l-Azetidine-2-carboxylate hydrolase from Pseudomonas sp. strain A2C was crystallized and diffraction data were collected to a resolution of 1.38 Å.
Keywords: l-azetidine-2-carboxylate hydrolase, Pseudomonas sp. strain A2C
Abstract
l-Azetidine-2-carboxylate hydrolase from Pseudomonas sp. strain A2C catalyzes a ring-opening reaction that detoxifies l-azetidine-2-carboxylate, an analogue of l-proline. Recombinant l-azetidine-2-carboxylate hydrolase was overexpressed, purified and crystallized using polyethylene glycol and magnesium acetate as precipitants. The needle-shaped crystal belonged to space group P21, with unit-cell parameters a = 35.6, b = 63.6, c = 54.7 Å, β = 105.5°. The crystal diffracted to a resolution of 1.38 Å. The calculated V M value was 2.2 Å3 Da−1, suggesting that the crystal contains one enzyme subunit in the asymmetric unit.
1. Introduction
l-Azetidine-2-carboxylate hydrolase (A2CH) from Pseudomonas sp. strain A2C catalyzes the hydrolytic opening of the azetidine ring of (S)-azetidine-2-carboxylic acid (AZC) and generates the product 2-hydroxy-4-aminobutyric acid (Gross et al., 2008 ▶). AZC, the substrate of the enzyme, is an analogue of l-proline and is biosynthesized by some plants such as lily of the valley. The incorporation of AZC into proteins as a substitute for l-proline causes protein misfolding and subsequent functional disruption. Therefore, AZC is highly toxic to most organisms. However, some bacteria have adapted to AZC by evolving a metabolic pathway that not only degrades the harmful AZC but also utilizes it as a sole source of nitrogen. One example of such bacteria is Pseudomonas sp. strain A2C, which was isolated from the soil beneath a lily of the valley plant. Pseudomonas sp. strain A2C contains a gene cluster composed of A2CH, an aminobutyrate transporter, a transaminase and hypothetical proteins. The product of the A2CH-catalyzed reaction can be further metabolized as a nitrogen source by proteins translated from other genes in the cluster.
The A2CH gene (GenBank ID 187942449) codes for 240 amino-acid residues. On the basis of amino-acid sequence analysis, A2CH has been shown to belong to the haloacid dehalogenase-like (HAD) superfamily, which includes haloacid dehalogenases, phosphoesterases, P-type ATPases and other hydrolases (Burroughs et al., 2006 ▶; see also Fig. 1 ▶). l-2-Haloacid dehalogenase (l-DEX), a member of the HAD superfamily, catalyzes the hydrolytic dehalogenation of l-2-haloacids [or (S)-2-haloacids] to produce d-2-hydroxy acids [or (R)-2-hydroxy acids] (Liu et al., 1995 ▶). The reaction begins with a nucleophilic attack on the C2 carbon of the substrate by an enzyme aspartate residue that resides near the N-terminus of the protein. This displaces the halogen atom and an ester intermediate is formed. The resulting ester intermediate is hydrolyzed by the attack of an activated water molecule. The crystal structures of several l-DEXs from different species have been solved and support this mechanism (Li et al., 1998 ▶; Ridder et al., 1999 ▶).
Figure 1.
Sequence alignment of A2CH with HAD superfamily enzymes using MUSCLE (Edgar, 2004 ▶). The regions that are not conserved have been excluded. The sequences used here were as follows: l-2-haloacid dehalogenase from Pseudomonas sp. YL (232 amino acids; L-DEX_PY_3122176), phosphoserine phosphatase from Methanococcus jannaschii (211 amino acids; SerB_Mj_14719643), phosphonoacetaldehyde hydrolase from Bacillus cereus (267 amino acids; phnX_Bc_48425373) and soluble epoxide hydrolase from Rattus norvegicus (554 amino acids; SEH_Rn_462371). The number in each sequence name indicates the GenBank ID. The capitalized letters in the Consensus line show the common residues. Conserved aliphatic, positively charged and Ser/Thr residues are shown as l, + and *, respectively. This alignment was produced using the program CHROMA (Goodstadt & Ponting, 2001 ▶).
A2CH showed no enzymatic activity with either (R)- or (S)-2-chloropropionic acid (Gross et al., 2008 ▶). While some important residues in l-DEX are conserved in A2CH, such as the aspartate nucleophile, other important residues are not. For example, the region of l-DEX believed to help stabilize the halide leaving group is not conserved in A2CH. In fact, A2CH appears to be highly evolved for reaction with AZC: even l-proline shows no demonstrable reactivity with A2CH. In order to understand what underlies the high substrate specificity of A2CH, the three-dimensional structure of the enzyme will prove crucial. Additionally, structural analysis of A2CH will provide insights into the reaction mechanism of this novel enzyme within the HAD superfamily. Here, we describe the crystallization and preliminary X-ray diffraction studies of A2CH from Pseudomonas sp. strain A2C.
2. Experimental procedure
2.1. Expression and purification
(S)-(−)-Azetidine-2-carboxylate was obtained from Sigma–Aldrich (St Louis, Missouri, USA). (S)-2-Hydroxy-4-aminobutyrate was obtained from Tokyo Chemical Industry Co. Ltd (Tokyo, Japan). All other chemicals were of guaranteed grade from Nacalai Tesque (Kyoto, Japan) and Wako Pure Chemical Industries (Osaka, Japan).
The recombinant plasmid pET29-A2CH was constructed as reported previously (Gross et al., 2008 ▶). Escherichia coli strain BLR (DE3) (Novagen, Madison, Wisconsin, USA) was transformed with the plasmid for A2CH overexpression. The recombinant cells were cultured in 1 l LB medium containing kanamycin (50 µg ml−1) at 310 K for 12 h until the OD600 reached 1.4. For induction, isopropyl β-d-1-thiogalactopyranoside (IPTG) was added to the culture to a concentration of 0.1 mM and the culture was incubated at 303 K for a further 12 h. The cells were harvested by centrifugation at 4400g for 15 min, suspended in 50 mM potassium phosphate pH 7.5 and then disrupted by sonication. The sonicated mixture was centrifuged at 5300g for 30 min to remove cell debris. The crude extract was treated with 1%(w/v) streptomycin sulfate and the supernatant was used for further purification. The protein solution was loaded onto a DEAE-Toyopearl (Tosoh, Tokyo, Japan) column equilibrated with 50 mM potassium phosphate pH 7.5. A2CH was eluted in the flowthrough fraction. The fraction was dialyzed against 5 mM potassium phosphate pH 7.5 and loaded onto a DEAE-Toyopearl column equilibrated with 5 mM potassium phosphate pH 7.5. The enzyme was eluted in the flowthrough fraction and dialyzed against 5 mM potassium phosphate pH 7.5. Finally, the dialyzed fraction was applied onto a Gigapite (Seikagaku, Tokyo, Japan) column equilibrated with 5 mM potassium phosphate pH 7.5. The flowthrough fraction was collected and concentrated to 20 mg ml−1 using a Microcon YM-10 membrane (Millipore, Billerica, Massachusetts, USA) at 12 000g. In this step, the buffer was exchanged to 10 mM HEPES–NaOH pH 7.5.
2.2. Enzyme assay
A2CH activity was assayed using the method described for the assay of proline reductase (Seto, 1979 ▶), with slight modifications as reported previously (Gross et al., 2008 ▶). A 250 µl reaction mixture consisting of 100 mM potassium phosphate buffer pH 8, 10 mM MgCl2, 20 mM dithiothreitol, 10 mM AZC and 1 µg purified enzyme was incubated at 303 K. The reaction was stopped by the addition of 50 µl 5% HClO4 solution. Protein debris was removed by centrifugation at 20 000g for 5 min and 1 ml o-phthalaldehyde mixture was added to 25 µl supernatant. Fluorescence was measured at 455 nm. (S)-2-Hydroxy-4-aminobutyrate (0–30 nmol) was used to generate a standard curve.
Gel-filtration analysis was performed to determine the holoenzyme molecular weight using a Superose 6 10/300 GL column (Amersham Biosciences, Piscataway, New Jersey, USA) eluted with 50 mM potassium phosphate pH 7 containing 200 mM NaCl. The molecular-mass marker used was Mw-Marker (Oriental Yeast Co. Ltd, Tokyo, Japan) and was composed of cytochrome c (12.4 kDa), myokinase (32 kDa), enolase (67 kDa), lactate dehydrogenase (142 kDa) and glutamate dehydrogenase (290 kDa).
2.3. Crystallization
Initial screening for crystallization conditions was performed with the Wizard I, II and III, Cryo I and II (Emerald BioSystems Inc., Washington, USA), Crystal Screen I and II and PEG/Ion Screen (Hampton Research, California, USA) crystallization kits. Protein dissolved in 10 mM HEPES–NaOH pH 7.5 was crystallized at 277 K by the sitting-drop vapour-diffusion method using 96-well plates. Crystals were obtained from condition No. 3 of Wizard III [20%(w/v) PEG 3350, 0.1 M magnesium formate]. Further optimization of the crystallization conditions was carried out using 24-well plates by the sitting-drop vapour-diffusion method at 277 K. Needle-shaped crystals were obtained using the following condition: a droplet consisting of a mixture of 2 µl protein solution and 2 µl reservoir solution was equilibrated against 1 ml reservoir solution [15%(w/v) PEG 3350, 0.1 M magnesium acetate and 0.1 M imidazole–HCl pH 8].
2.4. X-ray data collection
Prior to data collection, the crystal was soaked in reservoir solution containing 15%(v/v) glycerol as the cryoprotectant. The crystal was then flash-cooled in a cold nitrogen-gas stream at 100 K. Diffraction data were collected on a Rigaku Jupiter 210 CCD detector using synchrotron radiation of wavelength 1.00 Å on BL38B1 of SPring-8 (Hyogo, Japan). The collected data were processed with HKL-2000 (Otwinowski & Minor, 1997 ▶).
3. Results and discussion
A2CH was successfully overexpressed in E. coli and purified to homogeneity (Fig. 2 ▶). The specific activity was 5.5 µmol min−1 mg−1. The gel-filtration analysis showed that the molecular mass of A2CH was about 21 kDa. The molecular mass of A2CH was calculated from the primary structure to be 27 111 Da, suggesting that A2CH is a monomeric enzyme.
Figure 2.
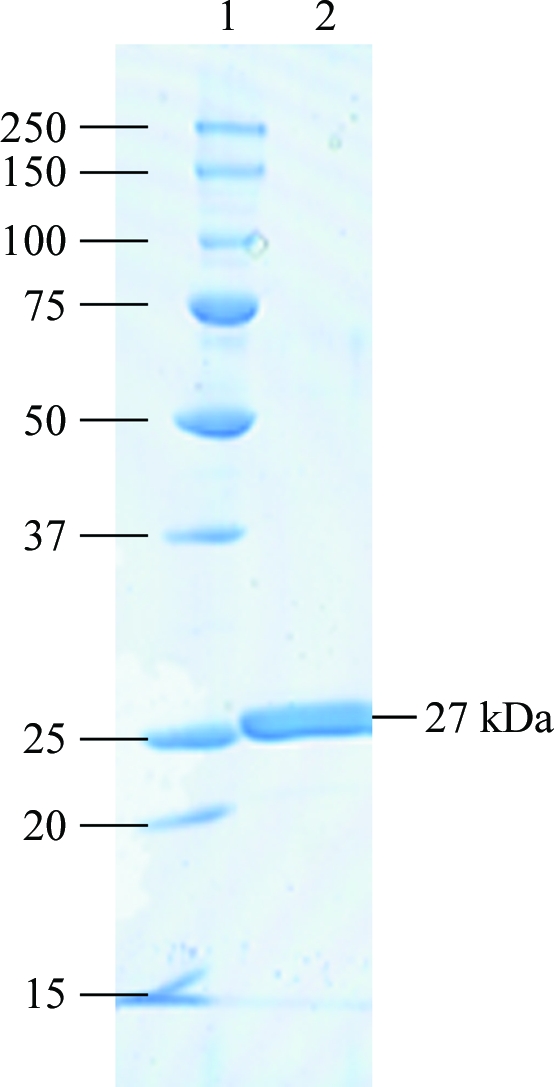
SDS–PAGE analysis of purified A2CH. Lane 1, molecular-mass markers (kDa); lane 2, 5 µg purified recombinant A2CH.
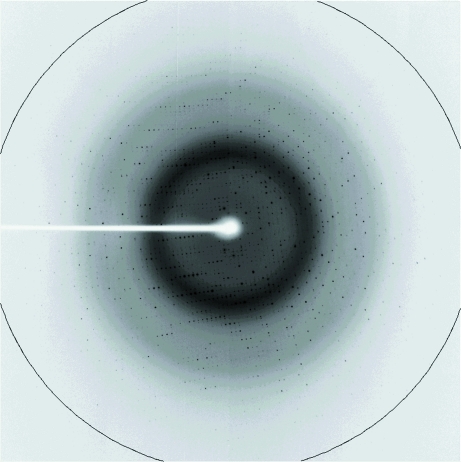
A2CH crystals appeared within a week (Fig. 3 ▶). Magnesium ions (Mg2+) and PEG were suitable precipitants for the crystallization of A2CH. The A2CH crystal yielded good diffraction images, as shown in Fig. 4 ▶. The crystal parameters and diffraction data statistics are summarized in Table 1 ▶. The crystal belonged to the monoclinic crystal system, space group P21, with unit-cell parameters as given in Table 1 ▶. The calculated unit-cell volume was 119 345.2 Å3. The crystal volume per protein molecular weight in the unit cell (V M) was calculated to be 2.2 Å3 Da−1, assuming the presence of one protein molecule in the asymmetric unit, with a solvent content of 44.1% (Matthews, 1968 ▶).
Figure 3.
Crystals of A2CH from Pseudomonas sp. strain A2C.
Figure 4.
X-ray diffraction pattern from a crystal of A2CH. The ring indicates 1.38 Å resolution.
Table 1. Data-collection and processing statistics.
Values in parentheses are for the highest resolution shell.
| No. of crystals | 1 |
| Beamline | SPring-8 BL38B1 |
| Wavelength (Å) | 1.0000 |
| Detector | Jupiter 210 CCD |
| Crystal-to-detector distance (mm) | 120 |
| Rotation range per image (°) | 1.00 |
| Total rotation range (°) | 192 |
| Exposure time per image (s) | 10.0 |
| Resolution range (Å) | 50–1.38 (1.43–1.38) |
| Space group | P21 |
| Unit-cell parameters (Å, °) | a = 35.6, b = 63.6, c = 54.7, β = 105.5 |
| Mosaicity (°) | 0.614 |
| Total No. of measured intensities | 183702 (12018) |
| Unique reflections | 47051 (4006) |
| Multiplicity | 3.9 (3.0) |
| Mean I/σ(I) | 26.5 (2.4) |
| Completeness (%) | 97.5 (83.6) |
| Rmerge† (%) | 3.9 (28.6) |
| Rmeas or Rr.i.m.‡ (%) | 4.5 (34.3) |
| Overall B factor from Wilson plot (Å2) | 31.5 |
R
merge = 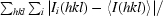
 , where I
i(hkl) is the intensity of the ith measurement of reflection hkl and 〈I(hkl)〉 is the mean value of I(hkl) for all i measurements.
, where I
i(hkl) is the intensity of the ith measurement of reflection hkl and 〈I(hkl)〉 is the mean value of I(hkl) for all i measurements.
R meas is the redundancy-independent R merge (Diederichs & Karplus, 1997 ▶).
We attempted to solve the structure of A2CH by molecular replacement using l-DEX as the search model, but failed. In order to determine the phase angle of the diffraction data, we are now preparing selenomethionine-incorporated A2CH for use in the single-wavelength anomalous dispersion method.
Acknowledgments
We thank Drs S. Baba and N. Mizuno of Japan Synchrotron Radiation Research Institute (JASRI) for their kind help with data collection.
References
- Burroughs, A. M., Allen, K. N., Dunaway-Mariano, D. & Aravind, L. (2006). J. Mol. Biol.361, 1003–1034. [DOI] [PubMed]
- Diederichs, K. & Karplus, P. A. (1997). Nature Struct. Biol.4, 269–275. [DOI] [PubMed]
- Edgar, R. C. (2004). Nucleic Acids Res.32, 1792–1797. [DOI] [PMC free article] [PubMed]
- Goodstadt, L. & Ponting, C. P. (2001). Bioinformatics, 17, 845–846. [DOI] [PubMed]
- Gross, C., Felsheim, R. & Wackett, L. P. (2008). J. Bacteriol.190, 4859–4864. [DOI] [PMC free article] [PubMed]
- Li, Y.-F., Hata, Y., Fujii, T., Hisano, T., Nishihara, M., Kurihara, T. & Esaki, N. (1998). J. Biol. Chem.273, 15035–15044. [DOI] [PubMed]
- Liu, J.-Q., Kurihara, T., Miyagi, M., Esaki, N. & Soda, K. (1995). J. Biol. Chem.270, 18309–18312. [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Ridder, I. S., Rozeboom, H. J., Kalk, K. H. & Dijkstra, B. W. (1999). J. Biol. Chem.274, 30672–30678. [DOI] [PubMed]
- Seto, B. (1979). Anal. Biochem.95, 44–47. [DOI] [PubMed]