Abstract
The TMPRSS2-ERG fusion, present in 50% of prostate cancers, is less common in prostatic intra-epithelial neoplasia (PIN), raising questions about whether TMPRSS2-ERG contributes to disease initiation. We identified the translational start site of a common TMPRSS2-ERG fusion and showed that transgenic TMPRSS2-ERG mice develop PIN, but only in the context of PI3-kinase pathway activation. TMPRSS2-ERG positive human tumors are also enriched for PTEN loss, suggesting cooperation in prostate tumorigenesis.
Recurrent gene fusions involving members of the ETS family of transcription factors occur frequently in human prostate cancer1. The TMPRSS2-ERG fusion gene, generated by an intrachromosomal deletion on chromosome 21 or by reciprocal translocation, is the most common rearrangement, found in ~50% of localized tumors1. The fusion is detected in essentially all the malignant cells within a focus of tumor, supporting a role in disease initiation. However, TMPRSS2-ERG is less commonly found in preneoplastic PIN lesions and, when present, is typically in PIN adjacent to fusion-positive tumor cells2. Potential explanations for the relatively low frequency of TMPRSS2-ERG in PIN versus cancer are: (i) TMPRSS2-ERG is not an initiating event, (ii) PIN may not be a reliable precursor lesion to invasive cancer, or (iii) PIN is molecularly heterogeneous but TMPRSS2-ERG driven PIN progresses rapidly to cancer.
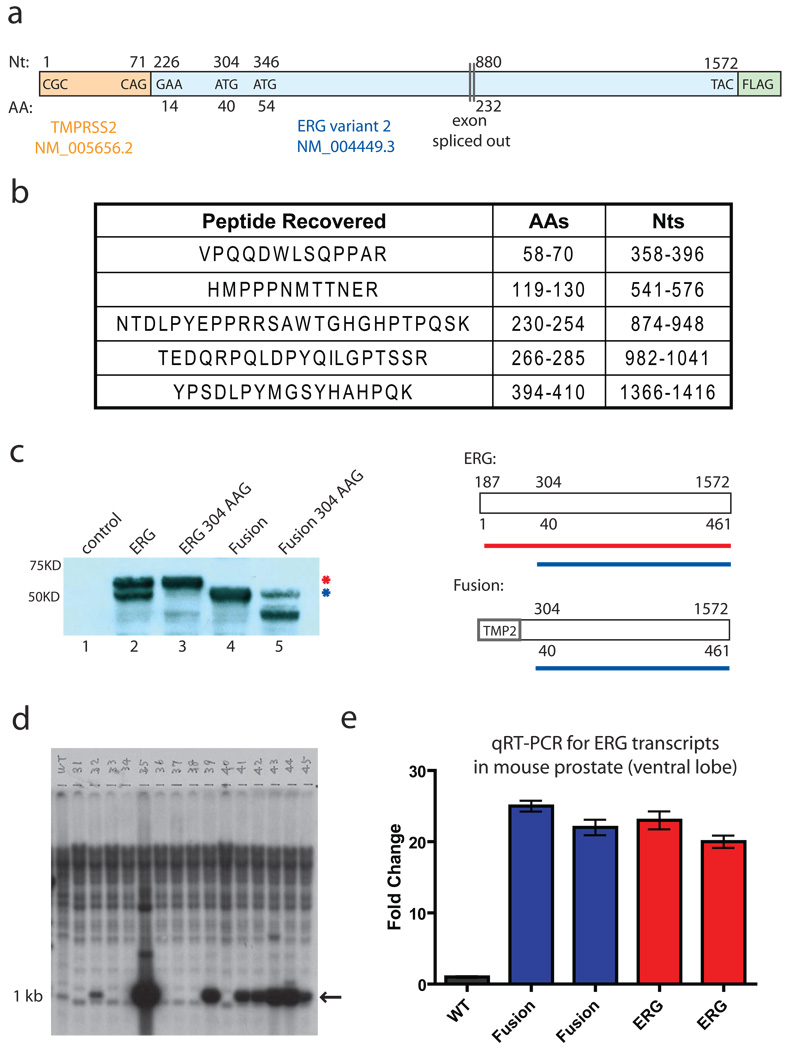
We addressed this question by generating transgenic mice expressing the TMPRSS2-ERG fusion in the prostate. Two ERG isoforms (variants 1 and 2) have been characterized, which differ in the N terminus as well as the presence of a downstream ERG exon due to alternative splicing. We designed our TMPRSS2-ERG construct (Fig. 1a) to re-create the most common translocation breakpoints3 and used the variant 2 ERG isoform based on exon array expression profiling data showing relative abundance of variant 2 in TMPRSS2-ERG positive human tumors4. Due to N-terminal truncation of ERG and fusion to non-coding TMPRSS2 sequence, the initiating methionine for ERG translation is removed, raising questions about the translational start site of TMPRSS2-ERG fusion mRNA. To address this issue, we conducted mass spectrometry analysis of the protein product of our TMPRSS2-ERG fusion (Fig. 1c, lane 4) and recovered five tryptic peptides. The most N-terminal peptide spanned amino acids 58–70 of ERG (nucleotides 358–396), narrowing down the possible start codon to the methionine encoded by nucleotide 304 (Met 40 in full length ERG) or to Met 54 at nucleotide 356 (Fig. 1b). Mutation of the ATG encoding Met 40 to AAG substantially impaired translation of the TMPRSS2-ERG protein (Fig 1c, compare lanes 4 and 5) and, in the context of full length ERG, eliminated expression of the faster migrating ERG isoform observed with wild-type ERG overexpression (Fig. 1c, compare lanes 2 and 3). These data implicate Met 40 as the principle start site for ERG expression in the TMPRSS2-ERG fusion as well as a potential alternative start site for wild-type ERG expression.
Fig. 1.
TMPRSS2-ERG fusion transcript produces a truncated ERG protein product. a, Diagram of TMPRSS2-ERG variant 2 construct. b, Tryptic peptides recovered from mass spectrometry performed on immunoprecipitated, gel-resolved protein from the fusion construct. c, ERG immunoblot of transiently transfected vector control, full length ERG, full length ERG with codon 304 mutation, fusion, or fusion with the codon 304 mutation. A diagram showing the protein products derived from the non-mutated constructs is shown on the right. d, Southern blot showing founder animals derived from blastocyst injections of ARR2PB-TMPRSS2-ERG construct. DNA was cut with BamHI and probed with a BamHI fragment that encodes the first 1050bp of the fusion. e, qRT-PCR using primers for the C-terminus of ERG on ventral prostates from a wildtype mouse, two TMPRSS2-ERG mice, and two full-length ERG transgenic mice. All Ct values were normalized to the Ct values of Nkx3.1 and fold change was calculated relative to the wildtype animal.
To create transgenic mice, we placed the TMPRSS2-ERG fusion construct under the control of the ARR2-Probasin promoter, used in previous models of PIN and prostate cancer5–7. Seventeen transgene-positive founder animals were generated by injection of the ARR2-PB-TMPRSS2-ERG construct into FVB blastocysts (Fig. 1d) and four demonstrated stable germline transmission of the transgene. Two lines (43 and 44) were expanded based on robust transgene expression at levels 20-fold greater than endogenous mouse Erg (Fig. 1e) and comparable to levels observed in an independently derived mouse expressing full length ERG variant 18. Prostates from TMPRSS2-ERG fusion mice from both lines were harvested at 2, 6, 12 and 14 months and compared with littermate controls. We found no histologic evidence of PIN or invasive cancer in 18 TMPRSS2-ERG mice examined (Fig. 2a). Although mild dysplasia was seen in some transgene-positive mice, these changes are commonly observed in wild-type mice and cannot be definitively attributed to transgene expression.
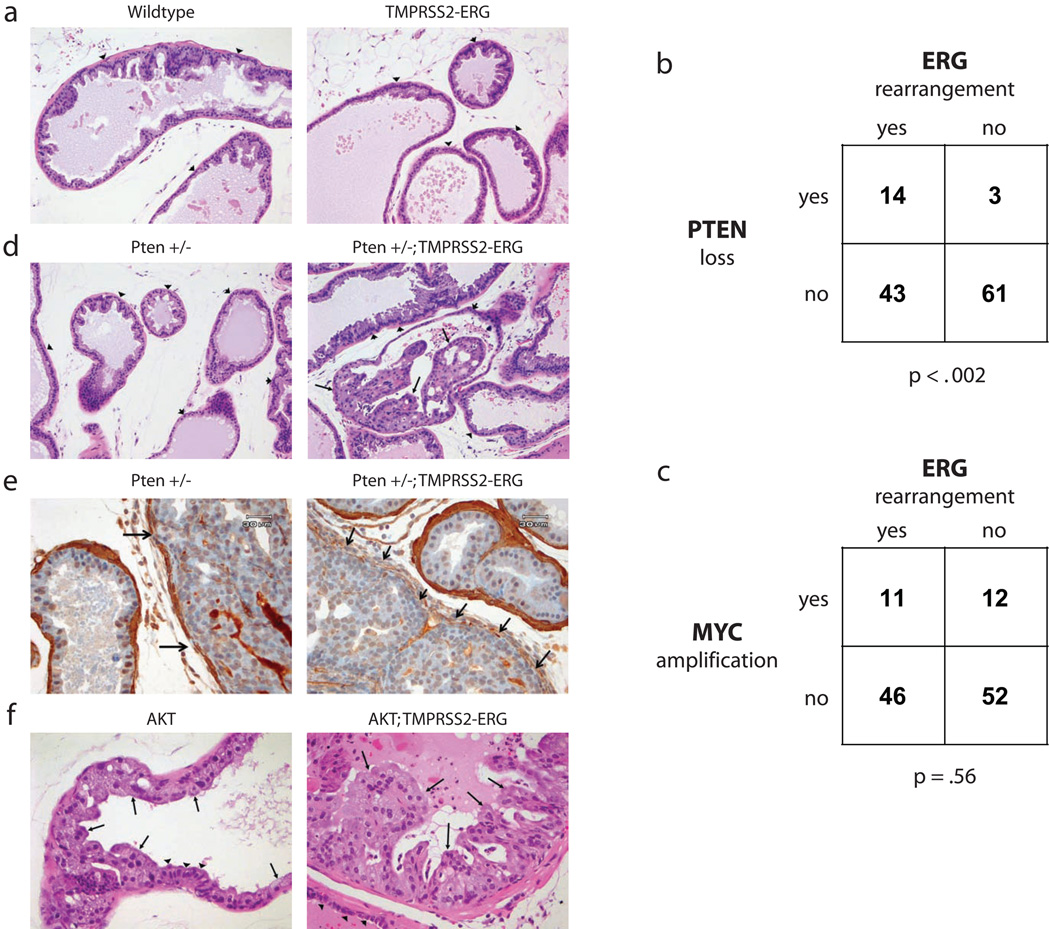
Fig. 2.
TMPRSS2-ERG fusion and PI3K pathway activation are cooperative events in prostate cancer. All pictures show the ventral prostate lobe. a, No PIN is noted in either wild type or TMPRSS2-ERG animals at 12 months of age (H&E, 200X). b, The proportion of human prostate cancers that show PTEN genomic loss, ERG rearrangement, both, or neither are shown (n = 121). PTEN loss was defined as having reduced copy number by RAE (heterozygous or homozygous loss; FDR q-value<6.93×10−8). ERG rearrangement was defined as the consequence of interstitial deletion by aCGH (q-value<6.93×10−8) and increased expression of ERG downstream of common translocation breakpoints (exons 5–8) relative to early ERG exons (exons 1–2) [>2 s.d. above the mean of normal samples (n=29) by exon expression array] The significance of the association between PTEN loss and ERG rearrangement is p-value<0.002 as determined by Fisher's exact test (right tail). c, The proportion of human prostate cancers that show MYC genomic amplification, ERG rearrangement, both, or neither are shown (n = 121). MYC amplification was defined as having increased copy number (q-value<1.2×10−7). ERG rearrangement and the significance of association (p-value=0.56) was defined as in Fig 2b. d, Normal prostate in Pten +/− and high grade PIN in Pten +/−; TMPRSS2-ERG mice at 6 months of age (H&E, 200X). Arrows indicate PIN and arrowheads show normal epithelium for comparison. e, Immunohistochemistry for smooth muscle actin in a Pten +/− and a Pten +/−; TMPRSS2-ERG mouse (400X). Compare disrupted and absent smooth muscle stroma around areas of possible microinvasion from a Pten +/−; TMPRSS2-ERG mouse (right column, arrows) to minimally attenuated smooth muscle sheath around area of high grade PIN in Pten +/− (left column, arrows) and to intact stroma of normal glands in both animals. f, Low grade PIN in AKT transgenic mice and high grade PIN in AKT; TMPRSS2-ERG double transgenic animals at 11 months of age (H&E, 400X). Arrows indicate PIN and arrowheads show normal epithelium for comparison.
These data suggest that TMPRSS2-ERG is insufficient to initiate prostate neoplasia and that cooperating oncogenic lesions are required. Two relatively common abnormalities in human prostate cancer are PTEN loss and MYC amplification, both of which play pathogenic roles in genetically engineered mouse models5,9. We therefore examined a cohort of TMPRSS2-ERG positive and negative human prostate tumors (n=121) for associated PTEN copy number loss or MYC amplification as measured by high resolution array-based comparative genome hybridization (aCGH). TMPRSS2-ERG status was determined based on interstitial deletion by aCGH and by exon array based mRNA expression profiling, which is highly correlated with fluorescent in situ hybridization analysis10. Although PTEN inactivation can also occur through loss of expression or point mutation, we determined PTEN status in these tumors by copy number alteration as this is a common mechanism of PTEN loss and provides robust evidence of gene inactivation. Tumors with PTEN loss were highly enriched for ERG rearrangement (p-value<0.002, Fisher’s one-sided exact test, Fig. 2b and Supplementary Tables 1–5), consistent with a similar association between reduced PTEN expression (measured by immunohistochemistry) and ERG fusion8. These studies are meaningful in light of a recent report that concurrent ERG rearrangement and PTEN loss in human prostate cancers predicts earlier disease recurrence11. In contrast, c-MYC amplification was not associated with ERG rearrangement (p-value=0.56, Fig. 2c). However, increased c-MYC mRNA levels, as measured by exon array, were associated with ERG rearrangement (p-value=0.0465, data not shown), consistent with recent evidence that c-MYC is a transcriptional target of TMPRSS2-ERG12. Taken together, these data establish that PTEN loss occurs in cooperation with TMPRSS2-ERG fusion, however ERG rearrangement does not result in selective pressure for MYC amplification.
To test the biological significance of the PTEN/TMPRSS2-ERG association in human tumors, we crossed our TMPRSS2-ERG mice with Pten +/− mice9. At six months of age, all compound mice examined (8 of 8)showed clear histologic evidence of PIN (Fig 2d). Notably, one Pten+/−; TMPRSS2-ERG animal had areas suggestive of microinvasive disease with attenuation of the periglandular smooth muscle stroma as well as stromal reaction, and inflammation around affected glands (Fig. 2e, Supplementary Figure 1). A focus of low grade PIN was observed in only 1 out of 8 Pten +/− littermate controls where the incidence of PIN at this age is reported to be about 10 percent9. To further explore the interaction with the PI3K pathway, we crossed TMPRSS2-ERG mice with prostate-specific AKT transgenic mice, which develop low grade PIN but do not progress to invasive cancer even at advanced age7. In contrast to 8 transgenic AKT littermate controls, 8 of 8 bigenic mice examined had developed more florid lesions by 10–12 months of age (Fig. 2f). We also crossed our TMPRSS2-ERG mice to ARR2PB-c-MYC (Hi-Myc) transgenic mice5 and found no evidence of cooperativity in 8 mice at 3 or 6 months, after which nearly all Hi-Myc mice have developed invasive cancer (Supplementary Figure 2). This result is consistent with the failure to detect significant coincidence of TMPRSS2-ERG fusion and c-MYC amplification in human tumors.
Our finding that TMPRSS2-ERG alone is insufficient to induce prostate neoplasia (or PIN) is in agreement with an independent study published in this issue8 but contrasts with two other reports that transgenic ERG expression induces PIN at 3–6 months of age13,14. These discrepancies are unlikely related to levels of transgene expression. All four models used the ARR2-Pb promoter, and our mice express the transgene at levels comparable to Carver et al. (Fig. 1e) and Klezovitch et al. (data not shown). Our mice differ in the inclusion of upstream untranslated TMPRSS2 sequences in the ERG fusion (which may impact translational efficiency, although we have no evidence for this based on transfection experiments) and the use of the variant 2 ERG isoform, which is an abundant allele in human tumors. The difference may lie in interpretation of mouse prostate histology, as the changes reported by Klezovitch et al. and by Tomlins et al. are subtle in comparison to the PIN phenotypes seen in other mouse prostate cancer models13,14. The higher frequency of microinvasion in the ERG;Pten +/− mice in the accompanying report may relate to mouse background because PI3K pathway activation induces a greater degree of proliferation, as measured by Ki-67 staining, in C57BL/6 (Carver et al.) relative to FVB (our model)15. Collectively, these data establish cooperativity between PI3K pathway activation and ERG aberration in induction of PIN but suggest that additional events are likely required for actual malignancy. Comprehensive human prostate cancer oncogenome studies are needed to fully annotate the frequency of alterations in these and other pathways.
Supplementary Material
Acknowledgements
This work was supported by a Ruth L. Kirschstein NRSA (7F32CA113132) to J.C.K. C.L.S. is an Investigator of the Howard Hughes Medical Institute and a Doris Duke Distinguished Clinical Scholar. B.S.T is a fellow of the Geoffrey Beene Cancer Research Center at MSKCC. We thank Kate Yen, Nicola Clegg, Yu Chen, Tambudzai Shamu, and the MSKCC CMG, Proteomics, and Molecular Cytology Core Facilities for technical help and/or discussions. We also thank Pier Paolo Pandolfi for sharing unpublished results and Valera Vasioukhin for providing transgenic prostate lobes.
Footnotes
Author contributions
J.C.K, J.X., J.W., D.H.L., B.S.C. and C.L.S. designed, created, and characterized the transgenic animals. S.S.C. and R.D.C provided the pathological analysis. J.X. and C.L.S. identified the potential start site. H.H, B.T., C.S., and W.L.G. performed and analyzed the CGH and exon array experiments. J.C.K. and C.L.S. wrote the initial draft of the manuscript and all authors contributed to the final version.
References
- 1.Kumar-Sinha C, Tomlins SA, Chinnaiyan AM. Nat Rev Cancer. 2008;8:497–511. doi: 10.1038/nrc2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perner S, et al. Am J Surg Pathol. 2007;31:882–888. doi: 10.1097/01.pas.0000213424.38503.aa. [DOI] [PubMed] [Google Scholar]
- 3.Tomlins SA, et al. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 4.Jhavar S, et al. J Mol Diagn. 2008;10:50–57. doi: 10.2353/jmoldx.2008.070085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ellwood-Yen K, et al. Cancer Cell. 2003;4:223–238. doi: 10.1016/s1535-6108(03)00197-1. [DOI] [PubMed] [Google Scholar]
- 6.Zhang J, Thomas TZ, Kasper S, Matusik RJ. Endocrinology. 2000;141:4698–4710. doi: 10.1210/endo.141.12.7837. [DOI] [PubMed] [Google Scholar]
- 7.Majumder PK, et al. Proc Natl Acad Sci U S A. 2003;100:7841–7846. doi: 10.1073/pnas.1232229100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carver BS, et al. Nat Genet. 2009 In Press. [Google Scholar]
- 9.Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi PP. Nat Genet. 1998;19:348–355. doi: 10.1038/1235. [DOI] [PubMed] [Google Scholar]
- 10.Attard G, et al. Oncogene. 2008;27:253–263. doi: 10.1038/sj.onc.1210640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoshimoto M, et al. Mod Pathol. 2008 [Google Scholar]
- 12.Sun C, et al. Oncogene. 2008;27:5348–5353. doi: 10.1038/onc.2008.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klezovitch O, et al. Proc Natl Acad Sci U S A. 2008;105:2105–2110. doi: 10.1073/pnas.0711711105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tomlins SA, et al. Neoplasia. 2008;10:177–188. doi: 10.1593/neo.07822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu Q, et al. Proc Natl Acad Sci U S A. 2007;104:17771–17776. doi: 10.1073/pnas.0708476104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.