Abstract
Platelet-derived growth factor (PDGF) is a pleiotropic protein with critical roles in both developmental as well as pathogenic processes. In the central nervous system specifically, PDGF is critical for neuronal proliferation and differentiation and has also been implicated as a neuroprotective agent. Whether PDGF also plays a role in synaptic plasticity, however, remains poorly understood. In the present study we demonstrated that in the rat hippocampal neurons PDGF regulated the expression of Arc/Arg3.1 gene that has been implicated in both synapse plasticity and long term potentiation. Relevance of these findings was further confirmed in vivo by injecting mice with intracerebral inoculations of PDGF, which resulted in a rapid induction of Arc in the hippocampus of the injected mice. PDGF induced long term potentiation in rat hippocampal slices, which was abolished by PDGF receptor-tyrosine kinase inhibitor STI-571. We also present evidence that PDGF-mediated induction of Arc/Arg3.1 involved activation of the MAPK/ERK (MEK) pathway. Additionally, induction of Arc/Arg3.1 also involved the upstream release of intracellular calcium stores, an effect that could be blocked by thapsigargin but not by EGTA. Pharmacological approach using inhibitors specific for either MAPK/ERK phosphorylation or calcium release demonstrated that the two pathways converged downstream at a common point involving activation of the immediate early gene Egr-1. Chromatin immunoprecipitation assays demonstrated the binding of Egr-1, but not Egr-3, to the Arc promoter. These findings for the first time, thus, suggest an additional role of PDGF, that of induction of Arc.
Keywords: Calcium Intracellular Release, ERK, Neurotrophic Factor, PI 3-Kinase, Protein Kinases, Arc, Egr, PDGF
Introduction
Members of the PDGF family are disulfide-bonded polypeptides that have multifunctional roles ranging from embryonic development to wound healing (1). PDGFs3 comprise of four polypeptide chains (A-D) that can form homo- or heterodimers (PDGF-AA, -BB, -AB, -CC, and -DD) and can also bind two types of receptors (PDGF-α and -β receptor) (2). Both of the receptors are structurally related and possess specific protein-tyrosine kinase domains (3). Ligand binding induces activation of the intracellular receptor kinases by formation of receptor dimers. Both PDGF-AA and -BB chains and their cognate receptors are widely expressed in the central nervous system (4, 5).
A number of studies have revealed the neuroprotective role of PDGFs that include inhibition of the N-methyl-d-aspartate (NMDA) receptor-dependent Ca2+ overload, leading to neuronal death (6–8), selective inhibition of NR2B-containing NMDA receptors, leading to potentiation of long term depression in hippocampal neurons(9), neuroprotection against human immunodeficiency virus protein toxicity (10), promotion of neuronal differentiation (11, 12), protection of hippocampal neurons against energy deprivation and oxidative injury (13), and modulation of synaptic transmission (14). Furthermore, the PDGF/PDGF-β receptor signal axis mediates dopamine receptor signals and inhibits γ-aminobutyric acid receptor activation (15, 16). In addition to PDGFs, PDGF-β receptor has also been implicated in neuroprotection after injurious events. For example, focal ischemia in rat brain causes a rapid increase in PDGF-B chain mRNA transcripts with a concomitant rise in PDGF-β receptor expression in the central nervous system (6).
Based on these vital roles of PDGF in the central nervous system, we sought to explore whether PDGF also was critical for long term potentiation, the most studied form of synaptic plasticity. Long term forms of learning and memory are dependent on rapid synthesis of new RNA and proteins in the cells (17). A set of immediate early genes including Arc/Arg3.1 (18), Homer (19), Narp (20), and cpg-2 (21) have been shown to play crucial roles in transcription-dependent plasticity. Regulation of Arc/Arg3.1 is very unique in that in stimulated neurons Arc mRNA had been shown to translocate from the nucleus to the dendrites where it gets translated and incorporated into the postsynaptic zone with other cytoskeletal proteins (22, 23). A critical role of Arc in synaptic plasticity has also been demonstrated in studies involving Arc/Arg3.1 knockdown, an effect that results in selective deficits in long term synaptic potentiation (24). Thus, Arc expression in the neurite has been strongly suggested as an index of synapse activation.
In addition to high frequency stimulation, neurotrophins such as brain-derived neurotrophic factors that exert multifunctional effects on the survival/development of central nervous system can also induce long term potentiation (25). Although PDGF shares functional and signaling similarities with neurotrophins, its role in long term potentiation and synaptic plasticity remains poorly resolved. In the present study we, thus, sought to explore the role of PDGF-B in the induction of synaptic plasticity with a focus on long term potentiation and the molecular mechanisms involved in the induction of Arc expression in the hippocampus. These findings can have possible implications in the development of central nervous system therapeutics targeted against neurodegenerative diseases.
EXPERIMENTAL PROCEDURES
Materials
Recombinant human PDGF was purchased from R&D Systems (Minneapolis, MN). Because PDGF, but not PDGF-AA, is implicated in preventing delayed neuronal death after forebrain ischemia in rats, especially in the hippocampus (26), where it is closely involved in the development of long term potentiation, in the present study we restricted studies to the role of PDGF and not the A chain. The phosphatidylinositol-3′ kinase (PI3K) inhibitor LY294002 and the MEK1/2 inhibitor U0126 were purchased from Calbiochem. STI-571, an inhibitor of PDGF receptor-tyrosine kinase, was obtained from Novartis, Basel, Switzerland. Dominant negative and constitutively active constructs of MEK were provided from Dr. Young Han Lee (Konkuk University, Korea). Chromatin immunoprecipitation assay kit was purchased from Upstate Biotechnology (Billerica, MA). Transcription inhibitor actinomycin D and the translation inhibitor actidione were purchased from Sigma.
Animals
Pregnant female Sprague-Dawley rats and C57BL/6N mice were purchased from Charles River Laboratories, Inc. (Wilmington, MA). All of animals were housed under conditions of constant temperature and humidity on a 12-h light, 12-h dark cycle, with lights on at 0700 h. Food and water were available ad libitum. All animal procedures were performed according to the protocols approved by the Institutional Animal Care and Use Committee of the University of Nebraska Medical Center.
Hippocampal Slices and Electrophysiological Recordings
Hippocampal brain slices were prepared from 15–35-day-old male Sprague-Dawley rats as previous described (27). Briefly, animals were anesthetized with isoflurane and decapitated. Brains were quickly removed from the cranial cavity and placed into ice-cold (4 °C), pre-oxygenated artificial cerebrospinal fluid. Hippocampi were dissected out, and transverse hippocampal slices (400 μm in thickness) were cut using a tissue chopper. Slices were kept in a humidified/oxygenated holding chamber at room temperature for at least 1 h before being transferred to a recording chamber. In the recording chamber, single hippocampal slices were submerged and continuously perfused (2.0 ml/min) with artificial cerebrospinal fluid containing 124.0 mm NaCl, 3.0 mm KCl, 2.0 mm CaCl2, 2.0 mm MgCl2, 1.0 mm NaH2PO3, 26.0 mm NaHCO3, and 10.0 mm glucose, equilibrated with 95% O2 and 5% CO2, pH 7.35–7.45. The temperature of the perfusate was maintained at 30 ± 1 °C with an automatic temperature controller (Warner Instrument Corp., Hamden, CT). Drugs were applied onto slices via incubation (1 h). Excitatory postsynaptic potentials (EPSPs) were elicited by a constant current stimulation (0.05 Hz, 50–400 μA) of Schaffer collateral-commissural axons using an insulated bipolar tungsten electrode. Intensity and duration of stimulation were adjusted to generate ∼30–40% of a maximal response. Evoked EPSPs were recorded with an Axopatch-1D amplifier (Axon Instruments, Inc., Union City, CA) in the CA1 dendrite field (stratum radium). Recordings were made with borosilicated glass microelectrodes with a tip diameter of 2.5–5.0 μm and a resistance of 1–5 megaohms when filled with artificial cerebrospinal fluid.
A 20-min control recording was conducted in each experiment once the adjustment of stimulation parameters was achieved. Each recording trial was an average of 3 consecutive sweeps. High frequency stimulation (100 Hz, 500 ms) was delivered twice in 20-s intervals at the same intensity as that employed in test pulses (low frequency). Electrical signals were filtered at 1 kHz and digitized at a frequency of 2.5 kHz using a Digidata 1320 interface (Axon Instruments, Inc.). Data were stored on a desktop PC and analyzed off-line using pCLAMP 10 software (Axon Instruments, Inc.). The initial slope of the EPSPs was analyzed and expressed in percentage of basal level (the average of initial slopes from the first 30 min was treated as 100%, i.e. the basal level). In bar graphs, the magnitudes of long term potentiation were quantified at the time point of 60 min after high frequency stimulation.
Microinjection
Microinjection was performed in male C57BL/6N mice, 12 weeks old, weighing 22–24 g. The mice were anesthetized with 2.5% isofluorane and placed in a stereotaxic apparatus for injection using the stereotaxic coordinates, AP-2.0 mm posterior, ML-1.6 mm lateral to midline, and DV-1.5 mm to bregma as described by Nunes-de-Souza et al. (28). All microinjections were performed using a 33-gauge stainless steel injector connected to a 10-μl syringe, which was operated by an influx pump. 1 μl solution was injected into the dorsal hippocampus at 0.15 μl per min. The concentration of PDGF used was 500 ng/μl, and vehicle was used as control. At the end of the injection the needle was kept in situ for a further 2 min. Three hours post-injection the mice were perfused and decapitated.
Immunostaining
For immunocytochemistry, cells were fixed in 4% paraformaldehyde followed by blocking with phosphate-buffered saline containing 10% bovine serum albumin. After blocking, cells were incubated at 4 °C overnight with different antibodies (Egr-1, Santa Cruz Biotechnology, Santa Cruz, CA). After washes cells, were incubated with the secondary goat anti-rabbit Alexa Fluor 488-conjugated antibody (1:500). For negative controls cells were treated as described above, except that the primary antibody treatment was omitted.
For immunohistochemistry, mice were perfused by transcardial perfusion using chilled 4% paraformaldehyde. Free-floating sections encompassing the entire hippocampus were sectioned at 40 μm on a cryostat. For Arc immunostaining, tissue sections were incubated with primary antibodies overnight at 4 °C. The Vector M.O.M immunodetection kit was used to avoid the cross-reaction between mouse tissue and the primary Arc antibody, which was generated in mice. Immunostaining was visualized by using 3,3′-diaminobenzidine as the substrate.
Cell Culture and Cell Lines
Primary rat neurons were prepared as described earlier with slight modifications (29). Briefly, hippocampus was dissected from embryonic day 18–19 Sprague-Dawley rats and dissociated with a mild mechanical trituration. Dissociated cells were seeded first in Dulbecco's modified Eagle's medium/F-12 supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. After 1 day, the cultures were supplemented with serum-free neurobasal medium containing B27 (50:1), 2 mm glutamax, 100 units/ml penicillin, and 100 μg/ml streptomycin. In all of the experiments cells were cultured for 7 days before treatment with PDGF or the inhibitors. The SH-SY5Y cell line was cultured following the protocols as our previous paper (10).
Western Blot Analysis
Treated cells were lysed using the Mammalian Cell Lysis kit (Sigma) and the NE-PER Nuclear and Cytoplasmic Extraction kit (Pierce). Cell lysates were subjected to the separation by 12% SDS-PAGE electrophoresis (about 20 μg of protein per well) and transferred to polyvinylidene difluoride membranes. Western blots were then probed with antibodies recognizing Arc (Santa Cruz,1:200), Egr-1 (Santa Cruz, 1:500), Egr-3 (Santa Cruz, 1: 500), phospho-extracellular signal-regulated kinase (ERK; Cell Signaling, 1:500), and ERK (Cell Signaling, 1:500). Signals were detected by chemiluminescence (Pierce). All experiments were repeated four times individually, and representative blots are presented in the figures.
Regulation of Egr-1
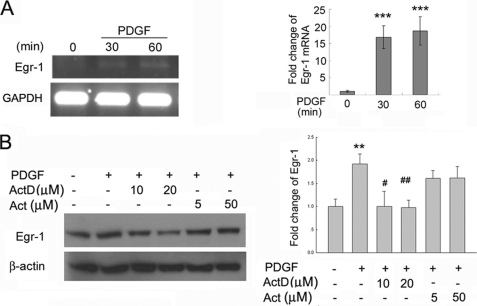
To examine whether PDGF-mediated regulation of Egr-1 occurred at the transcriptional and/or translational level, rat hippocampal neurons were pretreated with either actinomycin D (transcription blocker, at 10 and 20 μm) or with actidione (translational inhibitor, at 5 and 50 μm) followed by exposure to PDGF (20 ng/ml). The RNA and proteins extracted from rat hippocampus neurons were subjected to reverse transcription-PCR or Western blot, respectively. The sequences of rat Egr-1 and glyceraldehyde-3-phosphate dehydrogenase were as follows: rat Egr-1 sense primer, 5′-CCCAAACTGGAGGAGATGA-3′, and antisense primer, 5′-GGAGGCAGAGGAAGACGAT-3′; Rat glyceraldehyde-3-phosphate dehydrogenase sense primer, 5′- CCTTCATTGACCTCAACTAC-3′, and antisense primer, 5′-GGAAGGCCATGCCAGTGA-3′. Amplifications were carried out for 36 cycles of 94 °C for 40 s, 56 °C for 40 s, 72 °C for 1 min with final extension at 72 °C for 10 min.
Measurement of Free Intracellular Calcium
The changes in Ca2+ were monitored using Fluo-4/AM (Molecular Probes, Eugene, OR) dissolved in dimethyl sulfoxide. The rat primary hippocampal neurons in 35-mm culture dishes were rinsed twice with Bath solution (140 mm NaCl, 5 mm KCl, 1 mm CaCl2, 0.5 mm MgCl2, 10 mm glucose, 5.5 mm HEPES, pH 7.4), incubated in Bath solution containing 5 μm Fluo-4/AM with 5% CO2, 95% O2 at 37 °C for 40 min, rinsed twice with the Bath solution, mounted on a perfusion chamber, and scanned every second using confocal microscopy (X400) (Fluoview 300; Olympus). Fluorescence was excited at 488 nm, and the emitted light was read at 515 nm. All analyses of Ca2+ were processed at a single-cell level. To normalize for variations in initial fluorescence values, the values of the Ca2+ response were divided by the resting fluorescence value (calculated as the mean of at least three values before the application of PDGF).
Short Interfering RNA (siRNA) Transfection
siRNA target against PDGF-βR and Egr-1 were obtained from Dharmacon (Boulder, CO). SH-SY5Y cells were plated in a 24-well plate at a density of 4 × 104 cells per well 1 day before transfection. Before transfection, cell culture medium was replaced with 250 μl of pre-warmed culture medium. Two μl of Novagen Ribojuice siRNA transfection reagent (EMD Chemicals, Inc, Gibbstown, NJ) was then combined with 48 μl of serum-free RPMI 1640 medium (Invitrogen) for 5 min at room temperature. The Egr-1 siRNA was then added into the mixture described above to a final concentration of 150 nm. The siRNA and the reagent mixture were then incubated for 20 min at room temperature after which the combined mixture was added to the cells. The cell culture plate was shaken gently for 5 s and incubated for 24 h at 37 °C.
Chromatin Immunoprecipitation (ChIP) Assay
ChIP assay was performed according to the manufacturer's instructions (Upstate) with slight modifications. Briefly, after treatment of the cells, 18.5% fresh formaldehyde was added directly into the medium at a final concentration of 1% formaldehyde and incubated for 10 min at room temperature followed by quenching with 10× glycine. The cells were then scraped using 2 ml of pre-chilled phosphate-buffered saline containing a 1× protease inhibitor mixture. The cell pellet was harvested by spinning at 800 × g at 4 °C followed by adding lysis buffer (provided in the kit) to harvest nuclei. DNA was sheared by sonication. Fifty μl of the sheared cross-linked chromatin was then mixed with 20 μl of protein A magnetic beads and 5 μg of the corresponding antibody diluted in 450 μl of dilution buffer followed by incubation overnight at 4 °C. The magnetic beads binding antibody-chromatin complex was then washed with 0.5 ml each of a series of cold wash buffers in the order of low salt buffer, high salt buffer, LiCl buffer, and finally with Tris-EDTA buffer. The cross-linked protein-DNA complexes were reversed to free DNA by incubation at 62 °C for 2 h and purified using DNA purification spin columns following the manufacturer's instructions. Finally, the purified DNA was subjected to PCR to identify the promoter region containing GC-rich sequence estrogen response element (30). The sequence of the primers used to identify the promoter bound by transcription factors Egr-1/Egr-3 were as follows: sense, 5′-GAGAGGGCGCGGGCGGGCTCTGGCG-3′; antisense, 5′-CCGGCGGCTTTTTATGCTGCGCGGG-3′.
Statistical Analysis
Statistical analysis was performed using one-way analysis of variance with a post hoc Student's t test. Results were judged statistically significant if p < 0.05 by analysis of variance.
RESULTS
PDGF Induces Arc Expression in Neurons
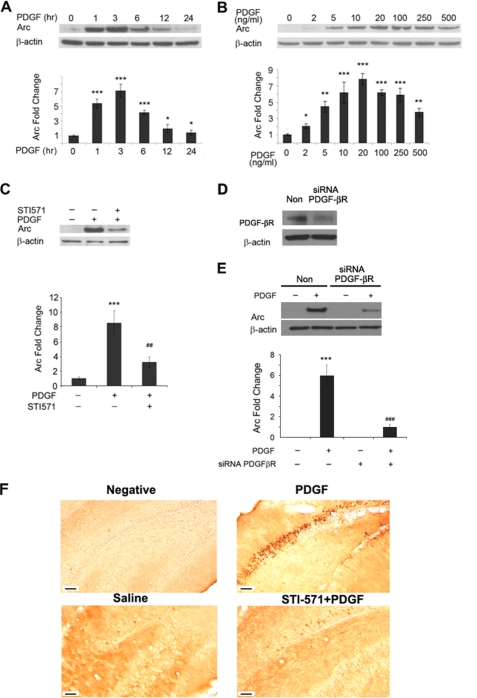
Because the expression of Arc/Arg3.1 gene is implicated in synapse plasticity and long term potentiation, we first wanted to investigate the effects of PDGF on Arc expression in primary rat hippocampal neurons to test whether PDGF plays a role in synaptic plasticity. To determine the optimal time of Arc induction, hippocampal neurons were treated with rhuman PDGF (20 ng/ml) for 1, 3, 6, 12, and 24 h followed by cell lysis and Western blot analysis. As shown in Fig. 1A, Arc induction was a rapid and early event with induction beginning as early as 1 h post PDGF treatment and reached a peak at about 2–3 h post stimulation. To optimize the dose of PDGF, hippocampal neurons were treated with varying doses of PDGF (2–500 ng/ml) for 2 h and subjected to Western blot analysis. As shown in Fig. 2, the dose-curve data demonstrated that PDGF concentrations as low as 5 ng/ml was able to induce Arc expression with peak induction at 20–100 ng/ml of PDGF. Pretreatment of hippocampal neurons with the receptor-tyrosine kinase inhibitor STI-571 (1 μm) for 1 h resulted in abrogation of PDGF-induced Arc expression (Fig. 1C), thus, underscoring the role of PDGF receptor-tyrosine kinase in this process. We acknowledge the fact that STI-571 is not a specific antagonist for PDGF-βR, as it is also known to inhibit PDGF-αR as well as other kinase activities. Hence, as an alternative approach we next sought to knock down PDGF-βR expression in neurons using the siRNA strategy. As expected, transfection of cells with PDGF-βR siRNA resulted in inhibition of receptor expression (Fig. 1D) and also abrogated the expression of PDGF-mediated induction of Arc (Fig. 1E), thereby confirming the role of PDGF-βR in this process.
FIGURE 1.
Time-course and dose-curve of PDGF-induced Arc expression in hippocampal neurons. A, the time course of PDGF-induced Arc expression in rat hippocampal neurons is shown. Arc expression reached peak 2–3 h post-PDGF treatment. *, p < 0.05; ***, p < 0.001 versus control group. B, shown is the dose-curve of PDGF-induced Arc expression 3 h post-treatment demonstrating that PDGF even at a concentration of 5 ng/ml was able to induce robust expression of Arc in rat hippocampal neurons. *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus control group. C, pretreatment of neurons with tyrosine kinase inhibitor-STI-571 was able to abrogate PDGF-induced Arc expression. ***, p < 0.001 versus control group; ##, p < 0.01 versus PDGF (20 ng/ml) group. D, Western blot analysis is shown of whole cell lysates from SH-SY5Y cells transfected with either PDGF-βR or nonsense (Non) siRNA using antibodies specific for PDGF-βR. E, transfection of SH-SY5Y cells with siRNAs specific for PDGF-βR abolished PDGF-mediated Arc expression. ***, p < 0.001 versus control group; ###, p < 0.001 versus PDGF (20 ng/ml) group. A representative immunoblot and the densitometric analysis from four separate experiments is presented. F, shown is PDGF-induced Arc expression in vivo. Adult mice were injected with either saline, PDGF, or STI-571 followed by PDGF, and 3 h later the animals were perfused and decapitated. Hippocampal sections were immunostained using the Arc antibody. Saline-injected animals demonstrated very low levels of Arc expression (bottom left panel). PDGF injections resulted in robust Arc expression in the hippocampus (top right panel). The right panels indicate higher magnifications of the boxed area in two respective pictures respectively. Scale bars indicate 50 μm. All the images are representative of three independent experiments.
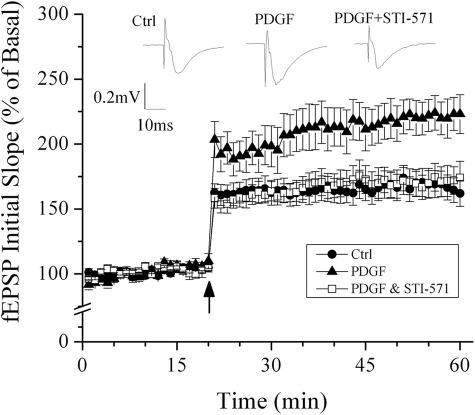
FIGURE 2.
Time course and average magnitude of long term potentiation recorded in the Schaffer-collateral to CA1 synapses from control slices (●), slices treated with PDGF (▴), and slices treated with PDGF+STI-571 (□). The graph plots the initial slope of the evoked EPSPs recorded from the CA1 dendrite field (stratum radium) in response to constant current stimuli. High frequency stimulation (100 Hz, 500 ms ×2) was delivered at the time indicated by an arrow. Note that PDGF induced an enhancement of long term potentiation and the blockade of PDGF-induced enhancement of long term potentiation by STI-571, Representative field EPSPs (fEPSPs) recorded from slices of control, treated with PDGF, and treated with PDGF+STI-571 are shown at the top of the figure. Ctrl, control.
To confirm the observed increase of Arc expression in hippocampus neurons in vitro, we sought to explore the expression of Arc in the brains of mice hippocampi injected with PDGF. Three hours post-injection of PDGF in the dorsal hippocampus, the mice were decapitated and subjected to the Arc immunostaining. Injection of PDGF resulted in a robust increase in Arc immunostaining in the mice hippocampi compared with the saline injected animals (Fig. 1F), which was abrogated by pretreatment with the tyrosine kinase inhibitor-STI-571.
PDGF-mediated Induction of Long Term Potentiation ex Vivo in Hippocampal Slices
Having determined that PDGF induced Arc expression and that Arc expression is implicated in long term potentiation, we next sought to test the effect of PDGF on long term potentiation. High frequency stimulation of Shaffer-collateral fibers produced robust long term potentiation recorded in CA1 region of non-treated (control) hippocampal slices. The average magnitude of long term potentiation measured 40 min after high frequency stimulation was 162.1 ± 9.9% of basal level (Fig. 2, n = 10). In contrast, incubation of hippocampal slices with PDGF produced an enhancement of long term potentiation with an average magnitude of 223.2 ± 14.9% that of basal level (Fig. 2, n = 9), in comparison with the long term potentiation magnitude recorded in control slices significantly (p < 0.01), suggesting an enhancement of long term potentiation by PDGF. To determine whether PDGF-induced enhancement of long term potentiation is mediated via PDGF receptors, we co-incubated hippocampal slices with PDGF and STI-571, a PDGF receptor-tyrosine kinase inhibitor. Our results showed that the average magnitude of long term potentiation recorded in slices co-incubated with PDGF and STI-571 was 174.1 ± 12.7% that of basal level (n = 6). Compared with the long term potentiation magnitude recorded in slices treated with PDGF, the difference was statistically significant (p < 0.05), indicating a blockade of PDGF-induced enhancement of long term potentiation by STI-571. Bath application of PDGF alone had no significant effect on basal synaptic transmission (n = 4, data not shown).
Involvement of Mitogen-activated Protein Kinase (MAPK)/ERK Pathway in PDGF-mediated Induction of Arc
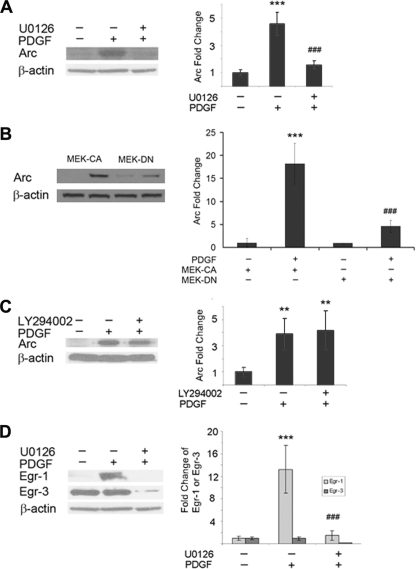
Several signal pathways have been implicated in PDGF induced biological responses including the MAPK/ERK, PI3K/Akt, and calcium release pathways (31, 32). To elucidate the roles of MAPK/ERK and PI3K pathways in PDGF-mediated Arc expression, we pretreated rat hippocampal neurons with either the PI3K inhibitor LY294002 (10 μm) or the MEK inhibitor U0126 (20 μm) for 1 h before exposing the neurons to PDGF for 30 min. PDGF-mediated induction of Arc was abrogated completely by pretreating the cells with the MEK inhibitor (Fig. 3A). To confirm the involvement of ERK pathway in PDGF-mediated Arc expression, neurons were transfected with constructs encoding either a dominant-negative (DN) or the constitutively active (CA) form of MEK. As shown in Fig. 3B, neurons transfected with the dominant-negative form of MEK failed to induce PDGF-mediated expression of Arc. Transfection of neurons with constitutively active MEK, on the other hand, resulted in PDGF-mediated induction of Arc expression, as expected (Fig. 3B). In contrast to the MEK inhibitor, PDGF-mediated induction of Arc was unaffected by the PI3K inhibitor (Fig. 3C).
FIGURE 3.
MAPK/ERK pathway is involved in PDGF-induced Arc expression. A, pretreatment of rat hippocampal neurons with MEK inhibitor (U0126, 20 μm) for 1 h resulted in inhibition of PDGF-mediated Arc expression is shown. ***, p < 0.001 versus control group; ###, p < 0.001 versus PDGF (20 ng/ml) group. B, dominant-negative (DN) MEK but not the constitutively active (CA) MEK resulted in abrogation of PDGF-mediated Arc expression. ***, p < 0.001 versus control group; ###, p < 0.001 versus PDGF (20 ng/ml) group. C, pretreatment of neurons with the PI3K inhibitor (LY294002, 10 μm) failed to inhibit PDGF-mediated Arc induction. **, p < 0.01 versus control group. D, exposure of rat hippocampal neurons to MEK inhibitor also resulted in abrogation of PDGF-mediated Egr-1 expression. ***, p < 0.001 versus control group; ###, p < 0.001 versus PDGF (20 ng/ml) group. Representative immunoblot and the densitometric analysis from four separate experiments is presented.
Based on the fact that the early growth response transcription factors Egr-1 and -3 play a role in Arc induction (30), and our findings that PDGF induced Arc expression, we next sought to examine the role of these transcription factors in PDGF-stimulated cells. As shown in Fig. 3C, similar to the up-regulation of Arc (Fig. 2), hippocampal neurons treated with PDGF for just 1 h demonstrated a dramatic up-regulation of Egr-1. Egr-3 expression, on the other hand, levels of which were high even in untreated cells, remained unchanged.
PDGF Regulated Arc Expression Involves Intracellular Calcium Transients
Because PDGF signaling also involves changes in calcium transients (33), we next wanted to investigate whether PDGF-induced Arc expression involved changes in calcium pools. We first assessed the effects of PDGF on primary hippocampal neurons using Ca2+-sensitive fluorophores. PDGF induced a rapid and sustained increase in intracellular calcium levels in hippocampal neurons. To determine whether the source of PDGF-induced Ca2+ transients was the intracellular (the endoplasmic reticulum (ER)) or the extracellular storage site for Ca2+, we preincubated cells with a blocker of ER Ca2+, thapsigargin or EGTA. The former specifically blocks the ER Ca2+-ATPase, causing a slow leak of Ca2+ from the store, thus depleting the store, whereas the latter works as a calcium chelator to deplete the calcium in media. Pretreatment of the cells with either thapsigargin (10 μm, for 2 h) or EGTA (200 μm, for 2 h) followed by subsequent exposure to PDGF blocked the majority of Ca2+ transients (Fig. 4A). Interestingly, however, although both thapsigargin and EGTA could block PDGF-induce Ca2+ transients, only thapsigargin but not EGTA was able to effectively block PDGF-induced Arc expression (Fig. 4B).
FIGURE 4.
Endogenous calcium is involved in the Arc expression induced by PDGF in rat hippocampal neurons. A, changes in intracellular [Ca2+]i levels in neurons after PDGF treatment were measured using the fluorescence intensity of Fluo-4 in a typical neuron. Pretreatment of cells with either EGTA or thapsigargin resulted in inhibition of PDGF-induced increase in intracellular Ca2+. B, pretreatment of rat hippocampal neurons with cells with either EGTA or thapsigargin resulted in decreased expression of Arc as well as Egr-1 but not Egr-3. The image is representative of at least three independent experiments. THA, thapsigargin. **, p < 0.01 versus control group; #, p < 0.05; ##, p < 0.01 versus PDGF (20 ng/ml) group.
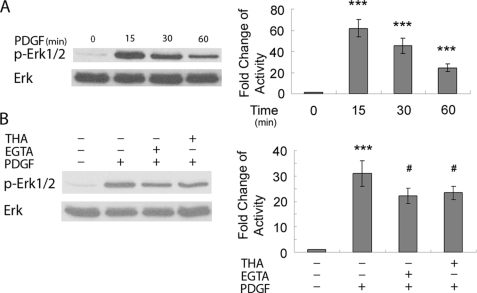
PDGF-induced Calcium Transients Lie Upstream of the MAPK/ERK Pathway
Having determined the abrogation of PDGF-induced Egr-1 expression by both the MEK inhibitor and calcium blocker thapsigargin, next we sought to further dissect the relationship between PDGF-induced calcium transients and activation of the MAPK/ERK pathway in rat hippocampal neurons. As shown in Fig. 5A, PDGF treatment resulted in time-dependent phosphorylation of ERK. Pretreatment of cells with the thapsigargin or EGTA significantly decreased PDGF-induced ERK phosphorylation (Fig. 5B). However, in the presence of ERK inhibitor U0126, PDGF was still able to elicit calcium response in rat hippocampal neurons (data not shown). Thus, in rat hippocampal neurons, MAPK/ERK inhibitor did not influence PDGF-induced Ca2+ release and reciprocally (data not shown), but calcium blockers decreased PDGF-induced ERK activation, thereby suggesting PDGF-induced Ca2+ release, which lies upstream of ERK pathway.
FIGURE 5.
PDGF-induced calcium transients lie upstream of MAPK/ERK pathway. A, exposure of rat hippocampal neurons to PDGF lead to time-dependent increase of ERK activation is shown. ***, p < 0.001 versus control group. B, pretreatment of rat hippocampal neurons with either EGTA or thapsigargin resulted in partially deactivation of ERK compared with PDGF treatment alone. ***, p < 0.001 versus control group; #, p < 0.05 versus PDGF (20 ng/ml) group. A representative immunoblot and the densitometric analysis from four separate experiments is presented. THA, thapsigargin.
PDGF-mediated Transcriptional Induction of Egr-1
The next step was to explore whether PDGF-mediated induction of Egr-1 expression occurred at the transcriptional and/or translational level. Rat hippocampal neurons were pretreated with either the transcription or translation inhibitor followed by exposure to PDGF for varying times. As shown in Fig. 6A, pretreatment of neurons with the transcription inhibitor, actinomycin D, abrogated PDGF-mediated induction of Egr-1. In contrast, pretreatment of cells with the translation inhibitor actidione did not exert any effect on PDGF-mediated induction of Egr-1 (Fig. 6B).
FIGURE 6.
PDGF regulates the expression of Egr-1 by regulating transcription of Egr-1 gene. A, PDGF induced expression of Egr-1 mRNA. A representative reverse transcription-PCR gel and the densitometric analysis of Egr-1 mRNA from four separate experiments is presented. ***, p < 0.001 versus control group. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. B, rat hippocampal neurons pretreated with the transcriptional inhibitor actinomycin D (Act D) resulted in inhibition of PDGF-mediated expression of Egr-1. Cells pretreated with the translational inhibitor Actidione (Act) failed to impact PDGF-mediated induction of Egr-1. **, p < 0.01 versus control group; #, p < 0.05; ##, p < 0.01 versus PDGF (20 ng/ml) group. Representative reverse transcription-PCR gel, immunoblot and the densitometric analysis from four separate experiments is presented.
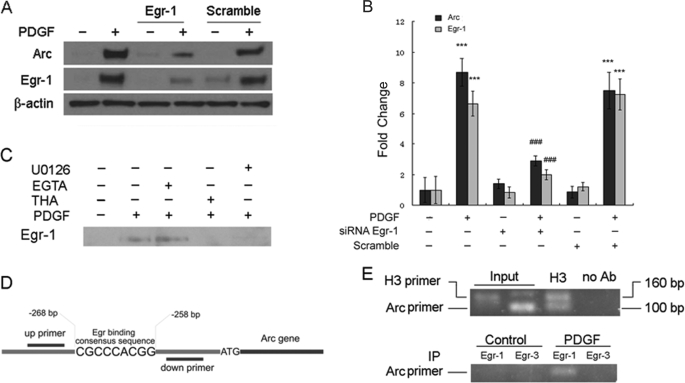
Egr-1 Translocation and Binding Are Critical for Arc Transcription
Because both the MAPK/ERK and calcium transients were found to regulate the expression of Egr-1 in PDGF-induced expression of Arc, the next step was to validate the role of Egr-1 in Arc expression. To confirm the role of Egr-1 in PDGF-induced Arc expression, Egr-1 expression in neuronal cell line SH-SY5Y was specifically down-regulated using specific siRNAs followed by assessment of PDGF-induced Arc expression. Treatment of SH-SY5Y cell Egr-1 siRNA (150 nm) blocked PDGF-induced expression of both Egr-1 and Arc (Fig. 7, A and B). Because Egr-1 activation involves its translocation to the nucleus, we next treated hippocampal neurons grown on tissue culture slides to PDGF for 1 h and subjected the cells to immunostaining using antibodies specific for Egr-1 and Egr-3. PDGF treatment resulted in nuclear translocation of Egr-1 but not of Egr-3 (data not shown). Furthermore, examination of nuclear extracts from PDGF-treated cells resulted in Egr-1 protein expression as evidenced by Western blot analysis (Fig. 7C). On the other hand, as expected, pretreatment of cells with either the MEK inhibitor U0126 or with thapsigargin led to inhibition of PDGF-induced nuclear translocation of Egr-1 (Fig. 7, D and E).
FIGURE 7.
Egr-1 is a transcription factor involved in PDGF-mediated induction of Arc/Arg3.1 gene in neurons. A, shown are SH-SY5Y cells transfected with 150 μm Egr-1 siRNA displayed decreased expression of PDGF-induced Egr-1 and Arc. B, densitometric analyses are shown of PDGF-mediated induction of Arc and Egr-1 in SH-SY5Y cells in the absence or presence of Egr-1 siRNA. ***, p < 0.001 versus control group; ###, p < 0.001 versus PDGF (20 ng/ml) group. A representative immunoblot and the densitometric analysis from four separate experiments is presented. C, Western blot analysis of the nuclear extracts from neurons treated with PDGF in the presence or absence of various inhibitors demonstrated that inhibitors that blocked PDGF-induced Arc expression were also able to inhibit PDGF-induced translocation of Egr-1. THA, thapsigargin. D, shown is a schematic illustration of the Egr-1 binding consensus sequence on the Arc/Arg3.1 promoter region. E, shown is a ChIP assay demonstrating the PDGF-mediated binding of Egr-1 but not Egr-3 to the Arc/Arg3.1 promoter. A representative PCR gel of the Arc promoter from four separate experiments is presented. IP, immunoprecipitate; Ab, antibody.
As final proof of involvement of Egr-1 in Arc expression, we sought to explore the binding of Egr-1 to the Arc promoter in its natural chromatin context to reveal active sites accessible to Egr-1. We investigated in vivo Arc promoter occupancy by ChIP assays. These experiments revealed an increased binding of Egr-1 but not Egr-3 to the Arc promoter in both rat primary neurons (Fig. 7E) and in the SH-SY5Y cells (data not shown) treated with PDGF.
DISCUSSION
The neurotrophic factor PDGF consists of a family of five dimeric ligands (PDGF-AA, -AB, -BB, -CC, and -DD) assembled from four gene products (PDGF A-D) that act via two classical receptor-tyrosine kinases, PDGF-αR (34, 35) and PDGF-β receptor (34, 36). Members of the PDGF family have multiple roles during embryogenesis and in a variety of pathological situations in the adult. Postnatally, PDGF-B expression is turned on in mammalian neurons, and PDGF-B protein is found in neurons broadly in the central nervous system in the adult primate brain (4, 37). Although this distribution likely reflects neuronal PDGF-B production, it is also possible that the neurons express PDGF receptors. Cultured rat neuronal cells have been shown to express PDGF-β receptor and respond to PDGF (38–40). PDGF pretreatment is known to attenuate excitotoxic death in cultured hippocampal neurons (7) and induce striatal neurogenesis in adult rats with 6-hydroxydopamine lesions (41). Another proposed mechanism by which PDGF mediates its neuroprotective effect is through the induction of neurite outgrowth, as demonstrated in a variety of in vitro neuronal systems, including fetal rat and human dopaminergic neurons (39), SH-SY5Y cells (42), and hippocampal HiB5 cells (43). The possibility has been raised that PDGF might act as a neuromodulator in the central nervous system. In the present study we demonstrate yet another novel function hitherto unidentified of PDGF; that of induction of Arc gene, a key player in long term potentiation in the central nervous system.
Previous work aimed at identifying the underlying mechanisms contributing to cytokine-induced long term potentiation or memory formation pointed to the role of immediate early genes as the key player in this process (44). Activation of these immediate early genes is thought to be essential for the long term synaptic plasticity (17, 45) and was a criteria used to distinguish long term versus short term plasticity. Because generation and maintenance of long term potentiation in the hippocampus is often accompanied with up-regulation of the immediate early gene Arc-1 (46, 47) and because PDGF exposure of rat hippocampal slices resulted in induction of long term potentiation, we sought to explore the mechanism of PDGF-mediated regulation of Arc-1/Arg3.1. Our findings demonstrated that PDGF was able to induce the expression of Arc/Arg3.1 in a concentration and time-dependent manner in rat hippocampal neurons. PDGF-induced Arc expression peaked at 3 h post-treatment and was sustained for up to 24 h after which the levels decreased to base line. Interestingly, PDGF-induced Arc expression could be remarkably blocked by pretreating the neurons with the PDGF receptor-tyrosine kinase inhibitor STI-571, thus underscoring the role of receptor engagement in this process. To evaluate the relevance of these cell culture studies in vivo, PDGF was injected directly into the adult mice hippocampi and examined for induced expression of Arc/Arg3.1 by immunostaining. PDGF treatment resulted in a rapid (3 h) up-regulation of Arc/Agr3.1 expression in the hippocampi of mice compared with the saline-injected controls. Positive immunostaining of Arc/Arg3.1 in the dendritic lamina of hippocampal neurons suggested the migration of Arc/Arg3.1 protein from the cell body to the synapse. These findings were in agreement with previous reports demonstrating localization of Arc/Arg3.1 protein in the post-synaptic regions after high frequency stimulation (48). Arc/Agr3.1 could, thus, play a role in reshaping of the neurosynapses, especially the smaller ones (48).
We next sought to explore the mechanism by which PDGF induced Arc/Arg3.1 expression. Published findings have suggested that the activation of MAPK/ERK signaling pathways play critical roles in both learning and memory and synaptic plasticity (49–53). Coincidently, the MAPK/ERK signaling pathways are also the key mediators of PDGF-induced proliferation and neurotrophic differentiation (54). Using the pharmacological approach involving the MAPK/ERK inhibitor U0126, we assessed the relationship between PDGF-induced ERK activation and Arc/Arg3.1 expression. PDGF-mediated induction of Arc/Agr3.1 expression in the hippocampal neurons involved activation of ERK, an effect that was abrogated by the ERK inhibitor. MAPK/ERK kinase signaling pathways are, thus, critical for PDGF-induced Arc/Arg3.1 expression. In contrast, PDGF-mediated activation of PI3K/Akt pathway was not involved the induction of Arc as the PI3K-specific inhibitor LY294002 was not able to abrogate PDGF-induced Arc/Arg3.1 expression.
It is well established that an increase in intracellular calcium is critical for N-methyl-d-aspartate receptor-dependent long term potentiation (55, 56) as has been demonstrated for brain-derived neurotrophic factor-induced long term potentiation, a process involving influx of extracellular calcium (57). In the current study exposure of neurons to PDGF resulted in increased intracellular levels of calcium, an effect that could be blocked by pretreating cells with EGTA and thapsigargin. Although both the agents were able to block PDGF-induced calcium surge, only thapsigargin and not EGTA was able to completely inhibit PDGF-induced Arc/Arg3.1 transcription, suggesting thereby that Ca2+ release from endoplasmic reticulum plays a critical role in this process.
To further dissect whether calcium release was critical for ERK activation, the effect of thapsigargin and EGTA on PDGF-mediated activation of ERK was examined (Fig. 6B). Both thapsigargin and EGTA was able to significantly decrease PDGF-mediated ERK activation. However, MEK inhibitor was also not able to abrogate calcium surge induced by PDGF, thus underscoring the fact that MAPK/ERK activation, which lies downstream of the calcium release pathway, and calcium release are both essential for PDGF-induced Arc/Arg3.1 expression. These findings are in agreement to reports demonstrating MAPK/ERK and Ca2+ pathways for various other synaptic plasticity genes (58, 59).
To further examine the downstream mediators of Arc gene expression by the MAPK/ERK and Ca2+ pathways, we focused our attention on the transcription factor Egr-1. Egr-1 is one of the important downstream factors that is regulated by both MAPK and Ca2+ pathways (60–62). In accordance with the previous studies on the induction of Egr-1 expression in rat mesangial cells (63) in the presence of PDGF, our findings also demonstrate PDGF-mediated regulation of Egr-1 in rat hippocampal neurons. Furthermore, this regulation by PDGF was at the transcriptional and not the translational level as pretreatment with the transcription inhibitor resulted in abrogation of PDGF-mediated induction of Egr-1.
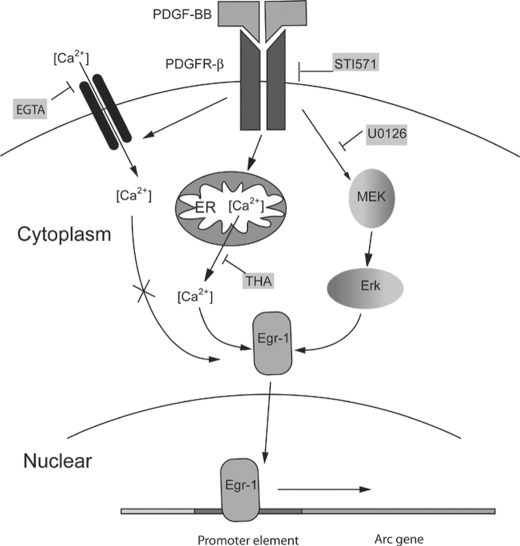
Previous studies by Abraham et al. (64) have demonstrated a correlation between Egr1 gene expression and stabilization of long term potentiation, and this has recently been corroborated in the gene mutant rat model (65). Our findings demonstrated that PDGF treatment of hippocampal neurons resulted in rapid induction of the transcription factor Egr-1, and both MEK inhibitor as well as the thapsigargin could effectively inhibit PDGF-mediated induction of Egr-1. In contrast, EGTA pretreatment of neurons failed to inhibit PDGF-induced expression of both Egr-1 and Arc/Arg3.1. Further corroboration of the role of Egr-1 in PDGF-induced Arc expression was carried out using the silencing RNA approach. Egr-1 siRNA- transfected cell lines failed to display PDGF-mediated induction of Arc/Arg3.1 expression (Fig. 7A). Furthermore, we were also able to demonstrate the binding of Egr-1 to promoter of the Arc/Arg3.1 gene using the ChIP assay. Egr-3, another member of the Egr-1 family of proteins that is also thought to contribute to Arc/Agr3.1 expression (30), was also tested for PDGF-mediated induction, translocation, and binding to the Arc promoter. PDGF exposure of neurons, however, was unable to induce the expression, translocation, and binding of Egr-3 to the Arc/Arg3.1 gene promoter. Taken together these findings identify Egr-1 as the junction transcriptional factor involved at the crossroads of two divergent pathways mediated by PDGF in the transcription of Arc/Arg3.1 as summarized in Fig. 8.
FIGURE 8.
Schematic illustration of signaling pathways involved in PDGF-mediated induction of Arc expression. After binding of PDGF to its cognate receptor-tyrosine kinase, there was an increase in intracellular calcium via either the influx of extracellular calcium or through the release of intracellular sources from the ER. Arc induction, however, is dependent only on the calcium released from the ER and not through the influx of extracellular calcium. PDGF can also result in activation of the MAPK/ERK pathway leading to downstream Arc expression. Both the pathways increased intracellular calcium, and the MAPK/ERK pathways converge downstream at the Egr-1 activation step, resulting in its nuclear translocation and subsequent binding to the Arc/Arg3.1 promoter, resulting in gene transcription. THA, thapsigargin.
Acknowledgments
We thank Novartis, Basel, Switzerland for providing STI-571. We thank from Dr. Young Han Lee (Konkuk University, Korea) for providing the dominant negative and constitutively active constructs of MEK.
This work was supported, in whole or in part, by National Institutes of Health Grants MH-068212, DA020392, DA023397, DA024442, and DA027729 (to S. B.).
- PDGF
- platelet-derived growth factor
- PDGF-R
- PDGF receptor
- ChIP
- chromatin immunoprecipitation
- ER
- endoplasmic reticulum
- EPSP
- excitatory postsynaptic potential
- ERK
- extracellular signal-regulated kinase
- MAPK
- mitogen-activated protein kinase
- MEK
- MAPK/ERK
- PI3K
- phosphatidylinositol 3′-kinase
- siRNA
- short interfering RNA.
REFERENCES
- 1.Heldin C. H., Westermark B. (1999) Physiol. Rev. 79, 1283–1316 [DOI] [PubMed] [Google Scholar]
- 2.Fredriksson L., Li H., Eriksson U. (2004) Cytokine Growth Factor Rev. 15, 197–204 [DOI] [PubMed] [Google Scholar]
- 3.Benito M., Lorenzo M. (1993) Growth Regul. 3, 172–179 [PubMed] [Google Scholar]
- 4.Sasahara M., Fries J. W., Raines E. W., Gown A. M., Westrum L. E., Frosch M. P., Bonthron D. T., Ross R., Collins T. (1991) Cell 64, 217–227 [DOI] [PubMed] [Google Scholar]
- 5.Gozal D., Simakajornboon N., Czapla M. A., Xue Y. D., Gozal E., Vlasic V., Lasky J. A., Liu J. Y. (2000) J. Neurochem. 74, 310–319 [DOI] [PubMed] [Google Scholar]
- 6.Egawa-Tsuzuki T., Ohno M., Tanaka N., Takeuchi Y., Uramoto H., Faigle R., Funa K., Ishii Y., Sasahara M. (2004) Exp. Neurol. 186, 89–98 [DOI] [PubMed] [Google Scholar]
- 7.Tseng H. C., Dichter M. A. (2005) Neurobiol. Dis. 19, 77–83 [DOI] [PubMed] [Google Scholar]
- 8.Ishii Y., Oya T., Zheng L., Gao Z., Kawaguchi M., Sabit H., Matsushima T., Tokunaga A., Ishizawa S., Hori E., Nabeshima Y., Sasaoka T., Fujimori T., Mori H., Sasahara M. (2006) J. Neurochem. 98, 588–600 [DOI] [PubMed] [Google Scholar]
- 9.Beazely M. A., Lim A., Li H., Trepanier C., Chen X., Sidhu B., Macdonald J. F. (2009) J. Biol. Chem. 284, 8054–8063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peng. F., Dhillon. N., Callen. S., Yao. H., Bokhari. S., Zhu. X., Baydoun. H. H., Buch. S. (2008) J. Neurovirol. 14, 62–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams B. P., Park J. K., Alberta J. A., Muhlebach S. G., Hwang G. Y., Roberts T. M., Stiles C. D. (1997) Neuron 18, 553–562 [DOI] [PubMed] [Google Scholar]
- 12.Erlandsson A., Enarsson M., Forsberg-Nilsson K. (2001) J. Neurosci. 21, 3483–3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng B., Mattson M. P. (1995) J. Neurosci. 15, 7095–7104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valenzuela C. F., Kazlauskas A., Weiner J. L. (1997) Brain Res. Brain Res. Rev. 24, 77–89 [DOI] [PubMed] [Google Scholar]
- 15.Oak J. N., Lavine N., Van Tol H. H. (2001) Mol. Pharmacol. 60, 92–103 [DOI] [PubMed] [Google Scholar]
- 16.Valenzuela C. F., Kazlauskas A., Brozowski S. J., Weiner J. L., Demali K. A., McDonald B. J., Moss S. J., Dunwiddie T. V., Harris R. A. (1995) Mol. Pharmacol. 48, 1099–1107 [PubMed] [Google Scholar]
- 17.Davis H. P., Squire L. R. (1984) Psychol. Bull. 96, 518–559 [PubMed] [Google Scholar]
- 18.Link W., Konietzko U., Kauselmann G., Krug M., Schwanke B., Frey U., Kuhl D. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 5734–5738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brakeman P. R., Lanahan A. A., O'Brien R., Roche K., Barnes C. A., Huganir R. L., Worley P. F. (1997) Nature 386, 284–288 [DOI] [PubMed] [Google Scholar]
- 20.Tsui C. C., Copeland N. G., Gilbert D. J., Jenkins N. A., Barnes C., Worley P. F. (1996) J. Neurosci. 16, 2463–2478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cottrell J. R., Borok E., Horvath T. L., Nedivi E. (2004) Neuron 44, 677–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steward O., Worley P. F. (2001) Neuron 30, 227–240 [DOI] [PubMed] [Google Scholar]
- 23.Steward O., Wallace C. S., Lyford G. L., Worley P. F. (1998) Neuron 21, 741–751 [DOI] [PubMed] [Google Scholar]
- 24.Guzowski J. F., McNaughton B. L., Barnes C. A., Worley P. F. (1999) Nat. Neurosci. 2, 1120–1124 [DOI] [PubMed] [Google Scholar]
- 25.Yin Y., Edelman G. M., Vanderklish P. W. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 2368–2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iihara K., Hashimoto N., Tsukahara T., Sakata M., Yanamoto H., Taniguchi T. (1997) J. Cereb. Blood Flow Metab. 17, 1097–1106 [DOI] [PubMed] [Google Scholar]
- 27.Xiong H., Baskys A., Wojtowicz J. M. (1996) Brain Res. 737, 188–194 [DOI] [PubMed] [Google Scholar]
- 28.Nunes-de-Souza R. L., Canto-de-Souza A., Rodgers R. J. (2002) Brain Res. 927, 87–96 [DOI] [PubMed] [Google Scholar]
- 29.Yao H. H., Ding J. H., Zhou F., Wang F., Hu L. F., Sun T., Hu G. (2005) J. Neurochem. 92, 948–961 [DOI] [PubMed] [Google Scholar]
- 30.Li L., Carter J., Gao X., Whitehead J., Tourtellotte W. G. (2005) Mol. Cell. Biol. 25, 10286–10300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang H., Bajraszewski N., Wu E., Wang H., Moseman A. P., Dabora S. L., Griffin J. D., Kwiatkowski D. J. (2007) J. Clin. Invest. 117, 730–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pratsinis H., Kletsas D. (2007) Eur. Spine J. 16, 1858–1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saqr H. E., Guan Z., Yates A. J., Stokes B. T. (1999) Neurochem. Int. 35, 411–422 [DOI] [PubMed] [Google Scholar]
- 34.Bergsten E., Uutela M., Li X., Pietras K., Ostman A., Heldin C. H., Alitalo K., Eriksson U. (2001) Nat. Cell Biol. 3, 512–516 [DOI] [PubMed] [Google Scholar]
- 35.Li X., Pontén A., Aase K., Karlsson L., Abramsson A., Uutela M., Bäckström G., Hellström M., Boström H., Li H., Soriano P., Betsholtz C., Heldin C. H., Alitalo K., Ostman A., Eriksson U. (2000) Nat. Cell Biol. 2, 302–309 [DOI] [PubMed] [Google Scholar]
- 36.Heldin C. H., Eriksson U., Ostman A. (2002) Arch. Biochem. Biophys. 398, 284–290 [DOI] [PubMed] [Google Scholar]
- 37.Sasahara A., Kott J. N., Sasahara M., Raines E. W., Ross R., Westrum L. E. (1992) Brain Res. Dev. Brain Res. 68, 41–53 [DOI] [PubMed] [Google Scholar]
- 38.Smits A., Kato M., Westermark B., Nistér M., Heldin C. H., Funa K. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 8159–8163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Othberg A., Odin P., Ballagi A., Ahgren A., Funa K., Lindvall O. (1995) Exp. Brain Res. 105, 111–122 [DOI] [PubMed] [Google Scholar]
- 40.Pietz K., Odin P., Funa K., Lindvall O. (1996) Neurosci. Lett. 204, 101–104 [DOI] [PubMed] [Google Scholar]
- 41.Mohapel P., Frielingsdorf H., Häggblad J., Zachrisson O., Brundin P. (2005) Neuroscience 132, 767–776 [DOI] [PubMed] [Google Scholar]
- 42.Hynds D. L., Burry R. W., Yates A. J. (1997) J. Neurosci. Res. 47, 617–625 [DOI] [PubMed] [Google Scholar]
- 43.Sung J. Y., Lee S. Y., Min D. S., Eom T. Y., Ahn Y. S., Choi M. U., Kwon Y. K., Chung K. C. (2001) J. Neurochem. 78, 1044–1053 [DOI] [PubMed] [Google Scholar]
- 44.Lanahan A., Worley P. (1998) Neurobiol. Learn Mem. 70, 37–43 [DOI] [PubMed] [Google Scholar]
- 45.Steward O., Worley P. (2002) Neurobiol. Learn. Mem. 78, 508–527 [DOI] [PubMed] [Google Scholar]
- 46.Guzowski J. F., Lyford G. L., Stevenson G. D., Houston F. P., McGaugh J. L., Worley P. F., Barnes C. A. (2000) J. Neurosci. 20, 3993–4001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Plath N., Ohana O., Dammermann B., Errington M. L., Schmitz D., Gross C., Mao X., Engelsberg A., Mahlke C., Welzl H., Kobalz U., Stawrakakis A., Fernandez E., Waltereit R., Bick-Sander A., Therstappen E., Cooke S. F., Blanquet V., Wurst W., Salmen B., Bösl M. R., Lipp H. P., Grant S. G., Bliss T. V., Wolfer D. P., Kuhl D. (2006) Neuron 52, 437–444 [DOI] [PubMed] [Google Scholar]
- 48.Kuhl D., Skehel P. (1998) Curr. Opin. Neurobiol. 8, 600–606 [DOI] [PubMed] [Google Scholar]
- 49.Frey U., Huang Y. Y., Kandel E. R. (1993) Science 260, 1661–1664 [DOI] [PubMed] [Google Scholar]
- 50.English J. D., Sweatt J. D. (1997) J. Biol. Chem. 272, 19103–19106 [DOI] [PubMed] [Google Scholar]
- 51.Rosenblum K., Futter M., Jones M., Hulme E. C., Bliss T. V. (2000) J. Neurosci. 20, 977–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Coogan A. N., O'Neill L. A., O'Connor J. J. (1999) Neuroscience 93, 57–69 [DOI] [PubMed] [Google Scholar]
- 53.Waltereit R., Dammermann B., Wulff P., Scafidi J., Staubli U., Kauselmann G., Bundman M., Kuhl D. (2001) J. Neurosci. 21, 5484–5493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Enarsson M., Erlandsson A., Larsson H., Forsberg-Nilsson K. (2002) Mol. Cancer Res. 1, 147–154 [PubMed] [Google Scholar]
- 55.Williams S., Johnston D. (1989) Neuron 3, 583–588 [DOI] [PubMed] [Google Scholar]
- 56.Mellor J., Nicoll R. A. (2001) Nat. Neurosci. 4, 125–126 [DOI] [PubMed] [Google Scholar]
- 57.Zheng F., Luo Y., Wang H. (2009) J. Neurosci. Res. 87, 380–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dolmetsch R. E., Pajvani U., Fife K., Spotts J. M., Greenberg M. E. (2001) Science 294, 333–339 [DOI] [PubMed] [Google Scholar]
- 59.Rosengart M. R., Arbabi S., Garcia I., Maier R. V. (2000) Shock 13, 183–189 [DOI] [PubMed] [Google Scholar]
- 60.Machado H. B., Vician L. J., Herschman H. R. (2008) J. Neurosci. Res. 86, 593–602 [DOI] [PubMed] [Google Scholar]
- 61.Moon Y., Yang H., Kim Y. B. (2007) Toxicol. Appl. Pharmacol. 223, 155–163 [DOI] [PubMed] [Google Scholar]
- 62.Mayer S. I., Thiel G. (2009) Eur. J. Cell Biol. 88, 19–33 [DOI] [PubMed] [Google Scholar]
- 63.Rupprecht H. D., Sukhatme V. P., Lacy J., Sterzel R. B., Coleman D. L. (1993) Am. J. Physiol. 265, F351–F360 [DOI] [PubMed] [Google Scholar]
- 64.Abraham W. C., Dragunow M., Tate W. P. (1991) Mol. Neurobiol. 5, 297–314 [DOI] [PubMed] [Google Scholar]
- 65.Lee J. L., Everitt B. J., Thomas K. L. (2004) Science 304, 839–843 [DOI] [PubMed] [Google Scholar]