BxlA from Streptomyces thermoviolaceus OPC-520 (molecular weight 82 kDa) was crystallized by the hanging-drop vapour-diffusion method at 289 K.
Keywords: BxlA, β-xylosidases, Streptomyces thermoviolaceus OPC-520
Abstract
BxlA from Streptomyces thermoviolaceus OPC-520, together with the extracellular BxlE and the integral membrane proteins BxlF and BxlG, constitutes a xylanolytic system that participates in the intracellular transport of xylan-degradation products and the production of xylose. To elucidate the mechanism of the hydrolytic degradation of xylooligosaccharides to xylose at the atomic level, X-ray structural analysis of BxlA was attempted. The recombinant BxlA protein (molecular weight 82 kDa) was crystallized by the hanging-drop vapour-diffusion method at 289 K. The crystals belonged to the monoclinic space group C2, with unit-cell parameters a = 142.2, b = 129.5, c = 101.4 Å, β = 119.8°, and contained two molecules per asymmetric unit (V M = 2.47 Å3 Da−1). Diffraction data were collected to a resolution to 2.50 Å and provided a data set with an overall R merge of 8.3%.
1. Introduction
Xylan is one of the major hemicellulose components of plant cells and is the second most abundant resource after cellulose. Streptomyces thermoviolaceus OPC-520 has a relatively high xylanase activity when grown in a medium containing xylan as a carbon source (Tsujibo et al., 1992 ▶). This bacterium secretes two types of xylanases in the presence of xylan: an acetyl xylan esterase and an α-l-arabinofuranosidase (Tsujibo et al., 1997 ▶). The xylan-degradation products (mainly xylobiose) produced by these enzymes are trapped by BxlE, transported into the cell by interaction with the integral membrane proteins BxlF and BxlG, and degraded to the final product xylose by an intracellular β-xylosidase (BxlA) (Tsujibo et al., 2001 ▶). Thus, the BxlA–E proteins constitute a xylanolytic system for the utilization of xylan. Genetic analysis of this bacterium revealed that the gene cluster consists of four different open reading frames organized in the order bxlE, bxlF, bxlG and bxlA. The gene cluster is transcribed as a polycistronic mRNA (Tsujibo et al., 2004 ▶).
The effective turnover of abundant organic molecules in the biosphere to reusable material is an important subject. Thus, as a part of studies of the applications of xylan, we have been studying the xylan-degradation mechanism by determining the crystal structures of the enzymes that constitute the xylanolytic system in Streptomyces thermoviolaceus OPC-520; the preliminary X-ray crystallographic analysis of BxlE has recently been reported (Seike et al., 2007 ▶). BxlA is an exopeptidase that hydrolyzes xylooligosaccharides but does not exhibit activity toward xylan (Tsujibo et al., 2001 ▶). Although BxlA plays an important role in the xylanolytic system, its catalytic mechanism is presently poorly understood at the atomic level. To elucidate the structure–function relationship of BxlA, we conducted an X-ray structural analysis of BxlA.
2. Methods and materials
2.1. Expression and purification
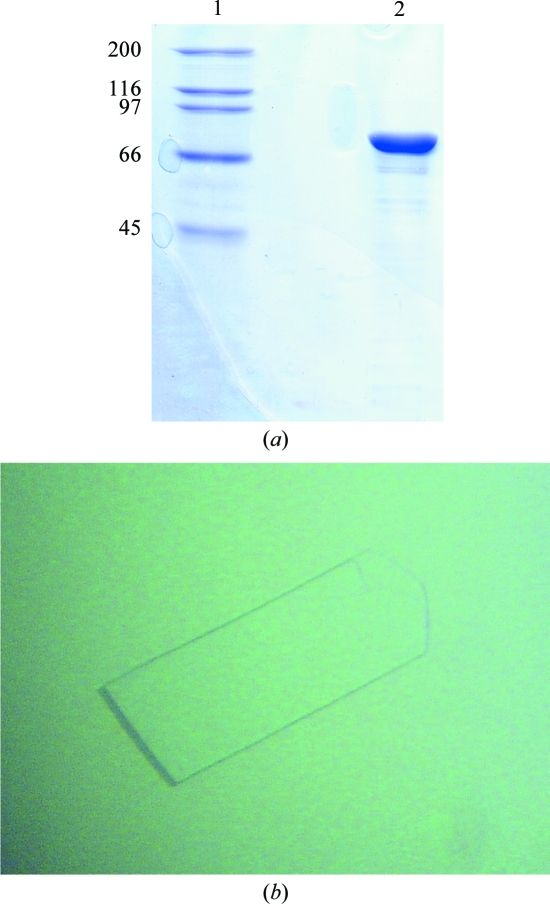
Cloning of the bxlA gene was performed according to a previous report (Tsujibo et al., 2001 ▶). The expression plasmid pGstBxlA coding for BxlA was constructed as follows. The oligonucleotide primers FwGstBxlA (5′-GAATTCATGACCACCGCCCCCTGGCAGGAT-3′) and RvGstBxlA (5′-GCGGCCGCTCAGTCCGCGGTGACAGTTCCGT-3′) were synthesized with EcoRI and NotI restriction sites (bold) to facilitate cloning in frame into the glutathione S-transferase (GST) fusion-protein expression vector pGEX-6P-1 (GE Healthcare). The DNA amplified by PCR was digested by EcoRI and NotI and the resulting fragment was inserted into the corresponding sites of pGEX-6P-1. Escherichia coli BL21 (DE3) cells harbouring pGstBxlA were induced with 0.01 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) at the mid-exponential growth phase (OD600 = 0.6) and incubated for a further 20 h at 310 K. Cells were harvested by centrifugation, washed with 50 mM Tris–HCl buffer pH 7.0 and resuspended in the same buffer. The cells were disrupted by sonication and the lysate was centrifuged at 18 000 rev min−1 for 60 min at 277 K. The supernatant was applied onto a Glutathione Sepharose 4B column (GE Healthcare) equilibrated with 50 mM Tris–HCl pH 7.0. After washing the column, PreScission protease (GE Healthcare) dissolved in the same buffer was loaded onto the column to remove the GST tag. BxlA was eluted from the column with the buffer. Fractions containing BxlA were concentrated to approximately 10–15 mg ml−1 using Centricon YM-50 centrifugal filters (Millipore). The purity of the final preparation was estimated to be >95% by SDS–PAGE (Fig. 1 ▶ a).
Figure 1.
(a) SDS–PAGE analysis of native BxlA. Lane 1, molecular-weight markers (kDa); lane 2, purified recombinant BxlA. (b) A single crystal of native BxlA. The crystal has approximate dimensions of 0.6 × 0.2 × 0.05 mm.
2.2. Crystallization
The size distribution of the protein molecule in solution was monodisperse, with a molecular weight of 82 kDa from dynamic light-scattering measurements. This protein has an α-helix and β-sheet structure according to a CD spectrum in the far-UV region.
An initial crystallization screen was conducted using the hanging-drop vapour-diffusion method at 289 K and crystallization kits from Hampton Research and Emerald BioSystems. No suitable crystals of BxlA alone were obtained. Therefore, cocrystallization was attempted with an inhibitor or substrate analogue and xylobiose was finally selected, despite being able to act as a substrate. Crystallization drops consisting of 2 µl protein solution and 2 µl reservoir solution were equilibrated against 0.5 ml reservoir solution. Thin plate-shaped crystals suitable for X-ray diffraction were obtained with a protein concentration of 14 mg ml−1 containing 10 mM xylobiose and reservoir solution comprised of 20%(w/v) PEG 3350, 0.2 M lithium sulfate and 0.1 M imidazole buffer (pH 5.2–5.5). Crystals grew within 1–2 months to maximum dimensions of approximately 0.6 × 0.2 × 0.05 mm (Fig. 1 ▶ b).
2.3. Data collection and processing
A BxlA crystal was transferred to cryoprotectant solution (reservoir solution containing 30% glycerol) prior to flash-cooling. Native data were collected at 100 K using Cu Kα radiation from an FR-E rotating-anode X-ray generator (Rigaku Corp.) equipped with CMF optics (Osmic Inc.) and an R-AXIS VII detector (the crystal-to-detector distance was 135 mm). The data within 180° of rotation were collected as 360 diffraction images using a 0.5° oscillation angle and an exposure time of 180 s. Data were processed with the CrystalClear program (Pflugrath, 1999 ▶). Two suitable derivatives were obtained by soaking the native crystals in solutions containing 1 mM K2PtCl4 for 2 d and 2 mM HgCl2 for 3 d. Data from derivatives were collected using the same conditions as for the native data.
3. Results and discussion
Recombinant BxlA protein was prepared in E. coli and crystallized using the vapour-diffusion method at 289 K. A total of 55 057 reflections were collected in the resolution range 25.58–2.50 Å with 99.9% completeness and an R merge of 8.3%. The BxlA crystals belonged to the monoclinic space group C2, with unit-cell parameters a = 142.2, b = 129.5, c = 101.4 Å, β = 119.8°. The V M value (the crystal volume per unit of protein molecular weight) was calculated to be 2.47 Å3 Da−1, indicating that two molecules formed the asymmetric unit, and a solvent content of about 50% was suggested from the V S value (Matthews, 1968 ▶). The number of molecules in the asymmetric unit was confirmed by cell-content analysis in the CCP4 program package (Collaborative Computational Project, Number 4, 1994 ▶). We applied the self-rotation function to check the NCS, but no peaks were found in the κ = 180° section on calculating the self-rotation function using data between 20 and 4 Å resolution. We attempted phase determination by the molecular-replacement method using the coordinates of maltose-binding protein (PDB code 1ex1; Varghese et al., 1999 ▶) as a search model. Structure determination of BxlA was also conducted using heavy-atom derivatives (Table 1 ▶). The X-ray structure analysis is promising for elucidation of the detailed mechanism of oligosaccharide degradation by BxlA at the atomic level.
Table 1. Statistics for the native, Pt-derivative and Hg-derivative BxlA diffraction data sets.
Values in parentheses are for the highest resolution shell.
| Native | Pt derivative | Hg derivative | |
|---|---|---|---|
| Space group | C2 | C2 | C2 |
| Unit-cell parameters | |||
| a (Å) | 142.2 | 143.1 | 141.4 |
| b (Å) | 129.5 | 129.8 | 130.1 |
| c (Å) | 101.4 | 101.9 | 100.3 |
| β (°) | 119.8 | 119.9 | 120.2 |
| Resolution range (Å) | 25.58–2.50 (1.71–1.65) | 25.12–2.50 (2.80–2.70) | 23.65–3.60 (3.73–3.60) |
| No. of unique reflections | 55057 | 54773 | 34786 |
| Average redundancy | 3.71 (3.41) | 3.12 (2.96) | 1.95 (1.79) |
| Completeness (%) | 99.9 (98.8) | 97.9 (93.7) | 97.5 (91.5) |
| Rmerge† | 0.083 (0.295) | 0.137 (0.390) | 0.195 (0.437) |
| Average I/σ(I) | 11.6 (3.9) | 6.7 (2.7) | 3.7( 1.7) |
R
merge = 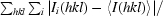
 , where Ii(hkl) and 〈I(hkl)〉 are the observed intensity and the mean intensity of related reflections, respectively.
, where Ii(hkl) and 〈I(hkl)〉 are the observed intensity and the mean intensity of related reflections, respectively.
References
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Pflugrath, J. W. (1999). Acta Cryst. D55, 1718–1725. [DOI] [PubMed]
- Seike, K., Sato, J., Tomoo, K., Ishida, T., Yamano, A., Ikenishi, S., Miyamoto, K. & Tsujibo, H. (2007). Acta Cryst. F63, 560–562. [DOI] [PMC free article] [PubMed]
- Tsujibo, H., Kosaka, M., Ikenishi, S., Sato, T., Miyamoto, K. & Inamori, Y. (2004). J. Bacteriol.186, 1029–1037. [DOI] [PMC free article] [PubMed]
- Tsujibo, H., Miyamoto, K., Kuda, T., Minami, K., Sakamoto, T., Hasegawa, T. & Inamori, Y. (1992). Appl. Environ. Microbiol.58, 371–375. [DOI] [PMC free article] [PubMed]
- Tsujibo, H., Ohtsuki, T., Iio, T., Yamazaki, I., Miyamoto, K., Sugiyama, M. & Inamori, Y. (1997). Appl. Environ. Microbiol.63, 661–664. [DOI] [PMC free article] [PubMed]
- Tsujibo, H., Takada, C., Tsuji, A., Kosaka, M., Miyamoto, K. & Inamori, Y. (2001). Biosci. Biotechnol. Biochem.65, 1824–1831. [DOI] [PubMed]
- Varghese, J. N., Hrmova, M. & Fincher, G. B. (1999). Structure, 7, 179–190. [DOI] [PubMed]