The expression, purification, preliminary crystallization and crystallographic analysis of phosphoketolase from L. lactis ssp. lactis (strain IL 1403) are reported.
Keywords: phosphoketolases, thiamine diphosphate-dependent enzymes
Abstract
Phosphoketolases are thiamine diphosphate-dependent enzymes which play a central role in the pentose-phosphate pathway of heterofermentative lactic acid bacteria. They belong to the family of aldehyde-lyases and in the presence of phosphate ion cleave the carbon–carbon bond of the specific substrate d-xylulose 5-phosphate (or d-fructose 6-phosphate) to give acetyl phosphate and d-glyceraldehyde 3-phosphate (or d-erythrose 4-phosphate). Structural information about phosphoketolases is particularly important in order to fully understand their mechanism as well as the steric course of phosphoketolase-catalyzed reactions. Here, the purification, preliminary crystallization and crystallographic characterization of d-xylulose 5-phosphate phosphoketolase from Lactococcus lactis are reported. The presence of thiamine diphosphate during purification was essential for the enzymatic activity of the purified protein. The crystals belonged to the monoclinic space group P21. Diffraction data were obtained to a resolution of 2.2 Å.
1. Introduction
Xylulose 5-phosphate phosphoketolase (EC 4.1.2.9) and fructose 6-phosphate phosphoketolase (EC 4.1.2.22) are key enzymes in the pentose-phosphate pathway of heterofermentative lactic acid bacteria and the d-fructose 6-phosphate (F6P) shunt of bifidobacteria. Phosphoketolases are thiamine diphosphate-dependent enzymes. In the presence of inorganic phosphate, these enzymes convert xylulose 5-phosphate (X5P) or fructose 6-phosphate (F6P) into acetylphosphate and glyceraldehyde 3-phosphate or erythrose 4-phosphate, respectively. To date, no structural data are available for any of the phosphoketolases. In addition to its particular significance in bacterial metabolism, phosphoketolase also seems to play a role in the virulence of microorganisms. A recent study demonstrated its adaptation role in full virulence of the insect-pathogenic fungus Metarhizium anisopliae (Duan et al., 2009 ▶).
The first phosphoketolase was extracted and purified from Lactobacillus plantarum (Heath et al., 1958 ▶). However, the first report of a phosphoketolase of known sequence was published much later and reported the isolation, purification and gene identification of the phosphoketolase from Bifidobacterium (Meile et al., 2001 ▶). Phosphoketolase genes from several microorganisms have subsequently been isolated and the correponding proteins have been expressed, i.e. for phosphoketolases from Lactobacillus pentosus (Posthuma et al., 2002 ▶), Leuconostoc mesenteroides (Lee et al., 2005 ▶), Lactobacillus paraplantarum (Jeong et al., 2007 ▶) and L. plantarum (Yevenes & Frey, 2008 ▶).
The mechanism of the reaction catalyzed by phosphoketolase is still unclear. Although it has been suggested that an α,β-dihydroxyethyl thiamine diphosphate (ThDP) intermediate is initially formed followed by its conversion into acetyl-ThDP (Merkler & Retey, 1981 ▶; Frey, 1989 ▶; Yevenes & Frey, 2008 ▶), there is still no direct evidence that supports acetyl-ThDP as an intermediate in phosphoketolase catalysis. Thus, crystal structures of phosphoketolase in complex with ThDP and specific substrates or inhibitors could significantly contribute to elucidating its reaction mechanism. It is also expected that such crystal structures will contribute to a better understanding of the role of the coenzyme-bound Mg2+ ion as well as of the steric course of the phosphoketolase-catalysed reaction (Merkler & Retey, 1981 ▶).
Here, we report the purification, crystallization and preliminary crystallographic analysis of phosphoketolase from Lactococcus lactis ssp. lactis IL 1403, a Gram-positive bacterium that is one of the most important microorganisms involved in the dairy industry.
2. Materials and methods
2.1. Cloning, expression and purification
The L. lactis phosphoketolase (ptk) gene was cloned from a previously reported genomic library of L. lactis ssp. lactis (strain IL 1403; Bolotin et al., 2001 ▶). The ptk gene (GenBank accession No. AE005176), which was reported as a hypothetical phosphoketolase gene, was amplified by PCR from a plasmid containing a genomic fragment, kindly provided by Alexei Sorokin, Jouy-en-Josas, France. The PCR fragment thus obtained was inserted into a pHAT2 prokaryotic expression plasmid. Recombinant phosphoketolase with an N-terminal His-tag fusion was overexpressed in Escherichia coli BL21 (DE3) RIL cells. The cells were grown at 298 K in Luria–Bertani medium containing 100 mg ml−1 ampicillin until an OD600 of 0.7 was reached. Protein expression was induced by the addition of 0.1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). Cell growth was continued at 298 K for 14 h after IPTG induction. The recombinant cells were harvested from the culture broth by centrifugation at 9000g for 10 min at 277 K and resuspended in buffer A (50 mM potasium phosphate pH 7.0, 300 mM NaCl, 20 mM imidazole, 0.2% Triton X-100, 1 mM DTT, 1 mM MgCl2, 1 mM ThDP with 1 mM phenylmethylsulfonyl fluoride as a protease inhibitor). The resuspended cells were disrupted using a French Press cell disrupter (Thermo Electron Corporation, USA) on ice. The cell debris was removed by centrifugation at 20 000g for 25 min at 277 K and the supernatant was filtered through a 0.45 µm filter. The filtrate was applied onto a HisTrap HP chromatography column (GE Healthcare, USA) equilibrated with buffer A. The bound protein was eluted with a linear gradient of 20–500 mM imidazole in buffer A. The homogeneity of the purified protein was assessed by 10% SDS–PAGE. The fractions were pooled, diluted in a 1:1 ratio with buffer A and concentrated using a Millipore centrifugal filter device (Amicon Ultra-4, 50 kDa cutoff; Millipore, USA). The concentrated protein was loaded onto a Superdex 200 16/60 column (GE Healthcare, USA) equilibrated with gel-filtration buffer [20 mM potasium monophosphate pH 7.0, 150 mM NaCl, 0.007%(w/v) β-octyl glucoside, 1 mM DTT, 1 mM MgCl2, 1 mM ThDP]. All protein-purification steps were performed at 277 K. The homogeneity of the purified protein was assessed by 10% SDS–PAGE. The purified protein was concentrated to 11 mg ml−1 using the same type of filter device from Millipore. The activities of the enzyme towards xylulose 5-phosphate and fructose 6-phosphate as substrates were determined as described in method A of Goldberg (1966 ▶) and by Orban & Patterson (2000 ▶). The protein concentration was determined using the Bradford assay (Bradford, 1976 ▶).
2.2. Crystallization and preliminary X-ray analysis
Initial crystallization experiments were performed at 293 K using the hanging-drop vapour-diffusion method in a 24-well Linbro plate (Hampton Research, USA) using the Crystal Screen, Crystal Screen 2 and PEG/Ion Screen kits from Hampton Research. The hanging drops were made up of 1 µl protein solution at 11 mg ml−1 protein concentration and 1 µl reservoir solution; the drops were equilibrated against 1 ml reservoir solution.
2.3. Data collection and processing
An optimized phosphoketolase crystal was picked up using an appropriate nylon loop, quickly soaked in cryoprotectant solution containing 25%(v/v) glycerol and flash-cooled in liquid nitrogen. A set of X-ray diffraction data was collected at 100 K to 2.2 Å resolution on beamline ID14-3 at the ESRF (Grenoble, France) with an oscillation angle of 0.5° and an exposure time of 5 s per image. The crystal-to-detector distance was set to 150 mm. Data were collected over a total of 185°. The data set was integrated and scaled using the programs XDS/XSCALE (Kabsch, 2010 ▶).
3. Results and discussion
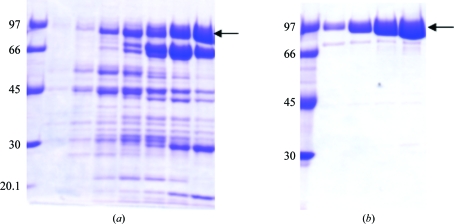
The presence of ThDP during purification was essential. In the absence of ThDP, which has been demonstrated to be an essential coenzyme of phosphoketolases (Heath et al., 1958 ▶; Goldberg, 1966 ▶; Meile et al., 2001 ▶), the purification procedure led to pure protein that was enzymatically inactive. Removal of imidazole after elution of phosphoketolase from an Ni-Sepharose affinity column also proved to be important, probably because imidazole can form a complex with the essential ThDP-bound Mg2+ ion, as noticed for other bacterial phosphoketolases (Yevenes & Frey, 2008 ▶). As shown in Fig. 1 ▶, purification of the prokaryotically expressed phosphoketolase produced a protein with high purity and a molecular weight that was in agreement with the theoretical value of the phosphoketolase amino-acid sequence (95 727 Da). The purified protein was active towards both xylulose 5-phosphate and fructose 6-phosphate as substrates; however, the specificity constant for the former was found to be significantly higher than that for the latter (manuscript in preparation).
Figure 1.
SDS–PAGE (10%) analysis of the phosphoketolase from L. lactis. (a) Fractions containing the phosphoketolase protein eluted from the HisTrap Ni2+ metal-affinity column. (b) Fractions containing purified protein eluted from a Superdex 200 gel-filtration column.
Optimization of the initial crystals was essential both to obtain single crystals and to improve the quality of the diffraction data. Tiny clustered plates were initially observed with a reservoir solution consisting of 20% PEG 3350 and 0.2 M NaSCN (PEG/Ion Screen condition No. 13) over a period of 4 d. A PEG fine screening was performed to improve the quality of the crystals (Fig. 2 ▶). Single crystals of reasonable diffraction quality were obtained when the temperature was reduced to 291 K and the PEG concentration was reduced to 17%.
Figure 2.
Crystal of recombinant phosphoketolase.
We tried using various additives to optimize the quality of the crystals but without visible improvements (data not shown). Further optimization of the crystals is in progress.
Based on the diffraction data set collected at 2.2 Å resolution the crystal belonged to the monoclinic space group P21, with unit-cell parameters a = 158.1, b = 141.5, c = 161.2 Å, β = 90.3°. Data-collection statistics and processing details are summarized in Table 1 ▶. Calculation of the Matthews coefficient (Matthews, 1968 ▶) indicated the presence of eight molecules in the asymmetric unit, with a corresponding V M value of 2.34 Å3 Da−1 and a predicted solvent content of 47.4%. Because the lack of sequence homology of phosphoketolase to known proteins, molecular replacement cannot be used to obtain initial phasing. Structure determination based on the existing data set as well as on those corresponding to heavy-atom and/or selenomethionine derivatives of phosphoketolase is under way.
Table 1. Diffraction data-collection statistics.
Values in parentheses are for the highest resolution bin.
| X-ray source | ID14-3 (ESRF, Grenoble) |
| Detector | MAR CCD detector (165 mm) |
| Wavelength (Å) | 0.931 |
| Temperature (K) | 100 |
| Crystal-to-detector distance (mm) | 150 |
| Exposure time (s) | 5 |
| Oscillation range (°) | 0.5 |
| Space group | P21 |
| Unit-cell parameters (Å, °) | a = 158.1, b = 141.5, c = 161.2, β = 90.3 |
| Resolution limit (Å) | 2.2 (2.3–2.2) |
| Completeness (%) | 96.5 (94.5) |
| No. of observations (overall/unique) | 1321732/346280 |
| Average multiplicity | 3.8 (3.4) |
| 〈I/σ(I)〉 | 8.9 (3.2) |
| Rsyn† (%) | 10.6 (39.3) |
| Rp.i.m.‡ (%) | 5.4 (27.0) |
| B factor from Wilson plot (Å2) | 30.1 |
R
sym = 100 × 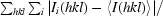
 , where I
i(hkl) is the intensity of the ith individual measurement of the reflection with Miller indices hkl and 〈I(hkl)〉 is the mean intensity of all measurements of I(hkl), calculated for I ≥ 3σ(I).
, where I
i(hkl) is the intensity of the ith individual measurement of the reflection with Miller indices hkl and 〈I(hkl)〉 is the mean intensity of all measurements of I(hkl), calculated for I ≥ 3σ(I).
R
p.i.m. = 100 × 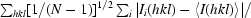
 , where I
i(hkl) is the intensity of the ith individual measurement of the reflection with Miller indices hkl and 〈I(hkl)〉 is the mean intensity of all measurements of I(hkl), calculated for I ≥ 3σ(I); N is the redundancy or multiplicity of the observed reflection (Weiss, 2001 ▶; Diederichs & Karplus, 1997 ▶).
, where I
i(hkl) is the intensity of the ith individual measurement of the reflection with Miller indices hkl and 〈I(hkl)〉 is the mean intensity of all measurements of I(hkl), calculated for I ≥ 3σ(I); N is the redundancy or multiplicity of the observed reflection (Weiss, 2001 ▶; Diederichs & Karplus, 1997 ▶).
Acknowledgments
We gratefully acknowledge access to the core facilities of the ZBM/LMB of the CAU and access to beamline ID14-3 at the ESRF (Grenoble). This research was supported by grants PN-II-ID-PCE-2007-1 No. 210/2007 from CNCSIS, PNCDI 02-2-PED-427 from BIOTECH and PN II No. 41038/2007 and CEEX 1/2005 from CNMP Romania.
References
- Bolotin, A., Wincker, P., Mauger, S., Jaillon, O., Malarme, K., Weissenbach, J., Ehrlich, S. D. & Sorokin, A. (2001). Genome Res.11, 731–753. [DOI] [PMC free article] [PubMed]
- Bradford, M. M. (1976). Anal. Biochem.72, 248–254. [DOI] [PubMed]
- Diederichs, K. & Karplus, P. A. (1997). Nature Struct. Biol.4, 269–275. [DOI] [PubMed]
- Duan, Z., Shang, Y., Gao, Q., Zheng, P. & Wang, C. (2009). Environ. Microbiol.11, 2351–2360. [DOI] [PubMed]
- Frey, P. A. (1989). Biofactors, 2, 1–9. [PubMed]
- Goldberg, M. L. (1966). Methods Enzymol.9, 515–520.
- Heath, E. C., Hurwitz, J., Horecker, B. L. & Ginsburg, A. (1958). J. Biol. Chem.231, 1009–1029. [PubMed]
- Jeong, D.-W., Lee, J. M. & Lee, H. J. (2007). J. Microbiol. Biotechnol.17, 822–829. [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Lee, J. M., Jeong, D.-W., Koo, O. K., Kim, M. J., Lee, J.-H., Chang, H. C., Kim, J. H. & Lee, H. J. (2005). Biotechnol. Lett.27, 853–858. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Meile, L., Rohr, L. M., Geissmann, T. A., Herensperger, M. & Teuber, M. (2001). J. Bacteriol.183, 2929–2936. [DOI] [PMC free article] [PubMed]
- Merkler, I. & Retey, J. (1981). Eur. J. Biochem.120, 593–597. [DOI] [PubMed]
- Orban, J. I. & Patterson, J. A. (2000). J. Microbiol. Methods, 40, 221–224. [DOI] [PubMed]
- Posthuma, C. C., Bader, R., Engelmann, R., Postma, P. W., Hengstenberg, W. & Pouwels, P. H. (2002). Appl. Environ. Microbiol.68, 831–837. [DOI] [PMC free article] [PubMed]
- Weiss, M. S. (2001). J. Appl. Cryst.34, 130–135.
- Yevenes, A. & Frey, P. A. (2008). Bioorg. Chem.36, 121–127. [DOI] [PubMed]