The crystallization and preliminary X-ray diffraction studies of DppA from P. pacifica SIR-I are reported.
Keywords: haloalkane dehalogenases, Plesiocystis pacifica SIR-I
Abstract
DppA from Plesiocystis pacifica SIR-I is a putative haloalkane dehalogenase (EC 3.8.1.5) and probably catalyzes the conversion of halogenated alkanes to the corresponding alcohols. The enzyme was expressed in Escherichia coli BL21 and purified to homogeneity by ammonium sulfate precipitation and reversed-phase and ion-exchange chromatography. The DppA protein was crystallized by the vapour-diffusion method and protein crystals suitable for data collection were obtained in the orthorhombic space group P21212. The DppA crystal diffracted X-rays to 1.9 Å resolution using an in-house X-ray generator.
1. Introduction
Haloalkane dehalogenases (EC 3.8.1.5) are α/β-hydrolase fold enzymes that catalyze the hydrolytic conversion of a broad spectrum of halogenated alkanes to the corresponding alcohols (Janssen, 2004 ▶). This reaction is of great environmental interest and haloalkane dehalogenases have therefore been extensively studied in recent years. Attempts have been made to enhance the speed of the catalytic reaction, to expand the substrate range and to increase the enantioselectivity (Bosma et al., 2002 ▶; Schanstra et al., 1996 ▶; Gray et al., 2001 ▶; Chaloupkova et al., 2003 ▶; Pries et al., 1994 ▶; Holloway et al., 1998 ▶). To date, only a handful of haloalkane dehalogenases have been crystallized (Franken et al., 1991 ▶; Newman et al., 1999 ▶; Marek et al., 2000 ▶; Stsiapanava et al., 2008 ▶; Mazumdar et al., 2008 ▶), of which the enzymes from Xanthobacter autotrophicus GJ10, Sphingomonas paucimobilis UT26 and Rhodococcus rhodochrous NCIMB13064 have been extensively investigated in several further studies. They all share the α/β-hydrolase fold, which is a hallmark of this class of enzymes (Ollis et al., 1992 ▶).
Plesiocystis pacifica SIR-1 is a myxobacterium which was isolated from the Japanese coast (Iizuka et al., 2003 ▶). The strain is Gram-negative, chemoheterotrophic and strictly aerobic (Iizuka et al., 2003 ▶). Its DNA sequence is available online at NCBI as a shotgun library (NCBI reference sequence NZ_ABCS00000000). The entry ZP_01908831.1 is tagged as a putative haloalkane dehalogenase with a calculated molecular weight of 32 kDa.
This haloalkane dehalogenase was chosen as it showed the highest sequence homology to another dehalogenase currently under study in an ongoing project and showed good alignment, with the residues of the catalytic triad (Asp-His-Asp) in reasonable positions.
2. Materials and methods
2.1. Protein expression and purification
The strain P. pacifica SIR-I was ordered from the German Collection of Microorganisms and Cell Cultures (DSMZ; DSM No. 14875) and was supplied as an actively growing culture on agar medium. Genomic DNA of P. pacifica SIR-I was amplified with the help of the GenomiPhi-Kit from GE Healthcare. The coding region of ZP_01908831.1 was cloned into a pET28a vector (without a His tag) and expressed in Escherichia coli BL21 Gold. The cells were grown at 310 K to an OD600 of 0.5, induced with IPTG (final concentration 0.1 mM) and incubated at 293 K overnight. Subsequently, the cells were harvested and resuspended in buffer A (10 mM Tris–HCl buffer pH 8). The cells were disrupted by adding lysozyme (incubation time of 1 h at 280 K on an orbital shaker) and further treatment with a French press (three rounds of disruption). The cell lysate was centrifuged at 8000g for 45 min to remove cellular debris.
Ammonium sulfate precipitation was carried out using 15%(w/v) ammonium sulfate in buffer A with an incubation time of 2 h at 280 K. Afterwards, the solution was centrifuged at 8000g for 30 min. The following purification steps were all performed using an ÄKTA Purifier (GE Healthcare). The cell lysate was applied onto a Butyl Sepharose column equilibrated with buffer A and eluted using a linear gradient from 15 to 0% ammonium sulfate. Fractions containing DppA were pooled and desalted by gel filtration (buffer A). Subsequently, the sample was applied onto a Q Sepharose ion-exchange column with buffer A, eluted using a linear gradient from 0 to 1 M NaCl and desalted by gel filtration (buffer A). The sample was then applied onto a high-resolution Resource Q ion-exchange column equilibrated with buffer A and eluted with a linear gradient from 0 to 1 M KCl. Finally, the protein solution was desalted (buffer A) and concentrated with Amicon Ultra spin columns (exclusion size 10 kDa) from Millipore.
2.2. Crystallization
Initial screening for crystallization conditions was performed by the sitting-drop vapour-diffusion method using a crystallization robot (HTPC, CyBio) and 96-well plates (CrystalQuick LP, Greiner Bio-One). The drops contained 0.3 µl protein solution (at 8 mg ml−1 in buffer A) and 0.3 µl reservoir solution and were equilibrated against 40 µl reservoir solution. From these initial screens (JBScreen Classic 1–10, Jena Bioscience) several hits were observed. The crystallization conditions were further optimized using 24-well crystallization plates (Greiner Bio-One) with the hanging-drop vapour-diffusion method. Each well contained 500 µl reservoir solution and the drop volume was a mixture of 1 µl protein solution and 1 µl reservoir solution.
2.3. Data collection and X-ray crystallographic analysis
For cryoprotection, the reservoir solution was mixed with 50%(v/v) PEG 400 to give a final concentration of 5%(v/v) PEG 400. Before the crystal was mounted, it was transferred into and soaked in this cryoprotectant for 30 s. The crystal was then flash-cooled to 110 K in a stream of nitrogen (Oxford Cryosystems).
Diffraction data were collected using a rotating-anode X-ray generator (MicroMax007, Rigaku) with Osmic multiple-layer optics (beam size 0.3 × 0.3 mm) and a CCD detector (Saturn 92, Rigaku).
The diffraction data were integrated and scaled using the software CrystalClear v.1.3.6 (Pflugrath, 1999 ▶). To obtain values of R p.i.m. and R r.i.m. the data were scaled with SCALA (Evans, 2006 ▶).
3. Results and discussion
The DppA protein was expressed in E. coli BL21 Gold and purified to homogeneity by ammonium sulfate precipitation and reversed-phase and ion-exchange chromatography. A final concentration of 8 mg ml−1 was used for crystallization experiments.
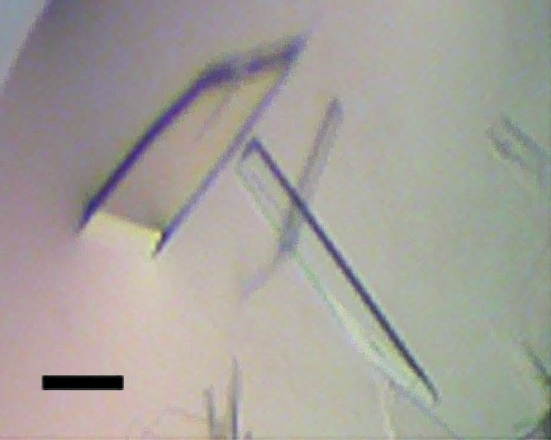
Crystals of DppA protein could be obtained with several precipitants from the initial screen using the vapour-diffusion method. The crystallization conditions were further optimized and tiny needles that were too small for data collection appeared after several days in most cases (0.1 M sodium acetate pH 4.6, 28–31% PEG 400, 0.1 M MgCl2; 0.1 M sodium phosphate–citrate pH 4.2, 0.2 M K2HPO4, 1.6 M NaH2PO4; 0.1 M Na HEPES pH 7.5, 1.8–2.2 M ammonium sulfate, 2% PEG 400; 0.1 M Na HEPES pH 7.5, 1.5–1.8 M ammonium sulfate, 0.2 M NaCl; 0.1 M Na MES pH 6.5, 1.8–2.0 M ammonium sulfate, 5% PEG 400; 0.1 M Na MES pH 6.5, 1.5–1.8 M ammonium sulfate). In one condition, comprised of 0.1 M Na MES pH 6.5, 1.8 M ammonium sulfate and 5% PEG 400, one crystal with an appropriate size for data collection (40 × 110 × 200 µm) was obtained after 4 d (Fig. 1 ▶).
Figure 1.
Crystals of the haloalkane dehalogenase DppA grown by the hanging-drop vapour-diffusion method. The crystal which was used for data collection (the leftmost crystal) has a thickness of approximately 40 µm. The solid bar represents 100 µm.
This crystal of DppA diffracted X-rays to a resolution of 1.95 Å on a rotating-anode X-ray generator. The integration limit was set to an average I/σ(I) of 2. The data statistics are reported in Table 1 ▶.
Table 1. X-ray data-collection statistics for DppA.
Values in parentheses are for the outer resolution shell.
| X-ray source | Rigaku MicroMax-007 |
| Detector | Saturn 92 |
| Wavelength (Å) | 1.5418 |
| Resolution range (Å) | 38.68–1.95 (2.02–1.95) |
| Space group | P21212 |
| Unit-cell parameters (Å) | a = 47.3, b = 108.5, c = 67.3 |
| Total reflections | 139903 |
| Unique reflections | 25613 |
| Completeness (%) | 98.5 (97.4) |
| Rmerge (%) | 12.0 (43.7) |
| Rr.i.m. (%) | 13.7 (57.9) |
| Rp.i.m. (%) | 6.4 (32.7) |
| Average I/σ(I) | 7.0 (2.3) |
| d-spacing (Å) | 2.02–1.95 |
| Mosaicity (°) | 1.00 |
| Multiplicity | 5.46 (4.13) |
| Wilson B factor (Å2) | 26.0 |
| No. of images | 258 |
The crystal of DppA belonged to the orthorhombic space group P21212 (No. 18), with unit-cell parameters a = 47.3, b = 108.5, c = 67.3 Å.
Assuming the presence of one molecule in the asymmetric unit, the Matthews coefficient V M was calculated to be 2.6 Å3 Da−1.
The structure could be solved by molecular replacement using the program Phaser (McCoy et al., 2007 ▶) with the haloalkane dehalogenase from X. autotrophicus GJ10 (PDB code 1edb; Verschueren et al., 1993 ▶), which has a sequence identity of 50%, as a search model. A unique solution was obtained with a monomer in the asymmetric unit with a likelihood score of 483.81 and a Z score of 21.51. Refinement is currently in progress.
References
- Bosma, T., Damborsky, J., Stucki, G. & Janssen, D. B. (2002). Appl. Environ. Microbiol.68, 3582–3587. [DOI] [PMC free article] [PubMed]
- Chaloupkova, R., Sykorova, J., Prokop, Z., Jesenska, A., Monincova, M., Pavlova, M., Tsuda, M., Nagata, Y. & Damborsky, J. (2003). J. Biol. Chem.278, 52622–52628. [DOI] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Franken, S. M., Rozeboom, H. J., Kalk, K. H. & Dijkstra, B. W. (1991). EMBO J.10, 1297–1302. [DOI] [PMC free article] [PubMed]
- Gray, K. A., Richardson, T. H., Kretz, K., Short, J. M., Bartnek, F., Knowles, R., Kan, L., Swanson, P. E. & Robertson, D. E. (2001). Adv. Synth. Catal.343, 607–617.
- Holloway, P., Knoke, K. L., Trevors, J. T. & Lee, H. (1998). Biotechnol. Bioeng.59, 520–523. [PubMed]
- Iizuka, T., Jojima, Y., Fudou, R., Hiraishi, A., Ahn, J.-W. & Yamanaka, S. (2003). Int. J. Syst. Evol. Microbiol.53, 189–195. [DOI] [PubMed]
- Janssen, D. B. (2004). Curr. Opin. Chem. Biol.8, 150–159.
- Marek, J., Vevodova, J., Kuta Smatanova, I., Nagata, Y., Svensson, L. A., Newman, J., Takagi, M. & Damborsky, J. (2000). Biochemistry, 39, 14082–14086. [DOI] [PubMed]
- Mazumdar, P. A., Hulecki, J. C., Cherney, M. M., Garen, C. R. & James, M. N. G. (2008). Biochim. Biophys. Acta, 1784, 351–362. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst.40, 658–674. [DOI] [PMC free article] [PubMed]
- Newman, J., Peat, T. S., Richard, R., Kan, L., Swanson, P. E., Affholter, J. A., Holmes, I. H., Schindler, J. F., Unkefer, C. J. & Terwilliger, T. C. (1999). Biochemistry, 38, 16105–16114. [DOI] [PubMed]
- Ollis, D. L., Cheah, E., Cygler, M., Dijkstra, B., Frolow, F., Franken, S. M., Harel, M., Remington, S. J., Silman, I., Schrag, J., Sussman, J. L., Verschueren, K. H. G. & Goldman, A. (1992). Protein Eng.5, 197–211. [DOI] [PubMed]
- Pflugrath, J. W. (1999). Acta Cryst. D55, 1718–1725. [DOI] [PubMed]
- Pries, F., van den Wijngaard, A. J., Bos, R., Pentenga, M. & Janssen, D. B. (1994). J. Biol. Chem.269, 17490–17494. [PubMed]
- Schanstra, J. P., Ridder, I. S., Heimeriks, G. J., Rink, R., Poelarends, G. J., Kalk, K. H., Dijkstra, B. W. & Janssen, D. B. (1996). Biochemistry, 35, 13186–13195. [DOI] [PubMed]
- Stsiapanava, A., Koudelakova, T., Lapkouski, M., Pavlova, M., Damborsky, J. & Kuta Smatanova, I. (2008). Acta Cryst. F64, 137–140. [DOI] [PMC free article] [PubMed]
- Verschueren, K. H. G., Kingma, J., Rozeboom, H. J., Kalk, K. H., Janssen, D. B. & Dijkstra, B. W. (1993). Biochemistry, 32, 9031–9037. [DOI] [PubMed]