Abstract
BACKGROUND:
The ability to digest lactose divides the world’s population into two phenotypes that may be risk variability markers for several diseases. Prebiotic effects likely favour lactose maldigesters who experience lactose spilling into their colon.
OBJECTIVE:
To evaluate the effects of fixed-dose lactose solutions on fecal bifidobacteria and lactobacilli in digesters and maldigesters, and to determine whether the concept of a difference in ability to digest lactose is supported.
METHODS:
A four-week study was performed in 23 lactose mal-digesters and 18 digesters. Following two weeks of dairy food withdrawal, subjects ingested 25 g of lactose twice a day for two weeks. Stool bifidobacteria and lactobacilli counts pre- and postintervention were measured as the primary outcome. For secondary outcomes, total anaerobes, Enterobacteriaceae, beta-galactosidase and N-acetyl-beta-D-glucosaminidase activity in stool, as well as breath hydrogen and symptoms following lactose challenge tests, were measured.
RESULTS:
Lactose maldigesters had a mean change difference (0.72 log10 colony forming units/g stool; P=0.04) in bifidobacteria counts compared with lactose digesters. Lactobacilli counts were increased, but not significantly. Nevertheless, reduced breath hydrogen after lactose ingestion correlated with lactobacilli (r=−0.5; P<0.001). Reduced total breath hydrogen and symptom scores together, with a rise in fecal enzymes after intervention, were appropriate, but not significant.
CONCLUSIONS:
Despite failure to achieve full colonic adaptation, the present study provided evidence for a differential impact of lactose on microflora depending on genetic lactase status. A prebiotic effect was evident in lactose maldigesters but not in lactose digesters. This may play a role in modifying the mechanisms of certain disease risks related to dairy food consumption between the two phenotypes.
Keywords: Fecal microflora, Lactose, Prebiotic
Abstract
HISTORIQUE :
La capacité de digérer le lactose divise la population mondiale en deux phénotypes qui peuvent risquer les marqueurs de variabilité de plusieurs maladies. Les effets prébiotiques favorisent probablement les personnes qui digèrent mal le lactose et subissent un épanchement de lactose dans le colon.
OBJECTIF :
Évaluer les effets de solutions de lactose à doses fixes sur les bifidobactéries et les lactobacilles fécales chez les personnes qui digèrent bien ou mal le lactose et déterminer si le concept d’une différence de capacité de digérer le lactose est étayé.
MÉTHODOLOGIE :
Les chercheurs ont effectué une étude de quatre semaines chez 23 personnes qui digèrent mal le lactose et 18 personnes qui le digèrent bien. Après deux semaines de sevrage des produits laitiers, les sujets ont ingéré 25 g de lactose deux fois par jour pendant deux semaines. Comme issue primaire, les chercheurs ont mesuré la numération de bifidobactéries et de lactobacilles dans les selles avant et après l’intervention. Comme issue secondaire, ils ont mesuré l’activité totale des anaérobies, des entérobactériacées, des bêta-galactosidases et des N-acétyl-bêta-D-glucosaminidases dans les selles, de même que l’hydrogène dans l’haleine et les symptômes après des tests de provocation par le lactose.
RÉSULTATS :
Les personnes qui digèrent mal le lactose ont une différence de changement moyenne (0,72 log10 unités formatrices de colonies/g de selle; P=0,04) dans les bifidobactéries par rapport à celles qui le digèrent bien. Les numérations de lactobacilles augmentaient, mais pas de manière significative. Néanmoins, une diminution d’hydrogène dans l’haleine après l’ingestion de lactose était corrélée avec les lactobacilles (r=−0,5; P<0,001). L’association d’une diminution d’hydrogène total dans l’haleine et des indices des symptômes, accompagnée d’une augmentation des enzymes fécaux après l’intervention, était appropriée, mais non significative.
CONCLUSIONS :
Malgré l’échec à obtenir une adaptation colique complète, la présente étude a fourni des données probantes démontrant un effet différentiel du lactose sur la microflore d’après le statut génétique de la lactase. Un effet prébiotique était évident chez les personnes qui digèrent mal le lactose, mais pas chez celles qui le digèrent bien. Ce phénomène peut contribuer à modifier les mécanismes de certains risques de maladie liés à la consommation de produits laitiers entre les deux phénotypes.
The ability (lactase persistent [LP]) or inability (lactase nonpersistent [LNP]) to digest lactose in adulthood is the most common known genetic polymorphism that divides humans into a dichotomous phenotype. The near-total loss of intestinal lactase by mid-childhood in the majority of LNP populations forces an altered pattern of food consumption and handling of lactose by this group (1,2). Because of symptoms incurred by intermittent consumption of lactose (mostly dairy foods [DFs]), lower quantities are generally consumed by LNP populations (3,4). However, regular consumption of lactose may lead to bacterial metabolism of nondigested lactose that spills into the lower intestine (prebiotic effect) leading to colonic adaptation, decreased symptoms and further increased intake and tolerability (5–8).
The overwhelming concentration of research on improving symptoms of lactose intolerance in LNP subjects may mask the significance of bacterial colonic adaptation. Over the past few decades, several authors have postulated that there are additional benefits to the interaction between undigested lactose and colonic bacteria (5,9–10). The best human model of the relevance of an adaptation effect on disease is afforded by studies on the protective effects of DFs (particularly milk consumption) against colorectal cancer. In this disease, a similar level of protection is apparently afforded both at high (western societies) and low (eastern societies) DF intakes in predominantly LP and LNP populations, respectively. In the latter’s case, part of the protection may relate to a prebiotic effect of lactose (11). Such a mechanism may also be relevant in other diseases (4).
Colonic spillage of lactose occurs at much lower intakes in LNP than LP subjects (12–14). To our knowledge, there is only a single study (14) that examined the quantity of lactose spill-age into the colon of LP subjects. The amount was found to be 4% to 8% of the ingested dose, representing approximately 2 g to 4 g of lactose per 1 L of milk consumed. Because LP subjects are more likely to reach such consumption levels, we were interested in whether this intake amount exerts measurable effects on colonic bacteria in this population. We hypothesized that lactose would selectively promote bacteria that are traditionally associated with health benefits (bifidobacteria and lactobacilli) in LNP subjects because more lactose spillover occurs in the colons of these individuals.
METHODS
Patients
Participants were recruited by advertisments and the establishment of a website. Men and women of different ethnic and racial backgrounds, who were between 18 and 49 years of age, were included. All procedures were performed without knowledge of the genetic status of the individual. Individuals with stable chronic conditions such as dyslipidemia and thyroid disorders requiring treatment were permitted to enrol. Subjects who were pregnant, had used antibiotics within the previous month, had acute or chronic digestive diseases including irritable bowel syndrome, or chronically used probiotics, nonsteroidal anti-inflammatory agents, narcotics or prokinetic drugs were excluded. The study was approved by the Research and Ethics Committee of the Sir Mortimer B Davis Jewish General Hospital (Montreal, Quebec). Informed, written consent was obtained from all participants, and the study was registered through the Protocol Registration System (www.ClinicalTrials.gov, ID NCT00599859).
Study outline
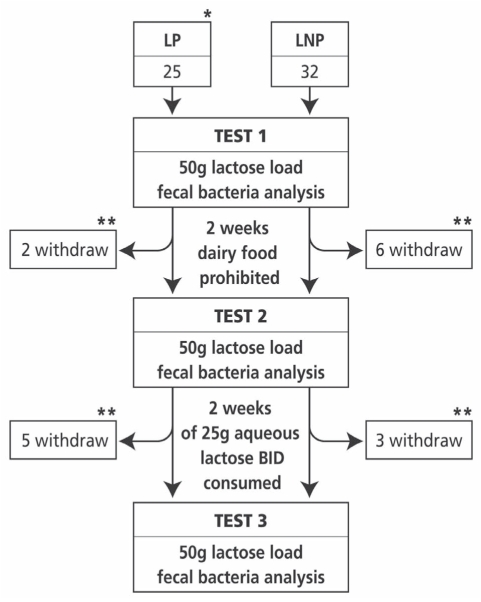
Qualified fasting participants presented to the laboratory between 08:00 and 10:00. Smoking and excessive activity were prohibited in preparation for breath hydrogen (BH2) testing. Subjects visited the laboratory on three occasions (Figure 1). The cross-sectional part of the study evaluated DF intake in both LNP and LP individuals based on a three-day recall questionnaire – the results of which are published elsewhere (15). The results of the primary objective (ie, whether lactose ingestion affects colonic bacteria differently in LNP and LP subjects) are reported in the present article. Blood was drawn at the first visit to determine genetic lactase status. All other data pertains to visits 2 (baseline) and 3 (follow-up).
Figure 1).
Diagram depicting the flow of recruited participants and their genetic/phenotypic classification. The number of participants at each phase is shown with a summary of the reasons for dropping out of the study. *Lactase nonpersistent (LNP) and lactase persistent (LP) status was defined by genetic analysis. **The reasons for dropping out were as follows: time constraints (seven LP, two LNP subjects); personal (one LNP subject); tests too symptomatic (four LNP subjects) and could not get accustomed to taking regular lactose after test 2 (two LNP subjects). BID Twice daily
To eliminate possible bacterial adaptation incurred with regular daily DF consumption, participants were asked to withhold consuming DFs for the duration of the study, except for a maximum of two ounces of cream (equivalent to 3 g of lactose) per day that could be used for coffee. This amount was previously shown (16) not to lead to clinical adaptation to lactose over a three-week period, with adaptation believed to be eliminated within the first week after cessation of DFs (5).
At baseline, participants underwent a lactose challenge test and were measured for both BH2 and symptoms over a 4.5 h period (see details below). In addition, they provided approximately 10 g of stool in a sterile container during the visit.
After the baseline visit, participants were given 25 g of lactose powder (McKesson, Canada) dissolved in water (addition of lemon juice was allowed) and instructed to consume the solution twice a day for the ensuing 14 days. Lactose was started at lower doses and was to be increased to the final dose over four days. Dose escalation was achieved by starting with 12.5 g per day for one day, 12.5 g twice a day for two days then 25 g plus 12.5 g morning and evening, respectively. On the fifth day, subjects consumed 25 g of lactose twice a day. Subjects who were unable to return for the follow-up test 15 days later were provided with additional lactose for a mean (±SD) of 16±3 days (median 15 days [range 12 to 24 days]) to sustain intake until testing. Eight participants withdrew after test 1 and a further eight withdrew after test 2 (Figure 1). All remaining participants claimed to have taken the assigned amounts of lactose. Unfortunately, the return of empty containers was inadvertently inappropriately recorded. On completion of this period, stool and lactose challenge tests were repeated.
Lactase genetic tests
Blood drawn during the first phase of the study was used to determine the predominant genetic polymorphism in the lactase promoter region that was analyzed. The polymorphism (C/T-13910) associates completely with the promoter region of the LP/LNP gene in the majority of European populations (17,18). DNA was prepared using a commercially available DNA isolation kit (Gentra Systems, USA). A real-time polymerase chain reaction assay based on fluorescence resonance energy transfer (19,20) was used. The LightCycler DNA Master Hybridization Probes kit (Roche Diagnostics, Germany) was used for analysis of the C/T genetic polymorphism. The C/C genotype of this polymorphism is associated with LNP individuals, the T/T genotype is associated with LP individuals and the heterozygote C/T is also considered to be LP, but with a reduced measured level of intestinal lactase (17). As such, the cohort was classified into two phenotypes: lactose digester (TT and CT) and maldigester (CC).
BH2 test
The BH2 test is the only clinical test that physiologically reflects metabolic and/or bacterial changes to dietary interventions and assesses colonic adaptation (5,21). It was included in the present study primarily as a measure of the adaptation process. Clinically, colonic adaptation includes improved symptoms, a measurable decrease in BH2 and an increase in fecal beta (β)-galactosidase (see below) on rechallenge after continued regular intake of lactose (5). Generally, the changes in BH2 following intervention are statistically significant.
Briefly, BH2 was measured in parts per million (ppm) using a validated hand-held hydrogen chemical sensor (EC60 gastrolyzer, Bedfont Scientific Ltd, United Kingdom) (22,23). Following a baseline measurement, subjects ingested 50 g of lactose mixed in water. BH2 was then remeasured at 15 min, 30 min, 45 min, 60 min, 75 min and 90 min intervals following ingestion, and every 30 min thereafter for a total of 270 min (4.5 h). The baseline value was subtracted from readings recorded at each subsequent time interval. In general, an acceptable baseline value is 20 ppm or lower. A definite positive value is defined as more than 20 ppm above baseline at any time point (21). Results of each BH2 measurement were summed to obtain a value for total breath hydrogen (TBH2).
Total symptom score
Symptoms were recorded on a four-point Likert scale at baseline and at 30 min intervals following ingestion of 50 g of lactose for 240 min. Bloating, gas and cramps were assigned a score of 0 if there were no symptoms, 1 for mild symptoms, 2 for moderate symptoms and 3 for severe symptoms. Diarrhea was scored as 0 (none) or 1 (present). The total symptom score (TSS) was obtained by summing all scores at each time point; the minimum possible score was 0, with a maximum possible score of 90 ([9×3×3]+9×1).
Stool bacterial counts
Participants encountered no problems with successfully providing stool samples during each visit. Samples were coded, placed on ice and were shipped to the microbiology laboratory (McGill University, Macdonald site, Montreal, Quebec) within 2 h. The samples were analyzed on the same day. No samples were sent on weekends.
A slightly modified method based on a previous report (24) on fecal bacteria testing was used. For quantification of total bifidobacteria, total lactobacilli, total anaerobes (bifidobacteria, lactobacilli, bacteroides and clostridia) and total enterobacteria, the fresh fecal samples (corrected to 100 g) were introduced into an anaerobic jar (BBL GasPak, Becton Dickinson, USA) containing a gas mixture. A 1.0 g sample of feces was removed and homogenized in 4 mL of prepared brain heart infusion broth supplemented with yeast extract and 5-cysteine hydrochloride. A serial dilution was made and subsequently plated onto Bifidobacterium (Beerens), Lactobacillus (Rogosa) agar, and Enterobacteriaceae (MacConkey) plates. The counts (colony forming units [CFU]) of total culturable anerobes were enumerated on reinforced Clostridial medium agar (Fisher Scientific, USA). The plates were incubated anerobically for two to three days at 37°C, with the exception of the MacConkey plates, which were incubated aerobically overnight at 37°C. All bacterial counts are reported as CFU/g of stool. The primary changes of interest were focused on bifidobacteria and lactobacilli because they are known to proliferate in vitro with regular lactose challenge; however, all four bacterial strains were analyzed and compared.
Stool enzyme activity
The bacterial lactase enzymes β-D-galactosidase (β-gal) and N-acetyl-β-D-glucosaminidase (NAG) were assessed as measures of stool enzyme activity. β-gal is a ubiquitous bacterial enzyme analogous to mammalian intestinal lactase, which increases with bacterial colonic adaptation (16,25). Measurement of fecal β-gal was performed using the O-nitrophenyl-β-D-galactopyranoside (ONPG) method (26). Briefly, 20 μL of stool in buffer was added to 480 μg of ONPG in sodium phosphate buffer (pH 7.0). The reaction was allowed to proceed at 45°C for 10 min. Sodium carbonate (1 M) was added to stop the reaction. Optical density at 420 nm was subsequently read and β-gal activity was reported as units/g of stool.
NAG is also a ubiquitous bacterial enzyme involved in mucus digestion and may exert bactericidal effects (27–29). Although NAG has not been previously evaluated in models of colonic adaptation, it is hypothesized that it may be a marker of bacterial response to lactose because of its role in mucus digestion. NAG was analyzed using a buffered sample of 0.1 mL of stool added to 1 mL of substrate. After incubation at 37°C for 30 min with 0.5 mL of sodium carbonate buffer to stop the reaction, the optical density was read at 405 nm (26), with NAG activity reported as units/mg of stool.
Statistical analysis
The demographics of the population are described using means and SDs for continuous variables and percentages with standard errors for categorical variables. Normally distributed data were obtained by log10 transforming the bacterial CFU/g of stool and enzyme activity.
The primary objective was to determine whether lactose consumption increases bifidobacteria and lactobacilli counts differentially in LNP subjects versus LP subjects. The respective change in scores (ie, follow-up minus baseline) were calculated and reported as mean change with 95% CIs and P values. The change in scores between follow-up and baseline for TBH2, TSS, log10(CFU/g stool) for Enterobacteriaceae and total anaerobic bacteria, log10(g/stool) β-gal and log10(units/mg stool) NAG are also described.
P<0.05 was considered to be statistically significant and P<0.1 was considered to be a trend. Because of the nature of the experiment, the analysis was limited to subjects who were measured at both baseline and follow-up. For missing BH2 values and TSS at any particular time point, data were assigned using the following algorithm (which should bias the results toward the null hypothesis): if data were available for time points immediately before and following the time point with missing data, the average of the two was used. If there were no data before or following the missing data time point (eg, baseline missing or 270 min value missing), the average of the scores for that time point from the other visits (for this sccenario, data from the visit four weeks before baseline were included) were used. For missing data regarding bacterial counts and enzyme analysis, the average score of the other visits was used. A sensitivity analysis for subjects with no missing data was also performed.
Sample size
Based on a previous study using 15 LNP and 15 LP subjects (30), the two exposure groups (LNP and LP) were expected to be of near equal size. Sample sizes were calculated setting alpha at 0.05 and power at 0.8.
The calculated SD of change for lactobacilli over time in the study by Ito and Kimura (31) was 0.79 (log10 units) using P=0.05 (the reported P value was less than 0.05; the calculation used in the present study represents a conservative approach when only pre and post SDs are provided, as in the Ito and Kimura study). A similar SD of change was assumed for the other primary outcome bifidobacteria count. There are no data to determine what a clinically relevant difference in bacterial counts should be. Therefore, a three-fold difference (0.48 log10 units) was arbitrarily chosen to be clinically relevant. It was estimated that 45 participants per group would be required to detect a statistically significant difference.
RESULTS
Participants were classified as LP or LNP based on the genetic lactase test. Although the study had initially intended to recruit 46 subjects per group, only a total of 57 were recruited because of logistical reasons. From the initial 57 participants, 16 did not complete the study (seven LNP subjects and nine LP subjects), leaving 41 with data for baseline and follow-up visits. The demographic characteristics of subjects who dropped out and participated are shown in Table 1. The distribution of participants and reasons for leaving the study are shown in Figure 1.
TABLE 1.
Demographics
|
Participating (genotype*) |
|||
|---|---|---|---|
| Dropouts (n=16) | LNP (CC) (n=23) | LP (CT/TT) (n=18) | |
| Male sex, n (%) | 8 (50) | 7 (30) | 8 (44) |
| Race, n (%) | |||
| African | 3 (19) | 5 (22) | 2 (11) |
| Asian | 1 (6) | 8 (35) | 0 (0) |
| Caucasian | 12 (75) | 10 (43) | 16 (89) |
| Genetics, n (%) | |||
| CC | 7 (44) | 23 (100) | 0 (0) |
| CT | 6 (38) | – | 10 (56) |
| TT | 3 (19) | – | 8 (44) |
| Age† | 29.1±8.1 | 32.1±8.7 | 26.3±6.5 |
| Body mass index†, kg/m2 | 22.7±3.1 | 23.8±4.3 | 22.8±2.4 |
| Daily lactose intake at study entry‡, g/day | 16.5 (0–60.1) | 11.0 (4.5–36.9) | 9.5 (0.4–26.7) |
Categorical variables are shown as a per cent of total within that group.
Refers to the polymorphism at position 13910 of the lactase promotor region;
Data for continuous measures are presented as mean ± SD;
Data presented as median (interquartile range).
Daily lactose intake is based on responses to a three-day recall questionnaire. LNP Lactase nonpersistent; LP Lactase persistent
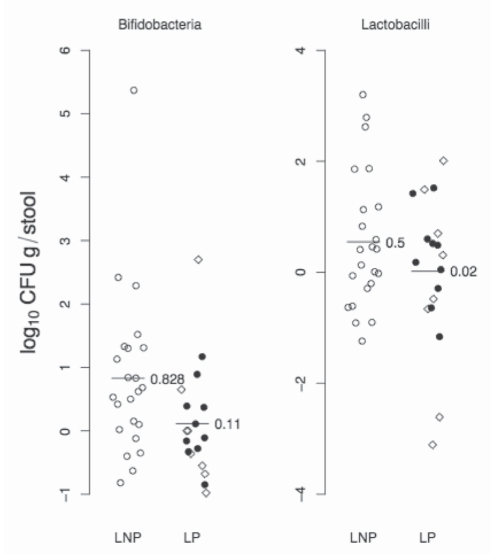
A scatter plot showing the mean change in primary outcomes (ie, bifidobacteria and lactobacilli counts) between tests 3 and 2 stratified according to LNP status is shown in Figure 2. The difference in the mean change in bifidobacteria counts (log10 CFU/g stool) for LNP subjects (genotype CC) versus LP subjects (genotypes TT and CT) was 0.72 (95% CI 0.03 to 1.04; P=0.04).
Figure 2).
Scatter plot showing the change in the primary outcomes: bifidobacteria and lactobacilli counts (log10 [colony forming units (CFU)/g stool]) between test 3 and test 2. Lactase nonpersistent (LNP) subjects are represented by open circles (genotype CC). Lactase persistent (LP) subjects are represented by solid circles (genotype TT) and open diamonds (genotype CT). In addition, the mean change for the respective LNP/LP groups is shown with a line and the exact value. No comparisons were made with test 1 (baseline)
As an exploratory posthoc analysis, the change in bifidobacteria counts were dichotomized to being positive based on thresholds of a 1log unit or greater change, and a 0.5log unit change. Of the LNP subjects, eight of 23 (35%) had a 1log unit change from baseline for bifidobacteria versus two of 18 (11%) for LP subjects (χ2=1.92, P=0.14 [Fisher’s exact test]). When a threshold of 0.5log units or greater was used, there were 14 of 23 (61%) LNP subjects with positive responses and four of 18 (22%) LP subjects (χ2=4.65, P=0.03 [Fisher’s exact test]).
In a similar analysis for lactobacilli, the difference in the mean change was 0.53log10 CFU/g stool (95%CI −0.30 to 1.36; P=0.20). At a threshold of 1log unit or greater change from baseline, there were seven of 23 (30%) positive responses in LNP subjects and four of 18 (22%) positive responses in LP subjects (χ2=0.05, P=0.73 [Fisher’s exact test]); the corresponding figures for a threshold of 0.5log units or greater change from baseline were nine of 23 (39%) for LNP subjects, and seven of 18 (39%) for LP subjects (χ2=0.09, P=1.0 [Fisher’s exact test]).
On posthoc analysis, a comparison of bifidobacteria and lactobacilli counts between LNP and LP individuals in tests 3 were 10.3log units versus 9.7log units (P=0.14), and 11.6log units versus 11.6log units (P=0.77), respectively.
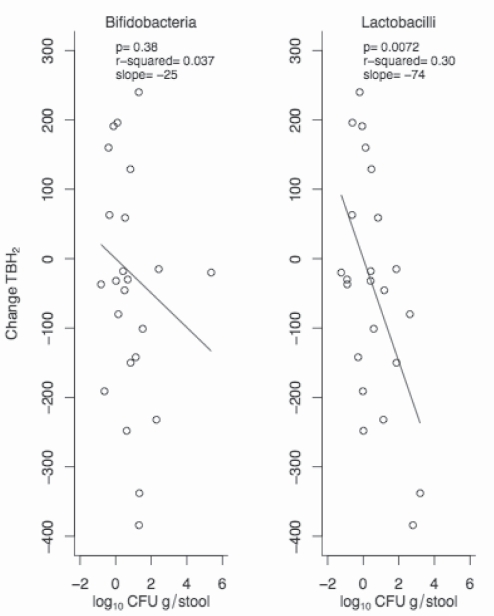
In a previous cross-sectional study (15), a very strong agreement (96.7% sensitivity and 92.6% specificity) between genetic classification and the results of the lactose challenge BH2 tests was found. To explore the suggestion from in vitro data that lactic acid-producing bacteria (bifidobacteria and lactobacilli) may be responsible for reduced BH2, the changes in TBH2 were plotted against the changes in bifidobacteria and lactobacilli in a posthoc analysis using only LNP subjects (Figure 3). The relationship was much stronger for lactobacilli than for bifidobacteria. LP subjects were not included because TBH2 was essentially zero at baseline and could not decrease further.
Figure 3).
Scatter plot showing the change in total breath hydrogen (TBH2) between test 3 and test 2, with the change in bacterial counts (colony forming units [CFU]) for the primary outcomes for bifidobacteria (left panel) and lactobacilli (right panel) in lactase nonpersistent subjects only (lactase persistent subjects have almost no TBH2 to begin with). The P value, r2 and slope are indicated in each panel. No comparisons were made with test 1 (baseline)
To explore whether there were differences in adaptability between LP heterozygotes (CT) and LP homozygotes (TT), the LP group was further investigated with additional posthoc analyses. There were no statistically significant differences in bifidobacteria counts between the CT and TT genotypes.
Table 2 shows the mean bacterial counts and mean fecal enzyme activities for LNP and LP subjects at baseline and follow-up. Changes between these two periods were evaluated in each individual with respect to the secondary comparisons of bacterial counts for Enterobacteriaceae and total anaerobic bacteria, β-gal and NAG stool enzymes, TBH2 and TSS (LNP versus LP). None of the comparisons achieved statistical significance, although there were the expected reductions in TBH2 and TSS after lactose challenge (TBH2: −57.9 ppm [95% CI −133.4 to 17.7]; P=0.13 and TSS: −4.6 units [95% CI −10.0 to 0.8; P=0.11]).
TABLE 2.
Counts and bacterial fecal enzymes for both lactase persistent (LP) and lactase nonpersistent (LNP) groups at baseline (test 2) and after 2 weeks of ingesting 25 g of lactose twice a day (test 3)
|
LP |
LNP |
|||
|---|---|---|---|---|
| Baseline | 2-week follow-up | Baseline | 2-week follow-up | |
| Bifidobacteria | 9.6±0.7 | 9.7±0.8 | 9.4±0.7 | 10.2±1.3 |
| Lactobacilli | 11.6±1.0 | 11.6±0.9 | 11.0±1.2 | 11.6±1.1 |
| Total anaerobes | 12.2±1.1 | 12.2±0.9 | 11.5±1.2 | 11.9±0.9 |
| Enterobacteriaceae | 7.0±1.1 | 6.7±1.1 | 6.8±1.2 | 6.9±1.1 |
| Beta-galactosidase | 2.0±0.3 | 1.9±0.3 | 1.8±0.4 | 1.9±0.4 |
| NAG | 1.9±0.2 | 1.8±0.2 | 1.8±0.2 | 1.8±0.2 |
Data presented as mean ± SD. Results are expressed as log10 colony forming units/g stool for all bacteria, log10units/g of stool for beta-galactosidase and log10 units/mg stool for N-acetyl-beta-D-glucosaminidase (NAG)
The results of the sensitivity analysis for the primary outcome using only complete case scenario data (n=25) found that the differences in the change scores for bifidobacteria was 0.72 (95% CI −01.4 to 1.58; P=0.10), and for lactobacilli was 0.08 (95% CI −1.42 to 1.59; P=0.91). These results are qualitatively similar to the results of the primary analysis.
DISCUSSION
The present pilot study supports the notion that prebiotic effects of lactose in humans occur predominantly in LNP rather than in LP individuals, even with high-dose lactose intake. While reduction in TBH2 following the adaptive period did not achieve the expected results in LNP subjects, a significant change was demonstrated in bifidobacteria in LNP compared with LP participants. The numerically reduced TBH2 observed in LNP was nevertheless closely linked with lactobacilli.
The concept of a prebiotic, nonabsorbed carbohydrate that selectively alters lower intestinal bacterial flora in a beneficial way to the host was first introduced by Gibson and Roberfroid (32), and altered to include detection of lactic acid bacteria in stool (33,34). In lactose-intolerant individuals, continued consumption of DFs can lead to adaptation (3,5–8,35–40), but not because of intestinal lactase induction (41) – both bifidobacteria and lactobacilli could be responsible (5,42,43). Also, lactose was shown to induce bifidobacteria proliferation in an in vitro model of the colon (44). Furthermore, lactose forms the backbone of some recognized prebiotics (45,46).
In the only other in vivo human study to evaluate fecal flora, Ito and Kimura (31) showed increased lactobacilli and bifidobacteria after a short exposure to lactose in lactose maldigesters. This study also found reduced bacteroides and Clostridium species. In our study, the mean change in bifidobacteria after a median of 15 days of lactose consumption was a 0.83 log10 units and approximately two-thirds of the subjects in the LNP group achieved 0.5log (three-fold) or greater change. It should be noted that this magnitude of change for bifidobacteria is similar to those reported with oligofructose (a recognized prebiotic) (47,48). Interestingly, reduction in BH2 was inversely correlated with lactobacilli in the present study. The effect on bifidobacteria in the LNP group was approximately seven-fold greater than that for the LP group, which showed minimal or virtually no change. Taken together, the in vitro studies and human observations provide support for a prebiotic effect of lactose, as redefined more recently (33,34), and a differential bifidogenic effect on LNP compared with LP phenotypes.
At the lactose doses we used (25 g twice a day), the expected spillage of 2 g/day to 4 g/day into the colon of LP individuals (14) may not have affected microflora in two weeks. We also examined whether C/T LP individuals fared any better with microbial effects and found none. These individuals have a 50% reduction in intestinal lactase (17) and might have been expected to spill more lactose than T/T LP individuals.
Several weaknesses in the present study need to be addressed. First, our results should be interpreted in light of the unanticipated reduction in study power to detect a three-fold change in bacterial counts. Nevertheless, the changes in bifidobacteria counts (from follow-up compared with baseline within groups) between LNP and LP subjects showed a greater difference than this. Therefore, the reason why outcome was not more dramatic may be related to the failure to induce complete adaptation in LNP participants. Because TBH2 was used as a control evaluating adaptation – a frequently observed phenomenon (5–8,31,35–40) – the question becomes why was this not achieved. Despite an attempt to use standard methodology in ascertaining consumption of lactose (ie, returned empty containers), we failed to properly record returns. Although historically suggested, we remain uncertain whether all participants explicitly followed the instructions. Second, we chose a period of 15 days of lactose consumption because it was previously shown that 16 days was adequate time for full adaptation to occur (5). However, this still may not have been enough time given the above outlined weaknesses. A longer period of lactose consumption, even with reduced adherence might have achieved the expected statistical significance in the reduction of BH2. Full adaptation may have enhanced bacterial effects. Finally, we recognize that due to the methods used to assess stool samples, deterioration in colony counts likely occurred (49); however, this was unavoidable, but due to the blinded nature of the study, the order and genetic status of the participants was unknown to those analyzing stool. As such, similar inaccuracies could have been incurred equally by both groups.
CONCLUSIONS
Despite limitations, the present study lends support to the concept that lactose exerts selective colonic prebiotic effects on LNP versus LP subjects. A bifidogenic effect of lactose is more evident in LNP individuals. Reduced TBH2 after lactose intervention is closely linked with metabolism by lactobacilli. Future studies should evaluate dose-time effects in these two populations and the specific species of bacteria affected. The contribution of host/intestinal bacterial interactions to disease is becoming more evident in some diseases (50,51). The possible modifying influence on the pathogenesis of lactose/lactase interactions should be taken into consideration.
Footnotes
FUNDING: This study was supported by a Grant in Aid from the Danone Institute of Canada, Quebec Branch. None of the authors have any conflicts of interests to declare.
COMPETING INTERESTS: None of the authors have financial associations with the granting agency.
REFERENCES
- 1.Maiuri L, Raia V, Potter J, et al. Mosaic pattern of lactase expression by villous enterocytes in human adult-type hypolactasia. Gastroenterology. 1991;100:359–69. doi: 10.1016/0016-5085(91)90203-w. [DOI] [PubMed] [Google Scholar]
- 2.Rossi M, Maiuri L, Fusco MI, et al. Lactase persistence versus decline in human adults: Multifactorial events are involved in down-regulation after weaning. Gastroenterology. 1997;112:1506–14. doi: 10.1016/s0016-5085(97)70031-3. [DOI] [PubMed] [Google Scholar]
- 3.Scrimshaw NS, Murray EB. The acceptability of milk and milk products in populations with high prevalence of lactose intolerance. Am J Clin Nutr. 1988;48S:1079. doi: 10.1093/ajcn/48.4.1142. (Abst) [DOI] [PubMed] [Google Scholar]
- 4.Shrier I, Szilagyi A, Correa JA. Impact of lactose containing foods and the genetics of lactase on diseases: An analytical review of population data. Nutr Cancer. 2008;60:292–300. doi: 10.1080/01635580701745301. [DOI] [PubMed] [Google Scholar]
- 5.Hertzler SR, Savaiano DA. Colonic adaptation to daily lactose feeding in lactose maldigesters reduces lactose intolerance. Am J Clin Nutr. 1996;64:232–36. doi: 10.1093/ajcn/64.2.232. [DOI] [PubMed] [Google Scholar]
- 6.Villar J, Kestler E, Castillo P, Juarez A, Menendez R, Solomons NW. Improved lactose digestion during pregnancy: A case of physiologic adaptation? Obstet Gynecol. 1988;71:697–700. [PubMed] [Google Scholar]
- 7.Szilagyi A, Salomon R, Martin M, Fokeeff K, Seidman E. Lactose handling by women with lactose malabsorption is improved during pregnancy. Clin Invest Med. 1996;19:416–26. [PubMed] [Google Scholar]
- 8.Pribila BA, Hertzler SR, Martin BR, Weaver CM, Savaiano DA. Improved lactose digestion and intolerance among African-American adolescent girls fed a dairy-rich diet. J Am Diet Assoc. 2000;100:524–8. doi: 10.1016/S0002-8223(00)00162-0. [DOI] [PubMed] [Google Scholar]
- 9.Uribe M, Marquez MA, Garcia-Ramos G, et al. Treatment of chronic portal-systemic encephalopathy with lactose in lactase-deficient patients. Dig Dis Sc. 1980;25:924–8. doi: 10.1007/BF01308043. [DOI] [PubMed] [Google Scholar]
- 10.Uribe-Esquivel M, Maran S, Poo JL, Munoz RM. In vitro and in vivo lactose and lactulose effects on colonic fermentation and portal-systemic encephalopathy parameters. Scand J Gastroenterol. 1997;32(Suppl 222):49–52. doi: 10.1080/00365521.1997.11720718. [DOI] [PubMed] [Google Scholar]
- 11.Szilagyi A, Nathwani U, Vinokuroff C, Correa JA, Shrier I. The effect of lactose maldigestion on the relationship between dairy food intake and colorectal cancer: A systematic review. Nutr Cancer. 2006;55:141–50. doi: 10.1207/s15327914nc5502_4. [DOI] [PubMed] [Google Scholar]
- 12.Oku T, Nakamura S, Ichinose M. Maximum permissive dose of lactose and lactitol for transitory diarrhea, and utilizable capacity for lactose in Japanese female adults. J Nutr Sci Vitaminol. 2005;51:51–7. doi: 10.3177/jnsv.51.51. [DOI] [PubMed] [Google Scholar]
- 13.Hertzler SR, Huynh B-CL, Savaiano DA. How much lactose is low lactose? J Am Diet Assoc. 1996;96:243–6. doi: 10.1016/S0002-8223(96)00074-0. [DOI] [PubMed] [Google Scholar]
- 14.Bond JH, Levitt MD. Quantitative measurement of lactose absorption. Gastroenterology. 1976;70:1058–62. [PubMed] [Google Scholar]
- 15.Szilagyi A, Shrier I, Chong G, et al. Lack of effect of lactose digestion status on quantitative fecal microflora. Can J Gastroenterol. 2009;23:753–9. doi: 10.1155/2009/693794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szilagyi A, Rivard J, Fokeeff K. Improved parameters of lactose maldigestion using lactulose. Dig Dis Sci. 2001;46:1509–19. doi: 10.1023/a:1010652223705. [DOI] [PubMed] [Google Scholar]
- 17.Enattah NS, Kuokkanen M, Forsblom C, et al. Correlation of intestinal disaccharidase activities with the C/T(13910) variant and age. World J Gastroenterol. 2007;13:3508–12. doi: 10.3748/wjg.v13.i25.3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuokkanen M, Enattah NS, Oksanen A, Savilahti E, Orpana A, Järvelä I. Transcriptional regulation of the lactase-phlorizin hydrolase gene by polymorphisms associated with adult-type hypolactasia. Gut. 2003;52:647–52. doi: 10.1136/gut.52.5.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nilsson TK, Johansson CA. A novel method for diagnosis of adult hypolactasia by genotyping of the −13910 C/T polymorphism with pyrosequencing technology. Scand J Gastroenterol. 2004;39:287–90. doi: 10.1080/00365520310008304. [DOI] [PubMed] [Google Scholar]
- 20.Chao CK, Sibley E. PCR-RFLP genotyping assay for a lactase persistence polymorphism upstream of the lactase-phlorizin hydrolase gene. Genet Test. 2004;8:190–3. doi: 10.1089/gte.2004.8.190. [DOI] [PubMed] [Google Scholar]
- 21.Romagnuolo J, Schiller D, Bailey RJ. Using breath tests wisely in a gastroenterology practice: An evidence-based review of indications and pitfalls in interpretation. Am J Gastroenterol. 2002;97:1113–26. doi: 10.1111/j.1572-0241.2002.05664.x. [DOI] [PubMed] [Google Scholar]
- 22.Metz G, Gassull MA, Leeds AR, Blendis LM, Jenkins DJ. A simple method of measuring breath hydrogen in carbohydrate malabsorption by end-expiratory sampling. Clin Sci Mol Med. 1976;50:237–40. doi: 10.1042/cs0500237. [DOI] [PubMed] [Google Scholar]
- 23.Van der Klei-van Moorsel JM, Douwes AC, van Oeveren JP. New principle for estimation of hydrogen in expired air. Eur J Pediatr. 1984;141:221–4. doi: 10.1007/BF00572764. [DOI] [PubMed] [Google Scholar]
- 24.Jung SJ, Houde R, Baurhoo B, Zhao X, Lee BH. Effects of galacto-oligosaccharides and a bifidobacteria lactis-based probiotic strain on the growth performance and fecal microflora of broiler chickens. Poultry Sci. 2007;87:1694–9. doi: 10.3382/ps.2007-00489. [DOI] [PubMed] [Google Scholar]
- 25.Flourie B, Briet F, Florent C, Pellier P, Maurel M, Rambaud JC. Can diarrhea induced by lactulose be reduced by prolonged ingestion of lactulose? Am J Clin Nutr. 1993;58:369–75. doi: 10.1093/ajcn/58.3.369. [DOI] [PubMed] [Google Scholar]
- 26.Yuen CT, Price RG, Chattagoon L, Richardson AC, Praill PF. Colorimetric assays for N-acetyl b-D glucosaminidase and b-D-galactosidase in human urine using newly-developed-nitrosyl substrates. Clin Chim Acta. 1982;124:195–204. doi: 10.1016/0009-8981(82)90387-4. [DOI] [PubMed] [Google Scholar]
- 27.Hussain AM, Daniel RCW, Frost AJ. The bactericidal effect of N-acetyl-β-D-glucosaminidase on bacteria. Vet Microbiol. 1992;32:75–80. doi: 10.1016/0378-1135(92)90008-h. [DOI] [PubMed] [Google Scholar]
- 28.Meslin JC, Andrieux C, Sakata T, et al. Effects of galacto-oligosaccharide and bacterial status on mucin distribution in mucosa and on large intestine fermentation in rats. Br J Nutr. 1993;69:903–12. doi: 10.1079/bjn19930090. [DOI] [PubMed] [Google Scholar]
- 29.Mayer C, Vocadio DJ, Mah M, et al. Characterization of a β-N-acetylhexosaminidase/β-glucosidase from Cellulomonas fimi. FEBS J. 2006;273:2929–41. doi: 10.1111/j.1742-4658.2006.05308.x. [DOI] [PubMed] [Google Scholar]
- 30.Szilagyi A, Malolepszy P, Hamard E, et al. Comparison of a real-time PCR Assay for lactase genetic polymorphism with standard indirect tests for lactose maldigestion. Clin Gastroenterol Hepatol. 2007;5:192–6. doi: 10.1016/j.cgh.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 31.Ito M, Kimura M. Influence of lactose on faecal microflora in lactose maldigesters. Microb Ecol Health Dis. 1993;6:73–6. [Google Scholar]
- 32.Gibson GR, Roberfroid MB. Dietary modulation of the human colonic microbiota: Introducing the concept of prebiotics. J Nutr. 1995;125:1401–12. doi: 10.1093/jn/125.6.1401. [DOI] [PubMed] [Google Scholar]
- 33.Gibson GR, Probert HM, Loo JV, Rastall RA, Roberfroid MB. Dietary modulation of the human colonic microbiota: Updating the concept of prebiotics. Nutr Res Rev. 2004;17:259–75. doi: 10.1079/NRR200479. [DOI] [PubMed] [Google Scholar]
- 34.Roberfroid M. Prebiotics: The concept revisited. J Nutr. 2007;137:830S–837S. doi: 10.1093/jn/137.3.830S. [DOI] [PubMed] [Google Scholar]
- 35.Habte D, Sterky G, Hjalmarsson B. Lactose malabsorption in Ethiopian children. Acta Paediatr Scand. 1973;62:649–54. doi: 10.1111/j.1651-2227.1973.tb17080.x. [DOI] [PubMed] [Google Scholar]
- 36.Sadre M, Karbasi K. Lactose intolerance in Iran. Am J Clin Nutr. 1979;32:1948–54. doi: 10.1093/ajcn/32.9.1948. [DOI] [PubMed] [Google Scholar]
- 37.Vesa TH, Korpela RA, Sahi T. Tolerance to small amounts of lactose in lactose maldigesters. Am J Clin Nutr. 1996;64:197–201. doi: 10.1093/ajcn/64.2.197. [DOI] [PubMed] [Google Scholar]
- 38.Suarez FL, Savaiano DA, Levitt MD. A comparison of symptoms after the consumption of milk or lactose-hydrolyzed milk by people with self-reported severe lactose intolerance. N Engl J Med. 1995;333:1–4. doi: 10.1056/NEJM199507063330101. [DOI] [PubMed] [Google Scholar]
- 39.Suarez FL, Savaiano DA, Arbisi P, Levitt MD. Tolerance to the daily ingestion of two cups of milk by individuals claiming lactose intolerance. Am J Clin Nutr. 1997;65:1502–6. doi: 10.1093/ajcn/65.5.1502. [DOI] [PubMed] [Google Scholar]
- 40.Szilagyi A, Malolepszy P, Yesovitch S, Nathwani U, Vinokuroff C, Cohen A. Inverse dose effect of pretest dietary lactose intake on hydrogen results and symptoms in lactase nonpersistent subjects. Dig Dis Sci. 2005;50:2178–82. doi: 10.1007/s10620-005-3028-4. [DOI] [PubMed] [Google Scholar]
- 41.Gilat T, Russo S, Gelman-Malachi E, Aldor TA. Lactase in man: A non adaptable enzyme. Gastroenterology. 1972;62:1125. [PubMed] [Google Scholar]
- 42.Jiang T, Savaiano DA. In vitro lactose fermentation by human colonic bacteria is modified by Lactobacillus acidophilus supplementation. J Nutr. 1997;1237:1489–95. doi: 10.1093/jn/127.8.1489. [DOI] [PubMed] [Google Scholar]
- 43.Jiang T, Savaiano DA. Modification of colonic fermentation by bifidobacteria and pH in vitro. Impact on lactose metabolism, short-chain fatty acid, and lactate production. Dig Dis Sci. 1997;42:2370–7. doi: 10.1023/a:1018895524114. [DOI] [PubMed] [Google Scholar]
- 44.Mäkivuokko HA, Saarinen MT, Ouwehand AC, Rautonen NE. Effects of lactose on colon microbial community structure and function in a four-stage semi-continuous culture system. Biosci Biotechno Biochem. 2006;70:2056–63. doi: 10.1271/bbb.60022. [DOI] [PubMed] [Google Scholar]
- 45.Bouhnik Y, Attar A, Joly FA, Riott M, Dyard F, Flourie B. Lactulose ingestion increases faecal bifidobacterial counts: A randomized double-blind study in healthy humans. Eur J Clin Nutr. 2004;58:462–6. doi: 10.1038/sj.ejcn.1601829. [DOI] [PubMed] [Google Scholar]
- 46.Jung SJ, Lee BH. Production and application of galacto-oligosaccharides from lactose by a recombinant β-galactosidase of Bifidobacterium infantis overproduced by pichia pastoris. Food Sci Biotechnol. 2008;17:514–8. [Google Scholar]
- 47.Gibson GR, Beatty ER, Wang X, Cummings JH. Selective stimulation of bifidobacteria in the human colon by oligofructose and inulin. Gastroenterology. 1995;108:975–82. doi: 10.1016/0016-5085(95)90192-2. [DOI] [PubMed] [Google Scholar]
- 48.Bouhnik Y, Raskine L, Simoneau G, Paineau D, Bornet F. The capacity of short-chain fructo-oligosaccharides to stimulate faecal bifidobacteria: A dose-response relationship study in healthy humans. Nutr J. 2006;5:8. doi: 10.1186/1475-2891-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ott SJ, Musfeldt M, Timmis KN, Hampe J, Wenderoth DF, Schreiber S. In vitro alterations of intestinal bacterial microbiota in fecal samples during storage. Diagn Microbiol Infect Dis. 2004;50:237–45. doi: 10.1016/j.diagmicrobio.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 50.Lakatos PL, Fischer S, Lakatos L, Gal I, Papp J. Current concept on the pathogenesis of inflammatory bowel disease-crosstalk between genetic and microbial factors: Pathogenic bacteria and altered bacterial sensing or changes in mucosal integrity take “toll”. World J Gastroenterol. 2006;28:1829–41. doi: 10.3748/wjg.v12.i12.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Szilagyi A. Risk modification of inflammatory bowel diseases by vitamin D, calcium and lactase/lactose interactions: Arguments based on relationships between IBD and colorectal cancer. In: Urster JO, editor. Research Trends in Nutrition for the Middle Aged and Elderly. New York: Nova Science; 2008. pp. 7–51. [Google Scholar]