The crystal structure of bilirubin oxidase (BOD) from M. verrucaria has been determined at 2.3 Å resolution using a merohedrally twinned crystal. BOD has four copper-coordination sites that are almost identical to those of other multicopper oxidases and is also very similar to them in overall structure.
Keywords: bilirubin oxidase, twinned crystals, multicopper enzymes, Myrothecium verrucaria
Abstract
Bilirubin oxidase (BOD), a multicopper oxidase found in Myrothecium verrucaria, catalyzes the oxidation of bilirubin to biliverdin. Oxygen is the electron acceptor and is reduced to water. BOD is used for diagnostic analysis of bilirubin in serum and has attracted considerable attention as an enzymatic catalyst for the cathode of biofuel cells that work under neutral conditions. Here, the crystal structure of BOD is reported for the first time. Blue bipyramid-shaped crystals of BOD obtained in 2-methyl-2,4-pentanediol (MPD) and ammonium sulfate solution were merohedrally twinned in space group P63. Structure determination was achieved by the single anomalous diffraction (SAD) method using the anomalous diffraction of Cu atoms and synchrotron radiation and twin refinement was performed in the resolution range 33–2.3 Å. The overall organization of BOD is almost the same as that of other multicopper oxidases: the protein is folded into three domains and a total of four copper-binding sites are found in domains 1 and 3. Although the four copper-binding sites were almost identical to those of other multicopper oxidases, the hydrophilic Asn residue (at the same position as a hydrophobic residue such as Leu in other multicopper oxidases) very close to the type I copper might contribute to the characteristically high redox potential of BOD.
1. Introduction
Bilirubin oxidase (BOD; EC 1.3.3.5), a member of the multicopper oxidase family, is found in Myrothecium verrucaria. It catalyzes the oxidation of bilirubin to biliverdin and then to a purple pigment in vitro (Shimizu, Kwon et al., 1999 ▶; Shimizu, Sasaki et al., 1999 ▶). In medical examinations of the liver, BOD is used for diagnostic measurement of the bilirubin concentration in serum (Kosaka et al., 1987 ▶; Shimizu, Kwon et al., 1999 ▶; Shimizu, Sasaki et al., 1999 ▶). Laccase, the most studied enzyme in this family, has great industrial potential in processes such as the decolorization of dyes, biopulping, organic synthesis and hair dyeing (Couto & Herrera, 2006 ▶).
Multicopper oxidases contain one type I, one type II and two (one pair of) type III coppers. The type II and III coppers form a trinuclear centre that reduces oxygen to water molecules and the type I copper functions as the electron mediator from the substrate to the trinuclear centre (Shleev et al., 2005 ▶; Enguita et al., 2004 ▶). Recently, BOD has been considered to be one of the best enzymes for reactions in biofuel cell cathodes because of its high reactivity at neutral pH (Cracknell et al., 2008 ▶; Tsujimura et al., 2001 ▶).
The structures of several multicopper oxidases, such as the CueO laccase from Escherichia coli (27% sequence identity; Roberts et al., 2003 ▶), the CotA laccase from Bacillus subtilis (35% sequence identity; Enguita et al., 2004 ▶), laccase from Trametes versicolor (Piontek et al., 2002 ▶) and ascorbate oxidase (Messerschmidt et al., 1992 ▶) have been reported. BOD is a monomeric protein (like other multicopper oxidases) with a molecular mass of 60 kDa. The structure of BOD has been predicted to be similar to those of other multicopper oxidases, but no crystal structure of BOD has been obtained.
In the present paper, we report the crystal structure of BOD. Blue bipyramid-shaped crystals of BOD were grown in MPD and ammonium sulfate solution using the hanging-drop vapour-diffusion method and were found to be merohedrally twinned in space group P63. The crystal structure of BOD was solved at 2.3 Å resolution by the SAD method using the anomalous diffraction of Cu atoms. The structure of BOD is similar to those of other multicopper oxidases such as the laccases CueO and CotA. The protein is folded into three domains and four copper-binding sites are located in domains 1 and 3. The copper-binding sites are almost identical to those in other multicopper oxidases.
2. Materials and methods
2.1. Materials
BOD from M. verrucaria (MT-1) was purified as described previously (Shimizu, Kwon et al., 1999 ▶; Shimizu, Sasaki et al., 1999 ▶). The sequence-database reference code for the protein is UniProtKB/Swiss-Prot Q12737. The sample was dialyzed against 20 mM Tris–HCl buffer pH 8.0 and further purified on a Mono Q column (Mono Q 5/50 GL, GE Healthcare, Buckinghamshire, England) equilibrated with 20 mM Tris–HCl pH 8.0 and eluted with a linear gradient of buffer containing 0–1 M KCl at a flow rate of 1.5 ml min−1. The purified sample was concentrated to about 10 mg ml−1 using Centricon-10 or Ultrafree-4 centrifugal filter units (Millipore, Bedford, Massachusetts, USA). The protein concentration was determined using a molar absorption coefficient at 600 nm of ∊600 = 5000 M −1 cm−1. Other chemicals were of guaranteed grade from Wako Pure Chemical Industries (Osaka, Japan), Nacalai Tesque (Kyoto, Japan) and Sigma–Aldrich Chemicals (St Louis, Missouri, USA).
2.2. Crystallization
The concentrated purified BOD sample (9.2 mg ml−1 in 10 mM Tris–HCl pH 8.0) was crystallized using the hanging-drop vapour-diffusion method. The solution in the crystallization drop was prepared on a siliconized cover glass by mixing identical volumes (3 µl + 3 µl) of protein solution and precipitant solution [10% 2-methyl-2,4-pentanediol (MPD), 1.44 M ammonium sulfate, 10% glycerol and 0.5 M KCl]. The droplets were equilibrated against 0.2–1.0 ml precipitant solution at 293 K. The obtained crystals were light blue in colour and bipyramidal in shape (spindle-shaped); the radius of the crystals was around 0.4 mm.
2.3. Data collection and processing
The crystal was mounted in a nylon CryoLoop (Hampton Research, Aliso Viejo, California, USA) and placed into liquid nitrogen. Cryoprotectant (10% MPD and 10% glycerol) was included in the precipitant solution used for crystallization. The X-ray fluorescence spectrum of the sample was measured for Cu atoms and the peak wavelength was determined to be 1.37751 Å. Diffraction data were collected in a cold nitrogen-gas stream (100 K) on an ADSC Quantum 4 CCD area detector (Rigaku/MSC, The Woodlands, Texas, USA) using synchrotron radiation at a wavelength of 1.37751 Å at the BL38B1 station of SPring-8 (Hyogo, Japan). The resulting data set was processed, merged and scaled using the HKL-2000 program package (HKL Research, Charlottesville, Virginia, USA) to a resolution of 2.5 Å (Table 1 ▶). Diffraction data from another crystal were collected at a wavelength of 1.0 Å and processed to a resolution of 2.3 Å in the same way (Table 2 ▶).
Table 1. Data-collection and phasing statistics.
Values in parentheses are for the highest resolution bin.
| Diffraction data | |
| X-ray source | SPring-8 (BL38B1) |
| Wavelength (Å) | 1.3775 |
| Detector | ADSC Quantum 4 CCD |
| Crystal system | Hexagonal |
| Space group | P63 (hemihedral twin) |
| Unit-cell parameters | a = b = 139.4, c = 135.4 |
| Resolution limits (Å) | 50–2.50 (2.59–2.50) |
| Measured reflections | 292856 (26721) |
| Unique reflections | 100078 (9897) |
| Completeness (%) | 98.1 (97.0) |
| Rmerge (%) | 4.7 (24.3) |
| Phasing | |
| Program used | SOLVE (SAD), RESOLVE |
| Resolution range (Å) | 20–2.90 (3.0–2.9) |
| Reflections used | 32824 (5803) |
| Completeness (%) | 99.4 (99.0) |
| I/σ(I) | 22.3 (3.0) |
| No. of Cu sites (in the asymmetric unit) | 8 |
| FOM | 0.24 (0.16) |
| FOM (RESOLVE) | 0.74 (0.42) |
Table 2. Data-collection and refinement statistics.
Values in parentheses are for the highest resolution bin.
| Diffraction data | |
| X-ray source | SPring-8 (BL38B1) |
| Wavelength (Å) | 1.00 |
| Detector | ADSC Quantum 4 CCD |
| Crystal system | Hexagonal |
| Space group | P63 (merohedral twin) |
| Unit-cell parameters (Å) | a = b = 139.7, c = 135.6 |
| Resolution limits (Å) | 50–2.30 (2.38–2.30) |
| Measured reflections | 381408 |
| Unique reflections | 66384 (6514) |
| Completeness (%) | 99.5 (98.4) |
| Rmerge (%) | 6.1 (41.4) |
| I/σ(I) | 30.2 (2.4) |
| Refinement | |
| Program used | phenix.refine |
| Resolution range (Å) | 33.6–2.30 (2.34–2.30) |
| Reflections used | 64543 (2852) |
| Completeness (%) | 96.7 (98.0) |
| Twin law | h, k, l and h, −h − k, −l |
| Twin fraction | 0.487 |
| Residues/waters (in the asymmetric unit) | 1026/317 |
| Cu atoms/sugar chains | 8/4 |
| Average B factor, protein/waters/Cu (Å2) | 51.7/43.8/38.2 |
| Bond length r.m.s.d. (Å) | 0.007 |
| Bond angles r.m.s.d. (°) | 1.450 |
| R factor (%) | 20.5 (29.9) |
| Rfree (%) | 26.3 (28.7) |
| Subunit Cα r.m.s. (Å) | 0.55 |
2.4. Phase determination and structure refinement
The initial phase determination as a single crystal (not as a twinned crystal) was performed using the SAD method implemented in the program SOLVE/RESOLVE (Terwilliger, 2003 ▶, 2004 ▶; Table 1 ▶). Using the eight Cu atoms found in the structure, a model of B. subtilis CotA laccase (PDB code 1gsk or 1uvw; Enguita et al., 2003 ▶, 2004 ▶) was roughly superimposed on the calculated electron-density map. The amino-acid sequence of CotA was changed to that of BOD and initial modelling was performed using the TURBO-FRODO program (AFMB, Marseille, France). Rigid-body refinement and restrained least-squares refinement were performed using the REFMAC program (Collaborative Computational Project, Number 4, 1994 ▶). Several rounds of refinement as a twinned crystal (with twin law h, −h − k, −l determined using phenix.xtriage), followed by manual model building using Coot (Emsley & Cowtan, 2004 ▶), were carried out using SHELXL (Sheldrick, 2008 ▶) and phenix.refine (Adams et al., 2002 ▶). Final twin refinement was performed using phenix.refine. The atomic coordinates of the BOD structure have been deposited in the Protein Data Bank (code 3abg).
2.5. Structure comparison with other multicopper oxidases
Structure-similarity searches using BOD as a query were performed using the program DaliLite (Holm et al., 2008 ▶; Holm & Park, 2000 ▶). Two multicopper oxidases, the laccases CueO and CotA, which were determined by DaliLite to have the same highest Z scores (46.0), were compared with BOD using the SSM algorithm implemented in Coot and the LSQKAB least-squares fitting in the CCP4 package.
2.6. Molecular-dynamics simulation of a BOD–mediator complex
The molecular dynamics of a BOD–mediator complex were calculated using GROMACS (Van Der Spoel et al., 2005 ▶) and the GMX force field. The coordinates of the mediator compound 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) (ABTS) were taken from the structure of the CotA–ABTS complex (PDB code 1uvw). A model of the BOD–ABTS complex was made by superimposing (using Coot) the main chains of BOD and CotA. Water molecules were added to the model and molecular-dynamics calculations were performed for 10.0 ps (5000 steps of 0.002 ps).
3. Results
3.1. Quality of the diffraction data
Although a clear diffraction pattern was observed for the BOD crystals, the graphs of the cumulative intensity distribution and the fourth moment of E calculated using the TRUNCATE program indicated the possibility of perfect twinning (data not shown). The program phenix.xtriage also suggested a high possibility of twinning with twin law (h, −h − k, −l) and 〈I 2〉/〈I〉2 = 1.615. Therefore, the data sets were processed as P63 (not P6322 as misleadingly presumed by visual examination of precession images from the data sets). Phase determination and initial refinement calculations could be carried out using P63 twinned data applied to a single-crystal (untwinned) model and statistics for the phase determination are shown in Table 1 ▶. Final twin refinement was calculated with the twin law (h, −h − k, −l) and the twin fraction was determined to be 0.487 (almost equal to 0.5). Statistics for the refinement are summarized in Table 2 ▶.
3.2. Quality of the final model
The complete BOD molecule comprises 572 amino-acid residues. However, residues 509–572 of molecule A and 519–572 of molecule B were not included in the final model because no clear interpretable electron density was observed for these residues. The asymmetric unit contained two molecules and the root-mean-square deviation (r.m.s.d.) between molecules A and B was 0.55 Å for all Cα atoms. Weak (the coordinate_sigma parameter was 0.5 in phenix.refine) noncrystallographic symmetry restraints for residues 1–508 were applied during refinement. Four carbohydrate chains in molecules A and B were added to residues Asn472 and Asn482. Two GlcNAcs and one mannose were added to Asn472 and two GlcNAcs were added to Asn482. Relevant refinement statistics are given in Table 2 ▶. The overall completeness, R factor and R free values were 96.7%, 0.205 and 0.263, respectively, for the data used in the refinement. For the highest resolution bin (2.34–2.30 Å), the completeness was 98.0% and the R factor and R free values were 0.299 and 0.287, respectively. Because twinned refinement can lead to artificially low R values, parts of the reported structure may not be free of significant errors. In the Ramachandran plot of the main-chain torsion angles, 61.9% of the residues are in the most favoured regions, with 97.5% of the residues lying within the generously allowed regions as defined in the program PROCHECK (Collaborative Computational Project, Number 4, 1994 ▶). The low percentage in the most favoured region is probably a consequence of the difficulty of refinement calculations using a twinned crystal.
3.3. Overall organization of the structure
Fig. 1 ▶ displays the overall structure of BOD (chain A in the asymmetric unit) as a ribbon representation. As in other multicopper oxidases, BOD is folded into three domains and the four metal-binding sites are located in the clefts formed by domains 1 and 3 (Fig. 1 ▶ a). The two multicopper oxidases CueO laccase and CotA laccase had the highest Z scores (46.0) from the DaliLite structure-similarity search server. The r.m.s.d.s calculated using the SSM superposition in Coot were 1.7 Å between BOD and CueO for the Cα atoms of 403 corresponding residues and 1.5 Å between BOD and CotA for 428 residues (Fig. 1 ▶ b).
Figure 1.
(a) Ribbon representation of the overall structure of BOD (molecule A). Domains 1, 2 and 3 are shown in light pink, light green and light blue, respectively. Type I Cu, type II Cu and type III Cu ions are depicted as purple, green and orange spheres, respectively. The figure was generated using MolScript (Kraulis, 1991 ▶) and Raster3D (Merritt & Murphy, 1994 ▶). (b) Cα plots of BOD (molecule A, red) and the laccases CueO (molecule A of 2yxw, blue; Kataoka et al., 2007 ▶) and CotA (molecule A of 1uvw, orange). The structures were superimposed using the Cα atoms using SSM superposition as implemented in the program Coot. The four Cu atoms of BOD, CueO and CotA are displayed in deep pink, cyan and yellow, respectively; an ABTS molecule bound to CotA is also shown in yellow. The figures were produced with MolScript and Raster3D.
3.4. Structure of the copper-binding sites
The type I Cu is coordinated by four amino-acid residues (His398, Cys457, His462 and Met 467) and the type II Cu is coordinated by two amino-acid residues (His94 and His401) (listed in Table 3 ▶). The type III copper coordination by six amino-acid residues (His136, His403 and His456; His134, His96 and His458) and a water molecule is shown in Fig. 2 ▶. Although a liganded water molecule was included between the two type III coppers in the model of molecule B, the B factor (40.2 Å2) of the O atom of the liganded water was considerably higher than that (26.6 Å2) in molecule A. The distances from the coppers to the liganded amino-acid residues in the molecule are summarized in Table 3 ▶. As shown in Fig. 2 ▶, clear electron density was observed for all four coppers in the 2F o − F c electron-density map and the anomalous map calculated using the diffraction data used for the SAD method.
Table 3. Copper–ligand distances (Å).
| Molecule A | Molecule B | CueO laccase (PDB code 2yxw) | CotA laccase (PDB code 1gsk) | |
|---|---|---|---|---|
| Type I Cu | ||||
| Cu(1)—His398 | 2.62 | 2.41 | 2.06 | 2.05 |
| Cu(1)—Cys457 | 2.33 | 2.26 | 2.16 | 2.20 |
| Cu(1)—His462 | 2.20 | 2.20 | 2.02 | 2.06 |
| Cu(1)—Met467 | 3.54 | 3.18 | 3.32 | 3.27 |
| Type II Cu | ||||
| Cu(4)—His94 | 2.06 | 2.06 | 1.96 | 1.85 |
| Cu(4)—His401 | 2.22 | 2.19 | 1.90 | 1.92 |
| Type III Cu | ||||
| Cu(2)—HOH | 2.29 | 2.67 | 1.98 | 2.11 |
| Cu(2)—His136 | 2.03 | 2.20 | 2.00 | 2.05 |
| Cu(2)—His403 | 2.07 | 2.21 | 2.03 | 2.05 |
| Cu(2)—His456 | 2.17 | 2.23 | 2.09 | 2.03 |
| Cu(3)—HOH | 2.52 | 2.49 | 1.99 | 2.19 |
| Cu(3)—His134 | 2.20 | 2.20 | 2.04 | 2.09 |
| Cu(3)—His96 | 2.13 | 2.37 | 2.03 | 1.85 |
| Cu(3)—His458 | 2.32 | 2.35 | 2.08 | 2.10 |
| Cu(2)—Cu(3) | 4.75 | 5.09 | 3.86 | 4.28 |
| Cu(2)—Cu(4) | 4.24 | 3.93 | 3.56 | 4.67 |
| Cu(3)—Cu(4) | 4.16 | 3.97 | 3.91 | 4.64 |
Figure 2.
Stereo diagrams (parallel eyes) depicting the copper-binding sites. Electron-density maps (2F o − F c, contoured at 1.5σ; blue) are shown for the type I Cu-binding site (a) and the type II and III Cu-binding sites (b). Anomalous maps (contoured at 6σ; red) were calculated using the diffraction data used for the SAD method and phases calculated using a final model. The final model is superimposed in a stick representation with atoms in standard colours for the Cu-coordinating residues and a water molecule (red spheres). Cu atoms are shown as small spheres coloured in the same colours as in Fig. 1 ▶. The figures were prepared with BobScript (Esnouf, 1999 ▶) and Raster3D.
3.5. Molecular-dynamics simulation for the BOD–mediator complex
A molecular-dynamics simulation was performed for the BOD–mediator (ABTS) complex (the initial model of BOD–ABTS was produced as described in §2.6). The average structure after the simulation shows the aromatic rings of the ABTS molecule and the Trp396 residue of BOD, indicating that the binding of the mediator molecule to BOD is stabilized by π–π interactions.
4. Discussion
In the study described here, the copper-bound structure of M. verrucaria BOD was determined at 2.3 Å resolution using merohedrally twinned crystals as the first demonstration of the structure of BOD. The type I Cu site of BOD has been reported to have a high redox potential compared with other multicopper oxidases with Met as an axial ligand (Tsujimura et al., 2005 ▶; Christenson et al., 2006 ▶). This study provides important insights regarding the characteristics of the redox potential of BOD.
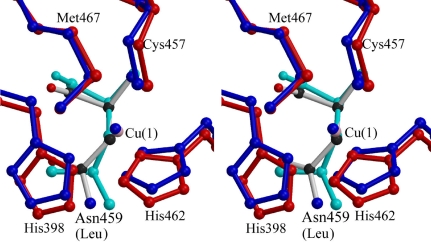
The overall structure of BOD was similar to those of two multicopper oxidases (CueO laccase from E. coli and CotA laccase from B. subtilis) deposited in the PDB, as shown in Fig. 1 ▶(b). The coordination of the type I Cu by four amino-acid residues (His398, Cys457, His462 and Met 467) and the coordination of the type II Cu by two amino-acid residues (His94 and His401) were almost the same as those in CueO (Fig. 3 ▶; Roberts et al., 2003 ▶). The coordination of the type III Cu by six amino-acid residues (His136, His403 and His456; His134, His96 and His458) and a water molecule was also similar to that in CueO. The distances between the Cu atoms and the liganded amino-acid residues in BOD, CueO and CotA are compared in Table 3 ▶. All bond lengths between coppers and liganded amino-acid residues in each molecule (A and B) are similar to those in CueO and CotA. While the coordination patterns of the principal residues are similar, BOD has a polar residue Asn459 at the opposite position to the axial ligand (Met467) of the type I Cu; the distance between Asn459 Nδ2 and the copper ion is 4.61 Å. CueO and CotA have nonpolar residues (Leu and Ile, respectively) at the position corresponding to Asn459 of BOD. The unique environment around the type I Cu might be responsible in part for the high redox potential. The redox potential of BOD is higher than those of CueO and CotA by 200–230 mV.
Figure 3.
Comparison of the type I copper-binding sites of multicopper oxidases. The Cu-coordinating residues and Cu atoms of two multicopper oxidases are shown as stereo diagrams. The four Cu atoms of BOD (red and pink) and CueO (blue) were superimposed using the LSQKAB program from the CCP4 package and the type I copper-binding sites are shown in the figure. The Asn (BOD) and Leu (CueO) residues close to the type I copper-coordinating site are also shown in standard colours and in cyan, respectively.
A molecular-dynamics simulation of the BOD–mediator (ABTS) complex suggested that the binding of the mediator molecule to Trp396 of BOD is stabilized by a π–π interaction. The corresponding Trp residue is not found in CotA or CueO. This coupling, which is not highly dependent on pH, might contribute to the higher activity at neutral pH of BOD.
Supplementary Material
PDB reference: bilirubin oxidase, 3abg
Acknowledgments
The synchrotron-radiation experiments were performed at BL38B1 of SPring-8 with the approval of the Japan Synchrotron Radiation Research Institute (JASRI).
References
- Adams, P. D., Grosse-Kunstleve, R. W., Hung, L.-W., Ioerger, T. R., McCoy, A. J., Moriarty, N. W., Read, R. J., Sacchettini, J. C., Sauter, N. K. & Terwilliger, T. C. (2002). Acta Cryst. D58, 1948–1954. [DOI] [PubMed]
- Christenson, A., Shleev, S., Mano, N., Heller, A. & Gorton, L. (2006). Biochim. Biophys. Acta, 1757, 1634–1641. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Couto, S. R. & Herrera, J. L. T. (2006). Biotechnol. Adv.24, 500–513.
- Cracknell, J. A., Vincent, K. A. & Armstrong, F. A. (2008). Chem. Rev.108, 2439–2461. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Enguita, F. J., Marcal, D., Martins, L. O., Grenha, R., Henriques, A. O., Lindley, P. F. & Carrondo, M. A. (2004). J. Biol. Chem.279, 23472–23476. [DOI] [PubMed]
- Enguita, F. J., Martins, L. O., Henriques, A. O. & Carrondo, M. A. (2003). J. Biol. Chem.278, 19416–19425. [DOI] [PubMed]
- Esnouf, R. M. (1999). Acta Cryst. D55, 938–940. [DOI] [PubMed]
- Holm, L., Kaariainen, S., Rosenstrom, P. & Schenkel, A. (2008). Bioinformatics, 24, 2780–2781. [DOI] [PMC free article] [PubMed]
- Holm, L. & Park, J. (2000). Bioinformatics, 16, 566–567. [DOI] [PubMed]
- Kosaka, A., Yamamoto, C., Morishita, Y. & Nakane, K. (1987). Clin. Biochem.20, 451–458. [DOI] [PubMed]
- Kataoka, K., Komori, H., Ueki, Y., Konno, Y., Kamitaka, Y., Kurose, S., Tsujimura, S., Higuchi, Y., Kano, K., Seo, D. & Sakurai, T. (2007). J. Mol. Biol.373, 141–152. [DOI] [PubMed]
- Kraulis, P. J. (1991). J. Appl. Cryst.24, 946–950.
- Merritt, E. A. & Murphy, M. E. P. (1994). Acta Cryst. D50, 869–873. [DOI] [PubMed]
- Messerschmidt, A., Ladenstein, R., Huber, R., Bolognesi, M., Avigliano, L., Petruzzelli, R., Rossi, A. & Finazzi-Agro, A. (1992). J. Mol. Biol.224, 179–205. [DOI] [PubMed]
- Piontek, K., Antorini, M. & Choinowski, T. (2002). J. Biol. Chem.277, 37663–37669. [DOI] [PubMed]
- Roberts, S. A., Wildner, G. F., Grass, G., Weichsel, A., Ambrus, A., Rensing, C. & Montfort, W. R. (2003). J. Biol. Chem.278, 31958–31963. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Shimizu, A., Kwon, J. H., Sasaki, T., Satoh, T., Sakurai, N., Sakurai, T., Yamaguchi, S. & Samejima, T. (1999). Biochemistry, 38, 3034–3042. [DOI] [PubMed]
- Shimizu, A., Sasaki, T., Kwon, J. H., Odaka, A., Satoh, T., Sakurai, N., Sakurai, T., Yamaguchi, S. & Samejima, T. (1999). J. Biochem.125, 662–668. [DOI] [PubMed]
- Shleev, S., Tkac, J., Christenson, A., Ruzgas, T., Yaropolov, A. I., Whittaker, J. W. & Gorton, L. (2005). Biosens. Bioelectron.20, 2517–2554. [DOI] [PubMed]
- Terwilliger, T. (2004). J. Synchrotron Rad.11, 49–52. [DOI] [PubMed]
- Terwilliger, T. C. (2003). Methods Enzymol.374, 22–37. [DOI] [PubMed]
- Tsujimura, S., Kuriyama, A., Fujieda, N., Kano, K. & Ikeda, T. (2005). Anal. Biochem.337, 325–331. [DOI] [PubMed]
- Tsujimura, S., Tatsumi, B., Ogawa, J., Shimizu, S., Kano, K. & Ikeda, T. (2001). J. Electroanal. Chem.496, 69–75.
- Van Der Spoel, D., Lindahl, E., Hess, B., Groenhof, G., Mark, A. E. & Berendsen, H. J. (2005). J. Comput. Chem.26, 1701–1718. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: bilirubin oxidase, 3abg