The results obtained demonstrate the great importance of solvent-inaccessible conserved hydrogen bonds between the Hfq monomers in the stabilization of the hexamer structure.
Keywords: Hfq, Sm-like proteins, tertiary protein structure, protein thermostability
Abstract
The bacterial Sm-like protein Hfq forms homohexamers both in solution and in crystals. The monomers are organized as a continuous β-sheet passing through the whole hexamer ring with a common hydrophobic core. Analysis of the Pseudomonas aeruginosa Hfq (PaeHfq) hexamer structure suggested that solvent-inaccessible intermonomer hydrogen bonds created by conserved amino-acid residues should also stabilize the quaternary structure of the protein. In this work, one such conserved residue, His57, in PaeHfq was replaced by alanine, threonine or asparagine. The crystal structures of His57Thr and His57Ala Hfq were determined and the stabilities of all of the mutant forms and of the wild-type protein were measured. The results obtained demonstrate the great importance of solvent-inaccessible conserved hydrogen bonds between the Hfq monomers in stabilization of the hexamer structure.
1. Introduction
In bacteria, Hfq protein acts as a global post-transcriptional regulator which binds small regulatory RNAs and promotes their interaction with mRNAs (Valentin-Hansen et al., 2004 ▶; Brennan & Link, 2007 ▶). It controls the expression of many genes by its action on mRNA translation, stability or polyadenylation (Zhang et al., 1998 ▶; Vytvytska et al., 2000 ▶; Hajnsdorf & Régnier, 2000 ▶; Sledjeski et al., 2001 ▶). Hfq is a small (70–110 amino-acid residues) thermostable protein which exists in a homohexameric form in solution (Brennan & Link, 2007 ▶; Zhang et al., 2002 ▶; Møller et al., 2002 ▶). A hexameric organization has also been observed in the crystal structures of Hfq from Staphylococcus aureus (Schumacher et al., 2002 ▶), the core part of Escherichia coli Hfq (Sauter et al., 2003 ▶) and Hfq from Pseudomonas aeruginosa (Nikulin et al., 2005 ▶). All of these proteins formed doughnut-shaped rings with outer and inner diameters of about 65 and 10 Å and a thickness of 25–30 Å.
Hfq belongs to the Sm/Sm-like protein family. This family includes eukaryotic Sm and Sm-like (Lsm) proteins and archaeal Lsm proteins (Wilusz & Wilusz, 2005 ▶). Eukaryotic Sm/Lsm proteins are involved in RNA processing in the cell (Kufel et al., 2004 ▶; Verdone et al., 2004 ▶). In crystals they form heptamers (Achsel et al., 1999 ▶; Mayes et al., 1999 ▶; Walke et al., 2001 ▶) or octamers (Naidoo et al., 2008 ▶), whereas in cytoplasm the Sm proteins are found as heterodimers or trimers and are only able to form heptamers in the presence of U-rich small nuclear RNAs (UsnRNAs; Achsel et al., 1999 ▶; Will & Lührmann, 2001 ▶).
Archaeal genomes usually encode one or two distinct Lsm proteins called Lsm1 and Lsm2 (Salgado-Garrido et al., 1999 ▶). Lsm3 proteins have only been identified in a few archaeal species (Mura et al., 2003 ▶; Kilic et al., 2006 ▶). Lsm1 is the most abundant of the archaeal species. Archaeal Lsm1 and Lsm2 proteins form stable homoheptamers, with the exception of Archaeoglobus fulgidus AF-Sm2, which can exist in hexameric or heptameric forms depending on the pH or the presence of RNA (Törö et al., 2001 ▶; Achsel et al., 2001 ▶; Kilic et al., 2006 ▶).
The Sm-protein family is characterized by a conserved motif of about 70 amino acids, which is called the Sm-domain. It is a β-barrel-type structure consisting of five β-strands, which are capped by an N-terminal α-helix (Fig. 1 ▶ a). The Sm-domain contains two conserved sequence motifs (Sm1 and Sm2) linked by a loop that differs in length and sequence depending on the species (Valentin-Hansen et al., 2004 ▶; Séraphin, 1995 ▶). Strands β1, β2 and β3 form the Sm1 motif and strands β4 and β5 constitute the Sm2 motif of the domain. The sequence of the Sm1 motif is conserved among all bacteria, archaea and eukarya (Kambach et al., 1999 ▶; Hajnsdorf & Régnier, 2000 ▶; Valentin-Hansen et al., 2004 ▶; Kilic et al., 2006 ▶). In contrast, the Sm2 motif has different consensus sequences in bacterial Hfq and eukaryal/archaeal Sm/Lsm proteins (Sauter et al., 2003 ▶). Analysis of the known crystal structures (Törö et al., 2002 ▶; Nikulin et al., 2005 ▶; Brennan & Link, 2007 ▶) has shown that the β-strands of the Sm2 motifs organize the protein ring by means of hydrogen bonds formed by the main-chain O and N atoms. The quaternary structure is additionally stabilized by contacts between strongly conserved amino acids. Previously, we have suggested (Nikulin et al., 2005 ▶) that the conserved YKHI consensus sequence of the Sm2 motif in Hfq should define its hexamer formation and that His57 could play a very important role in the stabilization of the hexamer structure. To prove this hypothesis, we mutated His57 in P. aeruginosa Hfq (PaeHfq) to alanine, threonine and asparagine, measured the stability of the wild-type hexamer and the obtained mutant forms and solved the crystal structures of PaeHfq with His57Thr and His57Ala mutations.
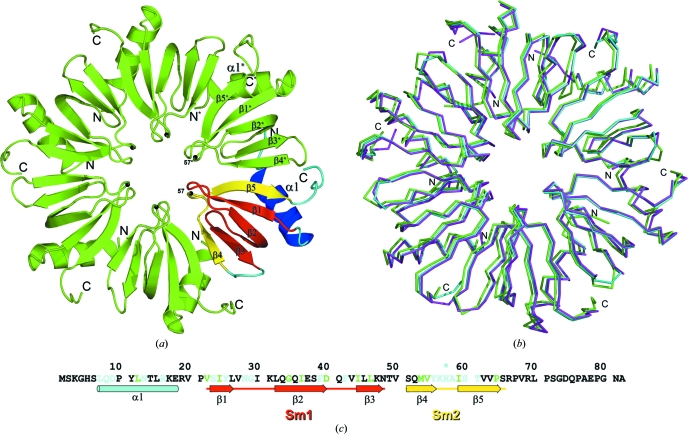
Figure 1.
(a) Overall structure of the Hfq hexamer from P. aeruginosa. One monomer is coloured according to the conserved sequence motifs: the Sm1 motif (β1, β2 and β3) is shown in yellow, the Sm2 motif (β4 and β5) in red and the N-terminal α1 helix in blue. The position of amino-acid residue 57 is shown by a black sphere. (b) Superposition of wild-type PaeHfq (cyan), H57T PaeHfq (magenta) and H57A PaeHfq (green). The Cα-atom r.m.s. deviations of H57T PaeHfq and H57A PaeHfq from the wild-type protein are 0.43 and 0.49 Å, respectively. (c) The amino-acid sequence of PaeHfq with corresponding secondary-structure elements. Amino-acid residues that are conserved in Lsm proteins from bacteria, archaea and eukarya are shown in green; those conserved in bacteria only are shown in cyan.
2. Materials and methods
2.1. Site-directed mutagenesis, gene expression and recombinant protein purification
To prepare mutant forms of PaeHfq, site-directed mutagenesis was carried out by PCR using oligonucleotides which contained the desired mutations. All of the mutants (hfqH57A, hfqH57N and hfqH57T) were constructed in two steps. In step 1, fragments carrying a mutation were amplified from pET22b(+)/Hfq DNA by PCR with the reverse primer 5′-CGGGATCCTCAAGCGTTGCCC-3′ and corresponding oligonucleotides for each fragment (H57A, 5′-GTTTACAAGGCGGCGATCTCC-3′; H57N, 5′-GTTTACAAGAACGCGATCTCC-3′; H57T, 5′-GTTTACAAGACCGCGATCTCC-3′). The fragments were then completed by PCR using the forward primer 5′-GGGAATTCCATATGTCAAAAGGGCAT-3′ and the PCR products obtained in step 1. The final PCR products were inserted into pET22b(+) plasmid DNA and verified by sequencing. All of the mutant proteins were purified as described previously (Nikulin et al., 2005 ▶).
2.2. Circular-dichroism (CD) measurements
CD measurements were performed on a Jasco J600 spectropolarimeter equipped with a Julabo F25 computer-controlled thermostat. All spectra and melting experiments were measured using a cell with a 0.1 mm path length. The melting experiments were performed by monitoring the change in ellipticity at 220 nm.
2.3. Crystallization and data collection
Protein crystals were obtained using the hanging-drop vapour-diffusion technique at 295 K. All drops were set up by mixing 2.0 µl protein solution (8 mg ml−1 protein, 100 mM NaCl, 50 mM Tris–HCl pH 8.0) with 2.0 µl reservoir solution (200 mM NH4Cl, 15% PEG MME 2000, 50 mM Tris–HCl pH 8.5, 20 mM CdCl2 or ZnCl2). Crystals appeared after 1 d and reached maximum dimensions of 300 × 100 × 50 µm within one week. Before freezing, the crystals were transferred to 15% PEG MME 2000, 15% PEG 400, 200 mM ammonium chloride, 50 mM Tris–HCl pH 8.5. X-ray diffraction data were collected from the crystals on EMBL beamline X12 (DESY, Hamburg) or the BL14.1 beamline at BESSY (Berlin) and were processed using XDS (Kabsch, 2010 ▶). Detailed data-collection statistics are given in Table 1 ▶.
Table 1. Data-collection and refinement statistics.
Values in parentheses are for the highest resolution shell.
| H57T PaeHfq | H57A PaeHfq | |
|---|---|---|
| Macromolecule details | ||
| PDB code | 3inz | 3m4g |
| No. of residues per monomer | 82 | 82 |
| Molecular assembly | Hexamer | Hexamer |
| Molecular weight of the hexamer (Da) | 54411 | 54489 |
| Data-collection statistics | ||
| Wavelength (Å) | 1.00 | 0.91841 |
| Resolution range (Å) | 30.0–1.7 (1.74–1.7) | 30.0–2.05 (2.16–2.05) |
| Space group | P21212 | P1 |
| Unit-cell parameters (Å, °) | a = 61.3, b = 71.2, c = 104.4, α = β = γ = 90 | a = 66.5, b = 66.6, c = 68.7, α = 91.8, β = 115.3, γ = 119.9 |
| Total reflections | 337107 (10581) | 234040 (34408) |
| Unique reflections | 50597 (2983) | 53606 (7783) |
| Redundancy | 6.7 (3.5) | 4.4 (4.4) |
| Completeness (%) | 94.1 (80.4) | 97.4 (96.5) |
| Rmerge (%) | 4.0 (36.2) | 5.5 (49.0) |
| Average I/σ(I) | 27.9 (3.9) | 14.4 (3.0) |
| Wilson B factor (Å2) | 31.2 | 32.9 |
| Refinement statistics | ||
| Resolution (Å) | 30.0–1.70 (1.74–1.70) | 30.0–2.05 (2.09–2.05) |
| Completeness (%) | 94.1 (80.4) | 97.4 (96.5) |
| Reflections | 50571 (2983) | 53561 (2704) |
| Test reflections | 2528 (131) | 2725 (127) |
| Rwork (%) | 0.149 (0.178) | 0.194 (0.296) |
| Rfree (%) | 0.218 (0.247) | 0.261 (0.393) |
| No. of waters | 379 | 331 |
| No. of ions | 11 | 18 |
| R.m.s. deviation from ideal geometry | ||
| Bonds (Å) | 0.010 | 0.005 |
| Angles (°) | 1.315 | 0.903 |
| Chirality (°) | 0.103 | 0.059 |
| Planarity (°) | 0.006 | 0.004 |
| Average B value (Å2) | ||
| Main chain | 31.39 | 45.24 |
| Side chain and water | 38.74 | 50.82 |
| MolProbity results | ||
| Ramachandran favoured (%) | 95.20 | 95.76 |
| Ramachandran allowed (%) | 98.74 | 99.88 |
| Ramachandran outliers (%) | 1.26 | 0.12 |
2.4. Structure determination and refinement
The protein structures were solved by the molecular-replacement method using the PHENIX package (Adams et al., 2002 ▶) with a hexamer of wild-type PaeHfq as the initial model (PDB code 1u1s; Nikulin et al., 2005 ▶). The simulated-annealing protocol following conventional residual refinement in combination with manual inspection in Coot (Emsley & Cowtan, 2004 ▶) was used to refine the model. Water molecules were introduced into the model using the ‘water pick’ function of Coot and the highest peaks in the F o − F c map were assigned to ions. At the final stage anisotropic ADP refinement of H57T PaeHfq was implemented, improving the R and R free factors from 0.199 and 0.244 to 0.149 and 0.218, respectively. The structure coordinates of H57A PaeHfq and H57T PaeHfq have been deposited in the Protein Data Bank (PDB codes 3inz and 3m4g, respectively).
3. Results and discussion
3.1. Crystal structures of H57A PaeHfq and H57T PaeHfq
The crystal structures of H57A PaeHfq and H57T PaeHfq were solved and refined to 2.05 and 1.7 Å resolution, respectively (Table 1 ▶). The substitutions did not change the overall shape of the hexamer or the conformations of the monomers (Fig. 1 ▶ b).
In the wild-type protein the side chain of His57 formed two hydrogen bonds to the main-chain O atoms of the adjacent monomer (Fig. 2 ▶ a). We supposed that the mutations would result in the disappearance of one or both of these hydrogen bonds. Indeed, the substitution of His57 by alanine led to a loss of the hydrogen bonds (Fig. 2 ▶ b). In contrast, the replacement of His57 by threonine gave rise to the formation of new hydrogen bonds between adjacent monomers that replaced those in the wild-type protein. Two water molecules acted as bridges connecting the hydroxyl of the threonine of one monomer to the main-chain carbonyl O atoms of Thr57 and Ile59 of another molecule (Fig. 2 ▶ c). Nevertheless, the compensation was not completely equivalent. In the wild-type protein one of the hydrogen bonds formed by His57 is inaccessible to solvent, whereas in H57T PaeHfq the water-bridge hydrogen bonds are accessible. In this case the protein atoms could easily form new hydrogen bonds to solvent. At higher temperature the water molecules could even escape from their sites. In this case, the hydroxyl group of Thr57 could be positioned at a short distance from the two carbonyl O atoms of the neighbouring monomer, which is not desirable. To prove this hypothesis, we measured the stability of the Hfq mutant proteins.
Figure 2.
The interface of two adjacent monomers in the PaeHfq hexamer. The main chains of the monomers are shown in green and yellow. Side chains are shown for residue 57 only. Hydrogen bonds are shown as dotted lines. (a) The wild-type PaeHfq crystal structure. (b) The H57A PaeHfq crystal structure. (c) The H57T PaeHfq crystal structure.
3.2. Stability of the Hfq mutant forms
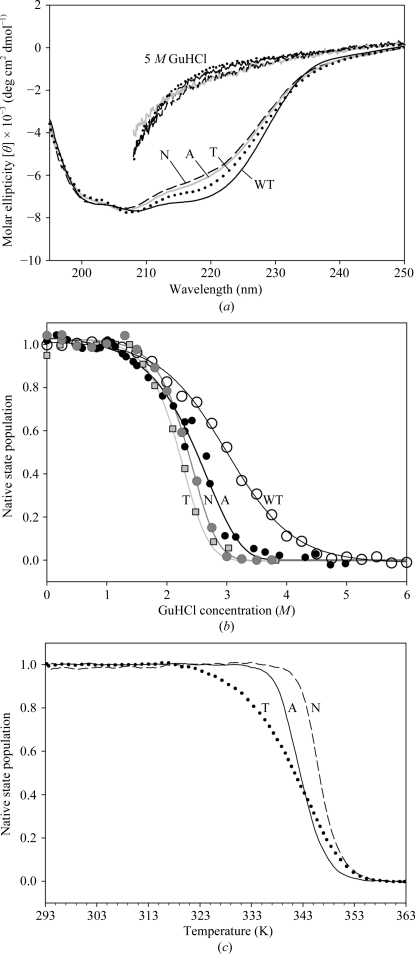
To evaluate the influence of the His57 substitutions on PaeHfq hexamer stability, CD spectra of the wild-type protein and its mutant forms were measured. At room temperature all these proteins had similar spectra corresponding to an α/β structure (Fig. 3 ▶ a). It was found that wild-type PaeHfq possesses extreme stability: its CD spectrum did not change during heating to 366 K or on the addition of urea up to 8 M. Difference scanning calorimetric experiments showed that the denaturation peak of wild-type PaeHfq appeared near 393 K (V. V. Filimonov, personal communication). Therefore, PaeHfq has one of the highest denaturation temperatures of known proteins (Tanaka et al., 2006 ▶). The secondary structure of wild-type PaeHfq, as well as those of its H57A, H57T and H57N mutants, was completely destroyed in the presence of 5 M GdnHCl (Fig. 3 ▶ a). The GdnHCl-induced unfolding of the proteins under equilibrium conditions demonstrated that all of the substitutions changed the stability of the protein considerably but in a similar way (Fig. 3 ▶ b).
Figure 3.
(a) CD spectrum of wild-type and mutant (H57A, H57T, H57N) PaeHfq proteins under nondenaturing conditions (lower lines) and in the presence of 5 M GdnHCl (upper lines). (b) Relative change of ellipticity at 220 nm during equilibrium unfolding of the proteins by GdnHCl. (c) Relative change of ellipticity at 220 nm during temperature unfolding of the mutant proteins in the presence of 1 M GdnHCl.
To reveal the difference in stability of the PaeHfq mutants, the relative changes in ellipticity at 220 nm were measured during temperature unfolding (Fig. 3 ▶ c). The presence of 1 M GdnHCl in the buffer was important in order to melt the proteins within the operating range of the spectropolarimeter. The H57N, H57A and H57T mutant forms of PaeHfq had melting temperatures of 346, 343 and 341 K, respectively, whereas wild-type PaeHfq retained its structure up to 366 K. Compared with the other mutants, the H57T PaeHfq had the lowest melting temperature, which was accompanied by a deterioration of melting-process cooperativity. The reason for this behaviour of H57T PaeHfq appears to be a consequence of the incorporation of water molecules between the side chain of the threonine and the main chain of the adjacent protein monomer as discussed above. In the H57N PaeHfq protein stereochemical analysis showed that the asparagine residue is able to organize a direct but water-accessible hydrogen bond to the main-chain atoms of the neighbouring monomer. Therefore, this substitution resulted in a decreased melting temperature for the mutant protein forms but did not lead to deterioration of the melting cooperativity.
Supplementary Material
PDB reference: P. aeruginosa Hfq, H57A mutant, 3inz
PDB reference: H57T mutant, 3m4g
Acknowledgments
The research was supported by the Russian Academy of Sciences, the Russian Federal Agency for Science and Innovation (02.740.11.0295), the Russian Foundation for Basic Research (10-04-00818) and the Program of the RAS on Molecular and Cellular Biology.
References
- Achsel, T., Brahms, H., Kastner, B., Bachi, A., Wilm, M. & Lührmann, R. (1999). EMBO J.18, 5789–5802. [DOI] [PMC free article] [PubMed]
- Achsel, T., Stark, H. & Lührmann, R. (2001). Proc. Natl Acad. Sci. USA, 98, 3685–3689. [DOI] [PMC free article] [PubMed]
- Adams, P. D., Grosse-Kunstleve, R. W., Hung, L.-W., Ioerger, T. R., McCoy, A. J., Moriarty, N. W., Read, R. J., Sacchettini, J. C., Sauter, N. K. & Terwilliger, T. C. (2002). Acta Cryst. D58, 1948–1954. [DOI] [PubMed]
- Brennan, R. G. & Link, T. M. (2007). Curr. Opin. Microbiol.10, 125–133. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Hajnsdorf, E. & Régnier, P. (2000). Proc. Natl Acad. Sci. USA, 97, 1501–1505. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kambach, C., Walke, S., Young, R., Avis, J., de La Fortelle, E., Raker, V., Lührmann, R., Li, J. & Nagai, K. (1999). Cell, 96, 375–387. [DOI] [PubMed]
- Kilic, T., Sanglier, S., Van Dorsselaer, A. & Suck, D. (2006). Protein Sci.15, 2310–2317. [DOI] [PMC free article] [PubMed]
- Kufel, J., Bousquet-Antonelli, C., Beggs, J. D. & Tollervey, D. (2004). Mol. Cell. Biol.24, 9646–9657. [DOI] [PMC free article] [PubMed]
- Mayes, A. E., Verdone, L., Legrain, P. & Beggs, J. D. (1999). EMBO J.18, 4321–4331. [DOI] [PMC free article] [PubMed]
- Møller, T., Franch, T., Hojrup, P., Keene, D. R., Bachinger, H. P., Brennan, R. G. & Valentin-Hansen, P. (2002). Mol. Cell, 9, 23–30. [DOI] [PubMed]
- Mura, C., Phillips, M., Kozhukhovsky, A. & Eisenberg, D. (2003). Proc. Natl Acad. Sci. USA, 100, 4539–4544. [DOI] [PMC free article] [PubMed]
- Naidoo, N., Harrop, S. J., Sobti, M., Haynes, P. A., Szymczyna, B. R., Williamson, J. R., Curmi, P. M. G. & Mabbutt, B. C. (2008). J. Mol. Biol.377, 1357–1371. [DOI] [PubMed]
- Nikulin, A., Stolboushkina, E., Perederina, A., Vassilieva, I., Blaesi, U., Moll, I., Kachalova, G., Yokoyama, S., Vassylyev, D., Garber, M. & Nikonov, S. (2005). Acta Cryst. D61, 141–146. [DOI] [PubMed]
- Salgado-Garrido, J., Bragado-Nilsson, E., Kandels-Lewis, S. & Séraphin, B. (1999). EMBO J.18, 3451–3462. [DOI] [PMC free article] [PubMed]
- Sauter, C., Basquin, J. & Suck, D. (2003). Nucleic Acids Res.31, 4091–4098. [DOI] [PMC free article] [PubMed]
- Schumacher, M. A., Pearson, R. F., Møller, T., Valentin-Hansen, P. & Brennan, R. G. (2002). EMBO J.21, 3546–3556. [DOI] [PMC free article] [PubMed]
- Séraphin, B. (1995). EMBO J.14, 2089–2098. [DOI] [PMC free article] [PubMed]
- Sledjeski, D. D., Whitman, C. & Zhang, A. (2001). J. Bacteriol.183, 1997–2005. [DOI] [PMC free article] [PubMed]
- Tanaka, T., Sawano, M., Ogasahara, K., Sakaguchi, Y., Bagautdinov, B., Katoh, E., Kuroishi, C., Shinkai, A., Yokoyama, S. & Yutani, K. (2006). FEBS Lett.580, 4224–4230. [DOI] [PubMed]
- Törö, I., Basquin, J., Teo-Dreher, H. & Suck, D. (2002). J. Mol. Biol.320, 129–142. [DOI] [PubMed]
- Törö, I., Thore, S., Mayer, C., Basquin, J., Séraphin, B. & Suck, D. (2001). EMBO J.20, 2293–2303. [DOI] [PMC free article] [PubMed]
- Valentin-Hansen, P., Eriksen, M. & Udesen, C. (2004). Mol. Microbiol.51, 1525–1533. [DOI] [PubMed]
- Verdone, L., Galardi, S., Page, D. & Beggs, J. D. (2004). Curr. Biol.14, 1487–1491. [DOI] [PubMed]
- Vytvytska, O., Moll, I., Kaberdin, V. R., von Gabain, A. & Bläsi, U. (2000). Genes Dev.14, 1109–1118. [PMC free article] [PubMed]
- Walke, S., Bragado-Nilsson, E., Séraphin, B. & Nagai, K. (2001). J. Mol. Biol.308, 49–58. [DOI] [PubMed]
- Will, C. L. & Lührmann, R. (2001). Curr. Opin. Cell Biol.13, 290–301. [DOI] [PubMed]
- Wilusz, C. J. & Wilusz, J. (2005). Nature Struct. Mol. Biol.12, 1031–1036. [DOI] [PubMed]
- Zhang, A., Altuvia, S., Tiwari, A., Argaman, L., Hengge-Aronis, R. & Storz, G. (1998). EMBO J.17, 6061–6068. [DOI] [PMC free article] [PubMed]
- Zhang, A., Wassarman, K. M., Ortega, J., Steven, A. C. & Storz, G. (2002). Mol. Cell, 9, 11–22. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: P. aeruginosa Hfq, H57A mutant, 3inz
PDB reference: H57T mutant, 3m4g