The Fab fragment of NC-1, a murine antibody that specifically recognizes the six-helix bundle core of HIV-1 gp41, has been crystallized in space group P3221. An X-ray diffraction data set was collected at 3.2 Å resolution and a clear molecular-replacement solution was obtained for solution of the structure.
Keywords: NC-1 antibody, HIV-1 gp41, Fab fragments
Abstract
NC-1 is a murine monoclonal antibody that specifically recognizes the six-helix bundle core of the human immunodeficiency virus type 1 (HIV-1) gp41. As such, it is a useful tool for probing gp41 conformations in HIV-1 membrane fusion. To establish the structural basis underlying the NC-1 specificity, X-ray crystallography was employed to solve its three-dimensional structure. To accomplish this, hybridoma-produced NC-1 antibody was first purified and digested with papain. Its Fab fragment was then purified using size-exclusion chromatography following Fc depletion using a Protein A affinity column. Finally, crystallization of NC-1 Fab was performed by the hanging-drop vapour-diffusion method and the protein was crystallized at pH 8.0 using PEG 6000 as precipitant. The results showed that the NC-1 Fab crystals belonged to the trigonal space group P3221, with unit-cell parameters a = b = 118.7, c = 106.0 Å. There is one Fab molecule in the asymmetric unit, with 67.5% solvent content. An X-ray diffraction data set was collected at 3.2 Å resolution and a clear molecular-replacement solution was obtained for solution of the structure.
1. Introduction
The envelope glycoprotein (Env) of human immunodeficiency virus type 1 (HIV-1) mediates virus–cell attachment and membrane fusion. Env is synthesized as a precursor (gp160) that is proteolytically processed to generate two noncovalently associated subunits: gp120 and gp41 (Kowalski et al., 1987 ▶). Binding of the surface subunit gp120 to cellular receptors (CD4 and a chemokine co-receptor such as CCR5 or CXCR4) induces a conformational change in the transmembrane subunit gp41 which ultimately leads to membrane fusion (Chan & Kim, 1998 ▶). Therefore, gp41 plays a major role in viral cell entry by mediating the fusion of viral and cellular membranes. A six-helix bundle (6-HB) core formed by the N- and C-terminal heptad-repeat regions (NHR and CHR, respectively) of gp41 has been identified as the fusion-active state and/or post-fusion conformation of gp41. In the 6-HB core of gp41 the three NHR helices form an interior parallel coiled-coil trimer, while the three CHR helices pack in the reverse direction into the three hydrophobic grooves on the surface of the NHR coiled-coil trimer (Chan et al., 1997 ▶; Tan et al., 1997 ▶; Weissenhorn et al., 1997 ▶).
NC-1 was generated and cloned from BALB/c mice immunized with the model polypeptide N36(L6)C34, which folds into a stable six-helix bundle (Jiang et al., 1998 ▶; Lu & Kim, 1997 ▶). NC-1 is a unique monoclonal antibody that specifically recognizes the conformational epitopes on the 6-HB core of gp41 and fails to react with the isolated N36 peptide or C34 peptide (Jiang et al., 1998 ▶). This conformation-dependent reactivity is dramatically reduced by point mutations within the N-terminal coiled-coil region of gp41 which impede formation of the gp41 core. Previous studies showed that NC-1 only bound to the surface of HIV-1-infected cells in the presence of soluble CD4 (Jiang et al., 1998 ▶). These results indicate that NC-1 is capable of reacting with gp41 in a conformation-specific manner and can therefore be used as a valuable conformational probe for studying the receptor-induced conformational changes in gp41 required for membrane fusion and HIV-1 infection and for screening for HIV-fusion inhibitors targeting gp41 (de Rosny et al., 2004 ▶; Dimitrov et al., 2005 ▶; Jiang et al., 1999 ▶; Liu, Zhao et al., 2003 ▶; Liu, Boyer-Chatanet et al., 2003 ▶; Sackett et al., 2006 ▶; Yang et al., 2000 ▶).
In this study, we report the crystallization of the NC-1 Fab fragment. A preliminary analysis of the X-ray diffraction data and a molecular-replacement solution are also presented.
2. Materials and methods
2.1. Production and purification of NC-1 and its Fab fragment
NC-1 antibodies were purified using Protein A affinity-column chromatography (HiTrap, GE Healthcare, USA) from the supernatant of cultured hybridoma cells expressing the murine monoclonal antibody NC-1. The supernatant was loaded onto the column and the column was washed with 20 mM Tris, 200 mM NaCl pH 8.0. The antibody fraction was eluted with 0.1 M glycine (Sigma–Aldrich, USA) pH 2.7. The eluate was neutralized immediately after elution using 1 M Tris–HCl pH 8.0.
The Fab fragment of NC-1 was prepared by limited digestion with papain (Sigma–Aldrich, USA). The reaction was carried out in 20 mM sodium phosphate buffer pH 7.4 containing 150 mM NaCl, 10 mM EDTA and 10 mM cysteine (Sigma–Aldrich, USA). The reaction was terminated by the addition of iodoacetamide (Sigma–Aldrich, USA) to a final concentration of 200 mM and the solution was dialyzed against 20 mM Tris–HCl pH 7.4 containing 150 mM NaCl. The papain digestion was monitored using reducing SDS–PAGE and prestained molecular-weight markers (Bio-Rad, USA). The purified Fab fragment was dialyzed against 10 mM Tris, 0.1 mM ZnCl2, 100 mM NaCl pH 7.5 and its purity was checked and confirmed by SDS–PAGE.
2.2. Crystallization
The NC-1 Fab was concentrated to 3.0 mg ml−1 using an Amicon centrifugal filter with a 30 kDa molecular-weight cutoff (Millipore, USA). Preliminary crystallization conditions were screened by the hanging-drop vapour-diffusion method using the Crystal Screen kits from Hampton Research (Aliso Viejo, California, USA). Hanging drops containing 1 µl protein solution and 1 µl crystallization solution were equilibrated against a reservoir containing 1 ml crystallization solution.
After optimization of initial crystallization conditions, 2 µl protein solution and 2 µl crystallization solution were mixed in a hanging drop and equilibrated against 1 ml crystallization solution in order to obtain crystals with larger sizes.
2.3. Data collection and processing
The NC-1 Fab crystal was cryoprotected using reservoir solution supplemented with 25%(v/v) glycerol and was flash-cooled in liquid nitrogen. X-ray diffraction data were collected at 100 K using synchrotron radiation on Stanford Synchrotron Radiation Lightsource (SSRL) beamline 7-1. Diffraction patterns were recorded on a MAR Q315R CCD detector with a crystal-to-detector distance of 500 mm. The oscillation angle was 0.5° and the exposure time was 30 s per frame. A total of 277 frames were collected. Diffraction data were processed using the HKL-2000 suite (Otwinowski & Minor, 1997 ▶).
2.4. Phasing by molecular replacement
The CNS program (Brunger, 2007 ▶) was used to solve the crystal structure of NC-1 Fab by molecular replacement. NC-1 is a mouse antibody that belongs to the IgG2a class with a κ light chain. Therefore, we chose the high-resolution (1.9 Å) Fab structure of a mouse IgG2a (PDB code 1kn2; D’Souza et al., 2002 ▶) as the starting model for molecular replacement. The cross-rotation function and translation function were calculated using data in the resolution range 20.0–4.0 Å. Rigid-body refinement (using VH, VL, CH and CL as independent segments) was performed in CNS to improve the molecular-replacement solution. Electron-density maps (both 2F o − F c and F o − F c difference maps) were calculated with σA weighting and bulk-solvent correction using CNS. Rigid-body refinement was also performed using CNS. Protein models and electron-density maps were visualized using O (Jones et al., 1991 ▶).
3. Results and discussion
3.1. Fab production and purification
After papain digestion, the NC-1 Fab was further purified by size-exclusion chromatography. Analysis of the purified NC-1 Fab by SDS–PAGE under reducing conditions revealed a doublet at ∼25 kDa which is characteristic of Fab fragments (Fig. 1 ▶). No contaminant protein bands were seen, indicating the high purity of the NC-1 Fab preparation.
Figure 1.
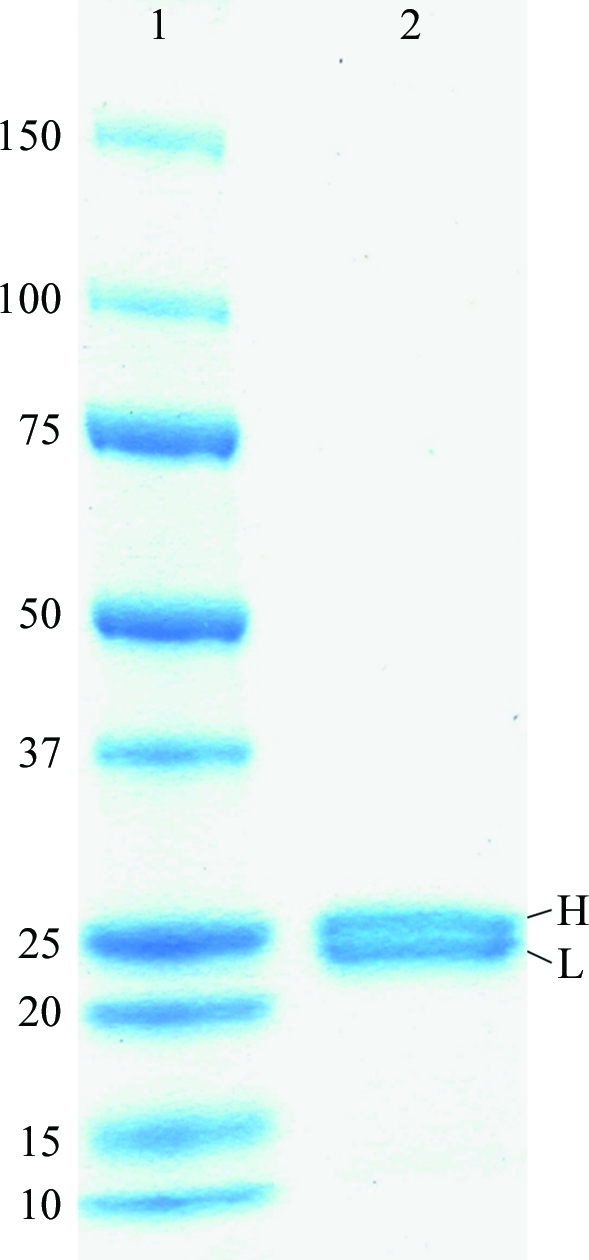
SDS–PAGE of the purified NC-1 Fab on a 12% gel under reducing conditions. Lane 1, molecular-weight markers (kDa). Lane 2, NC-1 Fab. The two bands at ∼25.2 and ∼24.0 kDa correspond to the heavy chain (H) and light chain (L) of the NC-1 Fab molecule, respectively.
3.2. Crystallization
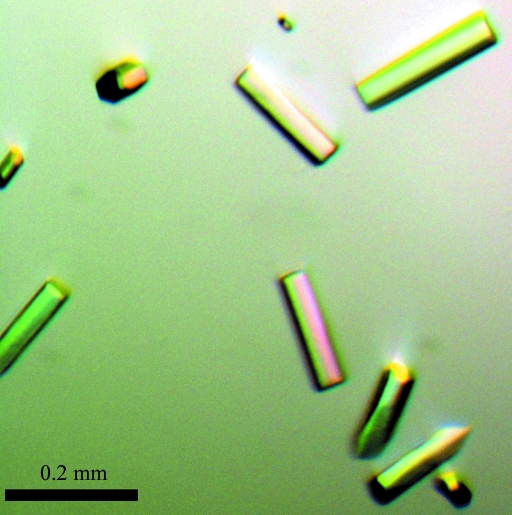
Initial crystals of the NC-1 Fab were obtained using Hampton Research Crystal Screen kits with PEG 6000 as precipitant. Further optimization of the crystal-growth conditions, including pH, protein concentration and precipitant concentration, resulted in crystals of diffraction quality (Fig. 2 ▶) which grew in 100 mM Tris–HCl pH 8.0 and 15%(w/v) PEG 6000 at room temperature. Crystals of the NC-1 Fab appeared within 2 d as thin rods and grew to maximum dimensions of ∼0.2 × 0.02 × 0.01 mm in a week. The crystals belonged to the trigonal space group P3221, with unit-cell parameters a = b = 118.7, c = 106.0 Å. The Matthews coefficient was found to be 3.8 Å3 Da−1, suggesting the presence of one Fab molecule in the asymmetric unit with a solvent content of 67.5%.
Figure 2.
Picture of NC-1 Fab crystals.
3.3. X-ray data collection and processing
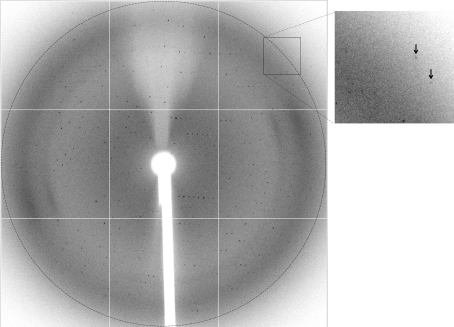
The NC-1 Fab crystals diffracted to a resolution beyond 3.0 Å (Fig. 3 ▶). Anisotropy was visually checked and was not significantly evident for reflections lower than 3.2 Å resolution. An X-ray diffraction data set was collected on Stanford Synchrotron Radiation Lightsource (SSRL) beamline 7-1. The crystal used for data collection was relatively small, forcing us to use 30 s exposures. Nevertheless, we collected a large angular range (138.5° in total) of data as evidenced by the data multiplicity. Crystal decay was evident to some degree for reflections beyond 3.2 Å, but was quite moderate for lower resolution shells. We therefore processed the data to 3.2 Å resolution using HKL-2000, which gave a mosaicity of 0.31°. Data-collection statistics are summarized in Table 1 ▶.
Figure 3.
A representative X-ray diffraction frame of the NC-1 Fab crystal recorded on a MAR Q315R CCD detector. The circle corresponds to a resolution of 2.98 Å. The two arrows in the enlarged box on the right indicate two reflections beyond 3.0 Å.
Table 1. Statistics of X-ray data collection.
Values in parentheses are for the highest resolution shell.
| Wavelength (Å) | 0.9794 |
| Resolution range (Å) | 20.0–3.2 (3.34–3.2) |
| Space group | P3221 |
| Unit-cell parameters (Å) | a = b = 118.7, c = 106.0 |
| No. of observed reflections | 119949 (11502) |
| No. of unique reflections | 14529 (1396) |
| Redundancy | 8.4 (8.2) |
| Completeness (%) | 94.4 (92.0) |
| Rmerge† (%) | 14.1 (49.7) |
| 〈I/σ(I)〉 | 18.7 (4.3) |
R
merge = 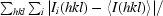
 , where I
i(hkl) is the ith observation of reflection hkl and 〈I(hkl)〉 is the weighted average intensity for all observations i of reflection hkl.
, where I
i(hkl) is the ith observation of reflection hkl and 〈I(hkl)〉 is the weighted average intensity for all observations i of reflection hkl.
3.4. The molecular-replacement solution
For clearer results in the cross-rotation and translation searches, the complementarity-determining region (CDR) residues (L24–L34, L50–L56, L89–L97, H26–H35, H50–H65 and H95–H102, where L is the light chain and H the heavy chain) and the linker peptides between the constant domains and the variable domains (L106–L115 and H112–H115) were excluded from the initial search model. The Fab constant and variable domains (designated CHCL and VHVL, respectively) were used as separate search models. The cross-rotation function gave a clear solution which was the highest peak in both the CHCL and VHVL cases using data in the resolution range 20.0–4.0 Å (Supplementary Table 1 ▶ 1). The top 10 peaks in cross-rotation searches were used in the subsequent translation search.
Based on the diffraction pattern alone, we were not able to distinguish between the enantiomorphic space groups P3121 and P3221. When we tried both of these space groups in molecular replacement, only the translation function in space group P3221 gave clear solutions for both CHCL and VHVL and these two solutions corresponded to the highest peaks in both cross-rotation and translation searches (Supplementary Table 1 ▶ 1), suggesting that the P3221 space group is the correct choice.
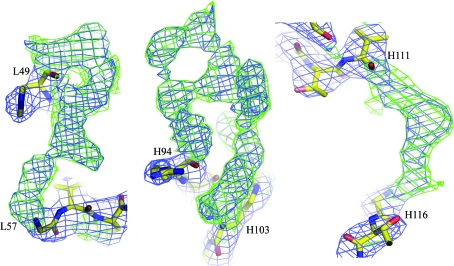
We performed the following to further confirm that the crystal belonged to space group P3221 and that these two solutions were the correct molecular-replacement solutions. Firstly, after rotation and translation in space group P3221 using the two solutions, CHCL and VHVL were combined to give the initial model. Visual inspection in O revealed reasonable crystal packing for the initial model with no molecular overlapping. Secondly, subsequent rigid-body refinement using the initial model lowered the R factor and R free to 39.4% and 38.7% from 44.4% and 44.0%, respectively, in the resolution range 20.0–3.2 Å with 8% of all the reflections randomly selected as the R free set. Finally, using this partial model we calculated σA-weighted 2F o − F c and F o − F c maps, both of which clearly showed distinguishable and continuous densities for all the missing CDR residues and the linker residues (Fig. 4 ▶).
Figure 4.
Representative electron densities for residues which were not included in map calculation, showing the densities for L50–L56 (L chain CDR2, left), H95–H102 (H chain CDR3, middle) and H112–H115 (linker between CH and VH, right). The 2F o − F c map is shown in blue and is contoured at 1.5σ. The F o − F c difference map is shown in green and is contoured at 2.0σ. Figures were prepared using PyMOL (DeLano, 2002 ▶).
In conclusion, we have crystallized the Fab fragment of the anti-HIV-1 antibody NC-1, which is unique in specifically recognizing the 6-HB core of HIV-1 gp41. We have collected a diffraction data set at 3.2 Å resolution. A clear molecular-replacement solution was obtained in space group P3221. Determination of the crystal structure of the NC-1 Fab fragment will lead to a better understanding of its specific binding to the gp41 core and in turn make it a valuable tool in probing gp41 conformations.
Supplementary Material
Supplementary material file. DOI: 10.1107/S1744309110019287/en5431sup1.pdf
Acknowledgments
We gratefully acknowledge the access to synchrotron radiation at Stanford Synchrotron Radiation Lightsource (SSRL) beamline 7-1. We thank Dr Hua Jing of the Scripps Research Institute for help with the X-ray data collection and processing. This project was supported in part by grants from the National Institutes of Health (GM071940 and AI069015 to ZHZ). LJ is partly supported by a grant from the UCLA AIDS Institute and the UCLA Center for AIDS Research (AI28697). SJ is supported by an NIH grant (AI46221) and CP is partly supported by a scholarship from the China Scholarship Council.
Footnotes
Supplementary material has been deposited in the IUCr electronic archive (Reference: EN5431).
References
- Brunger, A. T. (2007). Nature Protoc.2, 2728–2733. [DOI] [PubMed]
- Chan, D. C., Fass, D., Berger, J. M. & Kim, P. S. (1997). Cell, 89, 263–273. [DOI] [PubMed]
- Chan, D. C. & Kim, P. S. (1998). Cell, 93, 681–684. [DOI] [PubMed]
- DeLano, W. L. (2002). The PyMOL Molecular Viewer. http://www.pymol.org.
- Dimitrov, A. S., Louis, J. M., Bewley, C. A., Clore, G. M. & Blumenthal, R. (2005). Biochemistry, 44, 12471–12479. [DOI] [PMC free article] [PubMed]
- D’Souza, L. J., Gigant, B., Knossow, M. & Green, B. S. (2002). J. Am. Chem. Soc.124, 2114–2115. [DOI] [PubMed]
- Jiang, S., Lin, K. & Lu, M. (1998). J. Virol.72, 10213–10217. [DOI] [PMC free article] [PubMed]
- Jiang, S., Lin, K., Zhang, L. & Debnath, A. K. (1999). J. Virol. Methods, 80, 85–96. [DOI] [PubMed]
- Jones, T. A., Zou, J.-Y., Cowan, S. W. & Kjeldgaard, M. (1991). Acta Cryst. A47, 110–119. [DOI] [PubMed]
- Kowalski, M., Potz, J., Basiripour, L., Dorfman, T., Goh, W. C., Terwilliger, E., Dayton, A., Rosen, C., Haseltine, W. & Sodroski, J. (1987). Science, 237, 1351–1355. [DOI] [PubMed]
- Liu, S., Boyer-Chatenet, L., Lu, H. & Jiang, S. (2003). J. Biomol. Screening, 8, 685–693. [DOI] [PubMed]
- Liu, S., Zhao, Q. & Jiang, S. (2003). Peptide, 24, 1303–1313. [DOI] [PubMed]
- Lu, M. & Kim, P. S. (1997). J. Biomol. Struct. Dyn.15, 465–471. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Rosny, E. de, Vassell, R., Jiang, S., Kunert, R. & Weiss, C. D. (2004). J. Virol.78, 2627–2631. [DOI] [PMC free article] [PubMed]
- Sackett, K., Wexler-Cohen, Y. & Shai, Y. (2006). J. Biol. Chem.281, 21755–21762. [DOI] [PubMed]
- Tan, K., Liu, J., Wang, J., Shen, S. & Lu, M. (1997). Proc. Natl Acad. Sci. USA, 94, 12303–12308. [DOI] [PMC free article] [PubMed]
- Weissenhorn, W., Dessen, A., Harrison, S. C., Skehel, J. J. & Wiley, D. C. (1997). Nature (London), 387, 426–428. [DOI] [PubMed]
- Yang, X., Farzan, M., Wyatt, R. & Sodroski, J. (2000). J. Virol.74, 5716–5725. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material file. DOI: 10.1107/S1744309110019287/en5431sup1.pdf