Introduction
Much of what is currently understood about eukaryotic protein synthesis represents some very simple principles. DNA makes RNA makes protein. The genetic code accounts for the conversion of nucleic acid information into protein sequence information. The scanning hypothesis and the optimal sequence context around the initiating AUG define most of eukaryotic initiation using the predominant cap-dependent mechanism. The work of many has led to the type of initiation flow scheme shown in Fig. 1.
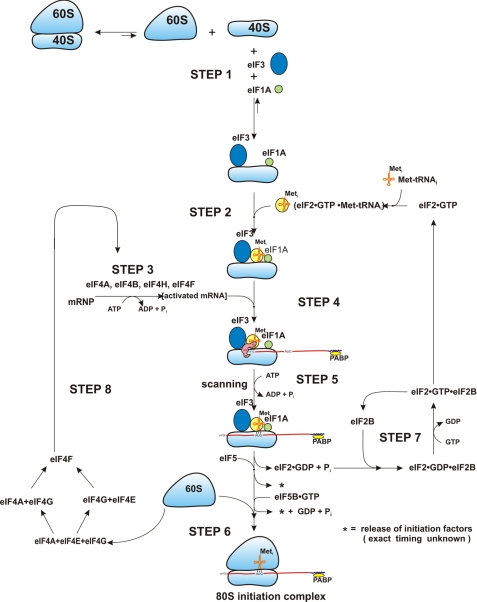
FIGURE 1.
The eukaryotic 80 S initiation pathway. A general scheme for the initiation steps in eukaryotic translation (m7G cap-dependent) is depicted. Elements that are uncertain are discussed in the text. This figure is adapted from Fig. 8 in Ref. 8.
Despite this detail, this picture is quite deceiving. First, there is more known than is depicted. Recent methodologies of cryo-electron microscopy and high field NMR combined with the almost miraculous crystal structures of bacterial ribosomes have added a considerable amount of information on the specificity and contact (working) surfaces of the ribosome and the translation factors that participate in protein biosynthesis. Second, methodologies are in place that allow for a more detailed analysis of the kinetic properties of the individual steps in this process. Finally, there are more and more examples emerging of elements influential in the regulation of translation as seen as post-translational modifications of the translation factors (or ribosomal proteins), non-coding RNAs, or cis-acting elements in the 5′- and 3′-UTRs2 of mRNAs that recruit trans-acting factors.
So where have we gone wrong? To begin with, much of the detail in Fig. 1 was determined in vitro with a series of partial confirmations in vivo, especially in the yeast system. As such, the major limitations to the pathway reflect the fact that most of the evidence used for formulation of the pathway was determined with artificial synthetic mRNAs, globin mRNA, or EMCV mRNA. It is usual to extrapolate from these experiments to more general mRNA translation; however, there are concerns with this approach. All of the in vitro systems function at a rate that is generally 5% (or less) of the in vivo rate, and there is rarely proof of turnover (i.e. that either a translation factor or the mRNA was used more than once). Second, a casual look at the pathway suggests that each translation initiation factor is used once, whereas available data indicate that multiple copies (or rounds of use) are required for one or more of the factors. Finally, each factor used in the in vitro experiments is present in the same stoichiometry. Numbers from mammalian systems and yeast indicate that this is clearly not the case, with the actual concentrations of different factors varying over a hundredfold range. However, the different concentrations of factors could reflect balancing concentration and binding constants or potential multiple turnovers. Thus, although the overall pathway shown in Fig. 1 may be correct, it certainly is not accurate in reflecting the full dynamics and kinetics of the initiation process.
Let us begin with “What's Right” with the model pathway shown in Fig. 1. For most mRNAs that are not regulated for expression, this pathway is accurate and reflects the translation of about 80% or more of the cellular mRNAs. Thus, changes that introduce secondary structure into the 5′-UTR reduce expression, as does an alteration of the nucleotide sequence around the initiating AUG (“the Kozak rules”) (1–3). The reduction in expression due to increased secondary structure can be reduced or eliminated by an increased concentration of eIF4A (4). Although not shown in Fig. 1, translation is very much dependent on the m7G cap at the 5′ end of the mRNA and recognition of this structure by eIF4E. Accessibility to this cap structure is the dominant determinant of the efficiency of an mRNA; this has been shown both experimentally and mathematically (5, 6). This establishes the competitive ranking of mRNAs for translation within the cell. Although not shown pictorially in step 3, this step is the conversion of an RNP complex with secondary and tertiary folding associated with a number of specific and nonspecific RNA-binding proteins into an activated mRNA, the place where the initial competition for translation begins (note that all mRNAs are initially visualized as messenger RNP particles that are about 50% RNA and 50% protein). From the numbers available, most cells contain more mRNA than is being translated, and thus, competition is a common element in translation.
A second element that seems correct is steps 7 and 8, which depict the recycling of eIF2 and eIF4F (7). These steps are the major sites for regulation of translation initiation. The control of eIF2 recycling is influenced by either phosphorylation of serine 51 in the α subunit of eIF2 or direct regulation of the activity of the exchange factor eIF2B. Reduction in ternary complexes, eIF2·GTP·Met-tRNAi, as a result of diminishing recycling seems to reduce the translation of all mRNAs equally. By contrast, regulation of the level of eIF4F activity, primarily via 4E-BP, which binds free eIF4E and thereby blocks the reconstitution of active eIF4F, accentuates the competition between mRNAs, more sharply favoring the more efficient mRNAs as the level of eIF4F activity is reduced (8).
A third step in Fig. 1 that is also correct is that the selection of the start codon is almost exclusively through the base paring of the anticodon of the Met-tRNAi and the start codon (additional effects via the purine at −3) (9). With this in mind, it then makes perfect sense that the ternary complex should arrive at the ribosome before the mRNA so that no matter where the AUG start codon is, the initiator tRNA would be in position to identify the start codon by base pairing.
Where is the pathway in Fig. 1 either incorrect or inaccurate? The pathway is ideally suited for the attachment of the first few ribosomes to an mRNA that begins as a messenger RNP. However, most of protein synthesis is accomplished by polysomes where multiple ribosomes are translating the mRNA. There is thus an apparent kinetic advantage for ribosomes that have just finished translating an open reading frame to undergo initiation on the same mRNA (the circular mRNA). This is envisioned both as a three-dimensional concept (the finishing ribosome is on average closer to the 5′ end of the mRNA than a 40 S subunit free in solution) and as a facilitated process (the interaction between the poly(A)-binding protein and eIF4F, which drives the circularization via protein/protein interactions). A second feature of the mRNA in polysomes is that, at least for the 5′-UTR and the coding region, there is likely very little protein present because it is removed by the scanning process or the process of elongation, although the protein content in the 3′-UTR may be unchanged. The absence of proteins should enhance placement of the mRNA into the mRNA track on the 40 S subunit. Thus, the pathway depicted for translation initiation may be correct for the first few rounds of translation but not for the more efficient rounds of expression from polysomes. Most importantly, the slow steps in the first initiation pathway may be decidedly different from polysome “reinitiation” and thus yield different predictions for the effect of restriction of ternary complexes or active levels of eIF4F.
Scanning
One of the oldest tenets of eukaryotic initiation is the scanning mechanism that incorporates the binding of the mRNA to the 5′ end of the mRNA by 43 S ribosomes (minimally 40 S·eIF3·eIF2·GTP·Met-tRNAi) and then subsequent movement of the ribosomal complex in a 3′ direction in search of the initiating AUG. Although much of the early work and various tests to evaluate the process were initiated by Dr. Marilyn Kozak (1), a number of other researchers have contributed to this work as well. The two most key features that evolved were that the recognition of the AUG start codon was exclusively via the codon/anticodon interaction between AUG and the anticodon of the initiator Met-tRNA (9) and that increases in secondary structure in the 5′-UTR lead to a decrease in the translational efficiency of the mRNA (10, 11). Kozak demonstrated that 40 S subunits bound near the m7G cap of the mRNA, and then in an ATP-dependent manner, migrated to the initiating AUG (1). To my knowledge, this is the only experiment that actually monitored the movement of the 43 S complex on the mRNA. Model studies with purified factors and synthetic mRNAs have shown that ATP is required for the binding of the 40 S ribosome to the initiating AUG (as a “toe print”); however, these studies do not distinguish whether the ATP requirement is for binding the mRNA to the 43 S complex (see step 3 in the pathway), for scanning of the mRNA (see step 5), or both (12, 13). In addition, it has not been clear which proteins are responsible for the ATP-dependent steps. Clearly eIF4A is sufficient in vitro because it is the only known protein added to assays that binds and hydrolyzes ATP. Other proteins have been suggested to participate in this process, but their role is not well defined among the three options (step 3, step 5, or both). Three RNA helicases have so far been implicated (Ded1, Dbp1, and Dhx29), but it is not clear whether the effect of these helicases reflects a specific protein requirement or a general level of RNA helicase activity such that a greater number of specific RNA helicases might make up for a deficiency in any one helicase (14).
The other question is how is scanning accomplished? The two extremes would be a processive movement where the 43 S complex slides along the mRNA continuously in a 5′ to 3′ direction in search of the initiating AUG and sensing each nucleotide along the way or the stochastic, random movement of the 43 S complex from the 5′ cap that on average moves 5′ to 3′ (although little steps in between may be in the 5′ to 3′ or 3′ to 5′ direction). Because DNA helicases are quite processive, unwinding thousands of nucleotides before releasing from the substrate, one could envision the placement of the enzyme on the surface of the 43 S ribosome so that the action of the helicase would pull the 43 S complex along the mRNA. However, none of the RNA helicases studied to date are similarly processive. In fact, most seem to affect the hydrolysis of only a single ATP and then are released from the substrate (at least as monitored by RNA duplex unwinding assays). Thus, even if the correct helicase had been defined as one of the four (including eIF4A) or a combination of all of them, the mechanism remains to be defined.
The above concerns address the simple “globin-like” mRNA. What happens when the mRNA has a long 5′-UTR or has a 5′-UTR that is GC-rich and by folding would appear to have numerous stem loops that must be melted? Could the scanning ribosome bypass such structures, perhaps by a ribosome-hopping mechanism, as noted in bacteria, or a shunting mechanism, as proposed for the adenovirus late 5′ leader (15, 16)? In this regard, the insertion of a simple but stable stem loop (ΔG > −30 kcal) appears to block the scanning 43 S ribosome. For mRNAs, with long and potentially structured 5′-UTRs, is there a different mechanism that employs similar features to the globin-like mRNA but that requires the recruitment of additional proteins for scanning (i.e. cis-acting sequences that recruit trans-acting factors)?
In all of this, one imagines a specific process based upon the need for an m7G cap for efficient initiation. However, for some mRNAs, there may be no factor requirement. Although this may be true for the HCV mRNA (see “Cellular IRES Elements” below), it is well known for the synthetic polymer poly(U), which served as the initial template for studies of elongation (the poly(U)-directed synthesis of polyphenylalanine).
Reinitiation
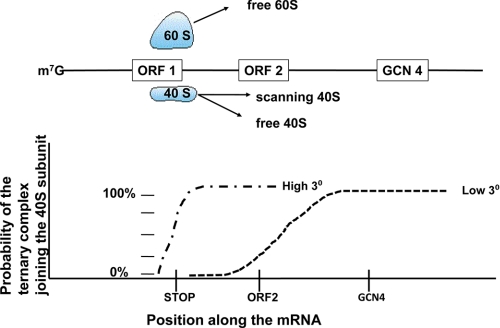
The best studied example of reinitiation is the synthesis of GCN4 in response to amino acid deprivation (17). The model developed to explain the regulation of GCN4 expression is that following the normal initiation of an upstream open reading frame as depicted in Fig. 1, the ribosome dissociates the 60 S subunit (and most of the 40 S subunits) at the termination codon, but 5–25% of the 40 S subunits remain bound to the mRNA and continue to scan in a 5′ to 3′ manner (Fig. 2). In the time period preceding arrival at the next start codon (Fig. 2, ORF2), at a minimum, a new ternary complex (eIF2·GTP·Met-tRNAi; Fig. 2, indicated by 3°) needs to be acquired. If this ternary complex is not acquired, then the 40 S subunit continues to scan to the start codon for GCN4. As this codon is considerably downstream of the ORF2 start codon, there is more time in which to acquire a ternary complex. Thus, the balance between recognizing the start codon at ORF2 or at GCN4 is governed by the kinetics of ternary complex acquisition. This process may be accentuated for GCN4 if there is sufficient secondary structure at the initiating AUG such that the 40 S subunit is likely to stall at that position.
FIGURE 2.
The process of reinitiation as seen in a model of the GCN4 mRNA. A ribosome is depicted having just completed the translation of the first small open reading frame (ORF1). At termination, the 60 S subunit is released, as are most, but not all, of the 40 S subunits. 40 S subunits that remain bound to the mRNA have a probability of acquiring a new ternary complex (3°) that is greatest under conditions of optimal protein synthesis (High 3°) but not under conditions of reduced protein synthesis (Low 3°). The probability of acquiring this second ternary complex is plotted below the schematic of the GCN4 mRNA. Initiation at ORF2 leads to termination and complete release of all subunits. Acquisition of a ternary complex 3′ of the AUG in ORF2 leads to the synthesis of GCN4 protein.
At a bare minimum, the following factors would be required: eIF2 as the ternary complex because only the tRNA anticodon will recognize the initiating AUG; eIF5, which serves as the GTPase-activating protein to trigger hydrolysis of the GTP in the ternary complex; and eIF5B, which permits the GTP-dependent joining with the 60 S subunit. Because both eIF1 and eIF1A function to open the mRNA-binding channel (presumably a requirement to allow for scanning) and eIF1A stabilizes the binding of the ternary complex, it is likely that these two proteins are also required (18).
As the GCN4 mRNA is already associated with the 40 S subunit, eIF3 would not be required to provide either a pool of free subunits for initiation (step 1) or a platform to assist the binding of the activated mRNA (step 4). However, eIF3 does stabilize the binding of the ternary complex to the 40 S subunit and may also play a role in the recruitment of other factors, either as an MFC or as individual factors. The eIF4 family of proteins (eIF4A, eIF4B, eIF4F, eIF4H) should not be required either for the activation of the mRNA or for the binding of the mRNA to the 40 S subunit. However, if the scanning described above is ATP-dependent, then either eIF4F or eIF4A (or perhaps a combination possibly including the RNA helicases described above) might be required as there has been no report of the 40 S subunit having either an ATP-binding site or the ability to scan. Once the initiating AUG has been identified, subsequent conversion to an 80 S initiation complex requires eIF5 and eIF5B as in the normal cap-dependent pathway (step 6). It remains to be determined whether this or some modified version of this scheme is responsible for the downstream initiations in the reinitiation pathway.
Individual Factors Versus the MFC
The original purification of mammalian and wheat germ translation factors sought to identify the smallest individual components required for initiation, elongation, or termination. The use of these proteins in model assays or partial assays then led to initiation schemes that depicted the addition or release of individual components at particular steps in the synthetic pathways. During purification, it was noted that a number of factors seemed to “stick” to other factors (eIF1A and eIF5B; eIF3 and eIF4B; eIF3 and eIF4F; eIF3 and eIF2; eIF4B and eIF4F; p67 and eIF2). The purification and subsequent analyses were consistent with individual translation factors binding to and being released from the 40 S subunit. This suggested that protein/protein interactions were occurring on the surface of the 40 S subunit and that these interactions were much tighter than initially realized. Thus, if protein A and protein B bound to each other with a Kd of 10−6 m in solution, that binding was probably 1–2 orders of magnitude tighter on the surface of the 40 S subunit.
With the development of the yeast system for factor analysis and the evolution of the “pulldown” assay, researchers have found complexes of greater size and number of components. The largest of these, the MFC, contains the ternary complex (eIF2·GTP·Met-tRNAi), eIF3, eIF1, and eIF5 (19). Although this and other complexes have been identified either by pulldown assays or during purification, there are still no kinetic data to confirm that such a complex(es) actually binds directly to the 40 S subunit.
Cellular IRES Elements
The groundbreaking studies of the Sonenberg (20) and Wimmer (21) groups on the IRES elements of poliovirus and EMCV, respectively, established the general principal that specific RNA sequences/structures exist that are capable of recruiting 40 S subunits. This led to the question as to whether there were cellular mRNAs that use a similar mechanism for expression. The answer is clearly yes, although which mRNAs truly contain an IRES element is still a topic for discussion (see the article by Wendy Gilbert in this series). However, the question has not been which mRNA, but rather what is the mechanism involved? At present, at least three distinct mechanisms have been proposed based upon the factors required for initiation in vitro. For viral elements, these are: eIF4E-independent but otherwise canonical initiation factors is required (i.e. poliovirus or EMCV); none of the eIF4 family of factors are required (i.e. HCV); and none of the canonical initiation factors are required (i.e. CrPV). Where studied, the cellular IRES elements all seem to fit the poliovirus model (22).
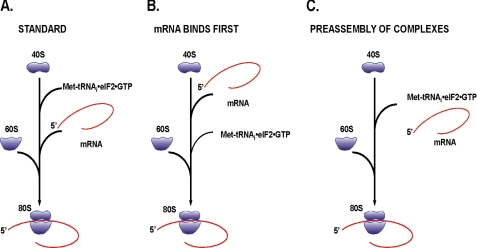
Although the above addresses the factor requirements, it does not delineate the mechanism involved in the process. With the identification of binding sites in the 5′-UTR of either poliovirus or EMCV mRNAs for eIF4F, it seemed that these structures are an alternative to the normal m7G cap that is recognized by the eIF4E subunit of eIF4F. If so, the initiation pathway might be identical to that shown in Fig. 1, with the caveat that a portion of the viral RNA functionally substitutes for the m7G cap. However, numerous publications have shown that when the levels of the ternary complex are reduced, the translation of EMCV mRNA is not altered. If the scheme in Fig. 1 is correct, the prediction is that all mRNAs will be reduced in expression to approximately the same degree. Because this is not what has been observed, are there alternate models that might explain this observation? Two alternative models to the standard scheme (Fig. 3A) are presented in Fig. 3, but there clearly could be more. The key feature is that some mechanism likely exists that would allow the IRES-containing mRNA to more competitively compete for the limiting ternary complex. The first model (Fig. 3B) proposes that the IRES-containing mRNA binds to the 40 S subunit before the binding of the ternary complex. In this way, the 40 S·mRNA complex could more efficiently bind the ternary complex (accentuated by codon/anticodon interactions and possible preferred 40 S confirmation). It is not clear whether the predominant effect would be on the forward reaction of binding the ternary complex or on the reverse reaction of releasing the ternary complex. In this way, 40 S·mRNA complexes would outcompete 40 S subunits. A second mechanism (Fig. 3C) proposes the formation of an MFC that would include the mRNA. Again, either through some interaction with the mRNA or a conformational change triggered by the mRNA, an MFC·mRNA outcompetes the free MFC for limiting ternary complexes. This competition feature is supported by the observation that under conditions of limiting eIF4F, the most competitive mRNAs are not reduced in expression, whereas the poorly competitive mRNAs see a drastic reduction in expression (8).
FIGURE 3.
Alternate pathways for initiation complex formation that are more competitive for limiting amounts of the ternary complex (eIF2·GTP·met-tRNAi). A shows the “normal, m7G cap-dependent” pathway for initiation where ternary complex binding precedes binding of the mRNA. B shows the possibility of the mRNA binding to the 40 S subunit prior to the binding of the ternary complex. C shows the possibility of an MFC preferentially sequestering both the mRNA and the ternary complex prior to both binding to the 40 S subunit. Options in either B or C would make translation of IRES-containing mRNAs less sensitive to the global reduction of ternary complexes.
The Protein Sediments with Ribosomes
Often, what appears to be a translational effect is further studied using sucrose gradients to determine whether protein (or mRNA) is undergoing a redistribution under a particular physiological condition. It is common that the protein of interest will sediment at a high molecular mass (in polysomes) when it is known that the purified protein is smaller (i.e. less than 10 S or 400 kDa). One needs to be cautious because ribosomes represent a considerable part of the soluble protein in a cell (15–25%), and any protein with a tendency to associate with other proteins may find a match in the ribosome. Also, for proteins that bind RNA, ribosomes/polysomes represent about 92% of the RNA in the cell, with most of the remainder being tRNA (perhaps because of its compact shape and small size, tRNA rarely binds to anything except the aminoacyl-tRNA synthetases). Thus, it is not surprising that most RNA-binding proteins are associated with ribosomes. There are also other cellular elements that may give the misleading impression of being in polysomes based solely upon their size (i.e. P bodies, cytoskeletal elements, etc.). Thus, there is a need to develop tests that are specific for polysomes and will distinguish them and other high molecular weight structures. Standard procedures in this regard include the use of puromycin, cycloheximide, RNase, or EDTA.
Homogeneous or Heterogeneous Populations?
Almost by definition, any high molecular protein complex is a mixture. This derives from the observation that on average, the protein synthesis error rate is 3–4 amino acids per 10,000. With an average of about 1,000,000 in molecular weight for proteins associated with each subunit, there should be 30–40 incorrect amino acids incorporated into each ribosomal subunit. The same is true for eIF3, which in mammals has a molecular weight of about 650,000 (in 13 subunits). By the same logic, eIF3 will contain 20–30 incorrect amino acids. If any of the mutant subunits fail to be incorporated into the larger complex, they are degraded.
However, the bigger question is “Are there complexes that lack a particular subunit function but have an mRNA-specific phenotype?” Two examples point to the existence of subpopulations of ribosomes. Fox and colleagues (23) have demonstrated the phosphorylation-dependent loss of ribosomal protein L13a from the ribosome as part of a silencing mechanism for the ceruloplasmin mRNA in response to interferon. The second is the characterization of the gene defect in X-linked dyskeratosis congenita as a pseudouridine synthase that modifies ribosomal RNA (24). The absence of this post-transcriptional modification yielded ribosomes that were defective in IRES-mediated translation. From just these two examples, one can clearly visualize the possible influence of ribosome modification as leading to either global reduction in translation, or perhaps more interestingly, the possibility of an mRNA-specific effect. One might thus anticipate studies of the consequences of phosphorylation of ribosomal protein S6 that is generally phosphorylated under conditions for optimal growth. Previous work indicated that there is no difference in the distribution of S6 phosphorylated ribosomes from non-phosphorylated ribosomes. In addition, in a yeast strain in which the target serine was mutated to alanine, the yeast grew like wild type (25). Both of these studies examined the influence on the major pathway of protein synthesis, the cap-dependent pathway. However, it is possible that alternate pathways (leaky scanning, reinitiation, IRES-mediated initiation, shunting) might be dramatically altered depending on S6 phosphorylation status. As these are minor pathways for translation initiation, the influence of this phosphorylation could easily be missed unless the cells/organism are/is examined under a variety of stress or developmental conditions.
Conclusion
In summary, there is more than enough work to be done over the next 20 years to keep a large cadre of researchers busy. Also, as is quite likely, there will be new observations from development, responses to stress, or disease that will provide new examples of post-translational regulation that alters translation, either globally or in an mRNA-specific manner, that will require the modification of exiting theories. Hopefully, this article, and the additional articles in this series, will provide a basis for better understanding the complexities of translational control.
Acknowledgments
The figures for this manuscript were prepared by Diane Baus. I thank Diane Baus and Dr. Lucas Reineke for critical reading of the manuscript and helpful comments.
This is the first article in the Thematic Minireview Series on Protein Synthesis. This minireview will be reprinted in the 2010 Minireview Compendium, which will be available in January, 2011.
- UTR
- untranslated region
- EMCV
- encephalomyocarditis virus
- RNP
- ribonucleoprotein
- IRES
- internal ribosome entry site
- MFC
- multifactor complex
- HCV
- hepatitis C virus.
REFERENCES
- 1.Kozak M. (1980) Cell 19, 79–90 [DOI] [PubMed] [Google Scholar]
- 2.Parkin N. T., Cohen E. A., Darveau A., Rosen C., Haseltine W., Sonenberg N. (1988) EMBO J. 7, 2831–2837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kozak M. (1986) Cell 44, 283–292 [DOI] [PubMed] [Google Scholar]
- 4.Svitkin Y. V., Pause A., Haghighat A., Pyronnet S., Witherell G., Belsham G. J., Sonenberg N. (2001) RNA 7, 382–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Godefroy-Colburn T., Thach R. E. (1981) J. Biol. Chem. 256, 11762–11773 [PubMed] [Google Scholar]
- 6.Godefroy-Colburn T., Ravelonandro M., Pinck L. (1985) Eur. J. Biochem. 147, 541–548 [DOI] [PubMed] [Google Scholar]
- 7.Mathews M. B., Sonenberg N., Hershey J. W. B. (eds) (2007) Translational Control in Biology and Medicine, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 8.Merrick W. C. (2003) Biochem. Mol. Biol. Educ. 31, 378–385 [Google Scholar]
- 9.Cigan A. M., Feng L., Donahue T. F. (1988) Science 242, 93–97 [DOI] [PubMed] [Google Scholar]
- 10.Kozak M. (1991) J. Biol. Chem. 266, 19867–19870 [PubMed] [Google Scholar]
- 11.Pelletier J., Sonenberg N. (1987) Biochem. Cell Biol. 65, 576–581 [DOI] [PubMed] [Google Scholar]
- 12.Dmitriev S. E., Pisarev A. V., Rubtsova M. P., Dunaevsky Y. E., Shatsky I. N. (2003) FEBS Lett. 533, 99–104 [DOI] [PubMed] [Google Scholar]
- 13.Pestova T. V., Kolupaeva V. G. (2002) Genes Dev. 16, 2906–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berthelot K., Muldoon M., Rajkowitsch L., Hughes J., McCarthy J. E. (2004) Mol. Microbiol. 51, 987–1001 [DOI] [PubMed] [Google Scholar]
- 15.Herr A. J., Nelson C. C., Wills N. M., Gesteland R. F., Atkins J. F. (2001) J. Mol. Biol. 309, 1029–1048 [DOI] [PubMed] [Google Scholar]
- 16.Yueh A., Schneider R. J. (2000) Genes Dev. 14, 414–421 [PMC free article] [PubMed] [Google Scholar]
- 17.Hinnebusch A. G. (2005) Annu. Rev. Microbiol. 59, 407–450 [DOI] [PubMed] [Google Scholar]
- 18.Passmore L. A., Schmeing T. M., Maag D., Applefield D. J., Acker M. G., Algire M. A., Lorsch J. R., Ramakrishnan V. (2007) Mol. Cell 26, 41–50 [DOI] [PubMed] [Google Scholar]
- 19.Singh C. R., Lee B., Udagawa T., Mohammad-Qureshi S. S., Yamamoto Y., Pavitt G. D., Asano K. (2006) EMBO J. 25, 4537–4546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pelletier J., Kaplan G., Racaniello V. R., Sonenberg N. (1988) Mol. Cell. Biol. 8, 1103–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jang S. K., Kräusslich H. G., Nicklin M. J., Duke G. M., Palmenberg A. C., Wimmer E. (1988) J. Virol. 62, 2636–2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elroy-Stein O., Merrick W. C. (2007) in Translational Control in Biology and Medicine (Mathews M. B., Sonenberg N., Hershey J. W. B., eds) pp. 155–172, Cold Spring Harbor Press, Cold Spring Harbor, NY [Google Scholar]
- 23.Mukhopadhyay R., Ray P. S., Arif A., Brady A. K., Kinter M., Fox P. L. (2008) Mol. Cell 32, 371–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoon A., Peng G., Brandenburger Y., Zollo O., Xu W., Rego E., Ruggero D. (2006) Science 312, 902–906 [DOI] [PubMed] [Google Scholar]
- 25.Kruse C., Johnson S. P., Warner J. R. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 7515–7519 [DOI] [PMC free article] [PubMed] [Google Scholar]