A hyperthermophilic archaeal Rieske-type [2Fe–2S] ferredoxin (ARF) from S. solfataricus P1 has been crystallized as a recombinant protein with a vector-derived long N-terminal extension region. The P43212 crystals of recombinant ARF diffracted to 1.85 Å resolution using synchrotron radiation.
Keywords: Rieske-type ferredoxins, [2Fe–2S] clusters, archaea, Sulfolobus solfataricus
Abstract
The hyperthermophilic archaeal Rieske-type [2Fe–2S] ferredoxin (ARF) from Sulfolobus solfataricus P1 contains a low-potential Rieske-type [2Fe–2S] cluster that has served as a tractable model for ligand-substitution studies on this protein family. Recombinant ARF harbouring a pET30a vector-derived N-terminal extension region plus a hexahistidine tag has been heterologously overproduced in Escherichia coli, purified and crystallized by the hanging-drop vapour-diffusion method using 0.05 M sodium acetate, 0.05 M HEPES, 2 M ammonium sulfate pH 5.5. The crystals diffracted to 1.85 Å resolution and belonged to the tetragonal space group P43212, with unit-cell parameters a = 60.72, c = 83.31 Å. The asymmetric unit contains one protein molecule.
1. Introduction
Proteins containing Rieske-type [2Fe–2S] clusters play essential functions in all three domains of life (Mason & Cammack, 1992 ▶; Berry et al., 2000 ▶; Link, 1999 ▶; Cramer et al., 2006 ▶). In contrast to regular plant-type and vertebrate-type [2Fe–2S] ferredoxins with complete cysteinyl ligations, the Rieske-type cluster has an asymmetric iron–sulfur core, with the Sγ atom of each of the two cysteine residues coordinated to one iron site and the Nδ atom of each of the two histidine residues coordinated to the other iron site (e.g. PDB entries 1rie, 1rfs, 1ndo, 1fqt, 1jm1, 1nyk and 2nuk; Iwata et al., 1996 ▶; Carrell et al., 1997 ▶; Kauppi et al., 1998 ▶; Colbert et al., 2000 ▶; Bönisch et al., 2002 ▶; Hunsicker-Wang et al., 2003 ▶; Kolling et al., 2007 ▶). With special interest in the critical functions of the histidine ligands in the biologically ubiquitous metallosulfur redox sites, the influence of substitution of each histidine ligand (His44, His64) by cysteine on the cluster assembly was addressed using archaeal Rieske-type ferredoxin (ARF; DDBJ/EMBL/GenBank accession No. AB047031; Cosper et al., 2002 ▶; Kounosu et al., 2004 ▶; Iwasaki et al., 2004 ▶, 2005 ▶, 2009 ▶; Dikanov et al., 2004 ▶) from the hyperthermophile Sulfolobus solfataricus strain P1 (DSM 1616T) as a tractable model. This 12 kDa protein (108 amino acids) contains the canonical cluster-binding motif -Cys42-Xaa43-His44-//-Cys61-Xaa62-Leu63-His64-, as found in bacterial low-potential Rieske-type ferredoxins involved in the biodegradation pathways of various alkenes and aromatic compounds (Link, 1999 ▶; Kounosu et al., 2004 ▶). Replacement of the His64 ligand by cysteine in ARF allowed the assembly of an oxidized [2Fe–2S] cluster with one histidine plus three cysteine ligands in the archaeal Rieske-type protein scaffold, whereas the replacement of the His44 ligand by cysteine caused the cluster insertion and/or assembly to fail (Kounosu et al., 2004 ▶). Replacement of three residues (His44, Lys45 and His64) in ARF (ARF-triple; H44I/K45C/H64C) by mimicking the mononuclear iron site in the Pyrococcus furiosus rubredoxin (Day et al., 1992 ▶; Adams et al., 2002 ▶) resulted in a rubredoxin-type mononuclear Fe(Cys)4 site in the Rieske-type protein scaffold (Iwasaki et al., 2005 ▶). In conjunction with the converse incorporation of an oxidized [2Fe–2S] cluster into the Clostridium pasteurianum rubredoxin polypeptide chain upon replacement of Cys42 by alanine, which normally accommodates a mononuclear Fe(Cys)4 site (Meyer et al., 1997 ▶), these results indicate the importance of the types and the spacing of ligands in the in vivo cluster-recognition/insertion/assembly in metallosulfur protein scaffolds (Iwasaki et al., 2005 ▶).
A deeper understanding of the design of the metal-binding site and evolutionary divergency from the same protein template to promote biological functionalities requires structural information at atomic resolution. Although recombinant ARF has been overproduced in Escherichia coli and can be obtained in appropriate forms for electron paramagnetic resonance and resonance Raman analyses in conjunction with stable-isotope (e.g. 2H, 15N and 57Fe) labelling and mutagenesis (Cosper et al., 2002 ▶; Kounosu et al., 2004 ▶; Iwasaki et al., 2004 ▶, 2005 ▶, 2009 ▶; Dikanov et al., 2004 ▶), no suitable crystals of wild-type ARF or its variants have been produced to date either in the absence or in the presence of a flexible hexahistidine tag at the N-terminus derived from a pET28a expression vector (usually cleaved off after purification by metal-affinity chromatography). Common methods of troubleshooting in this case may be to employ (i) a shorter (truncated) protein/domain fragment (as a flexible region, including a short histidine tag, can inhibit crystallization and may require cleaving off) or (ii) a (closely related) homologous protein from a different species for crystallization. Here, we present the crystallization of the recombinant wild-type ARF of S. solfataricus strain P1 using an alternative approach in a form suitable for X-ray studies and preliminary X-ray data analysis; although the role of a flexible affinity tag in protein crystallization has been the subject of considerable debate (Smyth et al., 2003 ▶; Carson et al., 2007 ▶; Smits et al., 2008 ▶; Backmark et al., 2008 ▶), we introduced an expression vector-derived long N-terminal extension plus a hexahistidine tag into the target protein of interest, which enabled a drastic improvement of its crystal quality. Thus, in some cases, this complementary strategy can reduce unfavourable interactions in protein crystallization and/or induce important crystal contacts and may aid a whole structurome-wide robust study on a selected organism with known genomic sequence and/or a target structural study of particular proteins, e.g. those of considerable biomedical and pharmaceutical interest.
2. Methods and results
2.1. Protein preparation and characterization
The arf gene coding for the archaeal Rieske-type ferredoxin (ARF) of S. solfataricus strain P1 (DSM 1616T) has been cloned and sequenced (DDBJ/EMBL/GenBank accession No. AB047031; Kounosu et al., 2004 ▶). The polymerase chain reaction (PCR) was carried out to amplify the arf gene using S. solfataricus P1 genomic DNA and the following oligonucleotide primers (designed based on the reported nucleotide sequence): ARF30aEcoF, 5′-GGG CCC GAA TTC ATG CTA GTC AGA GTG TCT AGC TTA-3′, and ARF30aXhoR, 5′-GGG CCC CTC GAG TTA AAT TTG TAT AAA AAT ATC TTT GCC-3′. The PCR product thus amplified was subcloned into an EcoRI/XhoI site in a pET30a vector (Novagen) and the nucleotide sequences were confirmed for both strands using an ABI PRISM 310 Genetic Analyzer automatic DNA sequencer (PE Biosystems) with a vector-specific T7 promoter and T7 terminator. The resultant vector was named pET30aARF. The resultant pET30aARF harbouring the arf gene was transformed into the host strain E. coli BL21-CodonPlus(DE3)-RIL (Stratagene). The transformants were grown overnight at 298 K in Luria–Bertani medium containing 50 µg ml−1 kanamycin and 0.2 mM FeCl3 and the recombinant holoprotein was overproduced with 1 mM isopropyl β-d-1-thiogalactopyranoside for 24 h at 298 K. The cells were pelleted by centrifugation and the recombinant ARF with a hexahistidine tag and a pET30a vector-derived long extension region at the N-terminus was purified at 293 K (Kounosu et al., 2004 ▶; Iwasaki et al., 2004 ▶); the proteolytic removal of the hexahistidine tag from the purified protein was omitted. The sample was further purified by gel-filtration chromatography (Sephadex G-75; Amersham Pharmacia Biotech) eluted with 10 mM HEPES–NaOH, 350 mM NaCl pH 7.5, concentrated to ∼17–18 mg ml−1 using a Centriprep-10 apparatus (Amicon) and stored frozen (193 K) until use.
The purified recombinant ARF contains 52 extra pET30a vector-derived residues (MHHHHHHSSGLVPRGSGMKETAAAKFERQHMDSPDLGTDDDDKAMADIGSEF) at the N-terminus which are absent from the S. solfataricus ARF sequence starting with Met1. Engineering of this N-terminal extension into the recombinant protein allowed not only rapid purification but also reproducible crystallization, presumably by inducing important crystal contacts and/or by changing the calculated pI from 8.14 to 6.14 (as predicted for the native and recombinant protein, respectively). The room-temperature visible absorption spectrum (measured using a Beckman DU-7400 spectrophotometer) of the recombinant ARF (data not shown) was identical to that of previously reported ARF samples obtained using a pET28aARF vector (based on a pET28a His-tag expression vector from Novagen; Kounosu et al., 2004 ▶), showing the presence of a Rieske-type [2Fe–2S] cluster.
2.2. Crystallization
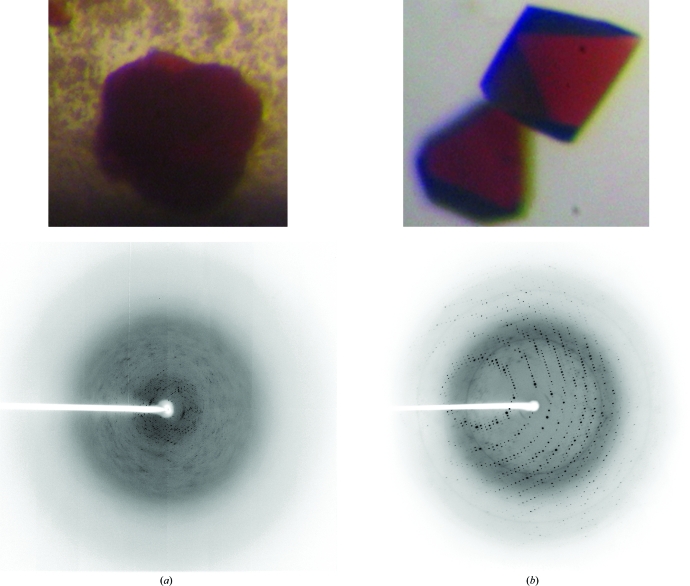
Preliminary screening took place by standard hanging-drop vapour diffusion in Linbro plates at 277 and 293 K using 0.5 ml reservoirs from commercially available sparse-matrix screening kits (Hampton Research Crystal Screens I and II and SaltRX and Qiagen JCSG+ and Protein Complex Suites). Dark reddish crystals were obtained in 1–3 d at 293 K (under oxygenic conditions without any reducing reagents) using three conditions. Optimized crystals (Fig. 1 ▶ b) were obtained by combining 1.0–2.0 µl protein solution with 1.0–2.0 µl reservoir solution consisting of 0.05 M sodium acetate, 0.05 M HEPES, 2 M ammonium sulfate pH 5.5. Drops were equilibrated against 0.5 ml reservoir solution and the crystals grew to maximal dimensions of approximately 0.25 × 0.25 × 0.3 mm in 2–4 d (Fig. 1 ▶ b, top). They were transferred into a cryoprotective solution containing 10%(v/v) glycerol in the same reservoir solution for flash-cooling in a cold nitrogen-gas stream. In comparison, the previous samples obtained using the pET28aARF vector (Kounosu et al., 2004 ▶), either with or without thrombin digestion of the pET28a vector-derived N-terminal extension region, had a significantly poorer reproducibility of crystallization (stacked cluster crystals were obtained only once in ∼6 months with reservoir solution containing 4 M sodium formate pH 8.8 and diffracted to 4.5 Å resolution using synchrotron radiation; Fig. 1 ▶ a).
Figure 1.
Typical crystals obtained using samples (a) before and (b) after engineering of a pET30a vector-derived N-terminal extension into the recombinant ARF (top) and the corresponding typical X-ray diffraction pattern, with a maximum resolution of (a) 4.5 Å (collected at a camera distance of 170 mm using a Jupiter210C CCD detector on SPring-8 beamline BL38B1) and (b) beyond 1.85 Å (collected at a camera distance of 150 mm using a Jupiter210C CCD detector on SPring-8 beamline BL32B2), respectively (bottom). The maximum dimensions of the dark reddish crystals in (b) are approximately 0.17 × 0.17 × 0.2 mm.
2.3. Crystallographic data collection and processing
X-ray diffraction data for recombinant ARF were collected from flash-cooled crystals using a Rigaku/MSC Jupiter210C CCD detector installed on the BL32B2 beamline at SPring-8, Japan. Data collection was performed at a wavelength of 1.0 Å with a total oscillation range of 180° and each diffraction image was taken with an oscillation angle of 1.0° and an exposure time of 10 s. The data were indexed and processed using HKL-2000 (Otwinowski & Minor, 1997 ▶). The crystals were found to diffract to 1.85 Å resolution (Fig. 1 ▶ b) and belonged to the tetragonal space group P41212 or its enantiomorph, with unit-cell parameters a = 60.72, c = 83.31 Å (Table 1 ▶). Assuming the presence of one protein molecule in the asymmetric unit, the Matthews coefficient was 2.1 Å3 Da−1, corresponding to a solvent content of 42% (Matthews, 1968 ▶).
Table 1. Data-processing statistics.
Values in parentheses are for the outer shell.
| Space group | P43212 |
| Unit-cell parameters (Å) | a = 60.72, c = 83.31 |
| Resolution range (Å) | 50–1.85 (1.92–1.85) |
| No. of measured reflections | 179961 |
| No. of unique reflections | 13760 (1203) |
| Completeness (%) | 98.7 (89.0) |
| Redundancy | 13.1 (8.9) |
| Rmerge† (%) | 3.9 (32.7) |
| 〈I/σ(I)〉 | 33.2 (3.2) |
| Mosaicity | 0.588 |
R
merge = 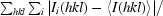
 , where I
i(hkl) and 〈I(hkl)〉 are the intensity of measurement i and the mean intensity for the reflection with indices hkl, respectively.
, where I
i(hkl) and 〈I(hkl)〉 are the intensity of measurement i and the mean intensity for the reflection with indices hkl, respectively.
Phase determination was successfully carried out by Fe-SAD using the programs SHELX (Sheldrick, 2008 ▶) and HKL2MAP (Pape & Schneider, 2004 ▶), which also established the tetragonal space group P43212. Construction, revision and analysis of atomic models using the ARF sequence are currently in progress.
Acknowledgments
We thank Dr Seiki Baba (SPring-8/JASRI) for obtaining the X-ray diffraction pattern shown in Fig. 1 ▶(a). This work was supported in part by The Japanese JSPS grants-in-aid 18608004 and 21659111 to TI. The diffraction data were collected on SPring-8 beamline BL32B2 with the approval of the Japan Synchrotron Radiation Research Institute (JASRI; Proposal No. 2010A1900).
References
- Adams, M. W. W., Jenney, F. E. Jr, Clay, M. D. & Johnson, M. K. (2002). J. Biol. Inorg. Chem.7, 647–652. [DOI] [PubMed]
- Backmark, A., Nyblom, M., Törnroth-Horsefield, S., Kosinska-Eriksson, U., Nordén, K., Fellert, M., Kjellbom, P., Johanson, U., Hedfalk, K., Lindkvist-Petersson, K., Neutze, R. & Horsefield, R. (2008). Acta Cryst. D64, 1183–1186. [DOI] [PubMed]
- Berry, E. A., Guergova-Kuras, M., Huang, L.-S. & Crofts, A. R. (2000). Annu. Rev. Biochem.69, 1005–1075. [DOI] [PubMed]
- Bönisch, H., Schmidt, C. L., Schäfer, G. & Ladenstein, R. (2002). J. Mol. Biol.319, 791–805. [DOI] [PubMed]
- Carrell, C. J., Zhang, H., Cramer, W. A. & Smith, J. L. (1997). Structure, 5, 1613–1625. [DOI] [PubMed]
- Carson, M., Johnson, D. H., McDonald, H., Brouillette, C. & DeLucas, L. J. (2007). Acta Cryst. D63, 295–301. [DOI] [PubMed]
- Colbert, C. L., Couture, M. M.-J., Eltis, L. D. & Bolin, J. (2000). Structure, 8, 1267–1278. [DOI] [PubMed]
- Cosper, N. J., Eby, D. M., Kounosu, A., Kurosawa, N., Neidle, E. L., Kurtz, D. M. Jr, Iwasaki, T. & Scott, R. A. (2002). Protein Sci.11, 2969–2973. [DOI] [PMC free article] [PubMed]
- Cramer, W. A., Zhang, H., Yan, J., Kurisu, G. & Smith, J. L. (2006). Annu. Rev. Biochem.75, 769–790. [DOI] [PubMed]
- Day, M. W., Hsu, B. T., Joshua-Tor, L., Park, J.-B., Zhou, Z. H., Adams, M. W. W. & Rees, D. C. (1992). Protein Sci.1, 1494–1507. [DOI] [PMC free article] [PubMed]
- Dikanov, S. A., Shubin, A. A., Kounosu, A., Iwasaki, T. & Samoilova, R. I. (2004). J. Biol. Inorg. Chem.9, 753–767. [DOI] [PubMed]
- Hunsicker-Wang, L. M., Heine, A., Chen, Y., Luna, E. P., Todaro, T., Zhang, Y. M., Williams, P. A., McRee, D. E., Hirst, J., Stout, C. D. & Fee, J. A. (2003). Biochemistry, 42, 7303–7317. [DOI] [PubMed]
- Iwasaki, T., Kounosu, A., Kolling, D. R. J., Crofts, A. R., Dikanov, S. A., Jin, A., Imai, T. & Urushiyama, A. (2004). J. Am. Chem. Soc.126, 4788–4789. [DOI] [PubMed]
- Iwasaki, T., Kounosu, A., Tao, Y., Li, Z., Shokes, J. E., Cosper, N. J., Imai, T., Urushiyama, A. & Scott, R. A. (2005). J. Biol. Chem.280, 9129–9134. [DOI] [PubMed]
- Iwasaki, T., Samoilova, R. I., Kounosu, A. & Dikanov, S. A. (2009). FEBS Lett.583, 3467–3472. [DOI] [PMC free article] [PubMed]
- Iwata, S., Saynovits, M., Link, T. A. & Michel, H. (1996). Structure, 4, 567–579. [DOI] [PubMed]
- Kauppi, B., Lee, K., Carredano, E., Parales, R. E., Gibson, D. T., Eklund, H. & Ramaswamy, S. (1998). Structure, 6, 571–586. [DOI] [PubMed]
- Kolling, D. J., Brunzelle, J. S., Lhee, S., Crofts, A. R. & Nair, S. K. (2007). Structure, 15, 29–38. [DOI] [PMC free article] [PubMed]
- Kounosu, A., Li, Z., Cosper, N. J., Shokes, J. E., Scott, R. A., Imai, T., Urushiyama, A. & Iwasaki, T. (2004). J. Biol. Chem.279, 12519–12528. [DOI] [PubMed]
- Link, T. A. (1999). Adv. Inorg. Chem.47, 83–157.
- Mason, J. R. & Cammack, R. (1992). Annu. Rev. Microbiol.46, 277–305. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Meyer, J., Gagnon, J., Gaillard, J., Lutz, M., Achim, C., Münck, E., Pétillot, Y., Colangelo, C. M. & Scott, R. A. (1997). Biochemistry36, 13374–13380. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Pape, T. & Schneider, T. R. (2004). J. Appl. Cryst.37, 843–844.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Smits, S. H. J., Mueller, A., Grieshaber, M. K. & Schmitt, L. (2008). Acta Cryst. F64, 836–839. [DOI] [PMC free article] [PubMed]
- Smyth, D. R., Mrozkiewicz, M. K., McGrath, W. J., Listwan, P. & Kobe, B. (2003). Protein Sci.12, 1313–1322. [DOI] [PMC free article] [PubMed]