 from the start of transcription. The remaining two elements were located at nucleotide positions −550 and −501 on the sense strand. In the Rbx1 promoter, two of the elements were on the reverse strand at nucleotide position −1780 and +24 from the start site of transcription. The other three elements were located at nucleotide position −2567, −507, and −491 on the sense strand. All constructs were transfected in Hepa-1 cells and analyzed for luciferase activity (Fig. 2, A and B, right panels). The Cul3 promoter construct containing no AREs, −203 to +55, showed very little luciferase activity when treated with either DMSO or t-BHQ. Interestingly, the Rbx1 promoter constructs located further upstream showed very low luciferase activity compared with the smaller constructs. It appears that Rbx1 promoter contains repressor element(s) in the promoter region between nucleotide −0.8 kb to −0.4 kb (Fig. 2B, right panel). This was evident from significant increase in basal expression from 0.4 kb promoter as compared with 0.8 kb promoter. The Cul3 and Rbx1 AREs that were most proximal to the start of transcription and expressing significant luciferase activity were mutated in their original construct, and designated as Mut. The mutated plasmids were also transfected in Hepa-1 cells and analyzed for luciferase activity in the absence and presence of t-BHQ to determine the role of individual AREs in expression and t-BHQ induction of the Cul3 and Rbx1 genes (Fig. 2, A and B, right panels). The results revealed that mutation of the ARE-5 in the Cul3 promoter and ARE-5 in the Rbx1 promoter resulted in the significant reduction in basal expression and abrogation of t-BHQ induction as compared with the wild type (p < 0.005). The Cul3 and Rbx1, WT, and Mut AREs were individually cloned in the pGL2 promoter vector (Fig. 3A), transfected in Hepa-1, and analyzed for luciferase activity to determine its role in t-BHQ induction of luciferase gene expression through the heterologous promoter (Fig. 3, B and C). The results demonstrated that Cul3 and Rbx1 WT ARE and not the mutants efficiently mediated expression and t-BHQ induction of luciferase gene expression.
FIGURE 2.
ARE sequence in the forward and reverse strand of the proximal promoters regulates expression and antioxidant induction of Cul3 and Rbx1 genes. A and B, deletion, mutagenesis, and transfection analysis. Serial deletions of mouse Cul3 and Rbx1 promoter separately attached to luciferase (Luc) reporter gene were transfected in Hepa-1 cells, treated with DMSO or 100 μm t-BHQ for 16 h, and analyzed for luciferase activity (right panels). Five putative ARE sequences for both Cul3 and Rbx1 are shown. ARE of interest was mutated within the original construct designated as WT and Mut. (left panels).
FIGURE 3.
Cul3 and Rbx1 WT AREs and not mutant AREs are inducible by antioxidants. A, ARE-luciferase expression in transfected cells. ARE-5 for both Cul3 and Rbx1 were separately attached to SV40 basal promoter hooked to luciferase reporter gene by cloning in vector pGL2 promoter, wild type and mutant sequences are shown. B and C, wild type and mutant Cul3 and Rbx1 AREs were transfected in Hepa-1 cells, treated with DMSO, or t-BHQ (100 μm for 16 h), and analyzed for luciferase activity. For all the above experiments, pGL2 empty vector was used as negative control. The results are expressed as fold increase in relative luciferase activity compared with untreated pGL2 transfection. The data shown are mean ± S.D. of three independent transfection experiments in A–C.
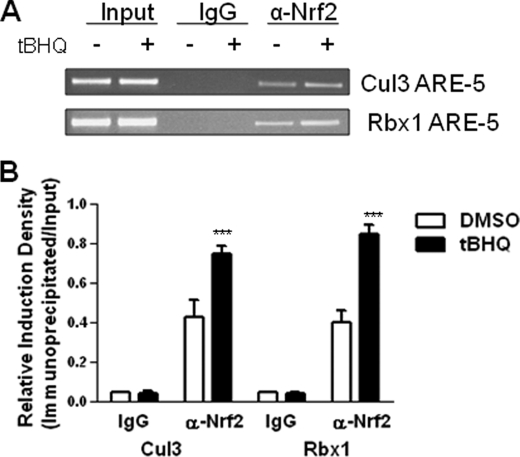
Antioxidant Increases in Vivo Binding of Nrf2 to Cul3 and Rbx1 ARE
ChIP assays were performed in Hepa-1 cells using an Nrf2-specific antibody and PCR primers covering the Cul3 ARE-5 and the Rbx1 ARE-5 regions in the respective promoters to determine the binding of Nrf2 to the AREs of the Cul3 and Rbx1 genes in DMSO and t-BHQ-treated Hepa-1 cells. The results demonstrated binding of Nrf2 to the Cul3 and Rbx1 gene promoters (Fig. 4A). The Nrf2 binding to the Cul3 and Rbx1 AREs was enhanced by 1.4-fold and 2.3-fold, respectively, in response to t-BHQ (p < 0.001) (Fig. 4B). These data indicate a specific interaction of Nrf2 to ARE-5 of the Cul3 and Rbx1 gene promoters, which is enhanced upon t-BHQ treatment. In similar experiments, Nrf2 failed to bind to ARE 1–4 in Cul3 and also in Rbx1 gene promoters (data not shown).
FIGURE 4.
Antioxidant increases binding of Nrf2 to Cul3 and Rbx1 ARE. A, ChIP assay. Hepa-1 cells were treated with 100 μm t-BHQ for 4 h, fixed with formaldehyde, cross-linked, and sheared the chromatin. The chromatin was immunoprecipitated with anti-Nrf2 antibody and control IgG. Nrf2 binding to Cul3 and Rbx1 promoter was analyzed by PCR with specific primers for Cul3 and Rbx1 ARE regions, respectively. Cul3 and Rbx1 ARE regions of Cul3 and Rbx1 promoter were also amplified from 5 μl of purified soluble chromatin before immunoprecipitation to show input DNA. B, densitometric analysis for ChIP assay. Relative Nrf2 binding to Cul3 and Rbx1 promoter was quantified. Signal intensity for PCR products was normalized to that of input for every sample. The experiment was repeated three times. The representative results are shown.
Nrf2 Mediates t-BHQ Induction of Cul3 and Rbx1 Gene Expression
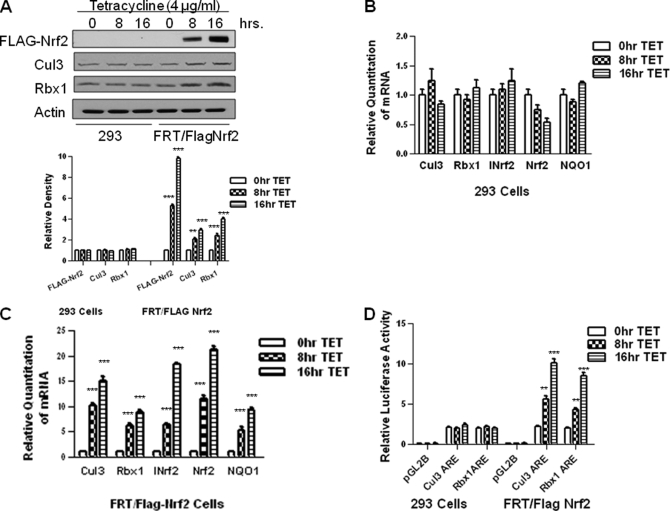
Oxidative and electrophilic stresses are known to induce the stability of Nrf2 that leads to nuclear accumulation, resulting in transcriptional activation of antioxidant and phase II drug-metabolizing enzyme genes, including NQO1 gene (14). Therefore, we evaluated the effect of increased and decreased expression of Nrf2 in regulation of Cul3 and Rbx1 gene expression. We used overexpression of Nrf2 and siRNA inhibition of Nrf2 to demonstrate a role of Nrf2 in mediated expression and t-BHQ induction of Cul3 and Rbx1 gene expression (Figs. 5 and 6). We also successfully established the Flp-In T-Rex 293 cell lines (293/FRT/FLAG-Nrf2) that upon stimulation with tetracycline showed a time-dependent increase in FLAG-Nrf2 protein (Fig. 5A) and RNA (Fig. 5, B and C). The time-dependent increase in FLAG-Nrf2, after stimulation with tetracycline, also led to increases in endogenous Cul3 and Rbx1 proteins. The real time-PCR analysis also revealed that tetracycline-induced overexpression of FLAG-Nrf2 led to time-dependent increases in Cul3 and Rbx1 gene expression. In the same experiment, the Nrf2 downstream gene NQO1 and INrf2 were also induced. The transfection of Flp-In T-Rex 293 or 293/FRT/FLAG-Nrf2 cells with Cul3 and Rbx1 ARE-Luc plasmid revealed time-dependent increases in luciferase gene expression upon stimulation with tetracycline (Fig. 5D).
FIGURE 5.
Overexpression of Nrf2 up-regulates endogenous and transfected Cul3 and Rbx1 gene expression. A, Western analysis of Flp-in T-REx 293 (293) cells or 293/FRT/FLAG-Nrf2 (FRT/FLAG-Nrf2) cells expressing tetracycline-induced FLAG-tagged Nrf2 were incubated with 4 μg/ml tetracycline (TET) for the indicated times. Cells were harvested, lysed, and probed with anti-FLAG, anti-Cul3, and anti-Rbx1. Anti-β-actin was used as loading control. Densitometry measurements of bands were quantitated and shown in graph blots below. B and C, Cul3 and Rbx1 gene expression was analyzed by real time-PCR. 293 cells or FRT/FLAG-Nrf2 cells were seeded in a monolayer and treated with 4 μg/ml tetracycline for indicated time points. RNA was extracted, converted to cDNA. 50 ng of cDNA was analyzed using primers and probes specific for Cul3 and Rbx1 mRNA. Tetracycline-induced Nrf2 expression was also confirmed using specific primers for exogenous Nrf2. NQO1 and INrf2 were used as positive control, respectively. GusB primers and probes were used as internal control. D, 293 cells or FRT/FLAG-Nrf2 cells were co-transfected with Cul3 or Rbx1 ARE plasmids and the internal control plasmid pRL-TK. Twenty-four hours after transfection, the cells were treated with 4 μg/ml tetracycline for 8 or 16 h. pGL2 vector was used as negative control. The cells were harvested and analyzed for luciferase activity. The data shown are mean ± S.D. of three independent transfection experiments.
FIGURE 6.
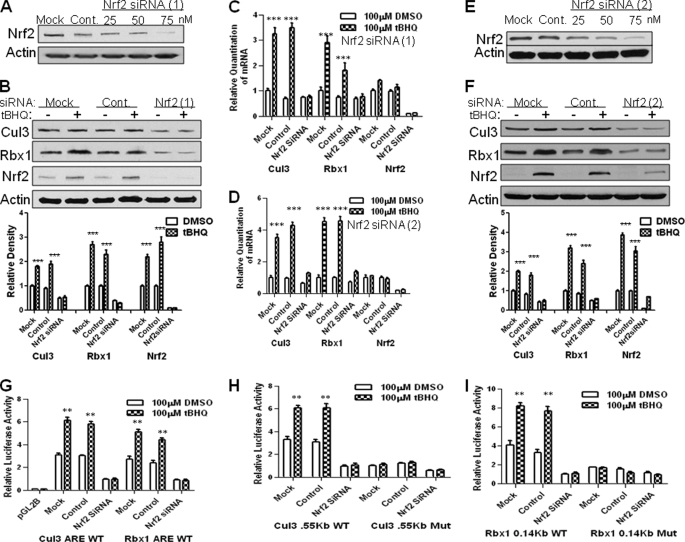
siRNA inhibition of Nrf2 decreases t-BHQ-inducible expression of Cul3 and Rbx1. A, Hepa-1 cells were transfected with control, 25, 50, or 75 nm of Nrf2 siRNA. Forty-eight hours after transfection, cells were harvested, lysed, and immunoblotted with anti-Nrf2 and actin antibodies. B, real time-PCR analysis of Hepa-1 cells transfected with control or Nrf2 siRNA. Twenty-four hours after siRNA transfection, cells were treated with 100 μm DMSO or t-BHQ. Cells were then harvested and total RNA was extracted and converted to cDNA. 50 ng of cDNA was analyzed for mRNA levels using Cul3 and Rbx1 primers and probes. Nrf2 was also probed for control. C, Western analysis of Cul3 and Rbx1 expression with Nrf2 siRNA. Hepa-1 cells were seeded and transfected with control or 75 nm of Nrf2 siRNA. Forty-eight hours after transfection, cells were harvested, lysed, and immunoblotted with anti-Cul3, anti-Rbx1, and anti-Nrf2 and anti-actin antibodies. Densitometry measurements of bands were quantitated and shown in graph blots to the right. D–F, A–C were repeated exactly as previously stated except with a different Nrf2 siRNA, the second Nrf2 siRNA will be referred to in the figures as Nrf2 siRNA (2). G–I, Hepa-1 cells were transfected with control or Nrf2 siRNA. Twenty-four hours after transfection, cells were transfected with Cul3 and Rbx1 promoters or Cul3 and Rbx1 WT and Mut. AREs. Cells were incubated for 16 h in the presence of DMSO or t-BHQ (100 μm). Transfected cells were harvested and analyzed for luciferase activity. The experiments were repeated three times, and the representative results are shown.
To further explore the role of Nrf2 in the t-BHQ-induced Cul3 and Rbx1 gene expressions, we transfected Hepa-1 cells with control or Nrf2 siRNA (Fig. 6, A--I) and analyzed for protein expression, gene expression, and luciferase activity. Real time-PCR analyses showed that Nrf2 siRNA, but not control siRNA, effectively inhibited Cul3 and Rbx1 mRNA expression (Fig. 6B). Western analysis showed that transfection of Hepa-1 cells with Nrf2 siRNA resulted in inhibition of Nrf2 and diminished t-BHQ induction of Cul3 and Rbx1 (Fig. 6C). In similar experiments, Nrf2 siRNA also inhibited Cul3 and Rbx1 WT AREs luciferase activity (Fig. 6G). Cul3 and Rbx1 promoters containing WT and Mutant AREs were also co-transfected in Hepa-1 cells with control or Nrf2 siRNA (Fig. 6, H–I). Nrf2 siRNA inhibited Cul3 and Rbx1 promoter luciferase activity. To make sure no off-target effects of a single Nrf2 siRNA were present, we transfected a different Nrf2 siRNA in Hepa1 cells (Fig. 6, D--F). The different Nrf2 siRNA showed similar results as the original siRNA causing decreased levels of Cul3 and Rbx1 mRNA expression and protein expression. The replacement of Hepa-1 with HepG2 cells also demonstrated Nrf2 mediated regulation of Cul3 and Rbx1 gene expression (data not shown).
Nrf2-mediated Up-regulation of Cul3 and Rbx1 Led to Increased Degradation of Nrf2
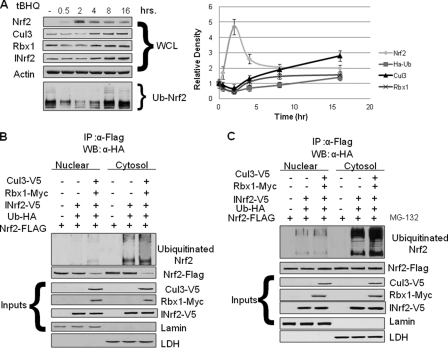
The treatment of Hepa-1 cells with antioxidant t-BHQ resulted in stabilization of Nrf2 that started within 0.5 h and peaked at 2 h after treatment (Fig. 7A). The Nrf2 levels declined at 4, 8, and 16 h after t-BHQ treatment. At 16 h, the Nrf2 levels were reduced to almost normal cellular levels. The stabilization of Nrf2 between 0.5 and 2 h led to increased expression of Cul3 and Rbx1 starting at 4 h and maximizing at 16 h. The ubiquitination of Nrf2 reduced at 0.5 and 2 h after t-BHQ treatment and then significantly increased at 8 and 16 h after t-BHQ treatment. In other words, t-BHQ-induced stabilization of Nrf2 was followed by increased expression of Cul3 and Rbx1 followed by increased ubiquitination and degradation of Nrf2. Our earlier published work has suggested that the Nrf2 is mostly degraded in cytoplasm as its degradation could be blocked in the presence of nuclear export inhibitor leptomycin B (31). However, we also found evidence that some of Nrf2 might also be degraded inside the nucleus (31). Next, we analyzed the cellular compartment-specific ubiquitination/degradation of Nrf2 in the absence and presence of proteasome inhibitor MG-132. Cytosol and nuclear extracts obtained from Nrf2-FLAG-transfected Hepa-1 cells were subjected to ubiquitination analysis (Fig. 7, B and C). The results demonstrate that overexpression of V5-Cul3, and Myc-Rbx1 leads to reduced levels of Nrf2 in both cytosol and nucleus confirming Cul3-Rbx1-mediated Nrf2 degradation (Fig. 7B, lower panel). However, enriching the ubiquitinated-Nrf2 indicated that most of the Nrf2 gets ubiquitinated in the cytosolic compartment (Fig. 7, B and C, upper panel). These results are complementary to our earlier published data and together conclude that Nrf2 ubiquitination and degradation mostly takes place in cytosol.
FIGURE 7.
Feedback loop between Nrf2 and Cul3-Rbx1. Activation of Nrf2 increases Cul3 and Rbx1 that degrades Nrf2. A, Hepa-1 cells were transfected with HA-Ub plasmid and treated with t-BHQ (100 μm) for the different time intervals. Whole cell lysates (WCL) were prepared and analyzed by Western blotting and probing with Cul3, Rbx1, and Nrf2 antibodies followed by β-actin antibody as loading control. INrf2 was used for positive control. In same experiment, whole cell lysates were immunoprecipitated (IP) with anti-Nrf2 antibody. Immunoprecipitates were analyzed by Western blotting and probing with anti-HA antibodies. Right panel, optical densities of Nrf2, Cul3, Rbx1, and ubiquitinated Nrf2 were normalized and plotted by time. The data presented are mean of three independent experiments. B and C, ubiquitination of Nrf2 in cytosol and nucleus. Hepa-1 cells were transfected with plasmids expressing Nrf2-FLAG, V5-INrf2, V5-Cul3, Myc-Rbx1, and HA-UB in the combinations as displayed. Cells were treated in the absence (B), or presence (C) of 10 μm MG-132 for 6 h. Cytosol and nuclear extracts were subjected to ubiquitination analysis similarly as in A. 50 μg of input extracts were probed with anti-V5, anti-FLAG, anti-Myc, anti-lamin B, and anti-lactate dehydrogenase antibodies.
DISCUSSION
Nrf2-mediated expression and coordinated induction of defensive genes, including detoxifying enzymes, is a mechanism of critical importance in protection against chemically induced oxidative stress and neoplasia (14). Therefore, the signals and mechanisms that regulate nuclear availability of Nrf2 are extremely important for the regulation of expression and induction of defensive genes (32). The regulation of Cul3 and Rbx1 is a very significant mechanism that controls Nrf2s abundance inside the nucleus. If Nrf2 is not tightly controlled, it can cause major problems within cells. Cul3 and Rbx1 regulation is also very important because they are responsible for the ubiquitination and degradation of Nrf2. To our knowledge, this is the first report demonstrating Nrf2s regulation of ubiquitin ligases.
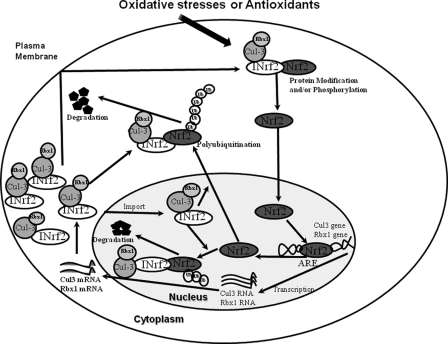
The results showed that antioxidant treatment induced the expression of Cul3 and Rbx1. This raised an interesting question regarding the mechanism of expression and antioxidant induction of Cul3 and Rbx1 and the in vivo role of increased Cul3 and Rbx1. The results also demonstrated the presence of a functional ARE on the forward and reverse strands of the Cul3 and Rbx1 promoters, respectively, able to bind Nrf2. The increase in expression of Nrf2 resulted in increased Cul3 and Rbx1 gene expressions. Mutations and deletions in the AREs and siRNA inhibition of Nrf2 significantly reduced both the expression and antioxidant induction of the Cul3 and Rbx1 genes. These experiments concluded that an ARE on the forward strand at nucleotide position −501 in the Cul3 promoter and an ARE on the reverse strand at nucleotide position +24 in the Rbx1 promoter and transcription factor Nrf2 regulated the expression and antioxidant induction of Cul3 and Rbx1 genes. Further studies showed that increased Cul3 and Rbx1 blocked Nrf2 activity by enhancing the ubiquitination and rapid degradation of Nrf2. In other words, Nrf2 induced Cul3 and Rbx1 for its own degradation. These results suggested the presence of a novel feedback autoregulatory loop between Cul3, Rbx1, and Nrf2 that controls cellular abundance of Cul3, Rbx1, and Nrf2 (Fig. 8).
FIGURE 8.
Autofeedback loop between Nrf2 and Cul3-Rbx1. A model that demonstrates an autoregulatory loop between Cul3-Rbx1 and Nrf2 is shown. The Nrf2 protein regulates the Cul3-Rbx1 genes at the level of transcription, and the Cul3-Rbx1 protein regulates the Nrf2 protein at the level of its activity.
The regulation of Cul3 and Rbx1 genes by the Nrf2 protein has an interesting consequence because Cul3 and Rbx1 protein can combine with Nrf2, via INrf2, and modulate down its activity as a transcription factor through degradation of Nrf2 (26). The regulation of Cul3 and Rbx1 is also important because there is a correlation of mutations within the ubiquitin-proteasome pathway and cancer (34). When Cul3 and Rbx1 protein is expressed in a cell, it blocks Nrf2 function, which results in less Nrf2 being made. Thus, the activity of Nrf2 and the levels of Cul3 and Rbx1 in a cell are kept in balance by the autoregulatory feedback loop. Factors that activate or inactivate either Cul3-Rbx1 or Nrf2 are expected to disrupt the autoregulatory loop with functional consequences. Xenobiotics and radiation that disturb this loop by dissociating Nrf2 from INrf2/Cul3-Rbx1 complex act to increase Nrf2 activity and Nrf2 downstream antioxidant gene expressions leading to protection and cell survival. Factors like mutations in Cul3 and Rbx1 can lead to their inactivation resulting in persistent nuclear accumulation of Nrf2 with adverse effects on cell survival.
In summary, Nrf2 serves as a sensor of oxidative and electrophilic stress. Nrf2 is translocated into the nucleus leading to activation of antioxidant genes that protect cells against adverse effects of chemical/radiation exposure. Nrf2 is then exported out of the nucleus, ubiquitinated, and degraded (33). Cul3 and Rbx1 are required for ubiquitination and degradation of Nrf2. Cul3 and Rbx1 are also capable of entering the nucleus to facilitate degradation of Nrf2 (21). A feedback autoregulatory loop between Cul3-Rbx1 and Nrf2 controls cellular abundance of Cul3, Rbx1, and Nrf2. Nrf2 regulates the expression and induction of Cul3-Rbx1 and their induction follow ubiquitination and degradation of Nrf2 and suppression of Cul3 and Rbx1 gene expressions. In other words, Nrf2 regulates Cul3 and Rbx1 by controlling its transcription and Cul3 and Rbx1 controls Nrf2 by facilitating its degradation.
Acknowledgments
We thank Dr. Suresh K. Niture and Dr. Emmanuel Unni, University of Maryland School of Medicine, Baltimore for helpful suggestions.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 ES012265.
- ROS
- reactive oxygen species
- Cul3
- Cullin 3
- Rbx1
- Ring box 1
- Nrf2
- NF-E2-related factor 2
- ARE
- antioxidant response element
- NQO1
- NAD(P)H:quinone oxidoreductase 1
- t-BHQ
- tert-butyl hydroquinone
- LMB
- leptomycin B
- LDH
- lactate dehydrogenase
- INrf2
- inhibitor of Nrf2 also known as Keap1
- ChIP
- chromatin immunoprecipitation
- siRNA
- short interfering RNA
- HA-Ub
- hemagglutinin-tagged ubiquitin
- ActD
- actinomycin D.
REFERENCES
- 1.Dhakshinamoorthy S., Long D. J., 2nd, Jaiswal A. K. (2000) Curr. Top. Cell Regul. 36, 201–216 [DOI] [PubMed] [Google Scholar]
- 2.Thelen M., Dewald B., Baggiolini M. (1993) Physiol. Rev. 73, 797–821 [DOI] [PubMed] [Google Scholar]
- 3.Breen A. P., Murphy J. A. (1995) Free Radic. Biol. Med. 18, 1033–1077 [DOI] [PubMed] [Google Scholar]
- 4.Bauer C. E., Elsen S., Bird T. H. (1999) Annu. Rev. Microbiol. 53, 495–523 [DOI] [PubMed] [Google Scholar]
- 5.Zhang D. D. (2006) Drug Metab. Rev. 38, 769–789 [DOI] [PubMed] [Google Scholar]
- 6.Kobayashi M., Yamamoto M. (2006) Adv. Enzyme. Regul. 46, 113–140 [DOI] [PubMed] [Google Scholar]
- 7.Copple I. M., Goldring C. E., Kitteringham N. R., Park B. K. (2008) Toxicology. 246, 24–33 [DOI] [PubMed] [Google Scholar]
- 8.Rushmore T. H., Pickett C. B. (1990) J. Biol. Chem. 265, 14648–14953 [PubMed] [Google Scholar]
- 9.Ikeda H., Nishi S., Sakai M. (2004) Biochem. J. 380, 515–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Venugopal R., Jaiswal A. K. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 14960–14965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang W., Jaiswal A. K. (2006) Free Radic. Biol. Med. 40, 1119–1130 [DOI] [PubMed] [Google Scholar]
- 12.Banning A., Deubel S., Kluth D., Zhou Z., Brigelius-Flohé R. (2005) Mol. Cell. Biol. 25, 4914–4923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim Y. J., Ahn J. Y., Liang P., Ip C., Zhang Y., Park Y. M. (2007) Cancer Res. 67, 546–554 [DOI] [PubMed] [Google Scholar]
- 14.Jaiswal A. K. (2004) Free. Radic. Biol. Med. 36, 1199–1207 [DOI] [PubMed] [Google Scholar]
- 15.Ramos-Gomez M., Kwak M. K., Dolan P. M., Itoh K., Yamamoto M., Talalay P., Kensler T. W. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 3410–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dhakshinamoorthy S., Jaiswal A. K. (2001) Oncogene 20, 3906–3917 [DOI] [PubMed] [Google Scholar]
- 17.Itoh K., Wakabayashi N., Katoh Y., Ishii T., Igarashi K., Engel J. D., Yamamoto M. (1999) Genes Dev. 13, 76–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang M. I., Kobayashi A., Wakabayashi N., Kim S. G., Yamamoto M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 2046–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang D. D., Lo S. C., Cross J. V., Templeton D. J., Hannink M. (2004) Mol. Cell. Biol. 24, 10941–10953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Furukawa M., Xiong Y. (2005) Mol. Cell. Biol. 25, 162–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niture S., Jaiswal A. K. (2009) J. Biol. Chem. 284, 13856–13868 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Geyer R., Wee S., Anderson S., Yates J., Wolf D. A. (2003) Mol. Cell. Biol. 12, 783–790 [DOI] [PubMed] [Google Scholar]
- 23.Sekhar K. R., Yan X. X., Freeman M. L. (2002) Oncogene 21, 6829–6834 [DOI] [PubMed] [Google Scholar]
- 24.Stewart D., Killeen E., Naquin R., Alam S., Alam J. (2003) J. Biol. Chem. 278, 2396–2402 [DOI] [PubMed] [Google Scholar]
- 25.Nguyen T., Sherratt P. J., Huang H. C., Yang C. S., Pickett C. B. (2003) J. Biol. Chem. 278, 4536–4541 [DOI] [PubMed] [Google Scholar]
- 26.Itoh K., Wakabayashi N., Katoh Y., Ishii T., O'Connor T., Yamamoto M. (2003) Genes Cells 8, 379–391 [DOI] [PubMed] [Google Scholar]
- 27.Bloom D. A., Jaiswal A. K. (2003) J. Biol. Chem. 278, 44675–44682 [DOI] [PubMed] [Google Scholar]
- 28.Maher J. M., Dieter M. Z., Aleksunes L. M., Slitt A. L., Guo G., Tanaka Y., Scheffer G. L., Chan J. Y., Manautou J. E., Chen Y., Dalton T. P., Yamamoto M., Klaassen C. D. (2007) Hepatology 46, 1597–1610 [DOI] [PubMed] [Google Scholar]
- 29.Wakabayashi N., Itoh K., Wakabayashi J., Motohashi H., Noda S., Takahashi S., Imakado S., Kotsuji T., Otsuka F., Roop D. R., Harada T., Engel J. D., Yamamoto M. (2003) Nat. Genet. 35, 238–245 [DOI] [PubMed] [Google Scholar]
- 30.Jain A. K., Jaiswal A. K. (2007) J. Biol. Chem. 282, 16502–16510 [DOI] [PubMed] [Google Scholar]
- 31.Jain A. K., Jaiswal A. K. (2006) J. Biol. Chem. 281, 12132–12142 [DOI] [PubMed] [Google Scholar]
- 32.Mohler J., Vani K., Leung S., Epstein A. (1991) Mech. Dev. 34, 3–9 [DOI] [PubMed] [Google Scholar]
- 33.Jain A. K., Bloom D. A., Jaiswal A. K. (2005) J. Biol. Chem. 280, 29158–29168 [DOI] [PubMed] [Google Scholar]
- 34.Mani A., Gelmann E. P. (2005) J. Clin. Oncol. 23, 4776–4789 [DOI] [PubMed] [Google Scholar]