Abstract
A characteristic feature of tissue resident human mast cells (MCs) is their hTryptase-β-rich cytoplasmic granules. Mouse MC protease-6 (mMCP-6) is the ortholog of hTryptase-β, and we have shown that this tetramer-forming tryptase has beneficial roles in innate immunity but adverse roles in inflammatory disorders like experimental arthritis. Because the key tissue factors that control tryptase expression in MCs have not been identified, we investigated the mechanisms by which fibroblasts mediate the expression and granule accumulation of mMCP-6. Immature mouse bone marrow-derived MCs (mBMMCs) co-cultured with fibroblast-like synoviocytes (FLS) or mouse 3T3 fibroblasts markedly increased their levels of mMCP-6. This effect was caused by an undefined soluble factor whose levels could be increased by exposing FLS to tumor necrosis factor-α or interleukin (IL)-1β. Gene expression profiling of mBMMCs and FLS for receptor·ligand pairs of potential relevance raised the possibility that IL-33 was a sought after fibroblast-derived factor that promotes tryptase expression and granule maturation via its receptor IL1RL1/ST2. MCs lacking IL1RL1 exhibited defective fibroblast-driven tryptase accumulation, whereas recombinant IL-33 induced mMCP-6 mRNA and protein accumulation in wild-type mBMMCs. In agreement with these data, synovial MCs from IL1RL1-null mice exhibited a marked reduction in mMCP-6 expression. IL-33 is the first factor shown to modulate tryptase expression in MCs at the mRNA and protein levels. We therefore have identified a novel pathway by which mesenchymal cells exposed to inflammatory cytokines modulate the phenotype of local MCs to shape their immune responses.
Keywords: Cell-Cell Interaction, Cytokine, Fibroblast, Mast Cell, Serine Protease
Introduction
Mast cells (MCs)2 are granulated cells of the myeloid lineage that reside within connective tissues (1). Although it has been known for some time that MCs complete their differentiation and granule maturation after they exit the bone marrow (2–4), the factors and mechanisms governing the final stages of their development remain poorly understood at the molecular level. Although it was originally proposed that the phenotype of a mature MC was irreversibly determined before its progenitor exits the bone marrow, it is now known that human and mouse MCs exhibit substantial plasticity in their development and that MCs can quickly alter the expression of their granule mediators in a cytokine-dependent manner (2, 5–9).
All human MCs contain abundant amounts of hTryptase-β (10–12), which is a tetramer-forming serine protease with tryptic-like substrate specificity (13). The ortholog of hTryptase-β is mouse MC protease (mMCP)-6 (14, 15). No human has been identified who lacks MCs, in part, because their tryptase-serglycin proteoglycan complexes are essential for combating bacterial and helminthic infections efficiently (16–18).
In the context of inflammatory arthritis, the number of MCs often increases >10-fold in the chronically inflamed joint (19). Their prominent roles in experimental arthritis and other MC-dependent inflammatory disorders have heightened interest in hTryptase-β and mMCP-6 (20, 21). Neutrophil accumulation and loss of aggrecan proteoglycans from cartilage are markedly reduced in mMCP-6-null C57BL/6 (B6) mice relative to (WT) B6 mice in two inflammatory arthritis models (20, 21). These in vivo studies provided the first direct evidence for a prominent involvement of MC-restricted tryptases in arthritis.
Given the growing evidence documenting the functional consequences of MC-restricted tryptases in health and disease, the factors and mechanisms that control the expression and granule accumulation of these neutral proteases need to be identified. Exposure of mouse bone marrow-derived MCs (mBMMCs) to IL-3 results in the transient expression of mMCP-6 mRNA (15, 22). Nevertheless, the amount of enzymatically active mMCP-6 protein in these nontransformed IL-3-developed cells is paltry relative to that of the mature MCs of the synovium, skin, and other connective tissues. The key factor that controls tryptase accumulation in MCs apparently does not originate from T cells because tryptase levels are not diminished in the synovial MCs of lymphocyte-deficient mice (23). Although the development of all MCs in vivo is highly dependent on mesenchymal cell-derived kit ligand (Kitl)/stem cell factor (1, 24, 25), exposure of mBMMCs to recombinant Kitl does not result in a significant increase in mMCP-6 mRNA and/or protein levels (26). Thus, the most important factor that regulates tryptase expression in tissue MCs remains obscure.
Synovial tryptase+ MCs reside in close proximity to fibroblast-like synoviocytes (FLS) (27). Likewise, the tryptase+ MCs in connective tissues are often in direct contact with fibroblasts and other mesenchymal cells. These observations prompted us to reexamine the interactions between MCs and mouse FLS and 3T3 fibroblasts vis à vis tryptase expression. We previously noted that in vivo-differentiated rat peritoneal and human lung MCs (as well as in vitro-differentiated mBMMCs) tightly adhere to mouse 3T3 fibroblasts (28–30) and rat chondrocytes (31). During co-culture with fibroblasts, immature mBMMCs develop a histochemical phenotype that is more similar to that of the mature MCs in the arthritic joint, due in part to increased expression of heparin-containing serglycin proteoglycans (30). The co-cultured mBMMCs also undergo granule maturation as evidenced by their marked increased granule accumulation of histamine and the exopeptidase carboxypeptidase A3 (32, 33). In support of these data, the cell granules become electron-dense at the ultrastructural level (34). The accumulated data led us to hypothesize that the FLS in synovial tissue elaborate a factor other than Kitl that is essential for inducing MC granule maturation into tryptase+ cells that resemble those in many connective tissues.
Here, we report that mouse FLS and 3T3 fibroblasts induce cultured mBMMCs to markedly increase their accumulation of enzymatically active mMCP-6. Although these mesenchymal cells constitutively produce the unknown MC regulatory factor, we discovered that its levels are markedly increased when these cells encounter cytokines that participate prominently in arthritis and other inflammatory disorders. Unexpectedly, we discovered that IL-33, a recently identified cytokine (35, 36) that induces MCs to exocytose a spectrum of inflammatory cytokines and chemokines (37), is an important fibroblast-derived factor in our in vitro system that induces tryptase accumulation. Finally, we show that the relevant receptor on the surface of the mBMMCs that recognizes mouse FLS- and 3T3 fibroblast-derived IL-33 is “IL-1 receptor-like 1” (IL1RL1; also known as ST2) (38). Consistent with our in vitro observations, mMCP-6 mRNA levels were significantly reduced in the MCs that reside in the joint tissues of IL1RL1-null mice.
EXPERIMENTAL PROCEDURES
Mice
WT B6 mice were obtained from The Jackson Laboratory. IL1RL1−/− (39) and mMCP-6-null (17, 21) B6 mice have been previously described. Experiments were conducted using animal protocols approved by the Animal Care and Use Committee of the Dana Farber Cancer Institute and Brigham and Women's Hospital.
mBMMC·FLS and mBMMC·3T3 Fibroblast Co-culture Systems
IL-3-dependent B6 mBMMCs and FLS were generated as described previously (40–42). B6 mBMMCs were chosen for these studies because the MCs in B6 mice cannot express the related tetramer-forming tryptase mMCP-7 due to a splice-site mutation in its gene (43, 44), thereby allowing easier interpretation of our enzymatic data. FLS were chosen for our co-culture studies because these cells represent a more physiologic population of nontransformed mesenchymal cells obtained from the ankle joint where mMCP-6+ MCs reside. Mouse 3T3 fibroblasts (line TIB 68, American Type Culture Collection, Manassas, VA) also were chosen because we have shown that this cell line induces WT mBMMCs to undergo granule maturation and because 3T3 fibroblasts are readily available, thereby allowing others to reproduce and extend our findings.
For 2-week co-culture experiments with FLS, 2 × 104 FLS were seeded into each 24-well plate. Forty eight h later, 1 × 105 IL-3/Kitl-generated WT mBMMCs that had been in culture for more than 8 weeks were seeded into the plates and allowed to directly contact the FLS monolayer. Alternatively, the mBMMCs were physically separated from the FLS during the co-culture by placing the mBMMCs into the upper chamber of a transwell culture dish with a membrane that contains 0.2-μm pores (Nalge Nunc International, Roskilde, Denmark). Half of the medium was changed every 4 days in these mBMMC·FLS co-cultures. Cells were enumerated by cytofluorometric staining, and microbead quantification after adherence to one another and/or the extracellular matrix was disrupted using trypsin or 10 mm EDTA (45). mBMMCs were identified by their surface expression of Kit and their unique morphologic features (e.g. presence of intracellular granules). Viability was determined by trypan blue exclusion.
For 3-week co-culture experiments with mouse 3T3 fibroblasts, 5 × 105 4-week IL-3-generated WT and mMCP-6−/− B6 mBMMCs were seeded into 35-mm culture dishes that in each instance contained a confluent monolayer of fibroblasts, as described previously (30). The resulting mBMMC·3T3 fibroblast co-cultures were maintained for 3 weeks in the presence of 50% WEHI-3 cell conditioned medium as a source of IL-3. Kitl was not added to the culture medium in these experiments. Because the entire conditioned medium was replaced every other day, only those mBMMCs that physically contacted the 3T3 fibroblast cell line were studied at the end of the co-culture in the experiments carried out with these cells.
Cytofluorometry
Cytofluorometric staining was performed as described previously (46). In brief, samples were washed with phosphate-buffered saline (PBS) supplemented with 2% fetal calf serum and then stained with appropriate antibodies and isotype controls. Cells were preincubated with an anti-CD16/CD32 antibody to avoid Fc-receptor-mediated staining. Intracellular staining was performed after fixation and permeabilization with Perm/Cytoperm solution (BD Biosciences), following the manufacturer's instructions. Cytofluorometric analyses were performed using an FACSDiva cytometer (BD Biosciences). Data were analyzed utilizing the Flow-Jo software package (Tree Star, Ashland, OR). The antibodies used in these experiments were CD117-Alexa Fluor 647 (Caltag, Carlsbad, CA), rat IgG-Alexa Fluor 647 (Caltag), IL1RL1-FITC (MD Bioscience, St. Paul, MN), affinity-purified rabbit anti-mMCP-6 antibody (47), and anti-rabbit-IgG-FITC (Jackson ImmunoResearch, West Grove, PA).
Immunocytochemical Staining of mBMMCs with Anti-mMCP-6 Antibody
The 3-week WT mBMMC fibroblast co-cultures were washed with serum-free RPMI 1640 medium and then exposed to trypsin for ∼5 min. The detached and separated cells were then placed on glass slides using a standard cytospin approach (5 min of centrifugation, 500 rpm). Immunocytochemical staining was performed as described previously (48)
SDS-PAGE Immunoblot Analysis
Mouse FLS were grown to confluence in 6-well plates. In some instances, the resulting cells were stimulated with 5 ng/ml mouse recombinant TNF-α and/or IL-1β (PeproTech, Rocky Hill, NJ) for 24 h before the cytokine-treated cells were placed in lysis buffer (25 mm Tris-HCl, pH 7.6, 150 mm NaCl, 10 mm EDTA, 1% Nonidet P-40, 0.1% SDS, 1% sodium deoxycholate, 5 mm phenylmethylsulfonyl fluoride, and 1× protease inhibitor mixture (Sigma)). Lysates were clarified by centrifugation at 9,500 × g for 10 min at 4 °C and then boiled in Laemmli sample buffer for 5 min at 95 °C. The proteins in the lysates were separated by SDS-PAGE using 15% acrylamide gels. After transfer to polyvinylidene difluoride membranes, the resulting protein blots were blocked for 30 min at room temperature with 5% milk proteins in PBS, washed in PBS, and incubated with anti-IL-33 antibody (1:5,000 dilution; MBL, Nagoya, Japan) or anti-β-actin antibody (1:5,000 dilution; BioLegend, San Diego) for 2 h at room temperature. The treated blots were washed three times for 5 min each in PBS and then incubated for 1 h with horseradish peroxidase-conjugated donkey anti-rabbit antibody (1:1,000 dilution; Jackson ImmunoResearch). After another three washes with PBS, the blots were developed using SuperSignal West Pico chemiluminescent substrate (Thermo Scientific, Waltham, MA). Densitometric analyses were performed by using NIH ImageJ. Each signal was evaluated in comparison with that of β-actin. Using a similar approach, lysates of WT and mMCP-6-null B6 mBMMCs before and after 3 weeks of co-cultured with mouse 3T3 fibroblasts were subject to SDS-PAGE immunoblot analysis by probing the protein blots with affinity-purified rabbit anti-mMCP-6 antibody directed against residues 160–178 in this tryptase (47).
Tryptase Biochemical Assay
Lysates were prepared from mBMMCs by sonication after normalization of lysate buffer volume based on cell numbers. All culture conditions contained growth factors for mBMMCs with >95% viability based on trypan blue exclusion. Tryptase enzymatic activity was quantified using the chromogenic tryptase/trypsin substrate S-2288 (DiaPharma, Westchester, OH), measuring absorbance at 405 nm after a 0.5–6-h incubation at room temperature. Tryptase activity was expressed as the amount of substrate cleaved relative to a standard curve performed with known amounts of recombinant hTryptase-β (R & D Systems, Minneapolis, MN) or pancreatic trypsin (Sigma).
Microarray Analyses of mBMMCs and 3T3 Fibroblasts
Total RNA was isolated from IL-3 differentiated mMCP-6-null B6 mBMMCs. mMCP-6-null B6 mBMMCs (17) were used in these microarray analyses rather than WT B6 mBMMCs because we concluded that the former cells probably would optimally express those surface receptors that control the expression of the tryptase to try to correct for their mMCP-6 deficiency created by our homologous recombination approach. Total RNA also was isolated from 3T3 fibroblasts. In all instances, the extracted RNA was purified using the RNeasy kit (Qiagen, Valencia, CA). The resulting RNA samples were further processed using the recommended protocols, hybridized to mouse 430_2.0 GeneChipsTM and read on a GeneChip scanner (Affymetrix, Santa Clara, CA) using GenePix Pro 4.1 software by the Arthritis Microarray Core Facility, Brigham and Women's Hospital and Harvard Medical School, Boston. Data filtering, transformation, and normalization were performed according to established protocols. Differential expression analyses were performed using GenePattern software from the Broad Institute.
Real Time-Quantitative-PCR (RT-qPCR) Assays
For RT-qPCR assays, total RNA was isolated from mBMMCs, mouse FLS, and ankle joints using the RNeasy minikit (Qiagen). FLS were stimulated with 5 ng/ml mouse recombinant TNF-α and/or IL-1β (PeproTech) for 24 h. To extract total RNA from mouse ankle joints, the skin around the ankle joints was removed, and tissue from the distal tibia to the mid paw was carefully collected to avoid bone marrow contamination. Harvested ankles were immersed in RNAlater (Qiagen) to minimize degradation of RNA. The samples were treated with proteinase K (55 °C, 15 min), and cells were disrupted with Buffer RLT lysis buffer (Qiagen). For the follow-up RT-qPCR assays, purified RNA was converted in each instance into cDNA using Quantitect reverse transcription kit (Qiagen). RT-qPCRs were then performed with SYBR Green Mastermix (SABiosciences-Qiagen, Frederick, MD) using primers for glyceraldehyde 3-phosphate dehydrogenase (GAPDH), mMCP-6, IL-33, and Kit (SABiosciences) on an Mx3000p PCR machine (Stratagene, La Jolla, CA). Relative expression was calculated using the comparative threshold cycle method. mMCP-6 and IL-33 mRNA levels were then normalized to that of the GAPDH or Kit transcript.
Histomorphometric Enumeration of Synovial MCs
Histomorphometric enumeration of MCs was performed in a blinded fashion as described previously (23). Briefly, synovial MCs were enumerated by counting the number of toluidine blue+ cells in synovial tissue surrounding ankle and tarsal joints in mid-sagittal hind paw sections. An eyepiece reticle (Leica Microsystems, Wetzlar, Germany) was used to define a unit of 0.04 mm2 restricted to within 200 μm of the synovial lining layer.
Statistical Analysis
p values were calculated using Student's t test in Prism software package 4.00 (GraphPad Software, San Diego). p values smaller than 0.05 were considered significant.
RESULTS
A Soluble FLS-derived Factor Induces Immature mBMMCs to Increase Their Expression of mMCP-6
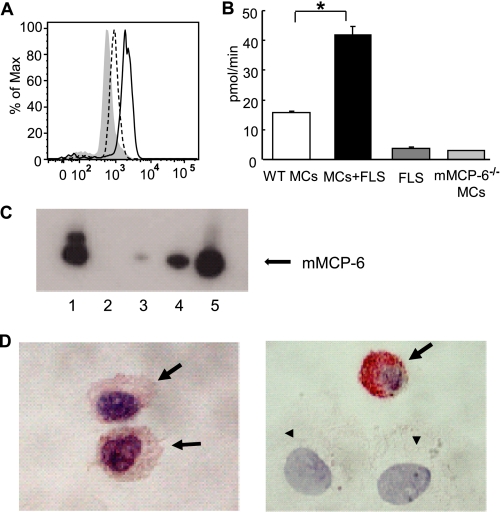
Using WT B6 mBMMCs (40), we examined whether FLS induced this nontransformed immature population of MCs to undergo granule maturation by measuring their levels of mMCP-6. After 2 weeks of co-culture with primary FLS, the granule content of mMCP-6 protein was quantified by intracellular cytofluorometry. In these studies, co-cultured mBMMCs exhibited a 2.5-fold increase in mMCP-6 protein relative to that of mBMMCs cultured in the absence of FLS (Fig. 1A). The tryptase activity of each sample was then quantified using the trypsin-susceptible chromogenic substrate S-2288. Consistent with the cytofluorometry results (Fig. 1A), 8-week mBMMCs co-cultured with FLS for an additional 2 weeks contained significantly more enzymatically active tryptase than mBMMCs maintained in the absence of FLS (Fig. 1B). In support of these data, the levels of mMCP-6 protein (Fig. 1C) and tryptase enzymatic activity (data not shown) increased over 10-fold (range 10–25-fold, n = 2) when 4-week WT mBMMCs were co-cultured with 3T3 fibroblasts for an additional 3 weeks in the absence of Kitl. In addition, mBMMCs before or after co-culture with fibroblasts were evaluated immunohistochemically for their mMCP-6 content. Consistent with the SDS-PAGE/tryptase biochemical assay data, mBMMCs in co-culture exhibited higher levels of mMCP-6 protein in their cytoplasmic granules than mono-cultured cells (Fig. 1D). Although the major tryptase present in B6 mBMMCs is mMCP-6, these cells also express mPrss31/tryptase-γ/transmembrane tryptase (22). To confirm that our tryptase functional measurements were not confounded by mPrss31, we quantified tryptase activity in mMCP-6−/−/mPrss31+/+ B6 mBMMCs co-cultured for 3 weeks with 3T3 fibroblasts; we found this activity was below our limit of detection (data not shown). Thus, mMCP-6 is the major tryptase in the secretory granules of the co-cultured B6 mBMMCs.
FIGURE 1.
Fibroblasts induce immature MCs to increase their levels of enzymatically active mMCP-6. A, intracellular cytofluorometric staining of Kit+ mBMMCs with anti-mMCP-6 antibody. B6 mBMMCs were collected after 14 days of co-culture with FLS (continuous line). Monoculture mBMMCs were maintained in parallel in medium supplemented with IL-3 and Kitl (dotted line). Gray shading shows control staining with anti-mMCP-6 antibody in tryptase-null (mMCP-6−/−/mMCP-7−/−) mBMMCs. B, tryptase enzymatic activities in mBMMCs co-cultured with FLS compared with control WT or mMCP-6−7− mBMMCs. As control, tryptase activities in FLS alone also are shown. IL-3 and Kitl (10 ng/ml, PeproTech) were added to all culture conditions. Results shown are means ± S.E. of data pooled from three independent experiments. *, p < 0.001 MCs + FLS co-culture versus MC monoculture condition. C, mMCP-6 protein levels in 4-week mBMMCs before and after an additional 3 weeks of co-culture with mouse 3T3 fibroblasts. Lysates were prepared from WT (lane 1) and mMCP-6−7− (lane 2) mBMMC fibroblast co-cultures and from WT monoculture mBMMCs maintained in parallel (lanes 3–5). Lanes 1 and 2 contain the protein content from ∼13,500 co-cultured mBMMCs in each instance. Lanes 3–5 contain the protein content from ∼15,000, 45,000, and 150,000 monocultured mBMMCs, respectively. As can be seen by comparing the data noted in lane 1 with that in lanes 3–5, co-cultured WT mBMMCs contain at least 10-fold more mMCP-6 protein on a per cell basis than non-co-cultured WT mBMMCs. The lack of an immunoreactive band in lane 2 documents the specificity of the anti-mMCP-6 antibody used in the SDS-PAGE immunoblot assay. The heterogeneous nature of the immunoreactive protein recognized by the anti-mMCP-6 antibody in this experiment is likely due to known variable glycosylation of the tryptase (75). Data are representative of two independent experiments. D, immunohistochemical staining with anti-mMCP-6 antibody of WT mBMMCs before (left panel) and after (right panel) co-culture with 3T3 fibroblasts. The alkaline phosphatase detection system confers a red color at sites of antibody binding. The slides were counterstained with hematoxylin. Fibroblasts (arrowheads) and MCs (arrows) in the right panel are labeled for clarity. Original magnitude, ×630.
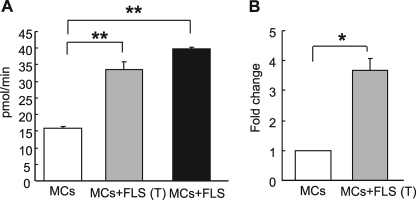
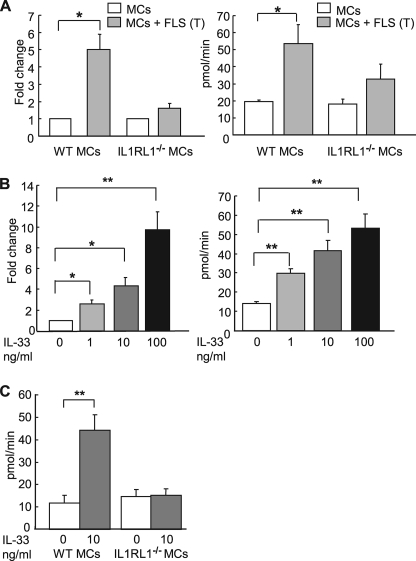
We extended these studies to assess whether or not the FLS-derived cytokine-like activity that promotes mMCP-6 expression requires direct cell-cell contact. For these studies, WT mBMMCs were allowed to physically contact FLS in the co-cultures. Alternatively, the two cell types were separated from one another using a porous membrane. Interestingly, mBMMCs that were prevented from contacting FLS using the transwell approach also demonstrated an elevation in tryptase enzymatic activity and mMCP-6 mRNA (Fig. 2, A and B).
FIGURE 2.
Soluble FLS-derived factor induces MCs to increase their tryptase activity and mMCP-6 mRNA levels. A, for co-culture, mBMMCs were allowed direct contact with FLS (MC+FLS) (black bar) or were co-cultured separately by a transwell membrane (MC+FLS (T)) (gray bar). B, mMCP-6 mRNA expression in mBMMCs after transwell co-culture with FLS (MC+FLS (T)) or after control mBMMCs culture. Data are presented as the fold induction of mMCP-6 mRNA after co-culture. The amount of mRNA in mBMMCs in control monoculture was defined as one. IL-3 and Kitl were added to all cultures in A and B. Results shown are the means ± S.E. of data pooled from three (A) and two (B) independent experiments. **, p < 0.01; *, p < 0.05.
Identification of Candidate Fibroblast-derived Soluble Mediators Using Microarray Approaches
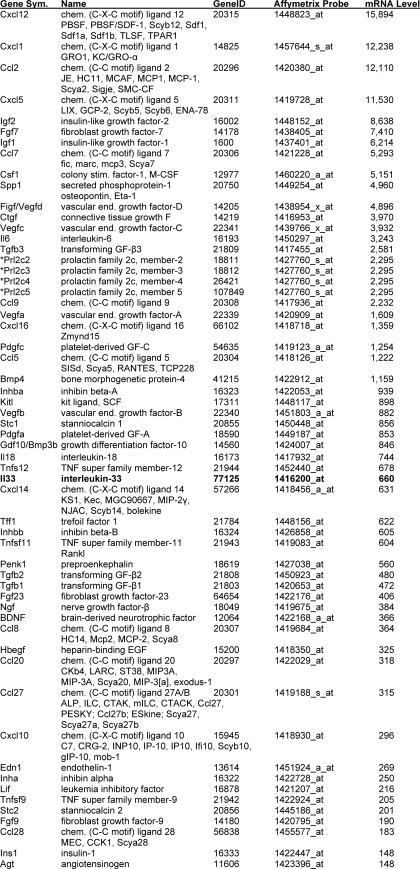
It is well known that fibroblasts express Kitl and that this cytokine promotes the development of MCs in vivo via its interaction with the tyrosine kinase receptor Kit on the surface of the MC-committed progenitor. Based on this example, we hypothesized that a similar cytokine·cytokine receptor mechanism probably promotes the FLS- and 3T3 fibroblast-dependent induction of mMCP-6 expression in mBMMCs. A high throughput microarray approach was therefore used to identify transcripts in mouse 3T3 fibroblasts that encode nearly every known protein, including 229 cytokines, chemokines, hormones, and growth and differentiation factors. We found that the transcripts that encode 59 of these 229 candidate proteins were constitutively expressed in mouse 3T3 fibroblasts at a level that exceeded 1% of the GAPDH and β-actin transcripts (Table 1 and supplemental Table 1). Kitl is a fibroblast-derived cytokine that constitutively regulates the development of MCs. As a validation of the microarray candidate approach, we noted that the Kitl transcript is the 24th most abundant transcript encoding a candidate protein that might regulate the co-cultured mBMMCs. In these microarray analyses, we also noted the prominent expression of the transcript that encodes the cytokine IL-33.
TABLE 1.
Constitutively expressed transcripts in mouse 3T3 fibroblasts that encode IL-33 and other candidate proteins that could regulate mMCP-6 expression in IL-3-developed mBMMCs
A microarray analysis of mRNA from mouse from 3T3 fibroblasts maintained in basal medium was carried out using an Affymetrix 430 2.0 GeneChip. The mRNA data for essentially every known protein can be found in supplemental Table 1. Noted below are the relative levels of the 58 most abundant transcripts that encode IL-33 and other candidate biologically active proteins that potentially could induce mBMMCs to increase their expression of mMCP-6. For reference, the arbitrary levels of the GAPDH and β-actin transcripts in these fibroblasts were 11,246–12,601 and 12,118–13,980 units, respectively. The levels of the noted candidate transcripts exceeded an arbitrary selected threshold of 1% of the levels of the transcripts that encode GAPDH and β-actin.
* Because the 1427760_s_at probe set on the GeneChip recognizes Prl2c2, Prl2c3, Prl2c4, and Prl2c5, it remains to be determined which prolactin family member is constitutively expressed in the fibroblasts.
We then searched for the transcripts that encode the receptors for our 59 candidate proteins in mMCP-6-null mBMMCs again using a high throughput microarray approach (Table 2 and supplemental Table 2). mBMMCs were found to contain abundant amounts of the transcript that encodes the IL-33 receptor IL1RL1. The levels of the IL1RL1 transcript in this population of MCs actually were greater than the levels of the Kit and GAPDH transcripts. Thus, the IL-33·IL1RL1 cytokine·cytokine receptor pair emerged as a prominent candidate signaling pathway for one of the soluble mediators elaborated by FLS and 3T3 fibroblasts that modulate mMCP-6 expression. Of note, others demonstrated a role for TGF-β-like cytokines in modulating mBMMC mMCP-6 mRNA levels (49). Indeed, our studies revealed the expression of TGF-β3 and its receptor in FLS and mBMMC, respectively (Tables 1 and 2). However, given the substantially higher mRNA expression level of IL1RL1 expression in mBMMC, we focused our attention on IL-33 and its receptor.
TABLE 2.
Constitutively expressed transcripts in IL-3-developed mBMMCs that encode receptors that recognize the fibroblast-derived factors noted in Table 1
A microarray analyses of IL-3-developed mMCP-6−7− mBMMCs was carried out using an Affymetrix 430 2.0 GeneChip. The mRNA levels in this MC population that encode essentially every mouse protein can be found in supplemental Table 2. Noted below are the mRNA levels of the five most abundant transcripts in mBMMCs that encode the receptors that recognize the fibroblast-derived biologically active proteins/ligands noted in Table 1. For reference, the arbitrary levels of the GAPDH transcript in these cells were 1600–5597 units.
Confirmation That FLS Express IL-33 and That mBMMCs Express IL1RL1
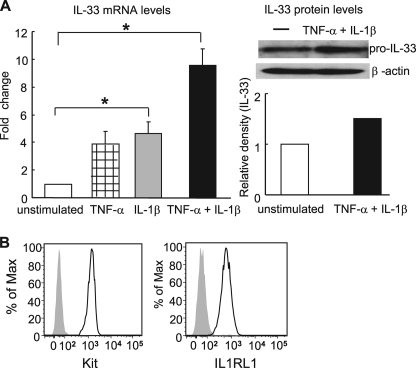
Having identified IL-33 as a potential candidate protein by which 3T3 fibroblasts might promote MC expression of mMCP-6, we proceeded to confirm the presence of this cytokine and its receptor in our FLS·mBMMC co-culture system. As seen in Fig. 3A, FLS constitutively expressed IL-33 mRNA and protein. Furthermore, the levels of this cytokine increased significantly in response to TNF-α and/or IL-1β. We next examined the presence of the IL-33 receptor on the surfaces of mBMMCs by cytofluorometry. As demonstrated previously (50, 51), we found that WT B6 mBMMCs expressed high levels of IL1RL1 protein on their plasma membranes in agreement with the microarray data (Table 2 and supplemental Table 2). Indeed, IL1RL1 staining intensity was similar to that of the abundantly expressed surface receptor Kit in this assay (Fig. 3B).
FIGURE 3.
FLS produce IL-33 and B6 mBMMCs express its receptor IL1RL1. A, IL-33 expression in FLS that were unstimulated (open bar) or were stimulated for 24 h with TNF-α (5 ng/ml) (striped bar), IL-1β (5 ng/ml) (gray bar), or TNF-α and IL-1β (black bar). Left panel, RT-qPCR measurement of IL-33 mRNA levels. Results shown are the means ± S.E. of data pooled from two independent experiments. *, p < 0.05 unstimulated versus IL-1β or TNF-α and IL-1β-stimulated FLS. Right panel, IL-33 protein expression followed by quantitative densitometry. Results shown are representative of data from two independent experiments. B, cell surface levels of IL1RL1 and Kit as assessed by cytofluorometry. Gray shaded area shows the staining of mBMMCs with isotype-matched antibody (left panel) or with an anti-IL1RL1 antibody in IL1RL1−/− mBMMCs (right panel).
FLS-derived IL-33 Regulates Tryptase Expression in mBMMCs
We next generated IL-3/Kitl driven mBMMCs from IL1RL1−/− and WT mice and examined mMCP-6 expression in these cells after they had been co-cultured with FLS. In contrast to WT mBMMCs, IL1RL1−/− mBMMCs exhibited no substantial change in mMCP-6 mRNA levels when co-cultured with FLS (Fig. 4A, left). In confirmatory studies, we examined the tryptase enzymatic activity in IL1RL1−/− mBMMC using the chromogenic assay. Tryptase activity was significantly increased in WT mBMMCs but not in IL1RL1−/− mBMMC in transwell co-culture with FLS (Fig. 4A, right). We next examined whether mouse recombinant IL-33 can directly induce tryptase expression in mBMMCs. For these studies, WT and IL1RL1-null mBMMCs were maintained in the presence of graded concentrations of recombinant IL-33 for 7 days. In these experiments, both mMCP-6 mRNA and functional tryptase were elevated in the WT mBMMCs by IL-33 in a dose-dependent manner (Fig. 4B). As anticipated, IL1RL1−/− mBMMCs were unresponsive to recombinant IL-33 (Fig. 4C).
FIGURE 4.
FLS-derived IL-33 promotes tryptase expression in mBMMCs. A, mMCP-6 mRNA expression (left panel) or tryptase enzymatic activity (right panel) in IL1RL1−/− or WT mBMMCs maintained in monoculture or co-cultured with FLS in transwell dishes. IL-3 and Kitl were added to all culture conditions. Results shown are the means ± S.E. of data pooled from two (left) and three (right) independent experiments. *, p < 0.05 monoculture versus co-culture WT mBMMC. B, mMCP-6 mRNA expression (left) and tryptase enzymatic activity (right) in B6 mBMMCs stimulated with graded concentrations of recombinant mouse IL-33 (R & D Systems) for 7 days. mMCP-6 mRNA expression was normalized to that of GAPDH. Results shown are the means ± S.E. of data pooled from three independent experiments. **, p < 0.01; *, p < 0.05. C, tryptase activities in IL1RL1−/− or WT mBMMCs stimulated with 10 ng/ml recombinant mouse IL-33 for 7 days. The mBMMCs in B and C were maintained in the presence of IL-3 and Kitl. Results shown are the means ± S.E. of data pooled from three independent experiments. **, p < 0.01.
From a technical standpoint, peripheral blood basophils are responsive to IL-3 and IL-33 (52, 53). Although mouse basophils express the tryptase family member mPrss34, they do not express mMCP-6 (54, 55). Nevertheless, basophils and basophil-committed progenitors are absent in the cell population obtained after unfractionated bone marrow cells are cultured >3 weeks in IL-3 enriched medium. To avoid potential confounding basophils in our studies, we utilized mBMMCs that had been in culture for at least 4 weeks in IL-3-enriched medium with or without Kitl. These findings established that FLS-derived IL-33 contributes to the regulation of mMCP-6 expression in MCs via the IL-33 receptor IL1RL1.
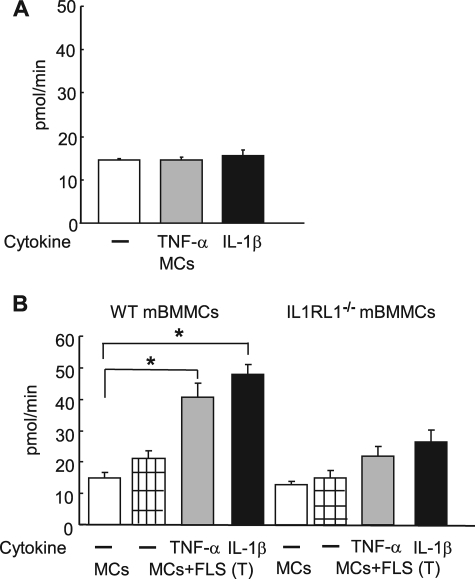
TNF-α and IL-1β Induce FLS to Increase Their Production of IL-33, Resulting in Augmented Tryptase Expression in Co-cultured mBMMCs
TNF-α and IL-1β are cytokines that participate in arthritis and other inflammatory disorders. mMCP-6 also plays a prominent role in two inflammatory arthritis models, promoting both disease intensity and cartilage injury. Having demonstrated a prominent role for the IL-33·IL1RL1 pathway in mMCP-6 induction in mBMMCs when these cells were co-cultured with FLS or 3T3 fibroblasts under basal conditions, we next examined the ability of TNF-α and IL-1β to modulate FLS regulation of the MC phenotype. Our rationale here was the marked increase in IL-33 mRNA levels observed in FLS when these mesenchymal cells encountered TNF-α and/or IL-1β (Fig. 3A). Indeed, exposure of mBMMCs·FLS co-cultures to TNF-α or IL-1β resulted in a prominent up-regulation of tryptase enzymatic activity. In contrast, these proinflammatory cytokines did not induce tryptase expression in mBMMCs in the absence of FLS (Fig. 5A). Additionally, the substantial increase of active tryptase enzymatic activity induced by TNF-α and IL-1β was remarkably abrogated in IL1RL1−/− mBMMCs (Fig. 5B). These observations demonstrate that the ability of FLS to impact the tryptase levels in MCs is augmented by stimuli typically derived from the TNF-α/IL-1β-expressing leukocytes that infiltrate tissues in inflammatory responses or potentially from MCs themselves in a local amplification loop (51).
FIGURE 5.
IL-33 secreted by TNF-α- or IL-1β-activated mouse FLS promotes a rapid increase in tryptase levels in WT mBMMCs. A, tryptase enzymatic activity in monoculture mBMMCs maintained in the presence of IL-3 and Kitl with or without 5 ng/ml TNF-α or IL-1β for 7 days. B, tryptase activity in WT (left) or IL1RL1−/− (right) mBMMCs maintained in monoculture or in transwell co-culture with FLS. Cells were cultured in the presence of IL-3 and Kitl with or without 5 ng/ml TNF-α or IL-1β (as labeled) for 7 days. Results shown are the means ± S.E. of data pooled from two independent experiments. *, p < 0.05 monoculture versus co-culture WT mBMMC in the presence of TNF-α or IL1β. p = not significant for IL1RL1−/− mBMMCs.
Decreased mMCP-6 mRNA Expression in IL1RL1−/− Synovial Tissue MCs
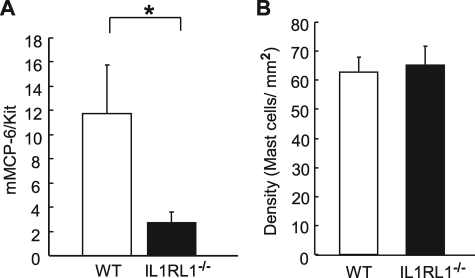
We next assessed the in vivo contribution of IL-33 to tissue MC tryptase levels. Because our previous studies demonstrated that synovial MCs express mMCP-6 (23), we examined the levels of this tryptase in the joint tissues of WT and IL1RL1−/− mice. Consistent with our in vitro data, mMCP-6 mRNA levels in synovial MCs of IL1RL1−/− mice were significantly decreased compared with that of WT mice (Fig. 6A). Because decreased mMCP-6 expression could result from a decreased tissue density of MCs or from less mMCP-6 expression in the MCs, we employed two approaches to control for decreased MCs as a trivial explanation for our findings. In the first method, we normalized our mMCP-6 mRNA levels to Kit mRNA levels because MCs are the major cell type in the joint that express Kit (Fig. 6A). In an independent approach, we quantified the density of MCs in the joints of WT and IL1RL1−/− mice. In these histomorphometric analyses, we found an equivalent density of MCs in both strains of mice (Fig. 6B).
FIGURE 6.
mMCP-6 mRNA expression is significantly decreased in synovial tissue MCs of IL1RL1−/− mice in vivo. A, mMCP-6 mRNA expression was quantified in joint tissues from WT (n = 4) or IL1RL1−/− (n = 6) ankles and then was normalized to that of Kit mRNA. *, p < 0.01. B, MC density was enumerated in ankle synovium from WT (n = 5) or IL1RL1−/− (n = 5) mice. Data are means ± S.E. Results are representative of two independent experiments.
DISCUSSION
In contrast to mucosal tissues, where chymase-expressing MCs predominate, the synovial sublining in mice and humans is studded with tryptase-expressing MC (56). This population of MCs increases in number in inflammatory arthritis. Because the phenotype of the MCs in synovial tissue is maintained in the absence of lymphocytes (23), we explored the mechanisms by which the mesenchymal cells in this tissue modulate MC maturation by examining the ability of FLS and a fibroblast cell line to regulate mMCP-6 expression in mBMMCs. Our initial studies revealed that FLS modulate mMCP-6 levels in cultured mBMMCs via an unknown soluble factor that is distinct from that of Kitl (Figs. 1 and 2). Microarray discovery approaches led us to hypothesize that IL-33 could be the relevant fibroblast-derived cytokine that regulates granule maturation of IL1RL1+ MCs (Tables 1 and 2 and supplemental Table 1). After having demonstrated that FLSs and 3T3 fibroblasts express IL-33 and that mBMMCs express its receptor (Fig. 3), recombinant IL-33 and IL1RL1-null mBMMCs were used in follow-up experimental approaches to show that this cytokine·receptor signaling pathway does indeed control the expression and granule accumulation of mMCP-6 in MCs in vitro (Fig. 4) and in vivo (Fig. 6). Genetic methods confirmed the contribution of IL-33 in modulating MC tryptase expression in vivo. Thus, these studies provide new insights beyond the biology of Kit/Kitl into mechanisms by which mesenchymal lineages in connective tissues regulate MC maturation.
IL-33 is a member of the IL-1 family of cytokines, and this factor was recently identified as a ligand for the orphan receptor IL1RL1 (36). IL-33 is known to be a potent activator of cytokine production in IL1RL1+ MCs (37, 50, 51, 57–59) as well as basophils (52, 53, 60), Th2 lymphocytes (36, 53), eosinophils (52, 61, 62), and myocytes (63, 64). Participation of IL-33 in disease pathophysiology has been uncovered in experimental arthritis (37, 65), infectious disease (66, 67), allergic inflammation (68–70), fibroproliferative disease (71), and cardiovascular disease (63, 64). This cytokine can have adverse or beneficial effects depending on the context in which it is expressed. IL-33 mRNA can be found in many organs, and there are presently 187 dbESTs in the human data base that originate from its gene (see GenBankTM Gene ID 9173 and UGID 130664). Relevant to our study, IL-33 is abundantly expressed in mesenchymal cells (e.g. smooth muscle cells, keratinocytes, and fibroblasts) (36), and this cytokine is elaborated by the synovial fibroblasts and vascular endothelial cells in the arthritic joint (37, 65, 72).
The demonstration that FLS-derived IL-33 participates in the granule accumulation of tryptase provides novel insight into a mechanism by which synovial tissue participates in disease pathophysiology. In the normal joint, FLS produce hyaluronan, lubricin, and other constituents of the normal joint fluid. However, these cells are relatively sparse (the compacted synovial lining is typically only 1–3 cells thick) and do not invade the cartilage in the joint or bone (73). FLS expand in number in the rheumatoid joint to form a hyperplastic lining frequently more than 10 cells thick. Previous studies revealed that these mesenchymal cells are key constituents in the pannus and that they are centrally involved in joint destruction. Because recent studies demonstrate that mMCP-6 contributes to inflammatory arthritis in multiple mouse model (20, 21), our observations now point to a previously unappreciated pathway by which FLS modulate disease activity indirectly by impacting the levels of proinflammatory tryptase in the synovium. Moreover, exposure of FLS to TNF-α or IL-1β induced in a rapid increase in the production of IL-33, accompanied by concomitant increases in MC tryptase expression (Fig. 5). Because TNF-α and IL-1β are prominent inflammatory mediators that originate from infiltrating leukocytes and tissue-resident synovial macrophages, our studies point to a pathway by which the synovial tissue amplifies these immune-derived signals to exacerbate joint inflammation via enhanced MC tryptase expression.
A number of factors (e.g. C5a, C3a, IgE/antigen, and IgG-containing immune complexes) induce varied populations of mouse and human MCs to coordinately release their preformed granule mediators and increase their expression of lipid and cytokine mediators. MCs can also be induced to produce cytokine mediators without emptying their granule contents when cellular activation occurs via a Toll-like receptor (74). Because IL-33 stimulates in vitro-differentiated human MCs to produce numerous cytokines and chemokines (57), the ability of this mesenchymal cell-derived factor to promote the granule accumulation of mMCP-6 was an unexpected finding. Indeed, we are unaware of another example where an exogenous factor stimulates MCs to increase their exocytosis of one class of mediators while promoting granule accumulation of another. Future investigations that are designed to deduce how IL-33·IL1RL1 signaling pathways differentially control which mediators accumulate and which are exocytosed from MCs should provide valuable information as to how this immune cell is regulated in the synovium and other connective tissue sites where IL-33 is present.
It deserves mention that the increase in tryptase expression that occurred when WT mBMMCs were co-cultured with FLS was not completely abrogated if IL1RL1−/− mBMMC was used (Fig. 4). This finding implies the presence of another soluble factor from FLS and fibroblasts that works in synergy with IL-33 to control mMCP-6 levels in MCs. Others have shown that TGF-β can regulate mMCP-6 mRNA levels in mBMMCs (49). As noted in Tables 1 and 2 and supplemental Tables 1 and 2, FLS constitutively express TGF-β3 and mBMMCs constitutively express its receptor Tgfbr1. Thus, TGF-β3 and its receptor are attractive candidates for further investigation in our co-culture system.
In summary, we demonstrate that IL-33 produced by mouse FLS and 3T3 fibroblasts promotes tryptase expression in mBMMCs and that synovium-derived IL-33 regulates mMCP-6 expression in the MCs that reside in joint tissues. Previous insights regarding mesenchymal cell-derived factors that regulate the granule phenotype of an MC have focused primarily on Kit·Kitl interactions. Our observations uncover a novel mechanism by which elaboration of IL-33 in the synovium can modulate MC tryptase expression. These observations delineate a means whereby FLS contribute to joint inflammation via enhancing MC tryptase expression. Our data raise the possibility that IL-33 regulation of IL1RL1+ MCs in other connective tissues contributes to beneficial and adverse roles of tryptases at those sites. MC-restricted tryptases have been implicated in numerous diseases and infections (16, 17). Thus, our observations point to further pathways by which tissues can impact and modulate MC-dependent immunity, inflammation, and connective tissue turnover.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants AI059746, AI065858, and HL036610. This work was also supported by research grants from The Cogan Family Foundation and the Harvard Club of Australia Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2.
- MC
- mast cell
- B6
- C57BL/6
- FLS
- fibroblast-like synoviocytes
- GAPDH
- glyceraldehyde 3-phosphate dehydrogenase
- IL
- interleukin
- IL1RL1
- IL-1 receptor-like 1
- Kitl
- kit ligand
- mMCP
- mouse MC protease
- PBS
- phosphate-buffered saline
- RT-qPCR
- real time-quantitative PCR
- TGF-β
- transforming growth factor-β
- TNF-α
- tumor necrosis factor-α
- WT
- wild type
- mBMMC
- mouse bone marrow-derived MC.
REFERENCES
- 1.Kitamura Y., Go S. (1979) Blood 53, 492–497 [PubMed] [Google Scholar]
- 2.Gurish M. F., Pear W. S., Stevens R. L., Scott M. L., Sokol K., Ghildyal N., Webster M. J., Hu X., Austen K. F., Baltimore D., et al. (1995) Immunity 3, 175–186 [DOI] [PubMed] [Google Scholar]
- 3.Gurish M. F., Austen K. F. (2001) J. Exp. Med. 194, F1–F5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gurish M. F., Tao H., Abonia J. P., Arya A., Friend D. S., Parker C. M., Austen K. F. (2001) J. Exp. Med. 194, 1243–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stevens R. L., Austen K. F. (1982) J. Biol. Chem. 257, 253–259 [PubMed] [Google Scholar]
- 6.Otsu K., Nakano T., Kanakura Y., Asai H., Katz H. R., Austen K. F., Stevens R. L., Galli S. J., Kitamura Y. (1987) J. Exp. Med. 165, 615–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanakura Y., Thompson H., Nakano T., Yamamura T., Asai H., Kitamura Y., Metcalfe D. D., Galli S. J. (1988) Blood 72, 877–885 [PubMed] [Google Scholar]
- 8.Ghildyal N., Friend D. S., Nicodemus C. F., Austen K. F., Stevens R. L. (1993) J. Immunol. 151, 3206–3214 [PubMed] [Google Scholar]
- 9.Friend D. S., Ghildyal N., Austen K. F., Gurish M. F., Matsumoto R., Stevens R. L. (1996) J. Cell Biol. 135, 279–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Irani A. A., Schechter N. M., Craig S. S., DeBlois G., Schwartz L. B. (1986) Proc. Natl. Acad. Sci. U.S.A. 83, 4464–4468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller J. S., Moxley G., Schwartz L. B. (1990) J. Clin. Invest. 86, 864–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vanderslice P., Ballinger S. M., Tam E. K., Goldstein S. M., Craik C. S., Caughey G. H. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 3811–3815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwartz L. B., Lewis R. A., Austen K. F. (1981) J. Biol. Chem. 256, 11939–11943 [PubMed] [Google Scholar]
- 14.Reynolds D. S., Stevens R. L., Lane W. S., Carr M. H., Austen K. F., Serafin W. E. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 3230–3234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reynolds D. S., Gurley D. S., Austen K. F., Serafin W. E. (1991) J. Biol. Chem. 266, 3847–3853 [PubMed] [Google Scholar]
- 16.Shin K., Watts G. F., Oettgen H. C., Friend D. S., Pemberton A. D., Gurish M. F., Lee D. M. (2008) J. Immunol. 180, 4885–4891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thakurdas S. M., Melicoff E., Sansores-Garcia L., Moreira D. C., Petrova Y., Stevens R. L., Adachi R. (2007) J. Biol. Chem. 282, 20809–20815 [DOI] [PubMed] [Google Scholar]
- 18.Huang C., De Sanctis G. T., O'Brien P. J., Mizgerd J. P., Friend D. S., Drazen J. M., Brass L. F., Stevens R. L. (2001) J. Biol. Chem. 276, 26276–26284 [DOI] [PubMed] [Google Scholar]
- 19.Crisp A. J., Chapman C. M., Kirkham S. E., Schiller A. L., Krane S. M. (1984) Arthritis Rheum. 27, 845–851 [DOI] [PubMed] [Google Scholar]
- 20.McNeil H. P., Shin K., Campbell I. K., Wicks I. P., Adachi R., Lee D. M., Stevens R. L. (2008) Arthritis Rheum. 58, 2338–2346 [DOI] [PubMed] [Google Scholar]
- 21.Shin K., Nigrovic P. A., Crish J., Boilard E., McNeil H. P., Larabee K. S., Adachi R., Gurish M. F., Gobezie R., Stevens R. L., Lee D. M. (2009) J. Immunol. 182, 647–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong G. W., Tang Y., Feyfant E., Sali A., Li L., Li Y., Huang C., Friend D. S., Krilis S. A., Stevens R. L. (1999) J. Biol. Chem. 274, 30784–30793 [DOI] [PubMed] [Google Scholar]
- 23.Shin K., Gurish M. F., Friend D. S., Pemberton A. D., Thornton E. M., Miller H. R., Lee D. M. (2006) Arthritis Rheum. 54, 2863–2871 [DOI] [PubMed] [Google Scholar]
- 24.Anderson D. M., Lyman S. D., Baird A., Wignall J. M., Eisenman J., Rauch C., March C. J., Boswell H. S., Gimpel S. D., Cosman D., et al. (1990) Cell 63, 235–243 [DOI] [PubMed] [Google Scholar]
- 25.Flanagan J. G., Leder P. (1990) Cell 63, 185–194 [DOI] [PubMed] [Google Scholar]
- 26.Gurish M. F., Ghildyal N., McNeil H. P., Austen K. F., Gillis S., Stevens R. L. (1992) J. Exp. Med. 175, 1003–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gruber B., Poznansky M., Boss E., Partin J., Gorevic P., Kaplan A. P. (1986) Arthritis Rheum. 29, 944–955 [DOI] [PubMed] [Google Scholar]
- 28.Levi-Schaffer F., Austen K. F., Caulfield J. P., Hein A., Bloes W. F., Stevens R. L. (1985) J. Immunol. 135, 3454–3462 [PubMed] [Google Scholar]
- 29.Levi-Schaffer F., Austen K. F., Caulfield J. P., Hein A., Gravallese P. M., Stevens R. L. (1987) J. Immunol. 139, 494–500 [PubMed] [Google Scholar]
- 30.Levi-Schaffer F., Austen K. F., Gravallese P. M., Stevens R. L. (1986) Proc. Natl. Acad. Sci. U.S.A. 83, 6485–6488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stevens R. L., Somerville L. L., Sewell D., Swafford J. R., Caulfield J. P., Levi-Schaffer F., Hubbard J. R., Dayton E. T. (1992) Arthritis Rheum. 35, 325–335 [DOI] [PubMed] [Google Scholar]
- 32.Serafin W. E., Dayton E. T., Gravallese P. M., Austen K. F., Stevens R. L. (1987) J. Immunol. 139, 3771–3776 [PubMed] [Google Scholar]
- 33.Dayton E. T., Pharr P., Ogawa M., Serafin W. E., Austen K. F., Levi-Schaffer F., Stevens R. L. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 569–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levi-Schaffer F., Dayton E. T., Austen K. F., Hein A., Caulfield J. P., Gravallese P. M., Liu F. T., Stevens R. L. (1987) J. Immunol. 139, 3431–3441 [PubMed] [Google Scholar]
- 35.Baekkevold E. S., Roussigné M., Yamanaka T., Johansen F. E., Jahnsen F. L., Amalric F., Brandtzaeg P., Erard M., Haraldsen G., Girard J. P. (2003) Am. J. Pathol. 163, 69–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmitz J., Owyang A., Oldham E., Song Y., Murphy E., McClanahan T. K., Zurawski G., Moshrefi M., Qin J., Li X., Gorman D. M., Bazan J. F., Kastelein R. A. (2005) Immunity 23, 479–490 [DOI] [PubMed] [Google Scholar]
- 37.Xu D., Jiang H. R., Kewin P., Li Y., Mu R., Fraser A. R., Pitman N., Kurowska-Stolarska M., McKenzie A. N., McInnes I. B., Liew F. Y. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 10913–10918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tominaga S., Yokota T., Yanagisawa K., Tsukamoto T., Takagi T., Tetsuka T. (1992) Biochim. Biophys. Acta 1171, 215–218 [DOI] [PubMed] [Google Scholar]
- 39.Townsend M. J., Fallon P. G., Matthews D. J., Jolin H. E., McKenzie A. N. (2000) J. Exp. Med. 191, 1069–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee D. M., Friend D. S., Gurish M. F., Benoist C., Mathis D., Brenner M. B. (2002) Science 297, 1689–1692 [DOI] [PubMed] [Google Scholar]
- 41.Lee D. M., Kiener H. P., Agarwal S. K., Noss E. H., Watts G. F., Chisaka O., Takeichi M., Brenner M. B. (2007) Science 315, 1006–1010 [DOI] [PubMed] [Google Scholar]
- 42.Razin E., Ihle J. N., Seldin D., Mencia-Huerta J. M., Katz H. R., LeBlanc P. A., Hein A., Caulfield J. P., Austen K. F., Stevens R. L. (1984) J. Immunol. 132, 1479–1486 [PubMed] [Google Scholar]
- 43.McNeil H. P., Reynolds D. S., Schiller V., Ghildyal N., Gurley D. S., Austen K. F., Stevens R. L. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 11174–11178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hunt J. E., Stevens R. L., Austen K. F., Zhang J., Xia Z., Ghildyal N. (1996) J. Biol. Chem. 271, 2851–2855 [DOI] [PubMed] [Google Scholar]
- 45.Rittner H. L., Mousa S. A., Labuz D., Beschmann K., Schäfer M., Stein C., Brack A. (2006) J. Leukocyte Biol. 79, 1022–1032 [DOI] [PubMed] [Google Scholar]
- 46.Nigrovic P. A., Gray D. H., Jones T., Hallgren J., Kuo F. C., Chaletzky B., Gurish M., Mathis D., Benoist C., Lee D. M. (2008) Am. J. Pathol. 173, 1693–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ghildyal N., Friend D. S., Stevens R. L., Austen K. F., Huang C., Penrose J. F., Sali A., Gurish M. F. (1996) J. Exp. Med. 184, 1061–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McNeil H. P., Frenkel D. P., Austen K. F., Friend D. S., Stevens R. L. (1992) J. Immunol. 149, 2466–2472 [PubMed] [Google Scholar]
- 49.Funaba M., Ikeda T., Murakami M., Ogawa K., Abe M. (2005) Cell. Signal. 17, 121–128 [DOI] [PubMed] [Google Scholar]
- 50.Moritz D. R., Rodewald H. R., Gheyselinck J., Klemenz R. (1998) J. Immunol. 161, 4866–4874 [PubMed] [Google Scholar]
- 51.Moulin D., Donzé O., Talabot-Ayer D., Mézin F., Palmer G., Gabay C. (2007) Cytokine 40, 216–225 [DOI] [PubMed] [Google Scholar]
- 52.Pecaric-Petkovic T., Didichenko S. A., Kaempfer S., Spiegl N., Dahinden C. A. (2009) Blood 113, 1526–1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smithgall M. D., Comeau M. R., Yoon B. R., Kaufman D., Armitage R., Smith D. E. (2008) Int. Immunol. 20, 1019–1030 [DOI] [PubMed] [Google Scholar]
- 54.Wong G. W., Yasuda S., Morokawa N., Li L., Stevens R. L. (2004) J. Biol. Chem. 279, 2438–2452 [DOI] [PubMed] [Google Scholar]
- 55.Ugajin T., Miyatani H., Demitsu T., Iwaki T., Ushimaru S., Nakashima Y., Yoshida Y. (2009) Intern. Med. 48, 693–695 [DOI] [PubMed] [Google Scholar]
- 56.Nigrovic P. A., Lee D. M. (2005) Arthritis Res. Ther. 7, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Allakhverdi Z., Smith D. E., Comeau M. R., Delespesse G. (2007) J. Immunol. 179, 2051–2054 [DOI] [PubMed] [Google Scholar]
- 58.Ho L. H., Ohno T., Oboki K., Kajiwara N., Suto H., Iikura M., Okayama Y., Akira S., Saito H., Galli S. J., Nakae S. (2007) J. Leukocyte Biol. 82, 1481–1490 [DOI] [PubMed] [Google Scholar]
- 59.Iikura M., Suto H., Kajiwara N., Oboki K., Ohno T., Okayama Y., Saito H., Galli S. J., Nakae S. (2007) Lab. Invest. 87, 971–978 [DOI] [PubMed] [Google Scholar]
- 60.Kroeger K. M., Sullivan B. M., Locksley R. M. (2009) J. Leukocyte Biol. 86, 769–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Suzukawa M., Koketsu R., Iikura M., Nakae S., Matsumoto K., Nagase H., Saito H., Matsushima K., Ohta K., Yamamoto K., Yamaguchi M. (2008) Lab. Invest. 88, 1245–1253 [DOI] [PubMed] [Google Scholar]
- 62.Cherry W. B., Yoon J., Bartemes K. R., Iijima K., Kita H. (2008) J. Allergy Clin. Immunol. 121, 1484–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sanada S., Hakuno D., Higgins L. J., Schreiter E. R., McKenzie A. N., Lee R. T. (2007) J. Clin. Invest. 117, 1538–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seki K., Sanada S., Kudinova A. Y., Steinhauser M. L., Handa V., Gannon J., Lee R. T. (2009) Circ. Heart Fail. 2, 684–691 [DOI] [PubMed] [Google Scholar]
- 65.Palmer G., Talabot-Ayer D., Lamacchia C., Toy D., Seemayer C. A., Viatte S., Finckh A., Smith D. E., Gabay C. (2009) Arthritis Rheum. 60, 738–749 [DOI] [PubMed] [Google Scholar]
- 66.Xu D., Chan W. L., Leung B. P., Huang F., Wheeler R., Piedrafita D., Robinson J. H., Liew F. Y. (1998) J. Exp. Med. 187, 787–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Humphreys N. E., Xu D., Hepworth M. R., Liew F. Y., Grencis R. K. (2008) J. Immunol. 180, 2443–2449 [DOI] [PubMed] [Google Scholar]
- 68.Pushparaj P. N., Tay H. K., H'ng S. C., Pitman N., Xu D., McKenzie A., Liew F. Y., Melendez A. J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 9773–9778 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 69.Kearley J., Buckland K. F., Mathie S. A., Lloyd C. M. (2009) Am. J. Respir. Crit. Care Med. 179, 772–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu X., Li M., Wu Y., Zhou Y., Zeng L., Huang T. (2009) Biochem. Biophys. Res. Commun. 386, 181–185 [DOI] [PubMed] [Google Scholar]
- 71.Tajima S., Bando M., Ohno S., Sugiyama Y., Oshikawa K., Tominaga S., Itoh K., Takada T., Suzuki E., Gejyo F. (2007) Exp. Lung Res. 33, 81–97 [DOI] [PubMed] [Google Scholar]
- 72.Carriere V., Roussel L., Ortega N., Lacorre D. A., Americh L., Aguilar L., Bouche G., Girard J. P. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 282–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee D. M., Kiener H. P., Brenner M. D. (2005) in Kelley's Textbook of Rheumatology (Harris E. D., Budd R. C., Firestein G. S., Genovese M. C., Sergent J. S., Ruddy S., Sledge C. B. eds) pp. 175–188, 7th Ed., W.B. Saunders/Elsevier, Philadelphia [Google Scholar]
- 74.Leal-Berumen I., Conlon P., Marshall J. S. (1994) J. Immunol. 152, 5468–5476 [PubMed] [Google Scholar]
- 75.Ghildyal N., Friend D. S., Freelund R., Austen K. F., McNeil H. P., Schiller V., Stevens R. L. (1994) J. Immunol. 153, 2624–2630 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.