Abstract
It is an unconventional gelsolin-related protein with four gelsolin-like (G) domains (G1 to G4), unlike typical gelsolin-related proteins with three or six G domains. GSNL-1 severs actin filaments and caps the barbed end in a calcium-dependent manner similarly to gelsolin. In contrast, GSNL-1 has different properties from gelsolin in that it remains bound to F-actin, and does not nucleate actin polymerization. To understand the mechanism by which GSNL-1 regulates actin dynamics, we investigated domain-function relationship of GSNL-1 by analyzing activities of truncated forms of GSNL-1. G1 plus the linker between G1 and G2 was sufficient for actin filament severing, whereas G1 and G2 were required for barbed-end capping. Actin severing activity of GSNL-1 was inhibited by phosphatidylinositol 4,5-bisphosphate (PIP2), and a PIP2-sensitive domain was mapped to G1–G2. At least two actin-binding sites were detected: a calcium-dependent G-actin-binding site in G1 and a calcium-independent G- and F-actin-binding site in G3–G4. These results reveal both conserved and different utilization of G domains between C. elegans GSNL-1 and mammalian gelsolin for actin-regulatory functions.
Actin filaments are relatively stable, and regulated disassembly of pre-existing filaments is often a critical process for remodeling of the actin cytoskeleton in a number of dynamic cellular events (1). Severing of actin filaments is an effective way to initiate filament disassembly. Gelsolin-related proteins (2–4) and actin-depolymerizing factor (ADF)/cofilin proteins (5–7) are the two major classes of actin-regulatory proteins that enhance actin filament turnover by severing and depolymerizing actin filaments. Gelsolin strongly severs actin filaments and caps the barbed ends in a calcium-dependent manner, while ADF/cofilin weakly severs actin filaments without capping filament ends in a calcium-independent manner. Thus, gelsolin and ADF/cofilin have different impacts on actin filament dynamics and distinct functions in regulating reorganization of the actin cytoskeleton.
Gelsolin has six repeats of homologous domains of 100 – 120 amino acids, which are designated as gelsolin-like (G) domains or segments (2–4). Despite the fact that the G domains of gelsolin have homologous sequences and three-dimensional structures, each G domain plays distinct roles in actin filament severing and capping. The most N-terminal G domain plus a linker sequence is sufficient for actin filament severing (8) and capping (9). The first and fourth G domains bind to actin at a cleft between subdomains 1 and 3 (10–13), while the second G domain binds to actin subdomain 2 that remains exposed on the surface of F-actin (12, 14, 15). Calcium-free inactive gelsolin has a tightly packed globular conformation with a number of interdomain interactions (16). Calcium activation of gelsolin induces large domain movements by dissociating some of the interdomain interactions and exposing actin-binding sites (12, 13, 17). Thus, each G domain of gelsolin has either direct or indirect roles in the calcium-dependent actin-regulatory activities.
G domains are found in many gelsolin-related proteins that have variable numbers of G domains ranging from two to six (1). However, roles of G domains of gelsolin-related proteins other than gelsolin in actin-regulatory activities are poorly understood. We have recently reported that gelsolin-like protein-1 (GSNL-1) from Caenorhabditis elegans is a novel gelsolin-related protein with four G domains (18). GSNL-1 is similar to gelsolin in that it severs and caps actin filaments. However, unlike gelsolin, GSNL-1 remains bound to actin filaments and does not nucleate actin polymerization (18). Three N-terminal G domains of GSNL-1 are homologous to the equivalent region of gelsolin, whereas the most C-terminal G domain of GSNL-1 is most closely related to the sixth G domain of gelsolin. Therefore, GSNL-1 lacks two G domains that are equivalent to fourth and fifth G domains of gelsolin. These unique properties of GSNL-1 prompted us to dissect functional domains of GSNL-1, as it can be a model to analyze how multiple G domains coordinate to regulate actin filament dynamics. We found that G domains of GSNL-1 have distinct roles in actin-regulatory activities with similarities and differences from those of gelsolin. These results provide interesting information on conservation and diversity of utilization of G domains in gelsolin-related proteins.
Experimental Procedures
Proteins and materials
Actin was purified from rabbit skeletal muscle acetone powder (Pel-Freeze Biologicals) as described (19). Pyrene-labeled actin was prepared as described (20). Bacterially expressed full-length GSNL-1 was purified as described previously (18). Phosphatidylinositol 4,5-bisphosphate (PIP2) (P-4516, Echelon Biosciences) was suspended in water at 1 mg/ml with brief sonication and stored at −20 °C.
Production of truncation mutants of GSNL-1
Fragments of GSNL-1 cDNA encoding truncated GSNL-1 sequences as shown in Fig. 1 were amplified by polymerase chain reaction from pGEX-GSNL-1 (18) as a template and cloned into pGEX-2T between the BamH I and Sma I sites using an Infusion PCR Cloning Kit (Clontech). The sequences of the inserts were verified by DNA sequencing. The Escherichia coli strain BL21(DE3) was transformed with the expression vectors and cultured in M9ZB (18.7 mM NaH4Cl, 22 mM KH2PO4, 42.3 mM Na2HPO4, 1 % Tryptone, 85.6 mM NaCl, 1 mM MgSO4, and 0.4 % glucose) containing 50 μg/ml ampicillin at 37 °C until A600 reached at 0.6 cm−1. Then, the cultures were cooled to room temperature, and expression was induced by adding 0.1 mM isopropyl-1-thio-β-D-galactopyranoside for 2 h at room temperature. The cells were harvested by centrifugation at 5000 × g for 10 min and homogenized by a French Pressure cell at 360–580 kg/cm2 in phosphate-buffered saline (137 mM NaCl, 2.7 mM KCl, 1.4 mM KH2PO4, 8 mM Na2HPO4). The homogenates were centrifuged at 20000 × g for 15 min, and the supernatants were applied to glutathione Uniflow (Clontech) column (1.5-ml bed volume). After washing with phosphate-buffered saline, 15 units of thrombin (Roche Applied Science) was added to cleave the N-terminal glutathione S-transferase (GST) tag, and GSNL-1 truncation mutants were eluted from the column. They were dialyzed against F-buffer (0.1 M KCl, 2 mM MgCl2, 20 mM HEPES-NaOH, pH 7.5) containing 50 % glycerol and stored at −20 °C. Protein concentrations were determined by densitometry of Coomassie blue-stained gels after SDS-PAGE using actin as a standard.
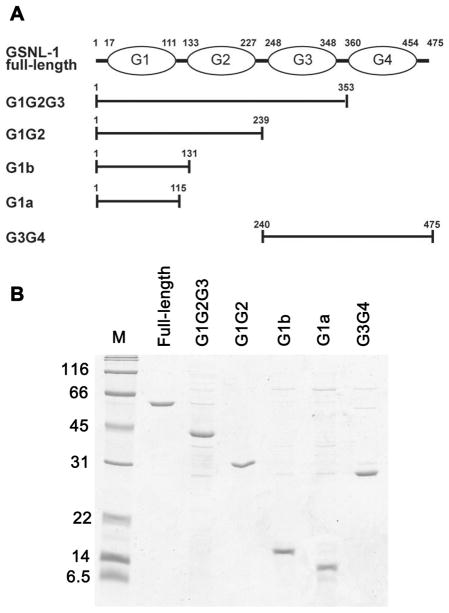
Figure 1.
Expression and purification of GSNL-1 variants. (A) Schematic representation of domain structure of full-length GSNL-1 (top). G domains are designated as G1 to G4. Positions of N- and C-termini of each domain are shown on top. Positions of amino acid sequence for each GSNL-1 fragments are shown. (B) Purified recombinant GSNL-1 variants (0.5 μg each) were subjected to SDS-PAGE and stained by Coomassie Brilliant Blue. Molecular mass markers (M) in kDa are shown on the left of the gel.
Labeling of actin by DyLight549
4 ml of polymerized actin at 4 mg/ml in a buffer containing 0.1 M KCl, 20 mM NaHCO3, 2 mM MgCl2, 0.2 mM ATP, and 0.2 mM dithiothreitol was mixed with 100 μl of 10 mg/ml DyLight 549 NHS-ester (Pierce/Thermo Scientific) in dimethylformamide and incubated overnight on ice. F-actin was sedimented by ultracentrifugation at 100,000 rpm for 1 hr by a Beckman TLA100.3 rotor, and suspended in 2 ml of G-buffer containing 0.2 mM CaCl2, 0.2 mM ATP, 0.2 mM dithiothreitol, and 2 mM Tris-HCl at pH 8.0. After brief sonication, it was dialyzed against G-buffer overnight to depolymerize actin and to remove unincorporated DyLight 549. Then, remaining F-actin was removed by ultracentrifugation at 100,000 rpm for 1 hr by a Beckman TLA100.3 rotor, and the supernatant was applied to gel filtration using Sephacryl S-300 (HiPrep 26/60, GE Healthcare) equilibrated with G-buffer. Fractions containing G-actin were pooled and concentrated by a Vivaspin 15R centrifugal concentrator (MWCO 10000) to ~40 μM actin. Concentration of actin was determined spectrophotometrically using an extinction coefficient of 0.63 mg−1 ml cm−1 at 290 nm. The molar ratio of the bound DyLight 549 to actin was 0.65, which was determined as manufacture’s instruction. DyLight 549-labeled G-actin was snap-frozen in small aliquots and stored at −80 °C.
Direct observation of actin filaments by fluorescence microscopy
Unlabeled G-actin was co-polymerized with DyLight 549-labeled G-actin at 2 μM total actin (20 % labeled) for 2 hr in F-buffer. Labeled actin was diluted to 0.4 μM in F-buffer with or without GSNL-1 variants in the presence of 0.1 mM CaCl2 or 5 mM EGTA and incubated for 5 min at room temperature. Then, the reactions were put on a nitrocellulose-coated coverslip, and immobilized actin filaments on the coverslip were observed by epifluorescence using a Nikon TE2000 microscope with a 60x Plan Apo objective (oil, numerical aperture of 1.4). Micrographs were captured by a SPOT RT Monochrome CCD camera (Diagnostic Instruments) and processed by IPLab (BD Biosciences) and Adobe Photoshop CS2.
Determination of critical concentration of actin
Varying concentrations of pyrene-labeled G-actin (20 % labeled) were polymerized overnight at room temperature in the presence of constant concentrations of GSNL-1 fragments in F-buffer containing 0.1 mM CaCl2 or 0.1 mM EGTA. Fluorescence intensity of pyrene (excitation at 366 nm and emission at 384 nm) at the steady state was measured with a Perkin-Elmer LS50B fluorescence spectrophotometer.
F-actin co-sedimentation assays
F-actin co-sedimentation assays were performed as described (21) with some modifications. Varying concentrations of GSNL-1 variants were added to 5 μM F-actin in F-buffer plus 0.2 mM dithiothreitol and 0.1 mM CaCl2 or 0.1 mM EGTA. After incubation for 1 hr at room temperature, the mixtures were ultracentrifuged by a Beckman TLA100 rotor at 436,000 × g for 15 min at 20 °C. Supernatant and pellet fractions were adjusted to the same volumes and subjected to SDS-PAGE and staining with Coomassie Brilliant Blue R-250 (National Diagnostics). Gels were scanned by an Epson Perfection V700 Photo Scanner at 300 dots per inch, and band intensity was quantified using Image J. To determine co-sedimentation of GSNL-1 variants with F-actin, experiments were performed in the absence of actin, and actin-independent sedimentations of GSNL-1 variants were quantified and subtracted from the experimental data in the presence of actin.
Non-denaturing polyacrylamide gel electrophoresis
Non-denaturing polyacrylamide gel electrophoresis to examine G-actin binding and PIP2 binding was performed as described (22, 23). GSNL-1 variants were incubated with G-actin or PIP2 in G-buffer for 30 min at room temperature. The samples were supplemented with 0.25 volume of a loading buffer (50 % glycerol, 0.05 % bromophenol blue) and electrophoresed using a Bicine/triethanolamine buffer system at 4 °C. The proteins were visualized by staining with Coomassie Brilliant Blue R-250 (National Diagnostics).
Analytical gel filtration chromatography
A Superdex 75 10/300 GL gel filtration column (24 ml bed, GE Healthcare) was equilibrated with F-buffer containing 0.2 mM dithiothreitol and 0.1 mM CaCl2, and proteins at 1 mg/ml (500 μl) were applied and eluted with the same buffer. The eluates were collected in fractions of 0.5 ml each, and analyzed by SDS-PAGE and staining with Coomassie Brilliant Blue R-250. Gels were scanned by an Epson Perfection V700 Photo Scanner at 300 dots per inch, and band intensity was quantified using Image J. Bovine serum albumin (66 kDa) (A8531, Sigma-Aldrich) and bovine carbonic anhydrase (29 kDa) (C7025, Sigma-Aldrich) were used as molecular mass standards.
Results
Construction of truncation mutants of GSNL-1 for domain mapping
To dissect functional domains of GSNL-1, we produced five truncation mutants containing different G domains as shown in Fig. 1. Each G domain was truncated from the C-terminus to generate G1G2G3, G1G2, and G1. We made two G1 variants with (G1b) or without (G1a) a linker between G1 and G2 (Fig. 1A) because a homologous region of gelsolin is implicated in actin filament severing activity (8, 24, 25). We also made G3G4, which lacks two N-terminal G domains. These GSNL-1 fragments were expressed in Escherichia coli as fusion proteins with GST and purified after removing the GST tag (Fig. 1B). We also tested bacterial expression of a mutant lacking only the C-terminal tail of 16 amino acids and a fragment containing G2G3G4, but these recombinant proteins were insoluble and not functionally tested in this study (our unpublished data).
Actin filament severing
GSNL-1 severs actin filaments in a Ca2+-dependent manner (18). F-actin severing activity of GSNL-1 truncation mutants were examined by direct microscopic observation of fluorescently labeled actin filaments. DyLight 549-labeled actin filaments were incubated with GSNL-1 variants in the presence or absence of calcium, immobilized on a nitrocellulose-coated coverslip, and observed by fluorescence microscopy. Control experiments of actin filaments with buffer only showed long filaments (Fig. 2A and B). At 20 nM of GSNL-1 mutants, G1G2G3 (Fig. 2E), G1G2 (Fig. 2G), and G1b (Fig. 2I) fragmented actin filaments in the presence of Ca2+ as well as full-length GSNL-1 (Fig. 2C) but not in the presence of EGTA (Fig. 2F, H, and J). Although G1a did not sever actin filaments at 20 nM (Fig. 2K and L), it exhibited severing activity at higher concentrations. G1a needed to be increased to 2–5 μM to achieve filament severing to a similar extent to 20 nM full-length GSNL-1 (data not shown), indicating that G1a was >100-fold weaker in actin severing than full-length GSNL-1, G1G2G3, G1G2, and G1b. It should be noted that the difference between G1a and G1b is the absence or presence of the G1–G2 linker sequence (16 amino acids) (Fig. 1A), indicating that this region is critical for actin filament severing. G3G4 did not sever actin filaments (Fig. 2M and N). These results indicate that G1 and the linker between G1 and G2 play critical roles in actin filament severing.
Figure 2.
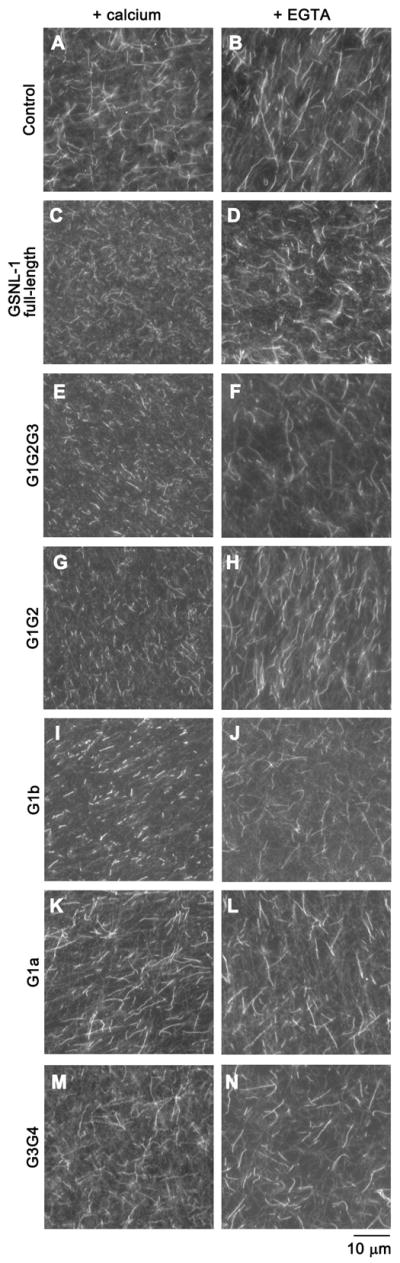
Direct observation of actin filament severing by fluorescence microscopy. DyLight 549-labeled actin filaments were incubated with buffer only (control) (A and B), 20 nM full-length GSNL-1 (C and D), 20 nM G1G2G3 (E and F), 20 nM G1G2 (G and H), 20 nM G1b (I and J), 20 nM G1a (K and L), or 20 nM G3G4 (M and N) in the presence of 0.1 mM CaCl2 (left column) or 5 mM EGTA (right column) for 5 min, and the filaments were observed by fluorescence microscopy. Bar, 10 μm.
Barbed-end capping
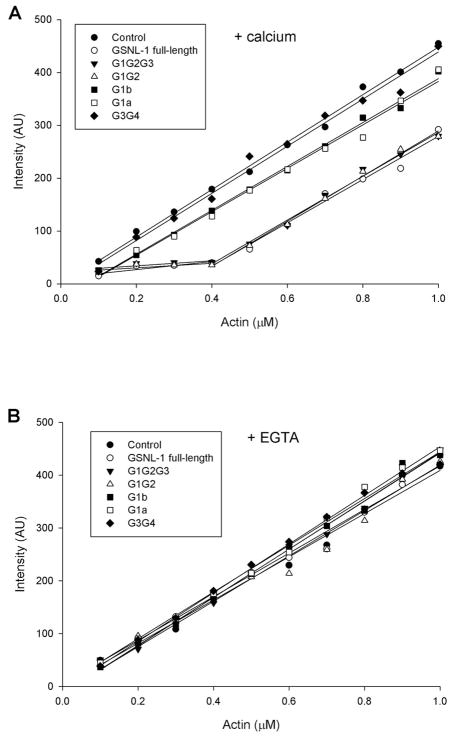
GSNL-1 caps the barbed end of actin filament in a Ca2+-dependent manner (18). Barbed-end capping activity was tested by effects of GSNL-1 truncation mutants on the critical concentration (Cc) of actin. Cc of actin with free ends is generally 0.1–0.2 μM. Since Cc at the barbed end (0.1 μM) is much lower than that at the pointed end (0.5 μM), Cc of barbed-end-capped actin will be near the Cc value at the pointed end (0.5 μM). In the presence of Ca2+, full-length GSNL-1 shifted Cc to ~0.4 μM (Fig. 3A, open circles) indicating that it capped the barbed end. G1G2G3 (Fig. 3A, closed triangles) and G1G2 (Fig. 3A, open triangles) similarly shifted Cc to ~0.4μM in the presence of Ca2+. However, G1a (Fig. 3A, open squares), G1b (Fig. 3A, closed squares), and G3G4 (Fig. 3A, closed diamonds) did not alter Cc. All tested GSNL-1 fragments did not alter Cc in the absence of Ca2+ (Fig. 3B). Thus, G1G2 is sufficient for Ca2+-dependent barbed-end capping. Lack of capping activity of G1a and G1b suggests that G2 is important for barbed-end capping.
Figure 3.
Effects of GSNL-1 variants on critical concentration of actin. Varying concentrations (0.1 – 1.0 μM) of 20 % pyrene-labeled actin were polymerized with or without 50 nM of GSNL-1 variants in the presence of 0.1 mM CaCl2 (A) or 5 mM EGTA (B). After overnight incubation, the pyrene fluorescence (arbitrary units) was measured and plotted as a function of total actin concentration.
G-actin binding
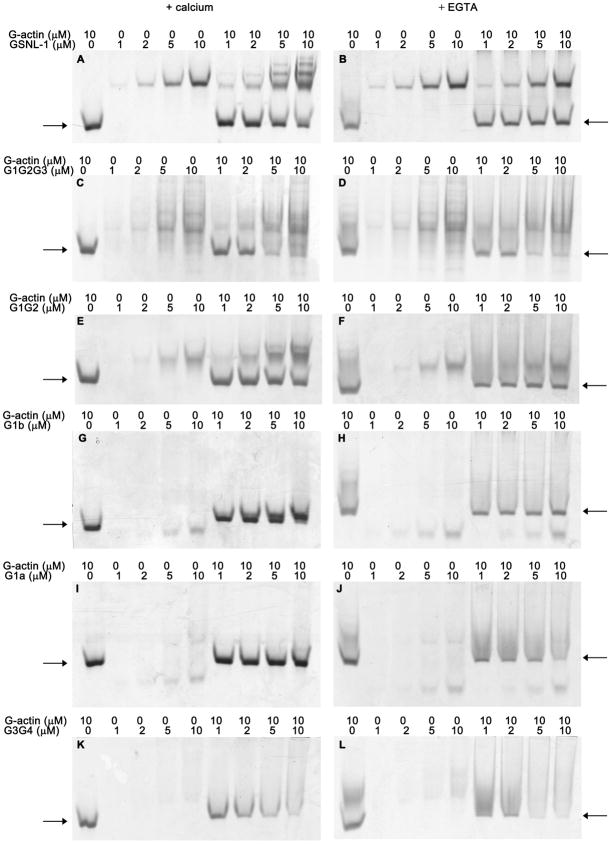
G-actin binding activity was examined by non-denaturing polyacrylamide gel electrophoresis. Full-length GSNL-1 binds to G-actin in the presence of calcium (18) and the complex of GSNL-1 and G-actin appeared as bands with slower mobility than those of GSNL-1 and actin (Fig. 4A). In the absence of calcium, no extra bands were detected in the mixtures of GSNL-1 and G-actin (Fig. 4B), indicating that this interaction is calcium-dependent. In the presence of calcium, all the tested GSNL-1 fragments bound to G-actin (Fig. 4C, E, G, I, and K). For G1G2G3 (Fig. 4C) and G3G4 (Fig. 4K), although bands of the complexes were not clearly detected, the band of actin was diminished when the concentrations of G1G2G3 or G3G4 were increased, suggesting that the complexes of G-actin and G1G2G3 or G3G4 migrated differently than G-actin alone but did not focus in distinct bands. For G1G2 (Fig. 4E) and G1b (Fig. 4G), bands of the complex appeared above the band of G1G2 or G1b by increasing the concentrations of the fragments. G1a did not apparently alter mobility of the actin band (Fig. 4I). However, the band of G1a, which was resolved in two bands, disappeared in the presence of G-actin, suggesting that the G1a-G-actin complex migrated at the same location as G-actin alone.
Figure 4.
Examination of G-actin binding by GSNL-1 variants by non-denaturing polyacrylamide gel electrophoresis. 10 μM G-actin and varying concentrations (0 – 10 μM) of full-length GSNL-1 (A and B), G1G2G3 (C and D), G1G2 (E and F), G1b (G and H), G1a (I and J), or G3G4 (K and L) were incubated alone or in mixtures in the presence of 0.1 mM CaCl2 (left column) or 0.5 mM EGTA (right column) for 30 min, and the samples were analyzed by non-denaturing polyacrylamide gel electrophoresis. Positions of free G-actin are indicated by arrows.
In the absence of calcium, G1G2G3 (Fig. 4D) and G3G4 (Fig. 4L) bound to G-actin, while G1G2 (Fig. 4F), G1b (Fig. 4H), and G1a (Fig. 4I) did not. G1G2G3 (Fig. 4D) and G3G4 (Fig. 4L) caused shifts in the band of actin in the presence of EGTA as well as in the presence of calcium (Fig. 4C and K). Therefore, G1G2G3 and G3G4 bound to G-actin in a calcium-independent manner. In contrast, no alterations were detected in the band patterns of G-actin and GSNL-1 fragments for mixtures of G-actin with G1G2 (Fig. 4F), G1b (Fig. 4H), or G1a (Fig. 4J) in the presence of EGTA. Thus, these GSNL-1 fragments bound to G-actin in a calcium-dependent manner. Overall, these results suggest that GSNL-1 has at least two G-actin-binding sites, a calcium-dependent site in G1 and a calcium-independent site in G3G4.
Native molecular mass
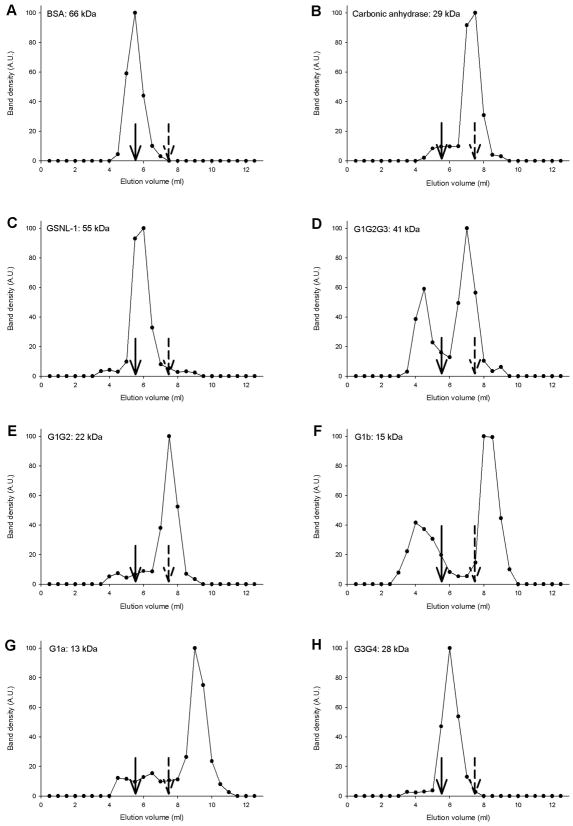
Our non-denaturing electrophoresis analysis of GSNL-1 variants and actin demonstrated that several variants were resolved in multiple bands as GSNL-1 variants themselves or in complex with actin (Fig. 4). We previously showed that multiple bands of the full-length GSNL-1:actin complex contain a 1:1 molar ratio of GSNL-1 and actin (18), but our analysis did not preclude the possibility that GSNL-1 and actin formed oligomers containing the same numbers of each molecule. We attempted to determine native molecular mass of the full-length GSNL-1:actin complex in G-buffer by gel filtration chromatography. However, our attempts failed due to non-specific interactions between the protein and the gel matrices under low ionic conditions in G-buffer (our unpublished data). Therefore, we estimated native molecular mass of GSNL-1 fragments alone in F-buffer in the presence of calcium by gel filtration chromatography using Superdex 75 (Fig. 5). As molecular mass standards, bovine serum albumin (BSA) (66 kDa) and carbonic anhydrase (CA) (29 kDa) were eluted at 5.5 and 7.5 ml, respectively (Fig. 5A and B). Full-length GSNL-1 (55 kDa) was eluted as a single peak at ~6.0 ml that is near but slightly slower than the peak of BSA (66 kDa) (Fig. 5C), indicating that GSNL-1 is a monomer. Similarly, G1G2 (Fig. 5E) and G1a (Fig. 5G) were eluted as single peaks that corresponded to elution volumes of monomers. G1G2G3 was eluted as two major peaks of estimated dimer (82 kDa) and monomer (41 kDa) (Fig. 5D), suggesting that multiple bands of G1G2G3 in non-denaturing electrophoresis (Fig. 4C and D) are partly due to mixed dimeric and monomeric states. G1b was also eluted as two peaks with one peak of large molecular mass and another peak that corresponded to monomer (15 kDa) (Fig. 5F). Since the peak of large molecular mass is broad and estimated to be much larger than calculated mass of G1b tetramer (60 kDa), this peak likely represents oligomers or aggregates of G1b. Interestingly, G3G4 was eluted as a single peak at 6.0 ml that is closer to BSA (66 kDa) than CA (29 kDa) (Fig. 5H), suggesting that G3G4 is a constitutive dimer (56 kDa). However, a smear appearance of G3G4 in non-denaturing electrophoresis (Fig. 4K and L) suggests that G3G4 is present as either monomer or dimer with variable conformational states under low ionic conditions in G-buffer.
Figure 5.
Estimation of native molecular mass of GSNL-1 variants by gel filtration chromatography. Bovine serum albumin (BSA) (A), carbonic anhydrase (CA) (B), GSNL-1 (C), G1G2G3 (D), G1G2 (E), G1b (F), G1a (G), or G3G4 (H) was applied to a Superdex 75 gel filtration column (24 ml bed) and eluted with F-buffer containing 0.1 mM CaCl2. The eluates were fractionated every 0.5 ml, and eluted proteins analyzed by SDS-PAGE and Coomassie blue staining. Density of the protein bands (arbitrary units) is plotted as a function of elution volume (ml). Arrows with solid lines and broken lines indicate peak positions of BSA (66 kDa) and CA (29 kDa), respectively. Molecular masses shown in the figure are calculated values for monomers.
F-actin binding
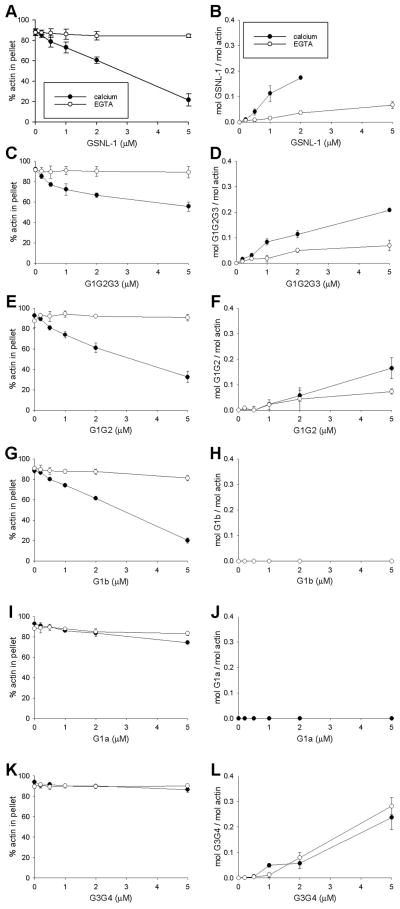
GSNL-1 binds to actin filaments in a calcium-dependent manner (18). F-actin binding of full-length GSNL-1 and GSNL-1 fragments was examined by co-sedimentation assays (Fig. 6). Full-length GSNL-1 co-sedimented with F-actin in a calcium-dependent manner as previously shown (Fig. 6B). We were not able to quantify co-sedimentation of GSNL-1 with actin at a high GSNL-1 concentration (5 μM) due to large deviations of the data. It also decreased pelletable actin in a calcium-dependent manner (Fig. 6A), which indicates an increase of fragmented short filaments. G1G2G3 had slightly weaker calcium-dependent actin fragmentation (Fig. 6C) and F-actin binding activities (Fig. 6D) than full-length GSNL-1 (Fig. 6A and B). G1G2 induced calcium-dependent actin fragmentation nearly as strong as full-length GSNL-1 (Fig. 6E) and very weak co-sedimentation with F-actin (Fig. 6F). Since co-sedimentation of G1G2 with F-actin was very little, it may simply represent its association with actin barbed ends but not with the side of filaments. Either G1b (Fig. 6H) or G1a (Fig. 6J) did not bind to F-actin. However, G1b (Fig. 6G), but not G1a (Fig. 6I), fragmented actin filaments as strongly as full-length GSNL-1 (Fig. 6A), which is consistent with the microscopic observations (Fig. 2). On the other hand, G3G4 co-sedimented with F-actin in the presence or absence of calcium (Fig. 6L) and showed no actin fragmentation activity (Fig. 6K). These results suggest that GSNL-1 has a calcium-independent F-actin binding site in G3G4. However, a regulatory site for calcium-sensitive F-actin binding activity still needs to be determined.
Figure 6.
Co-sedimentation assays of GSNL-1 variants with F-actin. 5 μM F-actin was incubated with varying concentrations (0 – 5 μM) of full-length GSNL-1 (A and B), G1G2G3 (C and D), G1G2 (E and F), G1b (G and H), G1a (I and J), or G3G4 (K and L) in the presence of 0.1 mM CaCl2 (closed circles) or 0.1 mM EGTA (open circles) for 1 hr and ultracentrifuged at 436,000 × g for 15 min. The supernatants and pellets were separated and analyzed by SDS-PAGE. Band intensities were quantified by densitometry, and actin-independent sedimentations of GSNL-1 variants were subtracted from the experimental data in the presence of actin. On the left column (A, C, E, G, I, and K), percentages of actin in the pellet fractions are plotted as a function of total concentrations of GSNL-1 variants. On the right column (B, D, F, H, J, and L), molar ratios of GSNL-1 variants to actin in the pellets are plotted as a function of total concentrations of GSNL-1 variants. Data are average ± standard deviations of three experiments.
Phosphoinositide binding
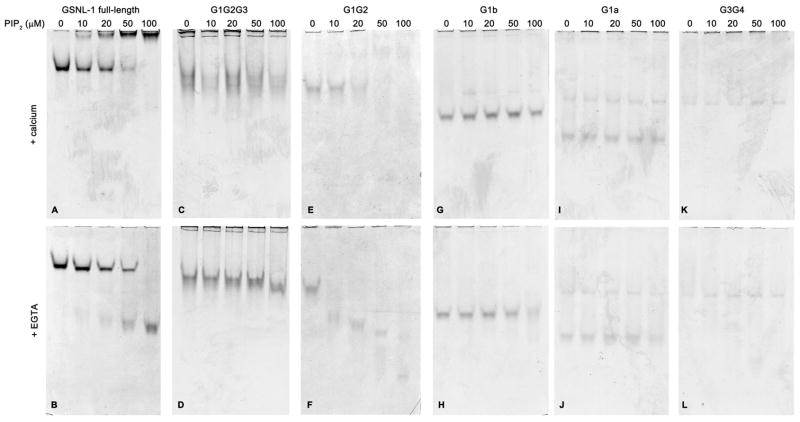
Gelsolin and gelsolin-related proteins directly bind to phospholipids including phosphatidylinositol 4,5-bisphosphate (PIP2) (26–28). By non-denaturing gel electrophoresis, we found that full-length GSNL-1 directly binds to PIP2 (Fig. 7A and B). By increasing the concentrations of PIP2, the band of GSNL-1 was shifted to an upper band in the presence of calcium (Fig. 7A) or a lower band in the presence of EGTA (Fig. 7B). Bands of G1G2G3 were not modified PIP2 in the presence or absence of calcium (Fig. 7C and D), suggesting that it did not bind to PIP2 or that the interaction was too weak to be detected by electrophoresis. In contrast, band shift of G1G2 occurred at much lower PIP2 concentrations than that of full-length GSNL-1 (compare Fig. 7E and A), suggesting that G1G2 binds to PIP2 with higher affinity than full-length GSNL-1. G1G2 bound to PIP2 with even higher affinity in the presence of EGTA (Fig. 7F) than in the presence of calcium (Fig. 7E). Thus, removal of G3 from G1G2G3 greatly augmented PIP2 binding, suggesting that G3 has an inhibitory role in PIP2 binding. G1b did not interact with PIP2 in the presence of calcium (Fig. 7G), while a weak band shift was detected for G1b in the presence of EGTA at 100 μM PIP2 (Fig. 7H). No PIP2-induced band shift was observed for G1a (Fig. 7I and J) or G3G4 (Fig. 7K and L), suggesting that these fragments did not bind to PIP2 or that the interactions were too weak to be detected by electrophoresis. These results indicate that G2 is necessary for PIP2 binding and that PIP2 binding is attenuated in G1G2G3.
Figure 7.
Examination of PIP2 binding by GSNL-1 variants by non-denaturing polyacrylamide gel electrophoresis. 5μM of full-length GSNL-1 (A and B), G1G2G3 (C and D), G1G2 (E and F), G1b (G and H), G1a (I and J), or G3G4 (K and L) were incubated with varying concentrations (0 – 100 μM) of PIP2 in G-buffer in the presence of 0.1 mM CaCl2 (A, C, E, G, I, and K) or 0.5 mM EGTA (B, D, F, H, J, and L) for 30 min, and the samples were analyzed by non-denaturing polyacrylamide gel electrophoresis.
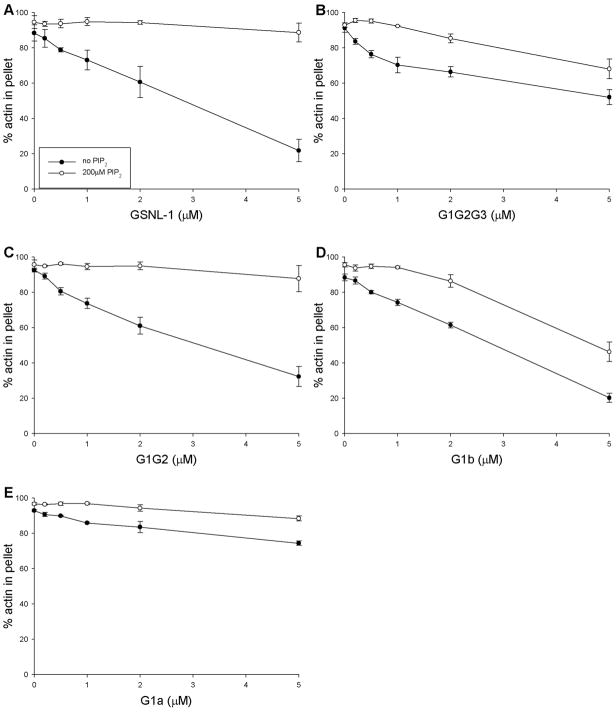
Binding of PIP2 to GSNL-1 inhibited its F-actin fragmentation activity as demonstrated by actin-sedimentation assays (Fig. 8). GSNL-1 fragmented actin filaments in the absence of PIP2 and decreased pelletable actin (Fig. 8A, closed circles). However, in the presence of 200 μM PIP2, full-length GSNL-1 did not show F-actin fragmentation (Fig. 8A, open circles). Similarly, activity of G1G2 was fully inhibited by PIP2 (Fig. 8C) as well as full-length GSNL-1. In contrast, activity of G1G2G3 (Fig. 8B), G1b (Fig. 8D), or G1a (Fig. 8E) was only weakly inhibited by PIP2 (Fig. 8B). These results are consistent with the PIP2-binding properties of GSNL-1 variants (Fig 7). Full-length GSNL-1 and G1G2 bound to PIP2 strongly and were effectively inhibited by PIP2, whereas G1b only weakly bound to PIP2 and were not strongly inhibited by PIP2. Activities of G1G2G3 and G1b was weakly inhibited by PIP2, whereas they did not show binding with PIP2 in non-denaturing electrophoresis, suggesting that binding of G1G2G3 or G1b to PIP2 was too unstable to be detected by electrophoresis. These results suggest that G2 is required for both PIP2 binding and PIP2 sensitivity.
Figure 8.
Effects of PIP2 on actin fragmentation by GSNL-1 variants. 5 μM F-actin was incubated with varying concentrations (0 – 5 μM) of full-length GSNL-1 (A), G1G2G3 (B), G1G2 (C), G1b (D), or G1a (E) in the presence of 0.1 mM CaCl2 with (open circles) or without (closed circles) 200 μM PIP2 for 30 min and ultracentrifuged at 436,000 × g for 15 min. The supernatants and pellets were separated and analyzed by SDS-PAGE. Band intensities were quantified by densitometry, and percentages of actin in the pellet fractions are plotted as a function of total concentrations of GSNL-1 variants. Data are average ± standard deviations of three experiments.
Discussion
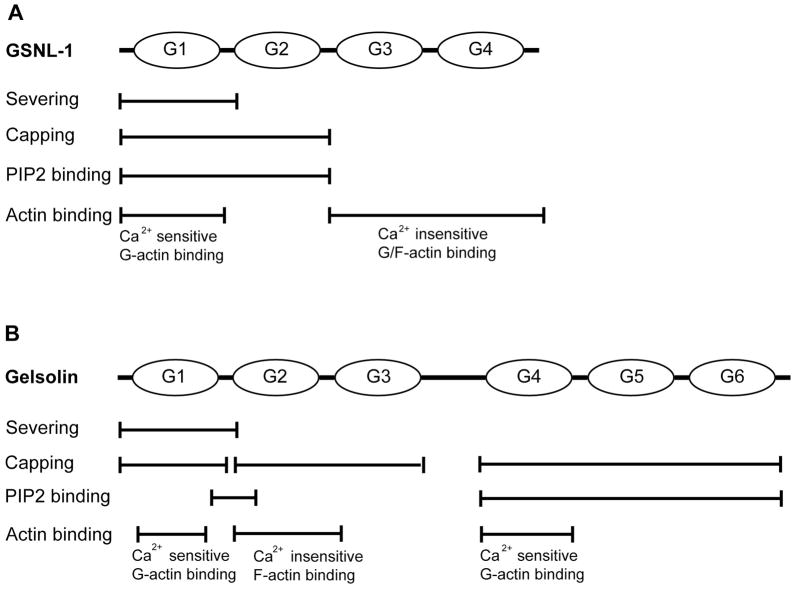
In the present study, dissection of functional domains of GSNL-1 identified distinct functions of the four gelsolin-like repeats and revealed similarities and differences between GSNL-1 and gelsolin in utilizing these domains to regulate actin dynamics (Fig. 9 and Table 1). Remarkably, only G1, the N-terminal G domain, plus a linker between G1 and G2 was sufficient for nearly full actin filament severing activity, whereas G2 was required for barbed-end capping and PIP2-binding (Fig. 9A). At least two actin-binding sites were identified: one in G1 that binds to G-actin in the presence of calcium and the other in G3G4 that binds to both G- and F-actin independent of calcium (Fig. 9A). These results show that the essential function of the N-terminal G domain (G1) in actin filament severing is conserved between GSNL-1 and gelsolin but roles of other G domains in barbed-end capping and PIP2 binding are diverged. Thus, GSNL-1 and gelsolin are conserved in the actin-filament severing and capping activities but with differential utilization of G domains.
Figure 9.
Comparison of functional domains between C. elegans GSNL-1 and mammalian gelsolin. (A) Functional domains of C. elegans GSNL-1 identified in this study. (B) Functional domains of mammalian gelsolin were adapted from Sun et al. (2) with modifications based on the following references: severing (8), capping (9, 31), PIP2 binding (32, 33, 36), calcium-sensitive G-actin binding in G1 (37), calcium-insensitive F-actin binding in G2 (31), and calcium-sensitive G-actin binding in G4 (38).
Table 1.
Summary of properties of GSNL-1 variants
| GSNL-1 | G1G2G3 | G1G2 | G1b | G1a | G3G4 | |
|---|---|---|---|---|---|---|
| Amino acid sequence | 1-475 | 1-353 | 1-239 | 1-131 | 1-115 | 240-475 |
| Filament severing | ||||||
| +Ca2+ | ++ | ++ | ++ | ++ | + | − |
| −Ca2+ | − | − | − | − | − | − |
| Capping | ||||||
| +Ca2+ | + | + | + | − | − | − |
| −Ca2+ | − | − | − | − | − | − |
| G-actin binding | ||||||
| +Ca2+ | + | + | + | + | + | + |
| −Ca2+ | − | + | − | − | − | + |
| F-actin binding | ||||||
| +Ca2+ | + | + | ± | − | − | + |
| −Ca2+ | − | − | − | − | − | + |
| PIP2 binding | ||||||
| +Ca2+ | + | − | + | − | − | − |
| −Ca2+ | + | − | + | ± | − | − |
| PIP2 regulation | ||||||
| +Ca2+ | ++ | + | ++ | + | + | ND |
G1 plus a linker between G1 and G2 (G1b) is the smallest GSNL-1 fragment that exhibits strong calcium-dependent actin-filament severing activity (Fig. 9A and Table 1). G1 without the G1–G2 linker (G1a) had only weak severing activity. Similar observations have been reported for gelsolin (Fig. 9B) (8). Kwiatkowski et al. demonstrated that a gelsolin fragment (PG160) containing G1 and a short G1–G2 linker sequence had nearly full severing activity while a shorter fragment (PG149) lacking the linker sequence had very weak severing activity (8). G1 of gelsolin binds to actin at a cleft between subdomains 1 and 3 (11). Amino acid sequence of the G1–G2 linker of gelsolin contains similarity with that of WASP homology 2 (29) and plays an important role in actin filament severing (8, 24, 25). The crystal structure of the gelsolin fragment PG160 in complex with G-actin shows that the G1–G2 linker makes a close contact with the surface of actin at residues 24 to 30 (29). Thus, the interaction between the G1–G2 linker and actin may promote or stabilize binding of G1 to actin and enhance its severing activity. Based on the sequence similarlity, G1 and the G1–G2 linker of GSNL-1 most likely bind to actin monomer in a similar manner to the equivalent region of gelsolin. The G1–G2 linker of G1b perhaps enhances binding to F-actin, but G1b did not co-sediment with F-actin (Fig. 6H). This might be due to instantaneous severing and disassembly of F-actin after association of G1b with F-actin. Therefore, role of the G1–G2 linker of GSNL-1 needs to be investigated with other techniques with higher temporal resolution. Functions of other G domains are different between GSNL-1 and gelsolin. G1b, G1G2, and G1G2G3 of GSNL-1 efficiently sever actin filaments in a Ca2+-dependent manner. In contrast, gelsolin fragment containing the three N-terminal G domains sever actin filaments independently of calcium, but gelsolin fragments containing G1 or G1G2 have calcium-sensitive severing activity (8, 30). Therefore, G3 of gelsolin masks the calcium-sensitivity, but this is not the case for GSNL-1. Atomic structures of gelsolin shows that G3 interacts with G1 in the absence of calcium and masks its actin binding site (16), but G3 is moved away from G1 when it binds to actin (12). Although intramolecular domain interactions in GSNL-1 are currently unknown, a mode of calcium-dependent conformational changes that involve G1 could be different between GSNL-1 and gelsolin.
G1G2 of GSNL-1 is the smallest fragment that has barbed-end capping activity (Fig. 9A and Table 1). G1a or G1b did not have capping activity, indicating that G2 is required for capping. In contrast, G1 and G2G3 of gelsolin can independently cap the barbed end (9, 31). Since G1 of gelsolin has only one actin binding site, Weber et al. (9) predicted that binding of gelsolin G1 to the barbed end of F-actin induces a conformational change that will slow down monomer dissociation from the end. Thus, G2 together with G1 of GSNL-1 may be required to cause such a change, or G2 may have an actin-binding site for a neighboring actin monomer at the barbed end, so that G1G2 can crosslink two monomers at the end to inhibit monomer dissociation. In either case, the difference in capping activity suggests that the actin-G1 interactions at the barbed end are somewhat different between GSNL-1 and gelsolin.
G1G2 of GSNL-1 is also the smallest fragment that binds to PIP2 (Fig. 9A and Table 1). Interaction of G1a or G1b with PIP2 was only detected by weak PIP2 inhibition of actin fragmentation by pelleting assays, suggesting that G2 is important for PIP2 binding. In gelsolin, a PIP2 binding site is located in residues 135-169 that encompass from the C-terminus of G1 to the N-terminus of G2 (Fig. 9B). This PIP2-binding site can be divided into two separate PIP2-binding sequences: P1 (residues 135-149) (32) and P2 (residues 150-169) (33). Comparison of the sequence of GSNL-1 and gelsolin shows that P2 is conserved in GSNL-1 but basic residues of gelsolin P1 are not conserved in GSNL-1. This suggests that the P2-like sequence in G2 of GSNL-1 is the primary PIP2-binding site. Since P2 of gelsolin is responsible for the PIP2-sensitivity (34), the P2-like sequence of GSNL-1 is predicted to be the PIP2-regulatory site.
The actin-regulatory activities of GSNL-1 are expected to be adapted to its biological functions. Messenger RNA of GSNL-1 is enriched in striated muscle in C. elegans (35). However, our preliminary studies indicate that a null mutation of gsnl-1 does not cause detectable changes in muscle actin organization under normal culture conditions (our unpublished data). Similarly, mammalian gelsolin is expressed in striated muscle and localizes to the thin filaments in myofibrils (1), but its role in muscle cells is not understood. Biochemical studies show that GSNL-1, as well as gelsolin, is regulated by calcium and PIP2, which are second messengers in a number of signaling pathways. Therefore, GSNL-1 might function in transient remodeling of the actin cytoskeleton upon signaling events during development and/or environmental stimuli. Our biochemical studies on GSNL-1 should provide basic knowledge to help further functional investigations of gelsolin-related proteins in C. elegans using genetic approaches.
Abbreviations
- ADF
actin-depolymerizing factor
- BSA
bovine serum albumin
- Cc
critical concentration
- CA
carbonic anhydrase
- G domain
gelsolin-like domain
- GSNL-1
gelsolin-like protein-1
- GST
glutathione-S-transferase
- PIP2
phosphatidylinositol 4,5-bisphosphate
Footnotes
This work was supported by grants from the National Institute of Health (R01 AR48615) and University Research Committee of Emory University to S. O.
References
- 1.Ono S. Mechanism of depolymerization and severing of actin filaments and its significance in cytoskeletal dynamics. Int Rev Cytol. 2007;258:1–82. doi: 10.1016/S0074-7696(07)58001-0. [DOI] [PubMed] [Google Scholar]
- 2.Sun HQ, Yamamoto M, Mejillano M, Yin HL. Gelsolin, a multifunctional actin regulatory protein. J Biol Chem. 1999;274:33179–33182. doi: 10.1074/jbc.274.47.33179. [DOI] [PubMed] [Google Scholar]
- 3.Silacci P, Mazzolai L, Gauci C, Stergiopulos N, Yin HL, Hayoz D. Gelsolin superfamily proteins: key regulators of cellular functions. Cell Mol Life Sci. 2004;61:2614–2623. doi: 10.1007/s00018-004-4225-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McGough AM, Staiger CJ, Min JK, Simonetti KD. The gelsolin family of actin regulatory proteins: modular structures, versatile functions. FEBS Lett. 2003;552:75–81. doi: 10.1016/s0014-5793(03)00932-3. [DOI] [PubMed] [Google Scholar]
- 5.Bamburg JR. Proteins of the ADF/cofilin family: essential regulators of actin dynamics. Annu Rev Cell Dev Biol. 1999;15:185–230. doi: 10.1146/annurev.cellbio.15.1.185. [DOI] [PubMed] [Google Scholar]
- 6.Bamburg JR, McGough A, Ono S. Putting a new twist on actin: ADF/cofilins modulate actin dynamics. Trends Cell Biol. 1999;9:364–370. doi: 10.1016/s0962-8924(99)01619-0. [DOI] [PubMed] [Google Scholar]
- 7.Ono S. Regulation of actin filament dynamics by actin depolymerizing factor/cofilin and actin-interacting protein 1: new blades for twisted filaments. Biochemistry. 2003;42:13363–13370. doi: 10.1021/bi034600x. [DOI] [PubMed] [Google Scholar]
- 8.Kwiatkowski DJ, Janmey PA, Yin HL. Identification of critical functional and regulatory domains in gelsolin. J Cell Biol. 1989;108:1717–1726. doi: 10.1083/jcb.108.5.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weber A, Pring M, Lin SL, Bryan J. Role of the N- and C-terminal actin-binding domains of gelsolin in barbed filament end capping. Biochemistry. 1991;30:9327–9334. doi: 10.1021/bi00102a027. [DOI] [PubMed] [Google Scholar]
- 10.Pope B, Way M, Weeds AG. Two of the three actin-binding domains of gelsolin bind to the same subdomain of actin. Implications of capping and severing mechanisms. FEBS Lett. 1991;280:70–74. doi: 10.1016/0014-5793(91)80206-i. [DOI] [PubMed] [Google Scholar]
- 11.McLaughlin PJ, Gooch JT, Mannherz HG, Weeds AG. Structure of gelsolin segment 1-actin complex and the mechanism of filament severing. Nature. 1993;364:685–692. doi: 10.1038/364685a0. [DOI] [PubMed] [Google Scholar]
- 12.Burtnick LD, Urosev D, Irobi E, Narayan K, Robinson RC. Structure of the N-terminal half of gelsolin bound to actin: roles in severing, apoptosis and FAF. EMBO J. 2004;23:2713–2722. doi: 10.1038/sj.emboj.7600280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robinson RC, Mejillano M, Le VP, Burtnick LD, Yin HL, Choe S. Domain movement in gelsolin: a calcium-activated switch. Science. 1999;286:1939–1942. doi: 10.1126/science.286.5446.1939. [DOI] [PubMed] [Google Scholar]
- 14.Van Troys M, Dewitte D, Goethals M, Vandekerckhove J, Ampe C. Evidence for an actin binding helix in gelsolin segment 2; have homologous sequences in segments 1 and 2 of gelsolin evolved to divergent actin binding functions? FEBS Lett. 1996;397:191–196. doi: 10.1016/s0014-5793(96)01086-1. [DOI] [PubMed] [Google Scholar]
- 15.McGough A, Chiu W, Way M. Determination of the gelsolin binding site on F-actin: implications for severing and capping. Biophys J. 1998;74:764–772. doi: 10.1016/S0006-3495(98)74001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burtnick LD, Koepf EK, Grimes J, Jones EY, Stuart DI, McLaughlin PJ, Robinson RC. The crystal structure of plasma gelsolin: implications for actin severing, capping, and nucleation. Cell. 1997;90:661–670. doi: 10.1016/s0092-8674(00)80527-9. [DOI] [PubMed] [Google Scholar]
- 17.Choe H, Burtnick LD, Mejillano M, Yin HL, Robinson RC, Choe S. The calcium activation of gelsolin: insights from the 3A structure of the G4–G6/actin complex. J Mol Biol. 2002;324:691–702. doi: 10.1016/s0022-2836(02)01131-2. [DOI] [PubMed] [Google Scholar]
- 18.Klaavuniemi T, Yamashiro S, Ono S. Caenorhabditis elegans gelsolin-like protein 1 is a novel actin filament-severing protein with four gelsolin-like repeats. J Biol Chem. 2008;283:26071–26080. doi: 10.1074/jbc.M803618200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pardee JD, Spudich JA. Purification of muscle actin. Methods Enzymol. 1982;85:164–181. doi: 10.1016/0076-6879(82)85020-9. [DOI] [PubMed] [Google Scholar]
- 20.Kouyama T, Mihashi K. Fluorimetry study of N-(1-pyrenyl)iodoacetamide-labelled F-actin. Local structural change of actin protomer both on polymerization and on binding of heavy meromyosin. Eur J Biochem. 1981;114:33–38. [PubMed] [Google Scholar]
- 21.Mohri K, Vorobiev S, Fedorov AA, Almo SC, Ono S. Identification of functional residues on Caenorhabditis elegans actin-interacting protein 1 (UNC-78) for disassembly of actin depolymerizing factor/cofilin-bound actin filaments. J Biol Chem. 2004;279:31697–31707. doi: 10.1074/jbc.M403351200. [DOI] [PubMed] [Google Scholar]
- 22.Safer D. An electrophoretic procedure for detecting proteins that bind actin monomers. Anal Biochem. 1989;178:32–37. doi: 10.1016/0003-2697(89)90351-5. [DOI] [PubMed] [Google Scholar]
- 23.Ono S, McGough A, Pope BJ, Tolbert VT, Bui A, Pohl J, Benian GM, Gernert KM, Weeds AG. The C-terminal tail of UNC-60B (ADF/cofilin) is critical for maintaining its stable association with F-actin and is implicated in the second actin-binding site. J Biol Chem. 2001;276:5952–5958. doi: 10.1074/jbc.M007563200. [DOI] [PubMed] [Google Scholar]
- 24.Southwick FS. Gain-of-function mutations conferring actin-severing activity to human macrophage Cap G. J Biol Chem. 1995;270:45–48. doi: 10.1074/jbc.270.1.45. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y, Vorobiev SM, Gibson BG, Hao B, Sidhu GS, Mishra VS, Yarmola EG, Bubb MR, Almo SC, Southwick FS. A CapG gain-of-function mutant reveals critical structural and functional determinants for actin filament severing. EMBO J. 2006;25:4458–4467. doi: 10.1038/sj.emboj.7601323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Janmey PA, Stossel TP. Modulation of gelsolin function by phosphatidylinositol 4,5-bisphosphate. Nature. 1987;325:362–364. doi: 10.1038/325362a0. [DOI] [PubMed] [Google Scholar]
- 27.Yin HL, Janmey PA, Schleicher M. Severin is a gelsolin prototype. FEBS Lett. 1990;264:78–80. doi: 10.1016/0014-5793(90)80769-f. [DOI] [PubMed] [Google Scholar]
- 28.Janmey PA, Matsudaira PT. Functional comparison of villin and gelsolin. Effects of Ca2+, KCl, and polyphosphoinositides. J Biol Chem. 1988;263:16738–16743. [PubMed] [Google Scholar]
- 29.Irobi E, Burtnick LD, Urosev D, Narayan K, Robinson RC. From the first to the second domain of gelsolin: a common path on the surface of actin? FEBS Lett. 2003;552:86–90. doi: 10.1016/s0014-5793(03)00934-7. [DOI] [PubMed] [Google Scholar]
- 30.Chaponnier C, Janmey PA, Yin HL. The actin filament-severing domain of plasma gelsolin. J Cell Biol. 1986;103:1473–1481. doi: 10.1083/jcb.103.4.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun HQ, Wooten DC, Janmey PA, Yin HL. The actin side-binding domain of gelsolin also caps actin filaments. Implications for actin filament severing. J Biol Chem. 1994;269:9473–9479. [PubMed] [Google Scholar]
- 32.Yu FX, Sun HQ, Janmey PA, Yin HL. Identification of a polyphosphoinositide-binding sequence in an actin monomer-binding domain of gelsolin. J Biol Chem. 1992;267:14616–14621. [PubMed] [Google Scholar]
- 33.Janmey PA, Lamb J, Allen PG, Matsudaira PT. Phosphoinositide-binding peptides derived from the sequences of gelsolin and villin. J Biol Chem. 1992;267:11818–11823. [PubMed] [Google Scholar]
- 34.Xian W, Janmey PA. Dissecting the gelsolin-polyphosphoinositide interaction and engineering of a polyphosphoinositide-sensitive gelsolin C-terminal half protein. J Mol Biol. 2002;322:755–771. doi: 10.1016/s0022-2836(02)00841-0. [DOI] [PubMed] [Google Scholar]
- 35.Fox RM, Watson JD, Von Stetina SE, McDermott J, Brodigan TM, Fukushige T, Krause M, Miller DM. The embryonic muscle transcriptome of Caenorhabditis elegans. Genome Biol. 2007;8:R188. doi: 10.1186/gb-2007-8-9-r188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feng L, Mejillano M, Yin HL, Chen J, Prestwich GD. Full-contact domain labeling: identification of a novel phosphoinositide binding site on gelsolin that requires the complete protein. Biochemistry. 2001;40:904–913. doi: 10.1021/bi000996q. [DOI] [PubMed] [Google Scholar]
- 37.Way M, Pope B, Gooch J, Hawkins M, Weeds AG. Identification of a region in segment 1 of gelsolin critical for actin binding. EMBO J. 1990;9:4103–4109. doi: 10.1002/j.1460-2075.1990.tb07632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pope B, Maciver S, Weeds A. Localization of the calcium-sensitive actin monomer binding site in gelsolin to segment 4 and identification of calcium binding sites. Biochemistry. 1995;34:1583–1588. doi: 10.1021/bi00005a014. [DOI] [PubMed] [Google Scholar]