Summary
Genome-scale studies have revealed extensive, cell type-specific co-localization of transcription factors, but the mechanisms underlying this phenomenon remain poorly understood. Here we demonstrate in macrophages and B cells that collaborative interactions of the common factor PU.1 with small sets of macrophage- or B celllineage-determining transcription factors establish cell-specific binding sites that are associated with the majority of promoter-distal H3K4me1-marked genomic regions. PU.1 binding initiates nucleosome remodeling followed by H3K4 monomethylation at large numbers of genomic regions associated with both broadly and specifically expressed genes. These locations serve as beacons for additional factors, exemplified by liver X receptors, which drive both cell-specific gene expression and signal-dependent responses. Together with analyses of transcription factor binding and H3K4me1 patterns in other cell types, these studies suggest that simple combinations of lineage-determining transcription factors can specify the genomic sites ultimately responsible for both cell identity and cell type-specific responses to diverse signaling inputs.
Introduction
The development of complex multicellular organisms involves hierarchically organized progenitor cells that ultimately give rise to terminally differentiated cell types with specialized functions. Cell fates are specified by lineage-determining transcription factors whose expression is often not limited to a single cell type (Tronche and Yaniv, 1992). Comparisons of the genome-wide binding patterns of different transcription factors in a variety of species and cell types have generated two major insights regarding transcription factor binding patterns: 1. Different factors in the same cell type tend to co-localize on a genome-wide scale (Chen et al., 2008; MacArthur et al., 2009). 2. The same factor in different cell types or at different stages of development exhibits different genome-wide binding patterns (Lupien et al., 2008; Odom et al., 2004; Sandmann et al., 2006).
Several mechanisms have been proposed to explain this phenomenon, including protein-protein interactions that enable ternary complex formation with DNA (Verger and Duterque-Coquillaud, 2002), pioneering factors that disrupt the closed nucleosome conformation and enable other factors to bind (Cirillo et al., 2002), cooperative binding of one or more factors to clustered sites that facilitates nucleosome displacement and stable binding of the factors involved (Boyes and Felsenfeld, 1996; Miller and Widom, 2003), and binding to chromatin marked in a cell type-specific manner by lysine 4-methylated histone H3 (H3K4me1/2) (Lupien et al., 2008), a sign of open chromatin that is correlated with activity of nearby genes (Heintzman et al., 2009). However, how transcription factors gain access to their eventual binding sites and the hierarchy of events that generate their cell type-specific binding and the associated epigenetic modification patterns have not previously been elucidated on a genome-wide scale.
The mammalian hematopoietic system represents a well-characterized model for the analysis of the combinatorial sets of transcription factors that orchestrate the development of distinct cell types from hematopoietic stem cells. Within the hematopoietic system, macrophages and B cells play essential and complementary roles in the innate and adaptive arms of the immune system. Recent studies suggest a model in which these cell types are derived from a lymphoid-primed multipotential progenitor (LMPP) that subsequently gives rise to common lymphoid progenitor (CLP) and granulocyte-macrophage progenitor (GMP) cells (Adolfsson et al., 2005) (Fig 1A). The Ets factor PU.1 is required for the generation of both GMP and CLP, and the later stages of macrophage and B cell development are additionally dependent on a number of cell type-restricted factors. Of these, AP-1 and C/EBP family factors are required for macrophage development and function (Friedman, 2007), while E2A, EBF1, Pax5 and Oct-2 play important roles in the development and function of B cells (Medina and Singh, 2005). In addition to its roles in hematopoietic development, recent evidence suggests that PU.1-bound sites in macrophages play a role in shaping the transcriptional response to inflammatory stimuli such as lipopolysaccharide (LPS), likely by generating cell type-specific regions of open chromatin that serve as beacons for the recruitment of transcriptional coactivators in response to stimuli (Ghisletti et al., 2010).
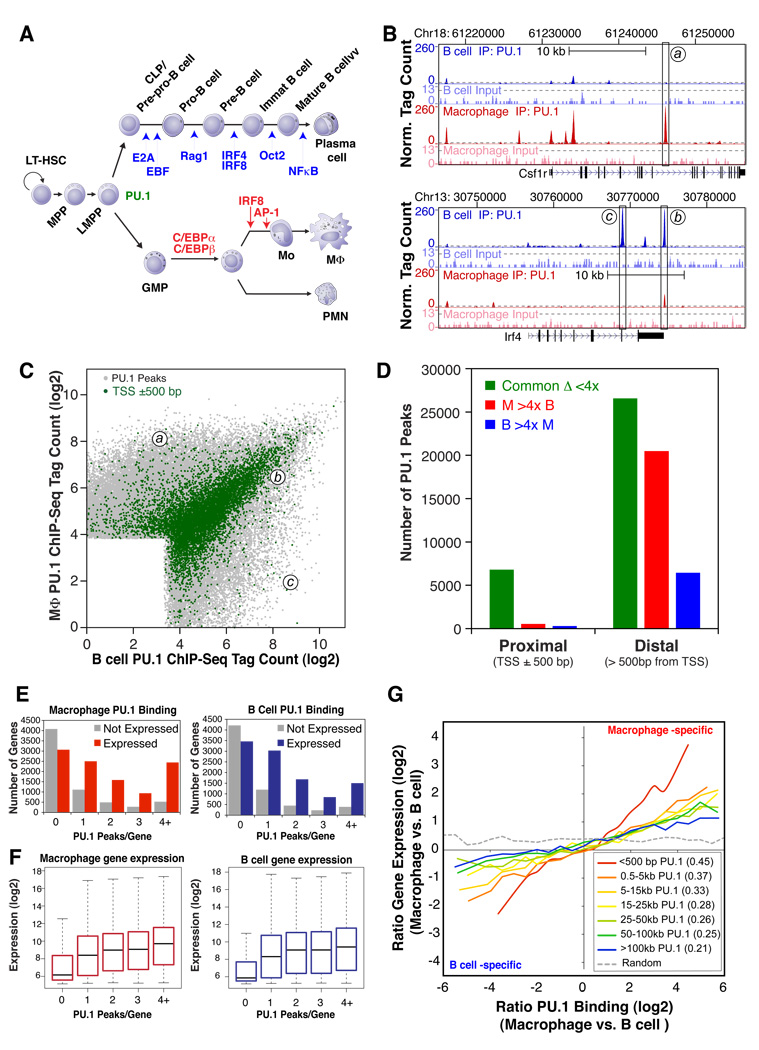
Figure 1. Identification and functional analysis of PU.1 binding sites in primary macrophages and splenic B cells.
(A) Simplified scheme for macrophage and B cell differentiation from hematopoietic stem cells, indicating genetically defined factors specific for B cells (blue) and macrophages (red) at the developmental transitions most severely affected by loss of a given factor. (B) UCSC Genome Browser image depicting PU.1 ChIP-Seq tags at the macrophage-specific Csf1r gene or the B cell-specific Irf4 gene. Dashed gray lines indicate an estimated 0.1% false discovery rate (FDR). Input DNA signal is shown 10-fold magnified. (C) PU.1-bound genomic sites are visualized by their respective normalized PU.1 ChIP-Seq tag counts (log2) within 200 bp of a given peak in macrophages and B cells. The coordinates of peaks a, b and c from panel 1B are indicated. Peak positions within 500 bp of a Refseq transcription start site are colored green. Jitter was added to the normalized tag counts to visualize otherwise overlapping data points. (D) Total number of common and cell-type specific PU.1-bound regions found in both promoter-proximal and distal genomic regions. Cell type-specificity was assigned to regions with 4-fold greater normalized tag counts in one cell type relative to the other. (E) Total number of genes with the specified number of PU.1 binding sites near their promoters. Genes were divided into subgroups of expressed and non-expressed genes based on their absolute normalized expression levels as measured by microarray hybridization (threshold: log2(microarray signal) = 6.5). (F) Distribution of the gene expression values for the combined subgroups of genes in the vicinity of the given number of peaks depicted in 1E. (G) Relationship between differential PU.1 binding and differential gene expression between macrophages and B cells. Subsets of PU.1 binding sites defined by their distance to the nearest TSS were sorted according to their difference in normalized tag counts between cell types. The moving average of the difference in gene expression values of the gene with the nearest TSS is reported relative to differential binding. The Pearson’s correlation coefficient for each group is reported in the insert (all p-values < 10−100).
To address the question of how lineage-determining transcription factors bind to genomic regions in a cell-specific manner, we investigated the genome-wide locations of PU.1 and the effects of loss or gain of other transcription factors on the PU.1 binding pattern in macrophages, B cells and different B cell progenitors. In addition, we assessed the impact of loss and gain of PU.1 on the cistrome of the myeloid-restricted transcription factor, C/EBPβ. Our results suggest that cell type-specific cistromes arise from collaborative interactions between small sets of lineage-determining factors that result in enhanced DNA binding, nucleosome remodelling and subsequent deposition of the epigenetic enhancer mark H3K4me1. The association of PU.1 and its collaborating factors at these genomic locations provides access points for the binding of additional transcription factors, which themselves do not appear to significantly contribute to shaping the overall master regulator genomic distribution, or the overall H3K4me1 pattern, but act to confer transcriptional functions to these distal sites.
Distinct PU.1 binding programs in macrophages and B cells
We initially performed chromatin immunoprecipitation-coupled deep sequencing (ChIP-Seq) (Barski et al., 2007) to define the PU.1 cistromes in mouse peritoneal macrophages and splenic B cells. These analyses identified 45631 and 32575 PU.1-bound genomic sites (FDR < 0.1%) in macrophages and B cells, respectively (Table S1), which included known PU.1 target sites (Figure S1A). Of these sites, 17130 were independently identified as bound by PU.1 in both cell types (Figure 1B and 1C, and Figure S1B). PU.1 was enriched at transcription start sites (TSS) relative to their genomic frequency but the majority of binding occurred at inter- and intragenic sites (Figure S1C).
Scatter plots of the tag counts around genomic peak coordinates enable comparative visualization of experimentally observed peaks and allow the overlay of other annotations present at these genomic coordinates. (For example, the tag counts associated with PU.1 peaks at the genomic coordinates a, b and c in Figure 1B are indicated in Figure 1C). Unexpectedly, more than 80% of the PU.1 sites localized to promoters (TSS ± 500 bp, green data points in Figure 1C) exhibited similar occupancy levels in both cell types, while differentially bound sites were mostly located in distal regions. (Figure 1C and 1D).
To examine the relationship of PU.1 binding sites and gene expression patterns, we performed transcriptome analysis of elicited macrophages and splenic B cells and correlated binding at each PU.1 site with the mRNA level of the closest gene. Out of 17004 genes analyzed, 58.0% and 54.9% were associated with at least one PU.1 peak in macrophages and B cells, respectively, with most of these genes exhibiting multiple peaks (Figure 1E). Genes associated with PU.1 binding were more likely to be expressed (Figure 1E) and the number of peaks per gene was positively associated with gene expression in both cell types (Figure 1F). Cell type-specific PU.1 binding was also correlated with cell type-specific gene expression (Figure 1G and Figure S1D). While this relationship was significant (Pearson correlation coefficient r2 = 0.30, p < 10−100), even cell type-specific genes often exhibited a mix of cell type-specific and commonly bound PU.1 peaks (e.g., Figure 1B, peak position (b)). The association between cell-specific PU.1 binding and cell-specific gene expression was strongest for peaks close to the TSS (Pearson correlation coefficient r2 = 0.45, p<10−100), however even the distantly located set of PU.1 peaks (>100 kb) showed correlation with vicinal gene expression (r2 = 0.21, p<10−100). This is in line with the known distinct roles of PU.1 in both transcription initiation and enhancer function (Fisher and Scott, 1998; Lichtinger et al., 2007).
PU.1 binds in the vicinity of other lineage-determining transcription factors
To gain insight into possible sequence determinants of the distinct PU.1 binding patterns observed in macrophages and B cells, we examined promoter-proximal and distal regions that were either commonly or cell type-specifically bound by PU.1 for enriched sequence elements by de novo motif analysis. The most enriched motifs were nearly identical in each of the different subsets and resembled the known PU.1 consensus element (Figure 2A and Figure S2A). The only exception was found at proximal promoter peaks common to both cell types, where PU.1 likely competes for binding with ETS factors, such as GABPα, at related ETS sites found in CpG islands (Xie et al., 2005). While nearly 60% of all PU.1 peaks contain either the PU.1 or GABP version of ETS motifs, the most enriched motif in the remaining peaks was a degenerate ETS core motif (RRGGAASY). Overall, ~85% of all PU.1 peaks contain an ETS site, strongly suggesting that PU.1 binds directly to the vast majority of its DNA target sites (Figure 2A and Table S2)
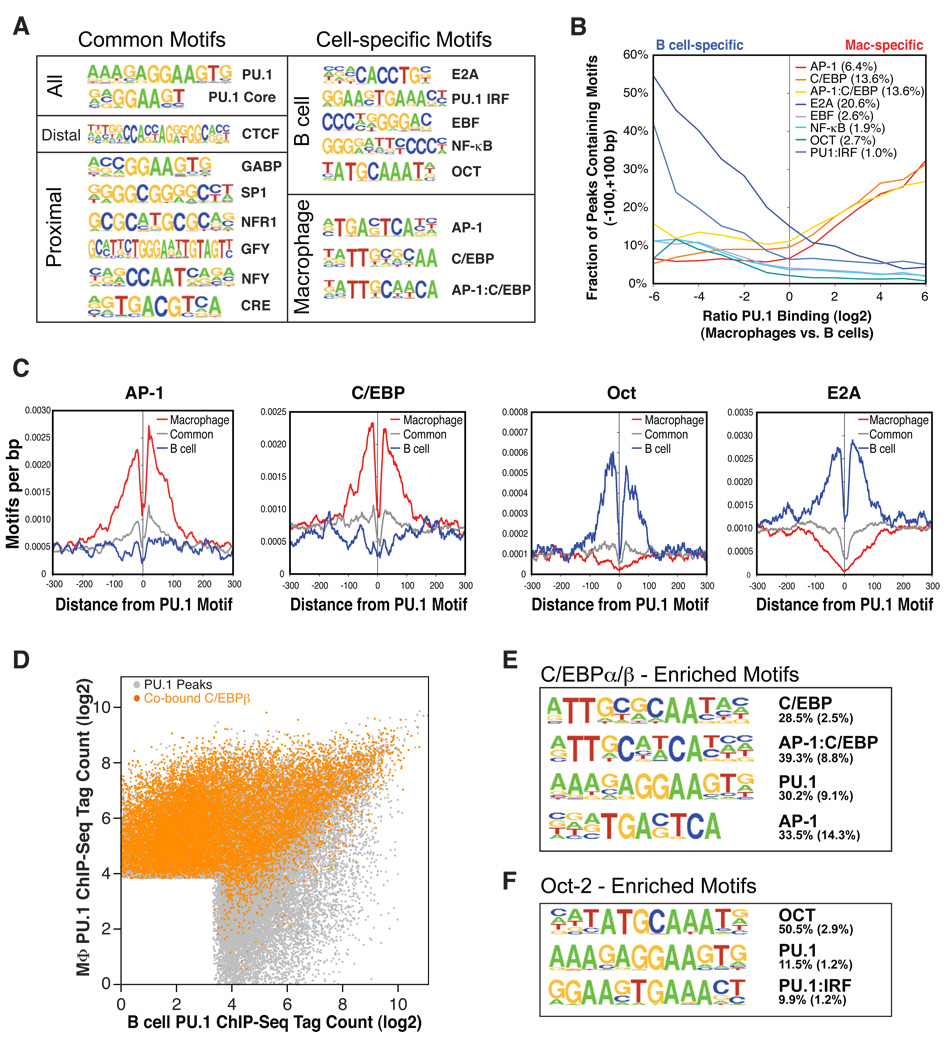
Figure 2. Transcription factors co-localize with PU.1 binding sites in macrophages and B cells.
(A) Sequence logos corresponding to enriched sequence elements identified by de novo motif analysis of promoter-proximal or inter-/intragenic PU.1 binding sites. Motifs at PU.1 sites common to macrophages and B cells were identified by comparing common peaks to randomly selected genomic regions. Motifs enriched at PU.1 sites specific to macrophages or B cells were discovered by directly comparing the sets of peaks that were exclusive to each cell type. Motif frequencies in different subsets of PU.1 peak regions are reported in Table S2. All motifs are enriched over background with p values < 10−100. (B) Frequencies of discovered motifs in PU.1 peaks (±100 bp) as a function of differential binding of PU.1 in macrophages and B cells. Expected motif frequencies based on 100k random genomic regions are reported in the legend. (C) Context-specific frequencies of AP-1, C/EBP, OCT, and E2A motifs in the vicinity of the central PU.1 motif found in PU.1 binding sites. Moving averages of motif frequencies within a 23 bp window are centered on distal macrophage-specific PU.1 sites (red), distal B cell-specific PU.1 binding sites (blue), and common distal PU.1 binding sites (gray) (>500 bp from the nearest TSS). (D) Differential PU.1 occupancy of PU.1 peaks as in Figure 1C; PU.1 peaks are colored orange if a C/EBPβ peak is located within 100 bp of the PU.1 peak. (E and F) Sequence logos are shown for the most highly enriched sequence motifs in C/EBPβ and Oct-2 binding sites, respectively. The fraction of peaks containing at least one instance of each motif within 100 bp of the peak center is given to the right of the motif with the expected frequency of the motif in 50k random regions given in parentheses.
In addition to the ETS motifs, common promoter-proximal PU.1-bound regions were highly enriched for motifs that have previously been shown to be generally enriched in promoters (Xie et al., 2005) (Figure 2A). Both promoter-proximal and promoter-distal commonly PU.1-bound regions exhibited enrichment for a PU.1-IRF composite, consistent with the ability of PU.1 and IRF4/8 to form a ternary complex on this class of composite sites in both cell types (Eisenbeis et al., 1993; Pongubala et al., 1992). Additionally, promoter-distal regions bound by PU.1 in both cell types were enriched for a CTCF motif, likely because CTCF-bound sites generally coincide with accessible chromatin as measured by DNase I hypersensitivity (Xi et al., 2007).
In striking contrast, macrophage-specific and B cell-specific PU.1-bound regions were significantly co-enriched for motifs for macrophage- and B cell-restricted, lineage-determining transcription factors, respectively (Figure 2A and 2B). This was evident to a similar degree in both promoter-proximal and promoter-distal regions (Table S2). Analysis of the spatial relationships between these co-enriched motifs and the PU.1 motif in each peak revealed that C/EBP and AP-1 motifs were highly enriched within 100 bp of macrophagerestricted distal PU.1 sites, while E2A, EBF, Oct and NF-κB motifs were correspondingly highly enriched near B cell-specific distal PU.1 sites (Figure 2B and 2C, Figure S2B). Conversely, the B cell-specific motifs were depleted in the vicinity of macrophage-specific PU.1 sites, while the regions around B cell-restricted PU.1 sites were depleted of macrophage-specific motifs (Figure 2C and Figure S2B). Secondary PU.1 motifs were also enriched within 100 bp of the central PU.1 motif, particularly at macrophage-specific and commonly bound sites (Figure S2B).
To confirm that the PU.1-co-enriched motifs were occupied by the associated cell type-restricted transcription factors, we carried out ChIP-Seq for C/EBPα and C/EBPβ in peritoneal macrophages and for Oct-2 in splenic B cells. For C/EBPα and C/EBPβ, which displayed an almost identical genomic binding pattern, we identified approximately 40000 binding sites (Table S1 and Figure S2C). Of these sites, 13840 (34.6%) were located within 100 bp of a PU.1-bound site, 60% of which were specific to macrophages, while only 1% were localized to B cell-specific PU.1 peaks (4× PU.1 tag difference) (Figure 2D). For Oct-2, the number of identified sites (1191) was smaller than for the C/EBPs, however the results complemented the above findings in that 596 (50%) of all Oct-2 sites were located within 100 bp of a PU.1 peak, of which 43% were B cell-specific and less than 1% macrophage-specific (4× PU.1 tag difference) (Figure S2D). C/EBP and Oct-2 binding sites exhibited similar relationships with gene expression as observed for PU.1 (Figure S2E). C/EBP- and Oct-2-bound regions were highly enriched for C/EBP and octamer motifs, respectively (Figure 2E). In addition, C/EBP-bound regions were co-enriched for a recently described C/EBP:AP-1 composite element (Cai et al., 2008) as well as canonical PU.1 and AP-1 motifs, whereas Oct-2-bound regions in B cells were highly enriched for a PU.1 as well as a PU.1 -IRF composite motif (Figure 2E).
Combinatorial interplay delineates stage-specific cistromes
The observation that PU. 1 binding occurred in the vicinity of motifs bound by lineage-restricted transcription factors prompted us to ask whether PU.1 binding to these sites was dependent on the presence of the respective factors. We addressed this question in B lineage progenitors devoid of both E2A and EBF (E2A−/−), EBF only (EBF−/−), or control cells that express both factors (Rag1−/−), which are arrested at successive stages in B cell development (Dias et al., 2008; Ikawa et al., 2004; Lin and Grosschedl, 1995; Mombaerts et al., 1992)(Figure 1A). ChIP-Seq for PU.1 in each of these cell populations yielded 44609, 36908 and 17210 peaks in E2A−/−, EBF−/− and Rag1−/− progenitors, respectively (FDR < 0.1%, Table S1). Surprisingly, despite the apparent loss in total number of PU.1 peaks comparing one developmental stage to the next, each successive stage was characterized by the appearance of PU.1 sites at new genomic positions that were not occupied by PU.1 in the previous progenitor stage. Using the PU.1 sites identified in E2A−/− cells as control, de novo motif analysis of the 3760, 2479, and 9504 PU.1 sites exhibiting 3× more tags in the EBF−/−, Rag1−/−, and mature B cells, respectively, revealed enrichment for distinct motifs for transcription factors expressed at respective stages of B cell development (Figure 3A and Table S3), demonstrating that PU.1 binding is dependent on the activity of other lineage-determining transcription factors. For example, co-enriched E2A motifs were gained comparing the PU.1 cistrome in EBF−/− cells to that in E2A−/− cells (Figure 3A). Furthermore, gained sites were primarily associated with genomic regions corresponding to B cell-specific PU.1 binding sites in mature splenic B cells, as exemplified by the reshaping of the PU.1 binding site pattern comparing CLP/pre-pro B (EBF−/− ) to pro-B cells (Rag1−/−) (red data points in Figure 3B).
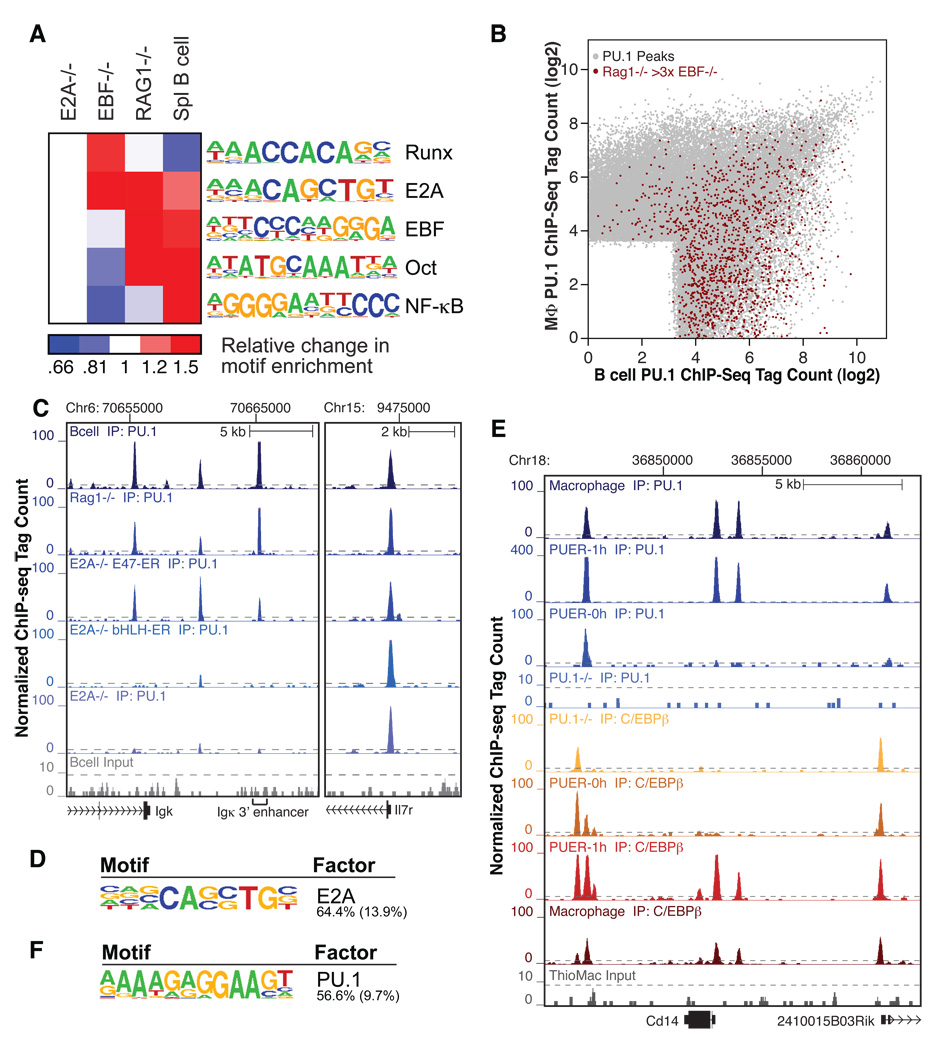
Figure 3. Co-operative interactions between PU.1 and cell type-restricted transcription factors define the macrophage and B cell-specific cistromes.
(A) Motifs in the vicinity of gained PU.1 sites during B cell development. Regions of 200 bp, centered on ChIP-Seq PU.1 peaks in the respective cell types were classified according to presence or absence of a given motif. Displayed is the ratio of the fraction of gained peaks (>3-fold tag count change) containing the motif relative to the fraction of unchanged peaks (<2-fold tag count change) containing the motif. Absolute peak numbers are provided in Supplemental Table S3. (B) Same plot as in Figure 1C, with PU.1 peaks seen specifically enriched in Rag1−/− vs. EBF−/−cells colored in red. (C) UCSC browser image of the PU.1 binding pattern at the IgK 3’ enhancer region. Top to bottom: B cells, Rag1−/− pro-B cells, E2A−/− CLP/pre-pro-B cells reconstituted with a conditional E2A (E47-ER) or a transactivation domain-deficient E2A (bHLH-ER) 6 h after tamoxifen treatment and E2A−/− CLP/pre-pro-B cells, as well as B cell ChIP input. The PU.1 site at the Il7r promoter is shown as a control for a PU.1 binding site that is invariant between the different cell types. (D) Top motif in the vicinity of PU.1 binding sites gained following reconstitution with full length E2A compared to constitutively bound PU.1 sites identified by de novo motif analysis. The fraction of PU.1 peaks containing at least one instance of the motif within 100 bp of the peak center in gained and constitutively bound peaks is given to the right and in parentheses, respectively. (E) UCSC browser tracks of PU.1 and C/EBPβ binding at the CD14 locus in primary macrophages, PU.1−/− cells and PUER cells without and 1 h after tamoxifen treatment. Top to bottom: PU.1 in macrophages, PUER cells after or without 1 h tamoxifen treatment and PU.1−/− cells (blue - dark to light), and C/EBPβ in PU.1−/− cells, PUER cells without or after 1 h tamoxifen treatment, and macrophages (brown to red). (F) Top motif enriched in the vicinity of gained C/EBPβ sites following activation of the PU.1-ER fusion protein compared to the constitutively bound sites identified by de novo motif analysis. The fraction of C/EBPβ peaks containing at least one instance of the motif within 100 bp of the peak center in gained and constitutively bound peaks is given to the right and in parentheses, respectively.
To directly test if the binding of PU.1 to B lineage-specific sites is dependent on E2A, we retrovirally transduced E2A–deficient CLP/pre-pro B cells derived from the bone marrow of E2A−/− mice (Ikawa et al., 2004) either with a tamoxifen-inducible E2A–estrogen receptor ligand binding domain fusion protein (E47-ER), or the analogous construct coding for a deletion mutant of E47 that retains the bHLH DNA binding and dimerization domain but lacks both activation domains (bHLH-ER) (Sayegh et al., 2003). Upon activation of the fulllength E47-ER fusion protein with tamoxifen for 6 hours, PU.1 binding increased at 3752 sites > 4× relative to the bHLH-ER control, as exemplified by the IgK 3’ enhancer (Figure 3C), and these sites were strongly co-enriched for an E2A motif (Figure 3D and Figure S3A). In contrast, the PU.1 cistrome in these cells (44609 peaks total) was not significantly altered by expression of the E2A deletion mutant lacking transcriptional activation domains (Figure 3C).
To conversely investigate whether PU.1 itself can also promote binding of one of its cistrome-associated factors, we assessed the genome-wide effects of inducing PU.1 activity on C/EBPβ binding in a PU.1-deficient (PU.1KO) myeloid progenitor cell line (Walsh et al., 2002). In the absence of PU.1, C/EBPβ exhibited a reduced overall binding pattern (22641 peaks), with loss of numerous binding sites associated with locations of PU.1 binding in primary macrophages, illustrated for the macrophage-specific CD14 gene (PU.1KO-Cebpb vs. Mac-Cebpb, Figure 3E). Motif analysis of the C/EBPβ-bound regions in PU.1−/− cells recovered C/EBP, C/EBP:AP-1, AP-1 and RUNX motifs, as well as a non-PU.1 Ets motif (AACAGGAAGT), but not the PU.1 binding motif found in primary macrophages (Figure S3G).
We next evaluated the consequences of PU.1 activity on C/EBPβ binding in PU.1−/−cells stably expressing a tamoxifen-responsive PU. 1-estrogen receptor ligand binding domain fusion protein (PUER) (Walsh et al., 2002). In this inducible system low levels of PU.1 activity and DNA binding are observed by ChIP-Seq in the absence of tamoxifen (8144 peaks total, exemplified in Figure 3E, PUER-PU.1–0h). This low level PU.1 activity was associated with partial recovery of C/EBPβ binding in the vicinity of some PU.1 binding sites (Figure 3E and Figure S3H, PUER- C/EBPβ-0h). Tamoxifen treatment induced PU.1 binding to a total of 37909 genomic regions after 1 h and resulted in a gain of 1710 C/EBPβ binding sites (>4× tags relative to the 0 h time point), restoring a large number of macrophage-specific PU.1 binding sites. The induced C/EBPβ binding regions were strongly enriched for a PU.1 motif (Figure 3F and Figure S3B) and were co-bound by PU.1 at over 75% of sites. Intriguingly, 57% of PU.1 binding sites observed in the absence of tamoxifen were co-bound by C/EBPβ, implying C/EBP and related factors play an important role in directing PU.1 localization when effective PU.1 concentrations are low.
PU.1 induces sequential nucleosome remodelling and histone H3K4 monomethylation
Recent studies have demonstrated that lineage-determining transcription factors preferentially associate with genomic regions marked by cell type-specific patterns of histone modifications, such as monomethylation of H3K4 (H3K4me1), that are suggested to signify accessible chromatin and/or enhancer-like elements (Heintzman et al., 2009). In addition, trimethylation of H3K4 (H3K4me3) marks active and/or poised promoters (Kim et al., 2005). We therefore profiled H3K4me1 and H3K4me3 in both macrophages and B cells using ChIP-Seq and found that the majority (>80%) of PU.1-bound sites were associated with either promoter-proximal H3K4me3 or distal H3K4me1 in both cell types. Cell-specific binding of PU.1 at distal sites was highly associated with cell-specific H3K4me1 (Figure 4A). Analysis of the genomic distribution of the H3K4me1 signal averaged for all distal PU.1 binding sites revealed a bimodal pattern with a pronounced reduction of signal centered over the PU.1 binding site (Figure 4B), analogous to that recently described for H3K4me1 around promoter-distal FoxA2- and STAT1-bound sites in mouse liver and HeLa cells, respectively (Robertson et al., 2008). Similarly, 80% of distal Oct-2 and C/EBP sites were marked by a corresponding pattern of cell-specific H3K4me1 (Figure 4B).
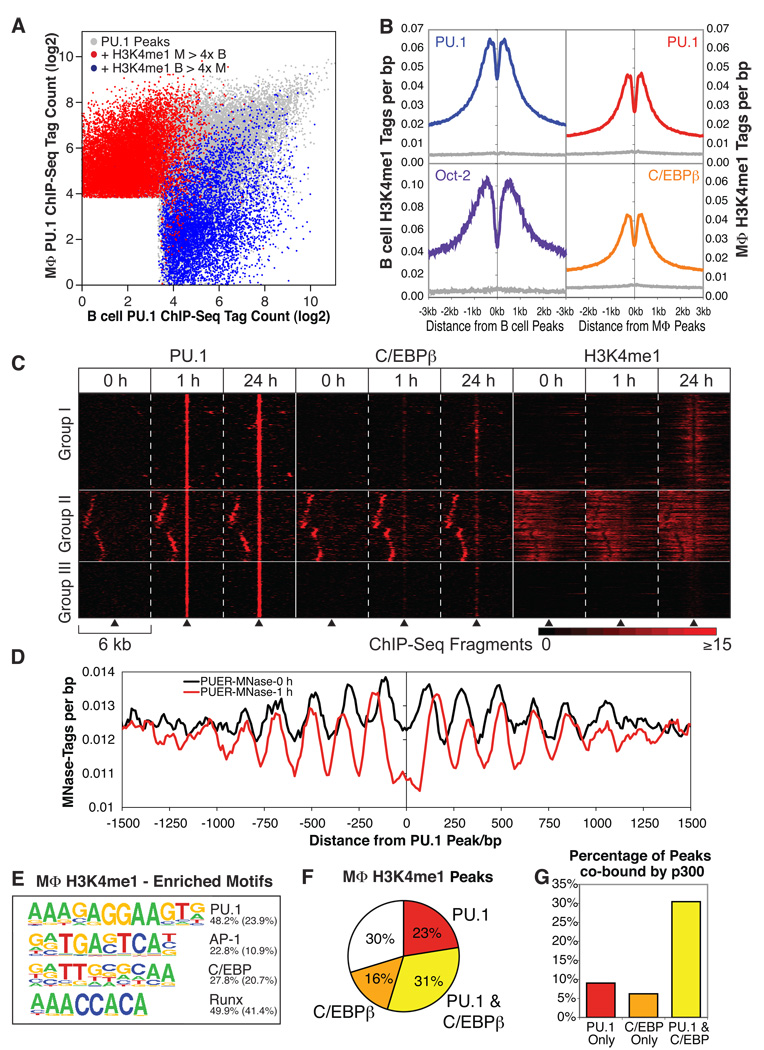
Figure 4. PU.1 and cooperating factors are required for histone modifications associated with enhancers.
(A) Same plot of PU.1 peaks as in 1C, peaks are colored red if the surrounding region (±500 bp) contains more than 4-fold more H3K4me1 tags in macrophages than in B cells. Peak positions fulfilling the same criteria for B cells versus macrophages are colored blue. (B) Cell type-matched cumulative normalized H3K4me1 ChIP-Seq and input sequencing tag counts per base pair are shown around distal peak positions (>3 kb from a TSS) of PU.1 (B cells and macrophages), C/EBPβ (macrophages), Oct-2 (B cells). (C) Temporal and spatial relationships of PU.1, C/EBPβ and H3K4me1 in PUER cells at different time points. Six kb-wide regions centered on genomic sites that gained PU.1 > 6-fold after 1 h tamoxifen were clustered according to their PU.1, C/EBPβ and H3K4me1 ChIP-Seq profiles at 0 h, 1 h or 24 h of tamoxifen treatment. Shown are representative sections (10 % of each group) of the resulting heat map. (D) Average nucleosome positions centered on induced PU.1 peaks before and after 1 h tamoxifen treatment as defined by MNase-Seq. (E) Sequence motifs associated with promoter-distal H3K4me1-marked 1 kb regions in macrophages. The top 4 motif results from de novo motif analysis are shown. The fraction of H3K4me1 marked regions containing at least one instance of each motif within 500 bp of the peak center is given to the right of the motif with the expected frequency of the motif in random regions in parentheses. (F) Pie chart depicting the overlap of PU.1 and C/EBPβ within 1 kb of focal H3K4me1 ChIP-Seq peaks. (G) Association of p300 with PU.1 and C/EBPβ co-bound sites. Percentages of transcription factor-bound regions co-bound by p300 are given. Absolute numbers were: PU.1 only, 21223 total, Co-bound by p300: 1921; C/EBPβ only, 20481 total, 1274 with p300; PU.1 & C/EBPβ, 13874 total, 4230 with p300. Peak positions for p300 were determined by analysis of p300 ChIP-Seq data for resting bone marrow-derived macrophages (Ghisletti et al., 2010).
These observations prompted us to address the question of whether H3K4me1 serves as a beacon to recruit PU.1 and collaborating transcription factors, and/or whether these factors are able to initiate the deposition of this mark. Using the PUER inducible system, we performed ChIP-Seq for H3K4me1 in PUER cells at 0, 1 and 24 h of tamoxifen treatment. Very little change in H3K4me1 signal was observed after 1 h (38 peaks gained), but a marked increase occurred at 3328 locations by 24 h (>4× tags, 15% of total H3K4me1 peaks present at 0 h). To evaluate the relationship between PU. 1 binding and H3K4me1, we selected a subset of 7428 highly induced PU.1 binding sites (>8-fold increase in tag count at 1 h) which exhibited less than 2 tags at 0 h. These binding sites could be classified into three groups, with representative peaks from each group shown in Figure 4C. The largest group, (Group I, 43%), consists of sites that gained significant H3K4me1 24 h after PU.1 activation. This gained H3K4me1 signal exhibited a bimodal spatial distribution identical to that observed for PU.1-associated H3K4me1 in macrophages and B cells (Figure S4A). Together with the fact that 90% of the sites that gain H3K4me1 are within a 1 kb window of a gained PU.1 site, these findings suggest that PU.1 binding is required to direct the local deposition of H3K4me1 at these sites. Group II (32%) consists of induced PU.1 binding sites that were marked by pre-existing H3K4me1. In this group, PU.1 binding initiated remodelling of the local H3K4me1-containing nucleosomes at 1 h and 24 h, again establishing a pattern identical to that in macrophages and B cells (Figure S4A). Group III (25%) consists of peaks that despite stable binding of PU.1 were not enriched for H3K4me1 at any time point, indicating that here, PU.1 binding is not sufficient to establish this mark. Increased recruitment of C/EBPβ to Groups I (37%) and II (30%) relative to Group III (11%) peaks (<100 bp) suggests that additional factors may be required for deposition of H3K4me1. Group I and II peaks, but not Group III peaks, were enriched for the 464 vicinal genes that were up-regulated greater than 4-fold in PUER cells following 24 h tamoxifen treatment (Weigelt et al., 2009) (Figure S4B).
To investigate whether the remodelling of H3K4me1-marked nucleosomes in the Group II peaks were consistent with changes in nucleosomal occupancy, we mapped nucleosome positions using MNase-Seq (Schones et al., 2008) in PUER cells at 0 h and 1 h after treatment with tamoxifen. Analysis of the nucleosome pattern before tamoxifen treatment at the 7428 regions where PU.1 is maximally gained at 1 h revealed a semi-regular pattern exposing the prospective PU.1 binding site on an expanded linker region between adjacent nucleosomes (Figure 4D). This pre-existing pattern is possibly due to low levels of PU.1 binding that are below detection by ChIP-Seq at the current sequencing depth, as it is possible to detect PU.1 at many of these sites in untreated PUER cells using qPCR-based ChIP assays (data not shown). Induction of PU.1 binding led to nucleosome remodelling across the entire set of induced sites, analogous to that suggested by the Group II H3K4me1-marked nucleosomes, resulting in further expansion of the linker region centered on the PU.1 binding site and compression of nucleosomes in either direction along the DNA for approximately 1 kb. In concert, these findings suggest a sequence of events at Group I sites in which PU.1 binding induces nucleosome remodelling, which is followed by monomethylation of H3K4.
Simple combinations of binding sites identify cell type-specific repertoires of H3K4me1-marked regions
The striking association of PU.1 and collaborating lineage-determining transcription factors with cell-specific H3K4me1 in macrophages and B cells led us to perform a de novo motif analysis of the approximately 20000 distal genomic regions that were focally marked by H3K4me1 in each cell type. This analysis revealed that these regions in macrophages were most highly enriched for PU.1, AP-1, C/EBP and RUNX motifs (Figure 4E), while H3K4me1-marked regions in B cells displayed maximal enrichment for PU.1, E2A, OCT, EBF and RUNX motifs (Figure S4C). This computational result is supported by ChIP-Seq data demonstrating that the majority (70%) of the H3K4me1-positive regions in macrophages were bound by PU.1 and/or C/EBPβ (Figure 4F) and is consistent with the finding that PU.1 (and likely C/EBP) binding can induce H3K4me1 deposition. Of note, analysis of the genome-wide location of p300 in resting macrophages (Ghisletti et al., 2010) indicates marked enrichment at genomic locations co-bound by PU.1 and C/EBPβ (Figure 4G).
To determine whether there is an analogous relationship between promoter-distal H3K4me1 and binding sites for transcription factors in other cell types, we performed de novo motif analysis on H3K4me1/H3K4me3 ChIP-Seq data sets from mouse embryonic stem cells (Meissner et al., 2008; Mikkelsen et al., 2007), liver (Wederell et al., 2008), human CD4+ T cells (Barski et al., 2007) and CD36+ erythrocyte precursors (Cui et al., 2009). Similar to the results in macrophages and B cells, H3K4me1-marked promoter-distal regions in these tissues were highly enriched for motifs for transcription factors required for the generation and maintenance of each cellular phenotype (Ivanova et al., 2006; Rothenberg and Taghon, 2005; Zaret et al., 2008) (Figure S4C). For example, H3K4me1-marked promoter-distal regions in ES cells were significantly enriched for binding sites for KLF4, OCT4, SOX2 and Esrrβ, factors that in combination are sufficient to reprogram somatic cells into induced pluripotent stem cells (Feng et al., 2009). In CD4+ T cells, this analysis revealed a strong association of H3K4me1-marked regions with Ets and Runx motifs, consistent with their known essential roles in T cell development and function (Figure S4C) (Rothenberg and Taghon, 2005). In this cell type, genome-wide data for nucleosome positions, DNase hypersensitivity and mononucleosomal ChIP-Seq for a wide range of histone modifications is available, allowing precise determination of nucleosome positions, chromatin accessibility and histone modifications (Barski et al., 2007; Boyle et al., 2008; Schones et al., 2008). Alignment of H3K4me1 ChIP-Seq, total nucleosome sequence tag positions and DNase I on the Ets motifs enriched in the H3K4me1-marked promoter-distal regions revealed nucleosome phasing around the Ets motif similar to the pattern we observed around PU.1 in PUER cells, which was accompanied by a sharp spike in DNase I hypersensitivity at the Ets site (Figure S4F). This result is in stark contrast to the continuous nucleosome distribution observed when aligning the data on the means of the tag distributions (Figure S4F) and establishes that the central gap in the H3K4me1 pattern over the transcription factor motif is reflective of the absolute nucleosome positions and represents the predominant DNase Iaccessible location at these promoter-distal sites.
PU.1 establishes cell type-specific cistromes for signal-responsive transcription factors
Given the large number of transcription factors that are known to play important roles in each cell type (e.g. greater than 350 transcription factors are transcribed in macrophages and ES cells, see Figure S4D), it was surprising to find that only small numbers of transcription factor motifs were highly overrepresented in the H3K4me1 patterns in the different cell types we analyzed (Figure S4C and S4E). To explore the relationship of transcription factors whose motifs are not enriched in H3K4me1-marked regions with H3K4me1-associated factors, the global H3K4me1 pattern, and cell type-specific gene expression, we initially focused on Liver X Receptors (LXRα and LXRβ). These oxysterolresponsive nuclear receptors play numerous roles central to macrophage function by regulating target genes involved in lipid metabolism, cell survival, and immunity (Rigamonti et al., 2008).
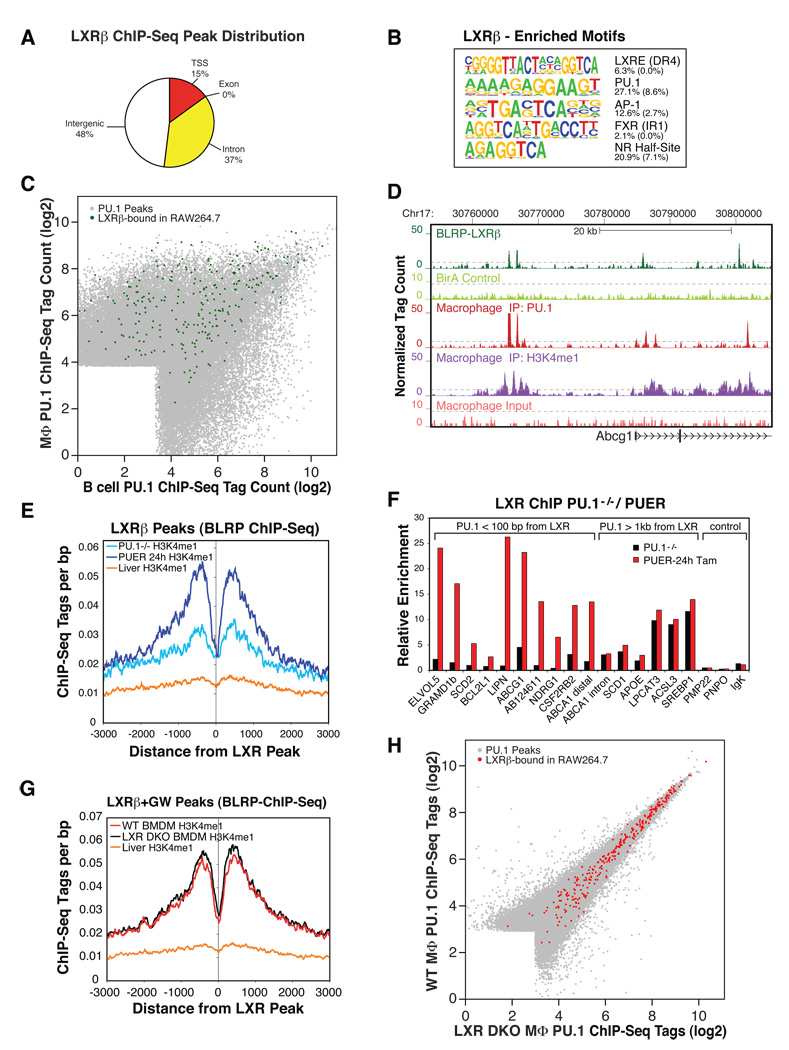
ChIP-Seq of in vivo biotin-tagged LXRβ in the murine macrophage RAW264.7 cell line (validated in Figure S5A and S5B) identified 664 confident LXR binding sites (FDR < 0.1%), 85% of which were distally located relative to the TSS of vicinal genes (Figure 5A). Conventional, antibody-based ChIP assays for endogenous LXRβ in primary macrophages confirmed 12 of 12 representative LXR binding sites (Figure S5C). While de novo motif analysis identified the consensus LXR response element (DR4) as the most highly enriched motif, PU.1 and AP-1 motifs were also highly co-enriched in LXRβ-bound regions (Figure 5B). In line with motif enrichment, 34% of LXRβ peaks were within 100 bp of a PU.1-bound site in primary macrophages (Figure 5C), exemplified for the established LXR target gene Abcg1 (Figure 5D). Additionally, macrophage LXR binding sites exhibited a macrophage-specific H3K4me1 signature, in contrast to the H3K4me1 pattern around the same sites in liver (Robertson et al., 2008), another tissue in which LXRs play key regulatory roles (Figure 5E and 5G).
Figure 5. PU.1 establishes part of the LXRβ cistrome and H3K4me1 pattern around LXRβ binding sites in macrophages, but not vice versa.
(A) Genomic annotation of high confidence LXRβ binding sites defined by ChIP-Seq of biotin-tagged LXRβ in RAW264.7 murine macrophages. (B) De novo motif analysis of LXR-bound regions in macrophages. The fraction of peaks containing at least one instance of each motif within 100 bp of the peak center is given to the right of the motif with the expected frequency of the motif in random regions in parentheses. (C) Macrophage and B cell PU.1 binding as in Figure 1C; PU.1 peak positions are colored green if an LXRβ peak is located within 100 bp of a PU.1 peak. (D) UCSC browser image of the LXR target gene locus ABCG1 depicting coordinate binding of LXR and PU.1, together with the associated H3K4me1 signature. The 2.5-fold magnified BirA control track denotes sequencing of a pulldown from formaldehyde-fixed, BirAtransgenic RAW264.7 devoid of BLRP-tagged proteins. (E) Cumulative H3K4me1 levels in PU.1−/− myeloid progenitors (PU.1−/−), PUER cells treated for 24 h with tamoxifen (PUER 24 h) or mouse liver as control (liver H3K4me1 ChIP-Seq data from (Robertson et al., 2008)) around LXRβ peak positions defined in RAW264.7 macrophages. (F) LXR binding at the indicated loci in PU.1−/− cells vs. PUER cells treated with tamoxifen for 24 h as compared by ChIP-qPCR. Values represent fold enrichment over ChIP with IgG control antibody. (G) Cumulative H3K4me1 levels in bone marrow macrophages derived from LXRα−/−/ LXRβ−/−mice (LXR DKO BMDM), wild type mice (WT BMDM) or liver (Robertson et al., 2008) as non-macrophage control, around LXRβ peak positions defined in RAW264.7 macrophages. (H) Scatter plot depicting PU.1 ChIP-Seq tag counts (log2) within 200 bp of combined genomic PU.1 peak positions defined in bone marrow-derived macrophages from wild type mice (WT MФ) or LXRα−/−/ LXRβ−/− mice (LXR DKO MФ). Genomic PU.1 peaks within 100 bp of an LXRβ peak in RAW264.7 macrophages are colored red.
To determine the influence of LXRs and PU.1 on each other’s DNA binding profiles, LXR binding in the absence or presence of PU.1 was compared in PU1−/− and tamoxifen-treated PUER cells, and conversely, PU.1 binding was assessed in wild type and in LXRα/β double-knockout bone marrow-derived macrophages. Quantitative ChIP analysis for LXRβ in PU.1−/− vs. tamoxifen-treated PUER cells demonstrated that LXRβ recruitment to sites with vicinal PU.1 binding (within 100 bp) was PU.1-dependent (Figure 5F). This was not the case for LXRβ binding sites that were not in close proximity to PU.1 (Figure 5F), indicating that PU.1 determines LXR binding in a spatially restricted manner. In concert with these findings, macrophage-specific LXRβ- bound regions displayed a significant PU.1-dependent gain in H3K4me1 signal comparing H3K4me1 ChIP-Seq profiles in PU.1−/− vs. tamoxifen-treated PUER cells (Figure 5E). Conversely, the PU.1 cistrome and H3K4me1 patterns around macrophage-specific LXRβ-bound sites were not significantly altered in LXRα/β double-knockout macrophages (Figure 5G and 5H), suggesting that LXRs do not contribute to defining the PU.1 cistrome or to establishing the H3K4me1 pattern at these regions.
PU.1 is required for LXR and TLR-dependent gene expression
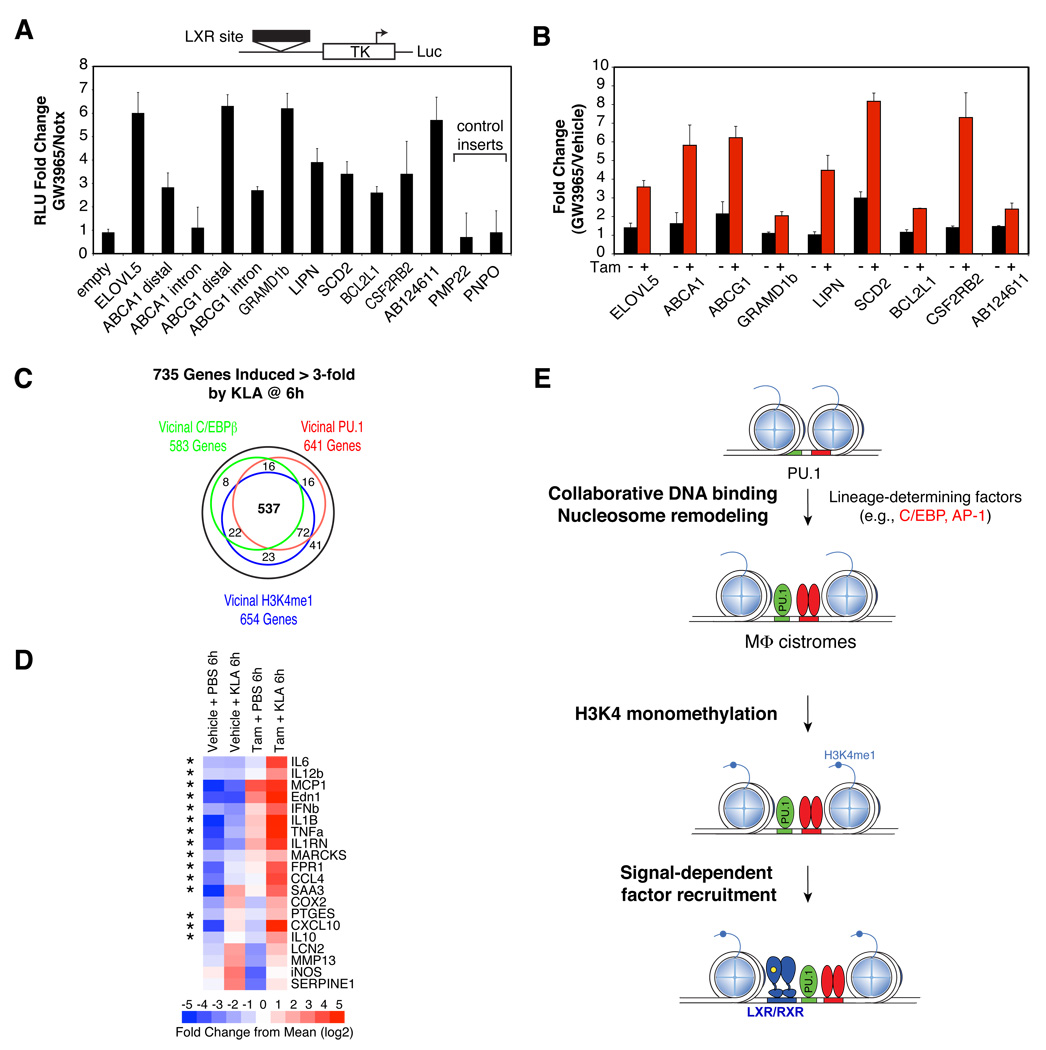
Although PU.1 binding sites are significantly correlated with expression of vicinal genes, (Figure 1E and 1F), an unbiased analysis of 71 sites in resting macrophages and B cells using a transient reporter gene assay revealed that a relatively small fraction (15%) conferred constitutive enhancer activity (Figure S6A). Given the finding that PU.1 participates in establishing the macrophage-specific LXR cistrome, it is possible that many of these sites might participate in defining the context-and signal-dependent transcriptional repertoire in each cell type. In support of this concept, regions containing both LXR and PU.1 binding sites exhibit significant sequence conservation in vertebrates, consistent with their potential importance as functional signal-responsive regulatory modules (Figure S5D). Functional analysis of representative LXR-PU.1 co-bound genomic regions vicinal to LXR target genes (as defined by their responsiveness to the synthetic LXR ligand GW3965 in primary macrophages, Figure S6B) by transient reporter gene analysis in RAW264.7 cells, demonstrated that 10 of 11 genomic regions tested exhibited ligand-dependent enhancer activity (Figure 6A). Furthermore, induction of LXR target genes in response to the LXR agonist GW3965 in PUER cells was markedly PU.1-dependent (Figure 6B).
Figure 6. PU.1 is necessary for LXR- and TLR4-dependent gene expression in macrophages.
(A) Relative enhancer activity of indicated LXR-bound regions inserted upstream of a minimal TK promoter driving luciferase expression in RAW264.7 macrophages. Data are presented as average fold change (±SEM) comparing 24 h treatment with 1 µM GW3965 vs. vehicle (DMSO) of three independent experiments performed in triplicate. (B) PUER cells were treated with tamoxifen or vehicle (EtOH) for 24 h, then with 1 µM GW3965 or vehicle (DMSO) for an additional 24 h. Average mRNA expression fold changes (±SEM) of the indicated genes as determined by quantitative real-time RT-PCR (qRT-PCR) comparing GW3965 versus vehicle in two independent experiments are shown, all expression changes were statistically significant (p<0.05, t-test). (C) Overlap of vicinal PU.1, C/EBPβ and H3K4me1 peaks in resting macrophages associated with genes that are induced more than 3-fold by KLA at 6 h. (D) PUER cells were treated with vehicle (DMSO) or tamoxifen for 24 h, then with vehicle (PBS) or KLA for 6 h. Messenger RNA levels for the indicated TLR4-responsive genes were determined by qRT-PCR and displayed as a heat map with absolute expression values ranging from low (blue) to high (red) compared to the mean level of expression for each gene. Asterisks denote genes whose KLA response is PU.1-dependent. (E) Schematic depicting the proposed model.
To test whether such PU. 1 dependence is a feature of other transcriptional programs defining macrophage-specific outputs, we assessed PU.1 dependence for TLR4 signaling responses in PUER cells. TLR4 ligation induces high magnitude changes in the expression of hundreds of genes in macrophages involved in innate immune responses through the activation of numerous signal-dependent transcription factors, including NF-κB and interferon regulatory factors (Lee and Kim, 2007). Transcriptome analysis of primary macrophages treated for 6 h with the specific TLR4 agonist Kdo2 lipid A (KLA) identified 735 genes induced more than 3-fold. PU.1 and C/EBPβ binding in resting macrophages occurred in the vicinity of 641 and 583 of these genes, respectively, and prior to KLA stimulation, these two factors co-localized, alone or together, with 537 of the 654 H3K4me1-marked regions associated with these genes (Figure 6C). Quantitative transcript analysis of 20 of these genes in the PUER system indicated that 14 required induction of PU.1 binding for qualitative or substantial quantitative responses to KLA (Figure 6D). Full responses of the remainder of these genes in untreated PUER cells indicated that the TLR4 signaling pathway is functional prior to activation of PU.1.
Discussion
Here we provide evidence that collaborative interactions between PU.1 and small sets of other lineage-determining transcription factors at closely spaced binding sites establish a large proportion of their respective macrophage and B cell-specific cistromes. The spatial distributions and frequencies of the motifs for each of these factors suggest these patterns arise from direct binding of each factor to its cognate DNA motif rather than tethering of one factor to the other. Conversely, the lack of a fixed distance between the motifs, their enrichment within 100 bp of each other and the nucleosome remodelling observed around sites where inducible PU. 1 binds in PUER cells, often in conjunction with C/EBPβ, is consistent with a mechanism in which concurrent binding of these factors enables effective competition with nucleosomes (Miller and Widom, 2003) to define cell type-specific binding patterns.
The finding that PU.1 binding leads to deposition of the H3K4me1 mark at promoter-distal sites that are correlated with cell type-specific patterns of gene expression (Heintzman et al., 2009) provides the initial evidence that PU.1 contributes to the ‘writing’ of this epigenetic signature on a genome-wide scale. Our data also indicate that PU.1 binding and nucleosome remodelling are not sufficient to result in the deposition of this mark in all cases (Figure 4C, Group III), suggesting that the mechanisms that ultimately lead to H3K4 monomethylation also require collaborative interactions with other transcription factors, such as C/EBPβ. Additionally, the motifs enriched in focally H3K4me1-marked promoter-distal regions in macrophages and B cells and the shape of the H3K4me1 patterns around the binding sites of three of these factors (C/EBPα/β, Oct-2) suggest that the other H3K4me1-associated factors are also involved in nucleosome displacement and targeting of H3K4me1 deposition around their binding sites. The observation that H3K4me1-marked regions in other tissues and cell types, including liver, erythrocytes, T cells and ES cells, also exhibit enrichment for limited sets of motifs for corresponding lineage determining factors (Figure S4C) suggests a general approach to the identification of such factors in distinct cell types, knowledge of which may facilitate cellular reprogramming efforts.
Taken together, our findings support a model in which two tiers of transcription factors cooperate to activate cis-active regulatory elements required for the development and function of macrophages and B cells (Figure 6E). The first tier consists of relatively small sets of lineage-determining factors, such as PU.1, C/EBPs and E2A, that act in a collaborative manner to delineate large sets of potential cis-regulatory elements in a cell type-specific fashion (exemplified for macrophages in Figure 6E). The specific attributes of these factors that enable this role remain to be defined, but presumably include expression levels and the ability to engage nucleosome remodeling factors and ultimately histone modifying enzymes that generate an accessible ‘proto-enhancer’ structure. Consistent with this, wild type E2A induces PU.1 binding at B cell-specific genomic sites that contain closely spaced PU.1 and E2A motifs, while a mutant form of E2A that is competent to bind to DNA but lacks transcriptional activation function does not (Figure 3C). This result highlights an important role for transactivation domains in enabling combinatorial transcription factor binding to chromatinized DNA at enhancer-like elements, in addition to their known roles in transcription initiation.
We provide further evidence that the priming of cis-active elements by lineage-determining factors is required for the subsequent binding of a second tier of factors, exemplified by LXRs (Figure 6E) and transcription factors that are activated by TLR4 signalling. This interpretation is supported by the recent observation that recruitment of p300 to new genomic locations in macrophages in response to TLR4 activation primarily occurs at regions exhibiting pre-existing H3K4me1 (Ghisletti et al., 2010), which from our analysis largely are sites of combinatorial interactions of PU.1, C/EBP and AP-1 transcription factors. Collectively, these sequential events lead to a functionally active enhancer capable of contributing to cell type-specific and/or signal-dependent gene expression, similar to the sequential steps leading to activation of the Csf1r locus during macrophage development (Krysinska et al., 2007). This model offers an explanation for the extensive genome-wide and cell type-specific co-localization of transcription factors observed in various previous studies (Chen et al., 2008; MacArthur et al., 2009), and provides insights into how simple combinations of lineage-restricted transcription factors on a genome-wide scale can specify promoter-distal cis-regulatory elements ultimately responsible for both cell identity and cell type-specific responses to diverse signaling inputs.
Experimental Procedures
Cell Isolation and Culture
Primary cells were isolated from male 6–8 week-old C57Bl/6 mice (Charles River Laboratories). Peritoneal macrophages were harvested by peritoneal lavage 3 days after i.p. injection of 3 ml thioglycollate, overnight culture and adherence selection. BMDM were generated as described (Valledor et al., 2004). Splenic B cells were isolated by magnetic depletion of CD43- and CD11b–expressing cells (Miltenyi). B220+ pro-B cells were magnetically enriched from bone marrow of Rag1 knockout mice and expanded for 10 days as described (Sayegh et al., 2005) with slight modifications. E2A−/−and EBF−/− pre-pro-B cells were cultured as described previously (Ikawa et al., 2004). PU.1−/− and PUER cells were propagated and the PU.1-ER fusion protein was activated with 100 nM 4-hydroxy-tamoxifen as described (Walsh et al., 2002).
Microarray Analysis
Gene expression profiling was performed in biological duplicates on 44K Whole Mouse Genome Oligo Microarrays (Agilent) according to the manufacturer’s instructions.
Retroviral Transduction
E47-ER-Tac and bHLH-ER-Tac retroviral constructs were described (Sayegh et al., 2003). Virus was generated by transfection of the constructs and packaging vector into HEK 293T cells. E2A–deficient hematopoietic progenitor cells were transduced as described previously (Quong et al., 1999). After 18 h, E47-ER and bHLH-ER were activated for 6 hr with 1 µM 4-hydroxy-tamoxifen and transduced cells were magnetically enriched (Miltenyi) for bi-cistronically co-expressed human CD25 (TAC antigen).
Chromatin Immunoprecipitation (ChIP)
ChIP was performed with 10–20×106 cells essentially as described (Metivier et al., 2003). Antibodies used: PU.1 (sc-352), C/EBPα (sc-61), C/EBPβ (sc-150), Oct-2 (sc-233) and pan-LXR (sc-1000) (Santa Cruz Biotech), H3K4me1 (ab8895) and H3K4me3 (ab8580) (Abcam) or LXRβ (PP-K8917-00, Perseus Proteomics).
MNase-Seq
Micrococcal nuclease digest and deep sequencing (MNase-Seq) was essentially performed as described (Schones et al., 2008).
High-Throughput Sequencing
Library preparation and high-throughput sequencing for 36 cycles was performed on Illumina Genome Analyzers I and II according to the manufacturer’s protocols (Illumina). The first 23–25 bp of each sequence tag were mapped to the mm8 assembly (NCBI build 36). Tag counts for each experiment were normalized to 107 specifically mapped tags.
Data Analysis
Peak finding and downstream data analysis was performed using HOMER, a software suite for ChIP-Seq analysis, which in part was created to support this study. Peaks were defined at a 0.1% estimated false discovery rate, indicated as a dashed line in genome browser images.
Supplementary Material
Acknowledgements
We thank Dr. Bing Ren, Dr. Michael Rehli and Dr. Constanze Bonifer for discussions and reading of the manuscript, Dr. Gary Hardiman and especially Colleen Ludka for assistance with Solexa sequencing, Rosa Luna for technical assistance, Dr. Young-Soo Kwon for suggestions and discussions and Lynn Bautista for assistance with manuscript preparation. SH was supported by an NIH postdoctoral training grant. These studies were primarily funded by NURSA consortium grant No DK62434 and further supported by NIH grants to CKG (HC088093, DK063491, CA52599), HS (P50 GM081892-01A1) and CM and a Foundation Leducq Transatlantic Network Grant to CKG. HS is an HHMI Investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
Sequencing and gene expression data are available in the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo) under the accession number GSE21512.
Detailed methods are described in the Supplemental Information.
References
- Adolfsson J, Mansson R, Buza-Vidas N, Hultquist A, Liuba K, Jensen CT, Bryder D, Yang L, Borge OJ, Thoren LA, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121:295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Boyes J, Felsenfeld G. Tissue-specific factors additively increase the probability of the all-or-none formation of a hypersensitive site. EMBO J. 1996;15:2496–2507. [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai DH, Wang D, Keefer J, Yeamans C, Hensley K, Friedman AD. C/EBP alpha:AP-1 leucine zipper heterodimers bind novel DNA elements, activate the PU.1 promoter and direct monocyte lineage commitment more potently than C/EBP alpha homodimers or AP-1. Oncogene. 2008;27:2772–2779. doi: 10.1038/sj.onc.1210940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9:279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- Cui K, Zang C, Roh TY, Schones DE, Childs RW, Peng W, Zhao K. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell. 2009;4:80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias S, Mansson R, Gurbuxani S, Sigvardsson M, Kee BL. E2A proteins promote development of lymphoid-primed multipotent progenitors. Immunity. 2008;29:217–227. doi: 10.1016/j.immuni.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenbeis CF, Singh H, Storb U. PU.1 is a component of a multiprotein complex which binds an essential site in the murine immunoglobulin lambda 2–4 enhancer. Mol Cell Biol. 1993;13:6452–6461. doi: 10.1128/mcb.13.10.6452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B, Jiang J, Kraus P, Ng JH, Heng JC, Chan YS, Yaw LP, Zhang W, Loh YH, Han J, et al. Reprogramming of fibroblasts into induced pluripotent stem cells with orphan nuclear receptor Esrrb. Nat Cell Biol. 2009;11:197–203. doi: 10.1038/ncb1827. [DOI] [PubMed] [Google Scholar]
- Fisher RC, Scott EW. Role of PU.1 in hematopoiesis. Stem Cells. 1998;16:25–37. doi: 10.1002/stem.160025. [DOI] [PubMed] [Google Scholar]
- Friedman AD. Transcriptional control of granulocyte and monocyte development. Oncogene. 2007;26:6816–6828. doi: 10.1038/sj.onc.1210764. [DOI] [PubMed] [Google Scholar]
- Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, Gregory L, Lonie L, Chew A, Wei CL, et al. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 2010;32:317–328. doi: 10.1016/j.immuni.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikawa T, Kawamoto H, Wright LY, Murre C. Long-term cultured E2A–deficient hematopoietic progenitor cells are pluripotent. Immunity. 2004;20:349–360. doi: 10.1016/s1074-7613(04)00049-4. [DOI] [PubMed] [Google Scholar]
- Ivanova N, Dobrin R, Lu R, Kotenko I, Levorse J, DeCoste C, Schafer X, Lun Y, Lemischka IR. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysinska H, Hoogenkamp M, Ingram R, Wilson N, Tagoh H, Laslo P, Singh H, Bonifer C. A two-step, PU.1-dependent mechanism for developmentally regulated chromatin remodeling and transcription of the c-fms gene. Mol Cell Biol. 2007;27:878–887. doi: 10.1128/MCB.01915-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MS, Kim YJ. Signaling pathways downstream of pattern-recognition receptors and their cross talk. Annu Rev Biochem. 2007;76:447–480. doi: 10.1146/annurev.biochem.76.060605.122847. [DOI] [PubMed] [Google Scholar]
- Lichtinger M, Ingram R, Hornef M, Bonifer C, Rehli M. Transcription factor PU.1 controls transcription start site positioning and alternative TLR4 promoter usage. J Biol Chem. 2007;282:26874–26883. doi: 10.1074/jbc.M703856200. [DOI] [PubMed] [Google Scholar]
- Lin H, Grosschedl R. Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature. 1995;376:263–267. doi: 10.1038/376263a0. [DOI] [PubMed] [Google Scholar]
- Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur S, Li XY, Li J, Brown JB, Chu HC, Zeng L, Grondona BP, Hechmer A, Simirenko L, Keranen SV, et al. Developmental roles of 21 Drosophila transcription factors are determined by quantitative differences in binding to an overlapping set of thousands of genomic regions. Genome Biol. 2009;10:R80. doi: 10.1186/gb-2009-10-7-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina KL, Singh H. Gene regulatory networks orchestrating B cell fate specification, commitment, and differentiation. Curr Top Microbiol Immunol. 2005;290:1–14. doi: 10.1007/3-540-26363-2_1. [DOI] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JA, Widom J. Collaborative competition mechanism for gene activation in vivo. Mol Cell Biol. 2003;23:1623–1632. doi: 10.1128/MCB.23.5.1623-1632.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombaerts P, Iacomini J, Johnson RS, Herrup K, Tonegawa S, Papaioannou VE. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- Odom DT, Zizlsperger N, Gordon DB, Bell GW, Rinaldi NJ, Murray HL, Volkert TL, Schreiber J, Rolfe PA, Gifford DK, et al. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303:1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pongubala JM, Nagulapalli S, Klemsz MJ, McKercher SR, Maki RA, Atchison ML. PU.1 recruits a second nuclear factor to a site important for immunoglobulin kappa 3' enhancer activity. Mol Cell Biol. 1992;12:368–378. doi: 10.1128/mcb.12.1.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quong MW, Harris DP, Swain SL, Murre C. E2A activity is induced during B-cell activation to promote immunoglobulin class switch recombination. EMBO J. 1999;18:6307–6318. doi: 10.1093/emboj/18.22.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigamonti E, Chinetti-Gbaguidi G, Staels B. Regulation of macrophage functions by PPAR-alpha, PPAR-gamma, and LXRs in mice and men. Arterioscler Thromb Vasc Biol. 2008;28:1050–1059. doi: 10.1161/ATVBAHA.107.158998. [DOI] [PubMed] [Google Scholar]
- Robertson AG, Bilenky M, Tam A, Zhao Y, Zeng T, Thiessen N, Cezard T, Fejes AP, Wederell ED, Cullum R, et al. Genome-wide relationship between histone H3 lysine 4 mono- and tri-methylation and transcription factor binding. Genome Res. 2008;18:1906–1917. doi: 10.1101/gr.078519.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg EV, Taghon T. Molecular genetics of T cell development. Annu Rev Immunol. 2005;23:601–649. doi: 10.1146/annurev.immunol.23.021704.115737. [DOI] [PubMed] [Google Scholar]
- Sandmann T, Jensen LJ, Jakobsen JS, Karzynski MM, Eichenlaub MP, Bork P, Furlong EE. A temporal map of transcription factor activity: mef2 directly regulates target genes at all stages of muscle development. Dev Cell. 2006;10:797–807. doi: 10.1016/j.devcel.2006.04.009. [DOI] [PubMed] [Google Scholar]
- Sayegh CE, Jhunjhunwala S, Riblet R, Murre C. Visualization of looping involving the immunoglobulin heavy-chain locus in developing B cells. Genes Dev. 2005;19:322–327. doi: 10.1101/gad.1254305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayegh CE, Quong MW, Agata Y, Murre C. E-proteins directly regulate expression of activation-induced deaminase in mature B cells. Nat Immunol. 2003;4:586–593. doi: 10.1038/ni923. [DOI] [PubMed] [Google Scholar]
- Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tronche F, Yaniv M. HNF1, a homeoprotein member of the hepatic transcription regulatory network. Bioessays. 1992;14:579–587. doi: 10.1002/bies.950140902. [DOI] [PubMed] [Google Scholar]
- Valledor AF, Hsu LC, Ogawa S, Sawka-Verhelle D, Karin M, Glass CK. Activation of liver X receptors and retinoid X receptors prevents bacterial-induced macrophage apoptosis. Proc Natl Acad Sci U S A. 2004;101:17813–17818. doi: 10.1073/pnas.0407749101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verger A, Duterque-Coquillaud M. When Ets transcription factors meet their partners. Bioessays. 2002;24:362–370. doi: 10.1002/bies.10068. [DOI] [PubMed] [Google Scholar]
- Walsh JC, DeKoter RP, Lee HJ, Smith ED, Lancki DW, Gurish MF, Friend DS, Stevens RL, Anastasi J, Singh H. Cooperative and antagonistic interplay between PU.1 and GATA-2 in the specification of myeloid cell fates. Immunity. 2002;17:665–676. doi: 10.1016/s1074-7613(02)00452-1. [DOI] [PubMed] [Google Scholar]
- Wederell ED, Bilenky M, Cullum R, Thiessen N, Dagpinar M, Delaney A, Varhol R, Zhao Y, Zeng T, Bernier B, et al. Global analysis of in vivo Foxa2-binding sites in mouse adult liver using massively parallel sequencing. Nucleic Acids Res. 2008;36:4549–4564. doi: 10.1093/nar/gkn382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigelt K, Lichtinger M, Rehli M, Langmann T. Transcriptomic profiling identifies a PU.1 regulatory network in macrophages. Biochem Biophys Res Commun. 2009;380:308–312. doi: 10.1016/j.bbrc.2009.01.067. [DOI] [PubMed] [Google Scholar]
- Xi H, Shulha HP, Lin JM, Vales TR, Fu Y, Bodine DM, McKay RD, Chenoweth JG, Tesar PJ, Furey TS, et al. Identification and characterization of cell type-specific and ubiquitous chromatin regulatory structures in the human genome. PLoS Genet. 2007;3:e136. doi: 10.1371/journal.pgen.0030136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3' UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, Watts J, Xu J, Wandzioch E, Smale ST, Sekiya T. Pioneer Factors, Genetic Competence, and Inductive Signaling: Programming Liver and Pancreas Progenitors from the Endoderm. Cold Spring Harb Symp Quant Biol. 2008 doi: 10.1101/sqb.2008.73.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.