Abstract
gp130 is the shared signal-transducing receptor subunit for the large and important family of interleukin 6-like cytokines. Previous x-ray structures of ligand-receptor complexes of this family lack the three membrane-proximal domains that are essential for signal transduction. Here we report the crystal structure of the entire extracellular portion of human gp130 (domains 1–6, D1–D6) at 3.6 Å resolution, in an unliganded form, as well as a higher resolution structure of the membrane-proximal fibronectin type III domains (D4–D6) at 1.9 Å. This represents the first atomic resolution structure of the complete ectodomain of any “tall” cytokine receptor. These structures show that other than a reorientation of the D1 domain, there is little structural change in gp130 upon ligand binding. They also reveal that the interface between the D4 and D5 domains forms an acute bend in the gp130 structure. Key residues at this interface are highly conserved across the entire tall receptor family, suggesting that this acute bend may be a common feature of these receptors. Importantly, this geometry positions the C termini of the membrane-proximal fibronectin type III domains of the tall cytokine receptors in close proximity within the transmembrane complex, favorable for receptor-associated Janus kinases to trans-phosphorylate and activate each other.
Keywords: Cell Surface Receptor, Crystallography, Jak Kinase, Receptor Structure-Function, Signal Transduction, Cytokine Receptor, Ectodomain, Fibronectin Type III Domains, gp130
Introduction
The cell surface receptor gp130 is a shared signaling receptor subunit for the interleukin 6 (IL-6)4-like cytokine family, which includes IL-6, IL-11, leukemia inhibitory factor (LIF), oncostatin M (OSM), and ciliary neurotrophic factor (CNTF) (1–4). This family of cytokines is involved in inflammatory and immune responses and also plays crucial roles in hematopoiesis, liver and neuronal regeneration, embryonic development, and fertility (1–4). Dysregulation of signaling contributes to diseases such as inflammatory bowel disease, osteoporosis, multiple sclerosis, multiple myeloma, and prostate cancer (1–4).
gp130 is the principal member of the “tall” cytokine receptor family, which also includes LIFR, OSM receptor, IL-12Rβ2, IL-27Rα, IL-31Rα, granulocyte colony-stimulating factor receptor, and leptin receptor (5). From sequence homology, the ectodomains of these receptors are predicted to be modular, containing 5–8 structural domains (6). gp130 has six domains: an N-terminal Ig-like domain (D1); a cytokine-binding module (D2–D3); and three contiguous, membrane-proximal, fibronectin type III (FNIII) domains (D4–D6) that define the tall receptor family.
Within this family, signaling occurs through two mechanisms. Binding of cytokines such as IL-6 or IL-11 to their specific cell surface receptor α-chains leads to gp130 homodimerization; binding of other cytokines of the IL-6 family can lead to heterodimerization of gp130 with a second signal-transducing tall receptor, such as LIFR or OSM receptor. Upon cytokine binding, the C-terminal FNIII domain “legs” of the tall receptors are brought into position on the cell surface with the correct proximity and orientation to allow trans-phosphorylation of receptor-associated Janus kinases (JAKs). Activated JAKs phosphorylate the receptors on specific tyrosine residues, creating docking sites for molecules with SH2 domains including the signal transducers and activators of transcription (STATs), which subsequently translocate to the nucleus to initiate the transcription of cytokine-responsive genes.
A variety of high resolution crystal structures of cytokine receptors have shown how tall receptors bind ligand and oligomerize with other signaling or non-signaling subunits through the membrane-distal region (7–12). Low resolution electron microscopy (EM) images showed that the legs approach each other close to the membrane in a number of signaling-competent gp130 complexes (5, 13, 14). It has also been established by mutagenesis that the legs of the receptor provide an essential geometry required for signaling (15–17). However, high resolution structural detail concerning these critical membrane-proximal FNIII domains is still lacking.
Here we report crystal structures of the entire extracellular portion of human gp130 (D1–D6) at 3.6 Å resolution and of the membrane-proximal FNIII domains alone (D4–D6) at 1.9 Å resolution. These structures together with previously published crystallographic and EM studies further our understanding of how the tall receptor family assemble high affinity signaling complexes.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Human gp130 D1–D6 (residues 1–597), bearing an N-terminal gp67 signal peptide, FLAG tag, and tobacco etch virus protease cleavage site, was expressed in Sf21 insect cells. Recombinant protein was purified using anti-FLAG M2-agarose (Sigma) and gel filtration chromatography. Human gp130 D4–D6 (residues 301–591) was expressed as an N-terminal His6 fusion in Escherichia coli BL21(DE3) cells and purified by sequential nickel affinity, anion exchange, and gel filtration chromatography. See Supplemental Methods for details.
Crystallization and Structure Determination
Crystallization was accomplished by hanging-drop vapor diffusion at 20 °C. Two crystal forms of gp130 D4–D6 were obtained from 12–20% polyethylene glycol 8000, 0.056 m NaCl, 0.1 m Tris-HCl, pH 8.5. Seleno-methionine (Se-Met)-substituted crystals were obtained under the same conditions. gp130 D1–D6 crystals were obtained from 1.5–2.0 m (NH4)2SO4, 0.1 m imidazole-malonate, pH 6.0.
Diffraction data were collected on beamlines MX1 or MX2 at the Australian Synchrotron. For D4–D6 structures, data were integrated and scaled using HKL2000 (18). A multiwavelength anomalous dispersion dataset (inflection point and remote) was collected from a Se-Met derivative for orthorhombic D4–D6 crystals. Four Se-Met sites were located using PHENIX (19), allowing phases to be calculated, and a model was built automatically, with manual editing performed in COOT (20). Refinement was performed using PHENIX and converged with Rwork = 0.177 and Rfree = 0.220 for data to 1.9 Å resolution. A native dataset was collected for monoclinic D4–D6 crystals and integrated to 3.1 Å. The 1.9 Å D4–D6 structure was used as a molecular replacement model using PHASER (21). Building and refinement were performed in COOT (20) and PHENIX (19), respectively. D1–D6 diffraction data were processed using XDS (22) and scaled with SCALA (23). A molecular replacement solution was found using PHASER (21), using our 1.9 Å structure of D4–D6 and gp130 D2–D3 from Protein Data Bank (PDB) entry 1P9M (11) as models. Refinement and building were performed in REFMAC (24) and COOT (20), respectively. Examples of electron density for 1.9 Å D4–D6 and 3.6 Å D1–D6 structures are shown in supplemental Fig. 1. Surface areas of the inter-FNIII domain interfaces were calculated using AREAIMol (23).
Fit of Crystal Structures into EM Density Map
We fitted our model structure into the EM map manually using PyMOL (25). Fitting refinement was performed in URO (26) using data from 260 to 30 Å.
RESULTS
Structure of the Membrane-proximal FNIII Domains of gp130
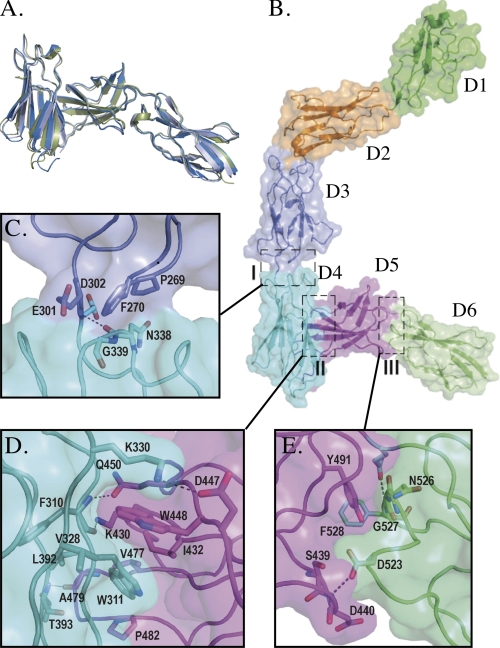
In the absence of structural information on the membrane-proximal domains of any tall receptor, we sought to structurally characterize this region of gp130. Two crystal forms of gp130 D4–D6 were obtained under similar crystallization conditions. Rod-shaped crystals diffracted to 1.9 Å and belonged to the space group P212121, with a single copy of the three-domain module in the asymmetric unit (supplemental Table 1). Plate-like crystals, space group C2, diffracted to 3.1 Å and contained two copies of D4–D6 in the asymmetric unit (supplemental Table 1), connected as a disulfide-linked dimer (Cys-395–Cys-395). As dimerization is not observed in the two other crystal structures reported here, we believe that this does not represent a biological interaction. As expected, each domain possesses the typical FNIII fold (27) (Fig. 1A, supplemental Fig. 3A). In D5, we observe a disulfide bond linking cysteines 436 and 444, as determined previously by mass spectrometry (28). The overall architecture of the three domains is completely maintained in both crystal forms (Fig. 1A), with r.m.s.d. for Cα atoms of the three different chains in the range 0.8–1.4 Å.
FIGURE 1.
Overall structure of gp130. A, overlay of the 1.9 and 3.1 Å (both chains) structures of D4–D6. B, the entire extracellular region of gp130 (D1 (green), D2 (orange), D3 (slate), D4 (cyan), D5 (magenta), D6 (lime)). Regions marked I, II, and III are shown in detail in parts C, D, and E, respectively.
Overall Structure of the Entire Extracellular Region of gp130
We also obtained crystals of the entire ectodomain of gp130 and were able to determine the structure by molecular replacement using our 1.9 Å structure of gp130 D4–D6 and the published structure of the gp130 D2–D3 domains (11). This was refined to 3.6 Å resolution with Rfree = 0.329 and Rwork = 0.261 (supplemental Table 1).
In the crystal structure, the ectodomain of gp130 adopts a wide open “C” shape, consisting of six well ordered domains (Fig. 1B). The orientation of D1 in the unliganded molecule is slightly different from that of D1 in the viral IL-6·gp130 tetramer (9) and IL-6·IL-6Rα·gp130 hexameric complex (11) (supplemental Fig. 2), but the structure of D1 itself is essentially unaltered (r.m.s.d. 1.0 Å). This suggests that when D1 participates in complex formation, a degree of flexibility in the D1–D2 linker allows a small conformational change to occur, enabling an oligomeric complex to assemble. The cytokine-binding module (D2–D3) is almost identical to published structures of this region with and without ligand (9–12) (r.m.s.d. 1.2 Å), confirming the view that binding of different ligands to this domain does not depend on conformational plasticity (12). The configuration of domains observed in the D4–D6 fragment is maintained in the complete ectodomain structure (r.m.s.d. 2.3 Å).
Inter-FNIII Domain Interactions
The D1/D2 and D2/D3 inter-FNIII domain interactions of gp130 have been detailed in published crystal structures (9–11). Here we focus on the previously unreported interfaces within the D3–D6 segment.
The linking segment between D3 and D4 is short (Glu-301 and Asp-302). The interface includes hydrophobic interactions between Pro-269 and Phe-270 of D3 and Asn-338 and Gly-339 of D4, respectively, and the linker region hydrogen-bonds to D4 through the backbone amide of Asp-302 and backbone carbonyl of Asn-338 (Fig. 1C). The interface has a limited total buried surface area of 467 Å2, suggesting that it may be flexible. This region is involved in crystal-packing interactions in which D3 and D5 clamp either side of D6 from a symmetry-related molecule (supplemental Fig. 4A), which may limit the movement of the domains. However, a similar interdomain interface is observed in other protein structures despite limited sequence homology. Structures of integrin β4 (liganded (29) and unliganded (30)) and type IIB receptor tyrosine phosphatase FNIII domains 2 and 3 (31), (supplemental Fig. 4B) have a similar buried surface area of ∼460 Å2 and align to D3–D4 with r.m.s.d. values in the range 1.1–1.4 Å, implying that this is a stable conformation.
The most notable feature of our gp130 structures is the unique orientation of D4 and D5 with respect to each other, which is observed in all crystal forms (Fig. 1, A and B). D4 connects to D5 by a five-residue linker, and the domains are oriented such that they lie across each other at an angle of ∼80°, resulting in an acute bend in the structure. The largely hydrophobic interface observed between D4 and D5 (Fig. 1D) is formed as a result of interactions between strands A, B, and G of D4 (i.e. the edge of the β-sandwich) packing against strands C, C′, and F of the four-strand sheet of D5, as well as FG loop (see supplemental Fig. 3B for strand labeling). The interaction interface is the most extensive within the gp130 molecule, with a total buried surface area of 1110 Å2. The key component of this interface is the burial of two tryptophan residues, one from D4 and one from D5, into hydrophobic regions on opposing domains (Fig. 1D). Trp-311 (D4) is packed against a hydrophobic region created by Leu-392, Ile-432, Val-477, and Pro-482, and Trp-448 (D5) is inserted into a hydrophobic pocket created by Trp-311, Val-328, Lys-330, and Ile-432. A limited number of direct polar contacts are also observed. The backbone carbonyl and amide groups of Phe-310 (D4) hydrogen-bond with the side chain of Lys-430 (D5) and the amide oxygen of Gln-450 (D5), respectively. Lys-430 and Gln-450 side chains also interact with each other. Lys-330 ϵ-NHζ (D4) forms a salt bridge with the side chain of Asp-447 (D5). The FG loop of D5 forms a type I β-turn and, within this, the backbone carbonyl atom of Ala-479 forms a hydrogen bond with the Oγ of Thr-393 in D4. It is interesting to note that Trp-311, Lys-330, and Trp-448 are completely conserved and Leu-392 and Ile-432 are identical or homologous among the tall receptor family (Fig. 1D, supplemental Fig. 3B). Other positions equivalent to those that contribute to the hydrophobic interface are also largely conserved as hydrophobic residues. The particular orientation of FNIII domains seen here has not been observed previously, but the size and hydrophobic nature of the interface suggest that it is rigid.
The D5/D6 interface (Fig. 1E) is stabilized by a network of interactions, including polar contacts (Ser-439 and Gln-494 of D5; Asp-523 and Asn-526 of D6) and hydrophobic interactions between Tyr-491 of D5 and Phe-528 of D6 and Tyr-491 and the backbone of residues 526–528. The side chain of Asp-523 (D6) interacts with the C-C′ loop of D5 to tether it to D6. The total buried surface area of 850 Å2 at the D5/D6 interface is larger than the buried surface of many other inter-FNIII domain interfaces (32) and implies a relatively inflexible junction.
Structural Model of the IL-6·IL-6Rα·gp130 Hexameric Complex and Fitting into Cryo-EM Map
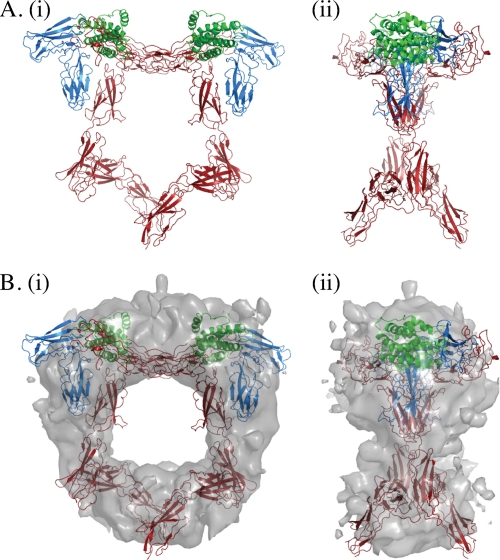
To gain insight into the conformation of the entire ectodomain within the context of a signaling complex, we combined the new structures described here with the published hexameric complex of IL-6·IL-6Rα·gp130 (11), which contains only D1–D3 of gp130. We overlaid the D2–D3 segment of gp130 within the hexamer with that of our unliganded D1–D6 gp130 structure (r.m.s.d. 1.2 Å). The general form of the modeled hexameric complex is of a pentagon (Fig. 2), in broad agreement with published electron micrographs (5, 13, 14). The membrane-proximal apex is open, with the C termini of the two gp130 molecules positioned to enter the membrane ∼65 Å apart. We then proceeded to fit this model into the only publicly available EM map of a gp130 complex, that of IL-11·IL-11Rα·gp130 (EMD-1223 (14)), which is structurally homologous to the IL-6·IL-6Rα·gp130 complex. There is remarkable agreement between our model and the published EM density (Fig. 2B), and the fit has a correlation coefficient of 0.59. Despite the rigid interfaces observed in our structure, one would expect a certain degree of flex for any molecule of such extended geometry. As we have four independent observations of D4–D6 (full-length D1–D6, the orthorhombic crystal of D4–D6, and two in the monoclinic crystal of D4–D6), we fitted these to the model by overlaying D4 of the different D4–D6 structures with D4 of the D1–D6 structure within our original model. Although the same D4/D5 and D5/D6 interfaces are maintained in these four views (Fig. 1, A and B), the intrinsic variability within the molecule is such that extension of the model with these different forms results in distances between the C termini of D6 between 65 and 30 Å (supplemental Fig. 5). The model in which the legs are 30 Å apart fits the EM map with a similar correlation coefficient of 0.61.
FIGURE 2.
Structural model of the extracellular IL-6·IL-6Rα·gp130 hexameric complex. A, gp130 D1–D6 was superimposed on the crystal structure of the IL-6·IL-6Rα·gp130 D1–D3 hexameric complex (11) by aligning gp130 D2–D3 of the two structures. i, “front” view; ii, “side” views following 90° rotation around the vertical axis of the front view shown in i. Green, IL-6; blue, IL-6Rα; red, gp130 D1–D6. B, fitting of the model into the EM map of Matadeen et al. (14). i, front view; ii, side view.
DISCUSSION
The distinguishing feature of the gp130 family of tall receptors is that the cytokine-binding/membrane distal half links to the membrane via membrane-proximal FNIII domains, D4–D6 in gp130. Mutagenesis studies have revealed a critical role for this region in receptor activation (15–17), but structural information on their disposition has been limited to low resolution EM data. Consistent with our structures, all electron micrographs proposed a C-shaped structure for gp130 in the complexed form, allowing the C-terminal regions of the ectodomains to come into close proximity, required for JAK transactivation. Superimposing our models of the hexameric IL-6·IL-6Rα·gp130 complex onto the cryo-EM map of Matadeen et al. (14) reveals that they have very similar dimensions, indicating that the conformations of liganded and unliganded gp130 are similar, and importantly, that the bent shape observed in the EM structures is not induced by ligand binding. There are some obvious discrepancies between the speculative fit of the legs in the low resolution EM studies and the detail of the crystal structures described here. In both the IL-11·IL-11Rα·gp130 and the gp130·LIFR·CNTFRα·CNTF structures, gp130 D4–D6 (and the homologous domains D6–D8 of LIFR) are modeled in a “beads on a string” arrangement, with the bend in the receptor required to form the C shape coming largely from a bend between D3 and D4. However, our structure reveals that the kink is not between D3 and D4, which are essentially linear, but between D4 and D5. In the structure of IL-6·IL-6Rα·gp130, the gp130 legs are modeled as asymmetrical, one with the similar linear arrangement of D4–D6 supposed in the other EM structures and one with a distinct bend between D4 and D5 (5). Although the authors commented that the asymmetry observed may be a preparation artifact and postulated the straight representation of the legs to be the more probable orientation, our crystal structure indicates that the bent conformation they observe accurately reflects the conformation of the signaling complex. They also present classes of molecule in which the legs are both symmetrical and bent, which match our model remarkably well (supplemental Fig. 5C).
The three key residues that maintain the D4/D5 interface, Trp-311, Lys-330, and Trp-448, are completely conserved in all members of the tall receptor family (Fig. 1D, supplemental Fig. 3B). Furthermore, positions equivalent to those that contribute to the buried hydrophobic regions are also largely conserved as hydrophobic residues. This implies that this interface, and therefore the unique orientation of these FNIII domains, is conserved throughout the tall receptor family. This is supported by EM images of the gp130·LIFR·CNTFRα·CNTF complex (13), in which both LIFR and gp130 adopt a very similar bent conformation in the leg region consistent with a bend between D4 and D5 (corresponding to D6 and D7 in LIFR).
The published EM models (5, 13, 14) show that the C termini of the ectodomains are in close proximity but do not agree on a common point of contact/crossover for the legs in the different gp130 complexes. Given our hypothesis that the arrangement of the legs will be similar across the entire family of tall receptors, it would also imply a common orientation of the legs of receptors with respect to each other within the signaling complexes. Repeating the fitting of models into all the EM data using our crystal structures will help clarify this.
Our model agrees with the paradigm that cytokine binding brings the C-terminal extracellular domains of two gp130 molecules into close proximity but does not resolve whether gp130 receptors interact directly in their membrane-proximal regions. Previous in vitro biochemical analyses using purified individual D4, D5, and D6 suggested that there could be a low affinity (KD > 5 μm) interaction between D4 domains (33). Our own work shows that even at concentrations of up to 250 μm, D4–D6 is monomeric in solution (supplemental Fig. 6). Isothermal titration calorimetry data presented by Boulanger et al. (11) using the IL-6·IL-6Rα·gp130 complex also shows that there is no gain in affinity of complex assembly when the legs are present as compared with when they are absent, which we interpret as indicating that there is no energetically favorable interaction within this region. In addition, in the three crystal forms, including one that is glycosylated, we do not see any signaling-competent crystal contacts between two D5 and/or D6 domains. Together this suggests that if an interaction occurs between these regions, it must be of extremely low affinity and induced by proximity.
There are reports that gp130 can exist as preformed inactive homodimers or as heterodimers with LIFR in the absence of ligand (34, 35). In the case of the erythropoietin receptor (EpoR), ligand-independent oligomerization is mediated by the transmembrane domain (36), and it is possible that any direct interaction between gp130 molecules also occurs in this region. Others have reported that binding of IL-6·IL-6Rα induces disulfide-linked homodimerization of gp130 (37). According to our model, disulfide bond formation would require substantial reorganization of gp130 if these were to form in the ectodomain; thus we suggest that a transmembrane or intracellular interaction is more likely.
JAKs, which are constitutively associated with the intracellular juxtamembrane region of cytokine receptors, are large molecules (∼1100 residues) containing four distinct globular domains. It is therefore possible that two JAK molecules could be close enough for transactivation without direct contact between terminal FNIII domains of the tall receptors as long as they are in reasonable proximity and in the correct orientation. The distance between the C termini of D6 domains is ∼30–65 Å in our modeled complexes of the IL-6·IL-6Rα·gp130 D1–D6 (Fig. 2, supplemental Fig. 5). The closest distance of 30 Å is comparable with that observed in the structure of the human Epo·EpoR complex (38), which also signals through JAK.
Collectively, the large volume of biochemical and structural studies on cytokine receptors has shown us that both proximity and orientation of the cytoplasmic receptor-bound JAKs are essential for receptor activation (see e.g. Refs. 39 and 40). The gp130 structures presented here reveal that the acute bend formed by D4 and D5 allows the ectodomain of gp130 to adopt a C shape, bringing the receptor complexes and their associated JAKs close enough for signaling without the need for major conformational change upon ligand binding. The unique configuration of the D4/D5 interface, which is conserved throughout the tall receptor family, predicts that this entire family may assemble functional cell surface complexes through this mechanism. It is apparent, however, that high resolution structures of full-length, liganded extracellular domains of complete receptor complexes and the entire extracellular, transmembrane, and intracellular domains of the receptors bound to JAKs will be required to further understand how cytokine-receptor assembly activates intracellular signaling processes. Our current effort lies in this direction.
Supplementary Material
Acknowledgments
We thank P. M. Colman for structural insight and continual support; O. B. Clarke for data collection; T. A. Willson for the gift of plasmids; and J. J. Babon, B. M. Smith, M. C. Lawrence, and J. M. Murphy for helpful discussions. Data were collected on the MX1 and MX2 beamlines at the Australian Synchrotron, Victoria, Australia. Crystallization trials were performed at the Bio21 C3 center.
This work was supported by the Australian National Health and Medical Research Council (Grants 257500, 280912, 487922, 461219, 461221, and 516730), the Australian Cancer Research Foundation, the Victoria Government Operational Infrastructure Scheme, and the Australian Government Independent Research Institutes Infrastructure Support Scheme.
The atomic coordinates and structure factors (codes 3L5H, 3L5I, and 3L5J) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Methods, Table 1, and Figs. 1–6.
- IL
- interleukin
- LIF
- leukemia inhibitory factor
- LIFR
- LIF receptor
- OSM
- oncostatin M
- CNTF
- ciliary neurotrophic factor
- CNTFR
- CNTF receptor
- JAK
- Janus kinase
- FNIII
- fibronectin type III
- EpoR
- erythropoietin receptor
- r.m.s.d.
- root mean square deviation
- EM
- electron microscopy
- Se-Met
- seleno-methionine
- STAT
- signal transducers and activators of transcription.
REFERENCES
- 1.Blanchard F., Duplomb L., Baud'huin M., Brounais B. (2009) Cytokine Growth Factor Rev. 20, 19–28 [DOI] [PubMed] [Google Scholar]
- 2.Heinrich P. C., Behrmann I., Haan S., Hermanns H. M., Müller-Newen G., Schaper F. (2003) Biochem. J. 374, 1–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heinrich P. C., Behrmann I., Müller-Newen G., Schaper F., Graeve L. (1998) Biochem. J. 334, 297–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taga T., Kishimoto T. (1997) Annu. Rev. Immunol. 15, 797–819 [DOI] [PubMed] [Google Scholar]
- 5.Skiniotis G., Boulanger M. J., Garcia K. C., Walz T. (2005) Nat. Struct Mol. Biol. 12, 545–551 [DOI] [PubMed] [Google Scholar]
- 6.Boulanger M. J., Garcia K. C. (2004) Adv. Protein Chem 68, 107–146 [DOI] [PubMed] [Google Scholar]
- 7.Tamada T., Honjo E., Maeda Y., Okamoto T., Ishibashi M., Tokunaga M., Kuroki R. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 3135–3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huyton T., Zhang J. G., Luo C. S., Lou M. Z., Hilton D. J., Nicola N. A., Garrett T. P. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 12737–12742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chow D., He X., Snow A. L., Rose-John S., Garcia K. C. (2001) Science 291, 2150–2155 [DOI] [PubMed] [Google Scholar]
- 10.Bravo J., Staunton D., Heath J. K., Jones E. Y. (1998) EMBO J. 17, 1665–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boulanger M. J., Chow D. C., Brevnova E. E., Garcia K. C. (2003) Science 300, 2101–2104 [DOI] [PubMed] [Google Scholar]
- 12.Boulanger M. J., Bankovich A. J., Kortemme T., Baker D., Garcia K. C. (2003) Mol. Cell 12, 577–589 [DOI] [PubMed] [Google Scholar]
- 13.Skiniotis G., Lupardus P. J., Martick M., Walz T., Garcia K. C. (2008) Mol. Cell 31, 737–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matadeen R., Hon W. C., Heath J. K., Jones E. Y., Fuller S. (2007) Structure 15, 441–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Timmermann A., Küster A., Kurth I., Heinrich P. C., Müller- Newen G. (2002) Eur. J. Biochem. 269, 2716–2726 [DOI] [PubMed] [Google Scholar]
- 16.Kurth I., Horsten U., Pflanz S., Timmermann A., Küster A., Dahmen H., Tacken I., Heinrich P. C., Müller-Newen G. (2000) J. Immunol. 164, 273–282 [DOI] [PubMed] [Google Scholar]
- 17.Hammacher A., Wijdenes J., Hilton D. J., Nicola N. A., Simpson R. J., Layton J. E. (2000) Biochem. J. 345, 25–32 [PMC free article] [PubMed] [Google Scholar]
- 18.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 19.Afonine P. V., Grosse-Kunstleve R. W., Adams P. D. (2005) CCP4 Newsletter, Vol. 42, Science and Technology Facilities Council (STFC) Daresbury Laboratory, Warrington, UK [Google Scholar]
- 20.Emsley P., Cowtan K. (2004) Acta. Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 21.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kabsch W. (1993) J. Appl. Crystallogr. 26, 795–800 [Google Scholar]
- 23.SERC Daresbury Laboratory (1994) Acta. Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 24.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta. Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 25.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA [Google Scholar]
- 26.Navaza J., Lepault J., Rey F. A., Alvarez-Rúa C., Borge J. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1820–1825 [DOI] [PubMed] [Google Scholar]
- 27.Leahy D. J., Hendrickson W. A., Aukhil I., Erickson H. P. (1992) Science 258, 987–991 [DOI] [PubMed] [Google Scholar]
- 28.Moritz R. L., Hall N. E., Connolly L. M., Simpson R. J. (2001) J. Biol. Chem. 276, 8244–8253 [DOI] [PubMed] [Google Scholar]
- 29.de Pereda J. M., Lillo M. P., Sonnenberg A. (2009) EMBO J. 28, 1180–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Pereda J. M., Wiche G., Liddington R. C. (1999) EMBO J. 18, 4087–4095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aricescu A. R., Siebold C., Choudhuri K., Chang V. T., Lu W., Davis S. J., van der Merwe P. A., Jones E. Y. (2007) Science 317, 1217–1220 [DOI] [PubMed] [Google Scholar]
- 32.Sharma A., Askari J. A., Humphries M. J., Jones E. Y., Stuart D. I. (1999) EMBO J. 18, 1468–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pflanz S., Kernebeck T., Giese B., Herrmann A., Pachta-Nick M., Stahl J., Wollmer A., Heinrich P. C., Müller-Newen G., Grötzinger J. (2001) Biochem. J. 356, 605–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tenhumberg S., Schuster B., Zhu L., Kovaleva M., Scheller J., Kallen K. J., Rose-John S. (2006) Biochem. Biophys. Res. Commun. 346, 649–657 [DOI] [PubMed] [Google Scholar]
- 35.Giese B., Roderburg C., Sommerauer M., Wortmann S. B., Metz S., Heinrich P. C., Müller-Newen G. (2005) J. Cell Sci. 118, 5129–5140 [DOI] [PubMed] [Google Scholar]
- 36.Constantinescu S. N., Keren T., Socolovsky M., Nam H., Henis Y. I., Lodish H. F. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 4379–4384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murakami M., Hibi M., Nakagawa N., Nakagawa T., Yasukawa K., Yamanishi K., Taga T., Kishimoto T. (1993) Science 260, 1808–1810 [DOI] [PubMed] [Google Scholar]
- 38.Syed R. S., Reid S. W., Li C., Cheetham J. C., Aoki K. H., Liu B., Zhan H., Osslund T. D., Chirino A. J., Zhang J., Finer-Moore J., Elliott S., Sitney K., Katz B. A., Matthews D. J., Wendoloski J. J., Egrie J., Stroud R. M. (1998) Nature 395, 511–516 [DOI] [PubMed] [Google Scholar]
- 39.Greiser J. S., Stross C., Heinrich P. C., Behrmann I., Hermanns H. M. (2002) J. Biol. Chem. 277, 26959–26965 [DOI] [PubMed] [Google Scholar]
- 40.Bravo J., Heath J. K. (2000) EMBO J. 19, 2399–2411 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.