Abstract
The majority of human skeletal dysplasias are caused by dysregulation of growth plate homeostasis. As TGF-β signaling is a critical determinant of growth plate homeostasis, skeletal dysplasias are often associated with dysregulation of this pathway. The context-dependent action of TFG-β signaling is tightly controlled by numerous mechanisms at the extracellular level and downstream of ligand-receptor interactions. However, TGF-β is synthesized as an inactive precursor that is cleaved to become mature in the Golgi apparatus, and the regulation of this posttranslational intracellular processing and trafficking is much less defined. Here, we report that a cysteine-rich protein, E-selectin ligand–1 (ESL-1), acts as a negative regulator of TGF-β production by binding TGF-β precursors in the Golgi apparatus in a cell-autonomous fashion, inhibiting their maturation. Furthermore, ESL-1 inhibited the processing of proTGF-β by a furin-like protease, leading to reduced secretion of mature TGF-β by primary mouse chondrocytes and HEK293 cells. In vivo loss of Esl1 in mice led to increased TGF-β/SMAD signaling in the growth plate that was associated with reduced chondrocyte proliferation and delayed terminal differentiation. Gain-of-function and rescue studies of the Xenopus ESL-1 ortholog in the context of early embryogenesis showed that this regulation of TGF-β/Nodal signaling was evolutionarily conserved. This study identifies what we believe to be a novel intracellular mechanism for regulating TGF-β during skeletal development and homeostasis.
Introduction
The body size of mammalian vertebrates is specified by endochondral ossification. Therefore, it is not surprising that a majority of human skeletal dysplasias are caused by dysregulation of growth plate homeostasis. The growth plate is composed of differentiating chondrocytes that occupy histologically distinct zones: (a) the resting zone (RZ) is the reservoir of chondrocyte progenitors; (b) the proliferating zone (PZ) contains actively proliferating chondrocytes important in bone elongation; (c) the hypertrophic zone (HZ) contains ECM-rich terminally differentiated chondrocytes. Chondrocyte differentiation and homeostasis are precisely regulated by interaction of signaling pathways including bone morphogenetic protein (BMP), TGF-β, Wnt, Hedgehog, and parathyroid hormone–related protein (PTHrP). These signaling molecules act directly or indirectly on chondrocytes within the growth plate as well as on cells of the perichondrium and periosteum. Dysregulation of these signaling pathways is a major cause of defective growth plate homeostasis and skeletal dysplasias (1, 2).
In vertebrates, a major source of TGF-β production is the skeletal system, where it controls both cartilage and bone homeostasis. This notion has been demonstrated in multiple mouse genetic studies and human diseases. TGF-β is synthesized as an inactive precursor (proTGF-β), containing a prodomain (the latency associated peptide [LAP]) and a mature peptide. Two precursor molecules are dimerized via disulfide bonds and cleaved by proprotein convertases, usually furin, at a specific site between LAP and the mature peptide. After cleavage, the 2 parts of proTGF-β remain associated with each other as the small latent complex (SLC). The cleavage/maturation process occurs in the Golgi apparatus and/or ECM and is necessary for generation of the bioactive mature TGF-β ligand, which can then bind TGF-β receptors to trigger downstream Smad-dependent or -independent pathways (3, 4). The body generates much more proTGF-β than needed, so the limiting factor of TGF-β activity is not TGF-β synthesis, but the availability of mature TGF-β. In contrast to the extensively studied TGF-β/TGF-β receptor (TGF-β/TGF-βR) downstream signaling pathways, the regulation of TGF-β maturation and bioavailability is much less understood. However, this level of regulation is an important target for diseases of the skeleton that alter TGF-β signaling.
E-selectin ligand 1 (ESL-1) is a type I transmembrane sialoglycoprotein with a large, approximately 1,100-aa N-terminal extracellular/lumenal domain, which contains 16 cysteine-rich motifs (5). This structure is unique and highly conserved in ESL-1 orthologs from C. elegans to humans, implying its potential important function during evolution. ESL-1 was identified as the ligand for E-selectin in myeloid cells (6). In addition, ESL-1 interacts with FGF2 in vitro (5) and copurifies with TGF-β1 in a large protein complex (7), and accordingly, it has also been named as cysteine-rich FGF receptor 1 (CFR-1) and latent TGF-β complex protein 1 (LTCP-1). However, the function of ESL-1 has largely remained unknown.
Here, we studied an Esl1-knockout mouse that unexpectedly exhibited a skeletal dysplasia phenotype. We discovered that ESL-1 plays an important role by inhibiting TGF-β maturation/bioavailability via an intracellular mechanism that is distinct from previously characterized ones that act within the ECM environment or downstream of ligand-receptor interaction.
Results
Loss of ESL-1 causes skeletal dysplasia.
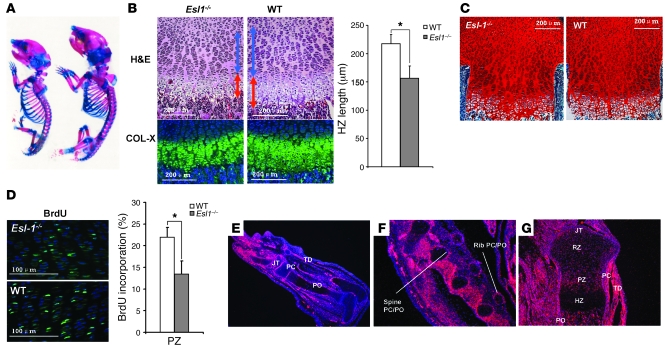
Because ESL-1 was identified as an E-selectin–interacting protein, its role in the regulation of leukocyte rolling was predicted (6). To test this, we generated Esl1–/– mice (Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI42150DS1) but did not find obvious defects in leukocyte rolling and blood count (data not shown and personal communication, K. Ley, La Jolla Institute for Allergy and Immunology, La Jolla, California, USA). Instead, loss of ESL-1 causes distinctive skeletal and growth defects. At birth, heterozygous mice were phenotypically normal, while Esl1–/– mice were approximately 30%–50% smaller than their WT littermates (Figure 1A). This was noted from E14.5 through maturity (Supplemental Figure 2). Skeletal preparations of Esl1–/– mice showed generalized shortening and thinning of all bony elements and distinctively narrow rib cages (Figure 1A). P1 Esl1–/– mice showed shortening of the growth plates that affected both the PZ and HZ, with decreased chondrocyte cell density and increased ECM deposition (Figure 1, B and C). The shortening of the growth plate was due in part to decreased chondrocyte proliferation, as quantified by BrdU incorporation assay (Figure 1D). These results suggest an important role of ESL-1 in skeletogenesis.
Figure 1. Esl1 expression pattern and skeletal dysplasia in Esl1–/– mice.
(A) Skeletal preparation (Alcian blue and alizarin red staining) of P3 WT (right) and Esl1–/– (left) mice. (B) H&E staining of P1 WT and Esl1–/– femur sections. Blue arrows indicate PZ; red arrows, HZ. The lengths of the HZ were separately confirmed by type X collagen (COL-X) immunofluorescence and quantified (n = 5). (C) Safranin O staining on P3 femur sections showing increased ECM deposition on mutant growth plate. (D) Esl1–/– chondrocytes in the P1 PZ of femurs show decreased proliferation compared with WT chondrocytes (n = 12) by BrdU incorporation assay. (E–G) In situ hybridization of Esl1 RNA probe in the skeletal tissues. At E16.5, Esl1 is highly expressed in perichondrium (PC), periosteum (PO), surface of joints (JT), tendons (TD) in the limb (E), PC and PO of rib and vertebrate (F). (G) In P1 limb, Esl1 is highly expressed in PC, PO, JT, and the PZ of cartilage, but less so in the RZ and HZ. *P < 0.05.
In human cells, an ESL1 splicing variant termed GLG2 was identified that encodes a protein with 24 additional amino acids in the C terminal of ESL-1 (8). However, the GLG2 isoform was found specifically in the primates but not in any non-primate species including mice, suggesting that ESL-1 but not GLG2 plays a major role in the molecularly conserved processes governing of skeletogenesis.
In addition to being widely expressed in various tissues including brain, kidney, intestine, etc., Esl1 is highly expressed in the skeletal system, including intervertebral discs at E12.5 (Supplemental Figure 3) and perichondrium and periosteum from E16.5 to P1, and shows increased expression in growth plate chondrocytes at P1 time points (Figure 1, E–G). Interestingly, the Esl1 expression pattern correlates well with TGF-βs expression patterns in all tested tissues in mouse embryos (9).
FGF signaling is not the major downstream target of ESL-1 during skeletal development.
The negative regulatory role of FGF signaling in chondrocyte proliferation is clearly demonstrated by activating mutations of FGFR3 in human achondroplasia and the phenotype of Fgfr3–/– and Fgf18–/– mice (10–12). Because of the interaction between ESL-1 and FGFs in vitro (5), we tested whether the skeletal phenotypes of Esl1–/– mice might be caused by elevated FGF activity. However, we detected similar levels of phosphorylated MEK1/2 (p-MEK1/2), the major downstream effectors of FGF signaling during chondrogenesis (13), in P3 rib cartilage of Esl1–/– mice versus WT littermates (Figure 2A). Furthermore, we tested potential FGF/ESL-1 in vivo interaction by generating Esl1–/–Fgfr3–/– mice. If loss of ESL-1 were to upregulate FGF activity in the skeleton, then loss of FGFR3 should compensate for this effect and rescue the Esl1–/– growth retardation. Instead, we found that the Esl1–/–Fgfr3–/– mice were similar to Esl1–/– littermates in size. Additionally, 4-week-old Esl1–/– and Esl1–/–Fgfr3–/– mice showed similar shortening of PZ and HZ in their growth plates (Figure 2, B and C, and Supplemental Figure 4). Hence, the Esl1–/– phenotype is dominant over the loss of Fgfr3–/– without evidence of epistasis, suggesting that FGF/FGFR3 signaling is not the major target of ESL-1 in cartilage.
Figure 2. FGF signaling is not the target of ESL-1 during skeletogenesis.
(A) p-MEK1/2 level during chondrogenesis is unchanged in P3 Esl1–/– versus WT rib cartilage (n = 3). Normalized results (to α-tubulin) are shown. (B) Four-week-old mice from Esl1–/– and Fgfr3–/– intercrosses. Esl1–/– growth retardation was not rescued by genetic inactivation of FGFR3. (C) H&E staining of growth plates from 4-week femurs of each genotype. Blue arrow indicates PZ; red arrow, HZ. Esl1–/–Fgfr3–/– (DKO) mouse growth plate is similar to that of Esl1–/– mice, suggesting absence of epistasis or rescue. The corresponding histogram of PZ and HZ lengths (n = 4) is shown. Scale bar: 100 μm. **P < 0.01.
ESL-1 upregulates TGF-β signaling in the growth plate.
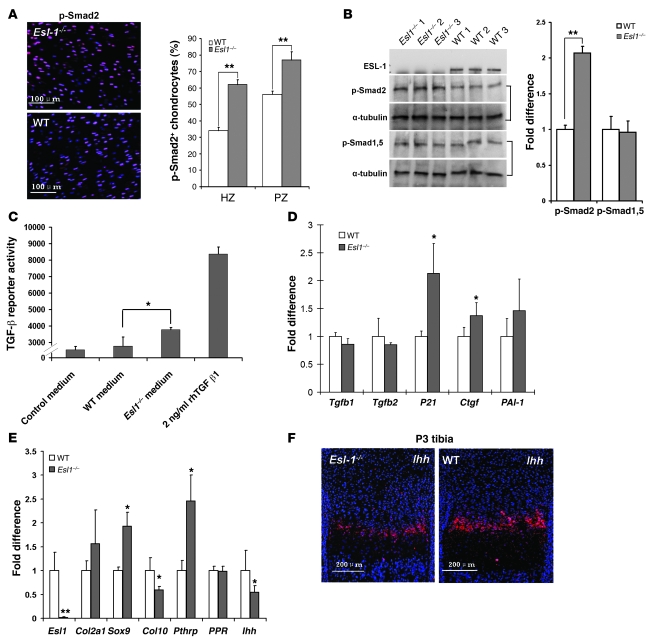
Because ESL-1 and TGF-β1 were found to copurify from a CHO cell line stably expressing TGF-β1 (7), we evaluated whether TGF-β signaling was perturbed in Esl1–/– growth plates. By immunofluorescence, we detected significantly increased numbers of phosphorylated-Smad2–positive (p-Smad2–positive) cells in the P1 Esl1–/– growth plate with α-tubulin or total Smad2 as a control (Figure 3A and Supplemental Figure 5). Western blot analysis confirmed that in P3 Esl1–/– rib cartilage protein, p-Smad2 was increased approximately 2-fold compared with that in WT mice, but the levels of phosphorylated Smad-1 and -5 were similar (Figure 3B), suggesting that TGF-β rather than BMP signaling was upregulated. Additionally, using a stable mink lung TGF-β reporter cell line (14), we found that conditioned medium from primary Esl1–/– chondrocyte culture exhibited higher TGF-β activity than that from WT chondrocytes (Figure 3C). Furthermore, in Esl1–/– mouse embryonic fibroblasts (MEFs), TGF-β downstream targets P21 (CDK inhibitor 1A), connective tissue growth factor (Ctgf), and plasminogen activator inhibitor 1 (PAI-1) were all upregulated, but Tgfb1 and Tgfb2 mRNA levels were unchanged (Figure 3D). These data suggest that loss of ESL-1 increased TGF-β signaling but not TGF-β expression and that the elevated TGF-β signaling may inhibit chondrocyte proliferation in the Esl1–/– growth plate.
Figure 3. TGF-β signaling is upregulated in Esl1–/– growth plates.
(A) p-Smad2–positive chondrocytes (red) merged with DAPI-stained (blue) chondrocytes are increased in P1 Esl1–/– femoral cartilage. n = 12. (B) The amount of p-Smad2 is increased in P3 Esl1–/– rib cartilage as revealed by Western blot, while the amount of p-Smad-1,5 is unchanged. Data were normalized to α-tubulin (n = 3). (C) Conditioned medium from Esl1–/– chondrocyte culture exhibits increased TGF-β activity compared with that from WT chondrocytes (n = 3). Numbers on y axis are direct reading from a illuminometer. Relative intensity from samples is shown. (D) qRT-PCR assays of TGF-βs and TGF-β downstream gene expression in MEFs. Tgfb1 and Tgfb2 are largely unchanged, but P21, Ctgf, and PAI-1 are all increased in Esl1–/– MEFs (n = 3). (E) qRT-PCR assays of cartilage differentiation markers. In P3 Esl1–/– rib chondrocytes (n = 5), expression of Sox9, Col2a1, and Pthrp is increased, while expression of Col10a1 and Ihh is decreased. PPR is unchanged. n = 5. *P < 0.05, **P < 0.01. (F) In situ hybridization with Ihh probe on P3 Esl1–/– and WT tibia.
We next evaluated the expression of chondrocyte markers by quantitative RT-PCR (qRT-PCR) of P3 rib cartilage RNA. Chondrocyte terminal differentiation markers, i.e., Col10a1 for hypertrophic chondrocytes and Indian hedgehog (Ihh) for prehypertrophic chondrocytes, were downregulated in Esl1–/– cartilage, while the markers for proliferating or resting chondrocytes, i.e., Sox9, Col2a1, and Pthrp, were increased in the mutant samples (Figure 3E). Furthermore, in situ hybridization showed that the Ihh-positive zone (preHZ) in Esl1–/– growth plate was narrower, with weaker signal intensity than in the WT sample (Figure 3F). During cartilage homeostasis, IHH and PTHrP cooperatively specify the transition of chondrocytes from proliferation to differentiation (15). IHH upregulates PTHrP through TGF-β, while PTHrP negatively regulates IHH expression (16). Moreover, loss of Ihh in mice leads to severely delayed proliferation and differentiation in growth plate chondrocytes. Therefore, the qRT-PCR data explain the Esl1–/– growth plate phenotype: the loss of ESL-1 augmented TGF-β signaling, which subsequently increased PTHrP expression and, accordingly, suppressed IHH expression in the negative feedback loop. This led to delayed chondrocyte proliferation and differentiation by shifting the IHH/PTHrP balance toward PTHrP. However, it is worth mentioning that PTHrP is commonly considered as a factor that maintains chondrocytes in an undifferentiated but proliferating mode. Hence, the decreased proliferation observed in Esl1–/– chondrocytes may be a result of other TGF-β downstream mechanisms outside the PTHrP/IHH axis. We also found that Sox9 and its direct downstream target Col2a1 are both increased in the mutant rib cartilage, consistent with previous observations that Hedgehog signaling is a negative regulator of Sox9 expression in the growth plate (17).
Gain of ESL-1 function in cartilage yields an growth plate phenotype opposite that resulting from loss of ESL-1 function.
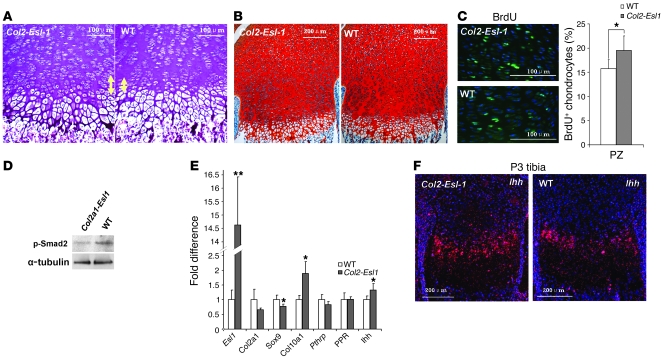
Since TGF-β can signal in both a paracrine and endocrine fashion, we overexpressed the Esl1 transgene using the Col2a1 cartilage-specific promoter in mice to determine whether ESL-1 acts directly on the cartilage via regulation of locally synthesized TGF-β. The Col2a1-Esl1 transgenic mice displayed increased proliferative chondrocytes, decreased ECM disposition, higher cell density in both the PZ and preHZ, and a longer preHZ (Figure 4, A–C). By Western blot analysis, decreased p-Smad2 was detected in the transgenic P3 rib cartilage, indicating downregulated TGF-β activity (Figure 4D). By qRT-PCR of transgenic rib cartilage (Figure 4E), we detected decreased Col2a1, Sox9, and Pthrp but increased Col10a1 and Ihh. These results are in contrast to the qRT-PCR data from the Esl1–/– cartilage. Additionally, increased Ihh expression in the preHZ was confirmed by in situ hybridization in P3 transgenic tibias (Figure 4F). Together, these data show that, as predicted, gain of ESL-1 expression within chondrocytes was sufficient to perturb a growth plate phenotype opposite to that caused by the generalized ESL-1 loss of function. Interestingly, the phenotype of Esl1 transgenic growth plate is reminiscent of that described in Ltbp3–/– mice. Latent TGF-β–binding protein 3 (LTBP3) regulates TGF-β secretion and bioavailability in bone and cartilage homeostasis. Loss of LTBP3 decreased TGF-β signaling and led to accelerated chondrocyte differentiation and hypertrophy (18). P3 Ltbp3–/– mutant mouse growth plates showed a longer hypertrophic chondrocyte zone and higher cell density in the PZ (Supplemental Figure 6A). p-Smad2–positive cells were also greatly decreased in the Ltbp3–/– growth plate chondrocytes (Supplemental Figure 6B). Together these data show that the decrease in TGF-β signaling observed in the 2 distinct genetic models of Col2a1-Esl1 transgenic and Ltbp3–/– mice caused similar growth plate phenotypes.
Figure 4. Gain of ESL-1 in the cartilage yields a growth plate phenotype opposite that resulting from the loss of ESL-1 function.
(A) H&E staining of P3 growth plates from WT and Col2a1-Esl1 transgenic mice (Tg). The Tg preHZ (yellow arrows) is longer than that of WT mice. (B) Safranin O staining on P3 Tg growth plate shows decreased ECM deposition and higher cell density. (C) The PZ of Tg femur shows increased BrdU-positive chondrocytes compared with that of WT littermates. (D) The amount of p-Smad2 is decreased in P3 Tg rib cartilage protein compared with that of WT littermates. (E) qRT-PCR of Tg P3 total cartilage RNA (n = 5) shows reduced Col2a1, Sox9, and Pthrp but increased Col10a1 and Ihh. PPR is unchanged. (F) In situ hybridization with Ihh probe on P3 Tg mice and WT tibia. *P < 0.05, **P < 0.01.
ESL-1 antagonizes TGF-β signaling in Xenopus laevis.
To further evaluate the antagonistic role of ESL-1 in the TGF-β signaling pathway, we performed several experiments using Xenopus laevis as a model system. Xenopus utilizes most of the signaling pathways found in mammals, and changes in these pathways lead to dramatic and distinctive embryonic phenotypes. For this reason, Xenopus embryos have been used extensively as a robust and sensitive system to explore functions of individual components of several signaling pathways. Although the role of TGF-β1–3 in Xenopus is not well defined, other TGF-β family members that share the same precursor-cleavage mechanism and downstream Smad transducers have been well studied in Xenopus. For example, it was shown that Xenopus nodal related 1–4 (Xnr1–4), members of the nodal subclass of TGF-β proteins, play major roles in Xenopus body axis formation utilizing well-defined TGF-β signaling pathways. We reasoned that if ESL-1 acts as an antagonist of Xnrs, overexpression of ESL-1 would lead to a phenotype similar to that of deficiency of one or more Xnr members (19, 20).
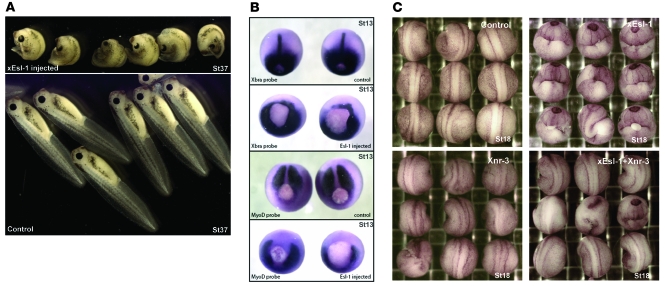
To this end, Xenopus Esl1 (xEsl1) mRNA was injected into the dorsal marginal zone of Xenopus embryos at the 2-cell stage (Supplemental Figure 7). We found that injected embryos at stage 37 exhibited a highly reproducible phenotype, characterized by severe trunk curvature and shortened axis (Figure 5A). At stage 17, the neural folds of injected embryos formed around the yolk plug but did not join together (Supplemental Figure 8). In contrast, when equal amounts of xEsl1 mRNA were injected into the ventral marginal zone, embryos developed normally and displayed no obvious abnormalities at gastrula, neurula, and tail bud stages, suggesting a highly specific dorsal function of ESL-1 (Figure 5A and Supplemental Figure 8). Furthermore, the phenotype of xEsl1 dorsal injection was dosage dependent (Supplemental Table 1). In addition, when mouse Esl1 mRNA was injected into Xenopus embryos, we observed phenotypes that were similar to, albeit less severe than, those with xEsl1 injections (data not shown). The phenotype of xEsl1-injected embryos strongly resembled that caused by deficiency of Xnr3, a TGF-β/Nodal member that specifies convergent extension movements. However, the xEsl1-injected embryos are distinct from Xnr1-, Xnr2-, or Xnr4-deficient embryos, whose phenotypes are considerably weaker than that resulting from Xnr3 deficiency (19). Thus, we decided to focus on the relationship between xEsl1 and Xnr3 in our studies. During body axis formation, Xnr3 expression is restricted to the organizer region, and it is essential for the expression of mesoderm markers such as Xbra and MyoD (19, 20). By in situ hybridization, the dorsal segment of the ring and the notochord showed reduced or absent expression of Xbra and MyoD in xEsl1-injected embryos (Figure 5B), indicating that the Xnr3 signal was markedly impaired by overexpression of xEsl1.
Figure 5. xEsl1 modulates Xenopus body axis formation.
(A) Morphological change of embryos (lateral view at stage 37) after injection at 2-cell stage. Upper panel: Embryos injected with xEsl1 mRNA (460 pg) in the dorsal marginal zone display shortened and curved body axis. Lower panel: Control embryos injected with xEsl1 mRNA (460 pg) in the ventral marginal zone show no change in phenotype. (B) The xEsl1 injection leads to changes of mesoderm markers detected by whole mount in situ hybridization (dorsal view at stage 13 [St13]). Top panel: Xbra was expressed around the blastopore and at notochord midline in the uninjected embryos. Second panel: Xbra expression was reduced or absent in xEsl1 mRNA–injected embryos. Third panel: MyoD was mainly expressed in presomatic plate in the uninjected embryos. Bottom panel: MyoD expression was absent in presomatic plate of xEsl1 mRNA–injected embryos. (C) Rescue of xEsl1-induced axis defects by coinjection with Xnr3 mRNA (dorsal view at stage 18). Embryos were injected at 2-cell stage in the dorsal marginal zone with the indicated mRNAs, xEsl1 (300 pg), Xnr3 (13 pg), or both. The coinjection of Xnr3 mRNAs rescued the curved body axis and open neural fold phenotypes of xEsl1 overexpression.
Rescue experiments were performed to explore the specific effect of ESL-1 on TGF-β regulation. We coinjected mRNAs of xEsl1 (300 pg) and Xnr3 (full-length pro-protein with LAP domain) (13 pg) into the dorsal marginal zone of 2-cell-stage embryos. Embryos were scored for abnormalities such as curved trunk and shortened axis. We found that coexpression of Xnr3 partially rescued the xEsl1-induced phenotype by approximately 50% on average in all independent tests (Figure 5C and Supplemental Table 2). The lack of full rescue may be partly explained by participation of additional TGF-β proteins in dorsal axis formation (19). In summary, the overexpression and rescue experiments showed that ESL-1 antagonism of TGF-β signaling is specific and evolutionarily conserved.
ESL-1 interacts with TGF-βs intracellularly and inhibits TGF-β/Smad signaling in a cell-autonomous fashion.
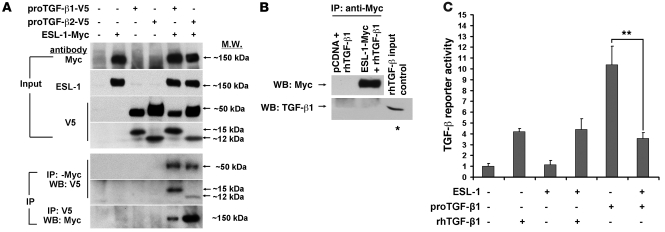
ESL-1 (LTCP-1) was copurified with proTGF-β1 in a large protein complex in the medium of CHO cells stably expressing TGF-β1 (7). In addition, although the majority of published data indicated that ESL-1 is localized in the Golgi apparatus, its alternatively glycosylated variant identified in epithelial cancer tissues was either on the cell surface or secreted (21), suggesting that the different subcellular localization of ESL-1 may be cell type specific. Therefore, possible mechanisms for ESL-1 inhibition of TGF-β may include: (a) intracellular inhibition of TGF-β bioavalibility, in a cell-autonomous fashion, or (b) extracellular interference with the interaction between secreted TGF-β and TGF-β receptors. To differentiate between these two possibilities, we first detected ESL-1 subcellular localization by immunostaining. We found that ESL-1 was restrictedly localized in the Golgi apparatus of most mouse tissues, e.g., in cartilage, and also in cell lines such as COS7 cells and MEFs, but it was detected at low levels in the ECM (see below). Second, by nonreducing Western blot assay on lysates from HEK293, HeLa, and COS7 cells cotransfected with proTGF-β1 and ESL-1, we found that ESL-1 was neither secreted with TGF-β nor bound to TGF-β covalently (Supplemental Figure 9). These data suggest that in these tested tissues and cell lines, ESL-1 is most likely to act intracellularly to regulate TGF-β bioavalibility. Hence, we next studied whether the intracellular ESL-1 can bind to TGF-β in a noncovalent manner. We coexpressed Myc-tagged ESL-1 and V5-tagged TGF-β1 or TGF-β2 in COS7 cells and performed coimmunoprecipitation assays. The anti-Myc antibody precipitates contained both the full-length proTGF-βs (~50 kDa) and mature TGF-β peptides (15 kDa for TGF-β1 and 12 kDa for TGF-β2), while anti-V5 antibody (TGF-β) precipitated ESL-1 as well (Figure 6A). However, when we incubated recombinant human TGF-β1 (rhTGF-β1, the mature TGF-β1 ligand) with the lysate of ESL-1-Myc–transfected cells, the rhTGF-β1 could not be coprecipitated with ESL-1 (Figure 6B). These data suggest that ESL-1 binds to TGF-β noncovalently within the cell and that LAP is required for this interaction.
Figure 6. ESL-1 can directly bind to TGF-β and inhibits TGF-β signaling.
(A) ESL-1 coimmunoprecipitates with both the proTGF-βs and TGF-β ligands. Transfection scheme is shown at the top. Note that anti-V5 antibody detects the V5-tagged proTGF-β1, proTGF-β2 (~50 kDa), and mature TGF-β peptides (TGF-β1: 15 kDa and TGF-β2: 12 kDa). The top 4 panels show Western blots of input lysates, and the bottom 3 show Western blot (WB) for immunoprecipitation. Antibodies used are listed on the left. ProTGF-β1 and proTGF-β2 and their mature ligands are all immunoprecipitated with Myc antibody, and vice versa, suggesting direct interaction between TGF-β and ESL-1. (B) ESL-1 cannot bind mature TGF-β without LAP. rhTGF-β1 (100 ng) was added to lysates of COS7 cells expressing ESL-1-Myc and then coimmunoprecipitated with anti-Myc antibody. The precipitates were analyzed with reducing Western blots. rhTGF-β1 (15 ng) was used as loading control (asterisk). (C) ESL-1 can effectively inhibit TGF-β activity from the plasmid-produced proTGF-β1 but cannot inhibit signaling from the exogenously added rhTGF-β1 (n = 3; **P < 0.01).
Because of the intracellular binding between TGF-β and ESL-1, we hypothesized that ESL-1 should regulate TGF-β in a cell-autonomous fashion. To test this, we expressed TGF-β1 and/or ESL-1 in COS7 cells, and tested the conditioned medium with the mink lung TGF-β reporter cells. In our study, ESL-1 could not alter the reporter activity induced by exogenous addition of rhTGF-β1. However, reporter activity was reduced by ESL-1 when proTGF-β1 was expressed endogenously from transfected plasmids (Figure 6C). These data suggest that ESL-1 inhibits TGF-β in a cell-autonomous fashion, instead of acting as a cell surface or matrix decoy receptor for TGF-βs.
ESL-1 blocks TGF-β maturation by furin proteases.
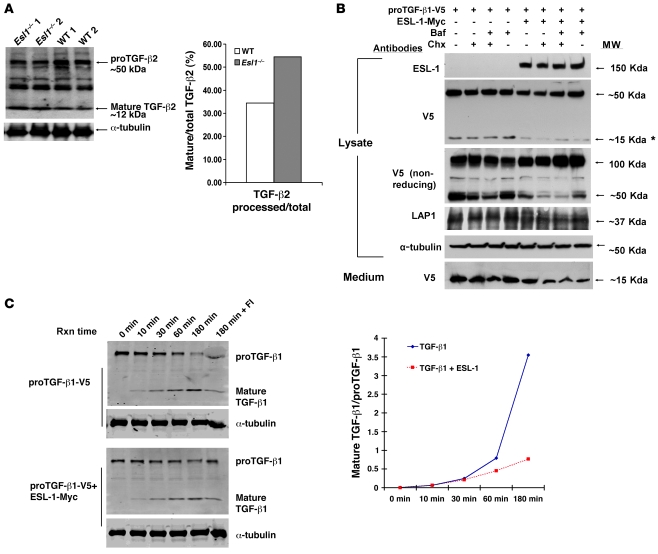
The Golgi apparatus is an important site for TGF-β maturation and secretion, i.e., the cleavage of TGF-β by furin proteases and the cosecretion of TGF-β with LTBPs (4, 22). The Golgi localization of ESL-1 and its functional inhibition of TGF-β signaling suggested that it may act at the level of TGF-β maturation. In fact, we found that TGF-β2, a predominant TGF-β family member in growth plate, was processed to a greater extent in Esl1–/– rib cartilage and also cultured Esl1–/– primary chondrocytes compared with WT counterparts (Figure 7A and Supplemental Figure 10). In contrast, activin βA, another TGF-β family member that is also expressed in growth plate and upstream of Smad2/3 signaling, was not processed differently between mutant and WT cartilage samples (Supplemental Figure 11). These data suggest that ESL-1’s effects on p-Smad2 activity in the growth plate homeostasis occur primarily via inhibition of TGF-β2 maturation.
Figure 7. ESL-1 inhibits TGF-β proteolytic processing.
(A) ESL-1 inhibits the intracellular cleavage and maturation of proTGF-β2 in vivo as shown by the relative abundance of mature TGF-β2 and proTGF-β2 on Western blot analysis of Esl1–/– versus WT cartilage lysates. α-Tubulin is used as a loading control. The ratio of mature TGF-β2 to total TGF-β2 is shown at right. (B) ESL-1 inhibits the intracellular cleavage of proTGF-β1 in vitro. The scheme of transfections and chemical treatments of COS7 cells is shown at the top. Primary antibodies and molecular weights are denoted. Both proTGF-β1-V5 (50 kDa) and the mature TGF-β1-V5 (15 kDa) can be detected by V5 antibody. Note that the amount of 15-kDa TGF-β1 (*) was significantly reduced in the presence of ESL-1. Under nonreducing condition, the presence of ESL-1 remarkably increased proTGF-β1 dimer (100 kDa). LAP1 (37 kDa) was decreased in the presence of ESL-1. α-Tubulin, as an internal control, was similar in all samples. Expression of ESL-1 reduced the secretion of mature TGF-β1 ligand found in medium. Baf, bafilomycin; Chx, cycloheximide. (C) ESL-1 inhibits furin processing of proTGF-β1 by in vitro furin assay. Western blot analysis of the furin reaction samples with V5 antibody. Reaction time (Rxn time), plasmids for transfections, and identity of the bands are noted. Furin inhibitor II (hexa-d-arginine) was added in the last sample (180 min + FI) to confirm the specificity of the furin reaction. The ratios of cleaved to uncleaved TGF-β1 at all time points is shown. The proportion of mature TGF-β1 ligand is greatly increased after 30 minutes incubation in the absence of ESL-1.
Furthermore, this finding was confirmed by a gain-of-function experiment in cell culture. COS7 cells were transfected with either proTGF-β1-V5 or proTGF-β1-V5/ESL-1-Myc. Cycloheximide or bafilomycin was added to minimize possible interference from overexpression of proteins and lysosomal processing. In the presence of ESL-1, decreased amounts of mature TGF-β peptide and LAP were detected in cell lysates. At the same time, increased amounts of the proTGF-β1 dimer (~100 kDa under nonreducing conditions) were detected in the lysate of ESL-1/TGF-β1 cotransfected cells. Less mature TGF-β1 was also detected in the culture medium of the cotransfected cells (Figure 7B). Similar trends were noted in cycloheximide- and/or bafilomycin-treated cells.
To test whether ESL-1 can specifically inhibit TGF-β maturation catalyzed by exogenously added furin protease, we incubated furin in a kinetic study with cell lysates from HEK293 cells transfected with either proTGF-β1-V5 or proTGF-β1-V5/ESL-1-Myc (Figure 7C). As expected, increasing ratios of ligand to proTGF-β1 were observed in both transfection schemes, with prolonged incubation showing furin-mediated cleavage of proTGF-β1 to mature TGF-β1. At shorter incubation time points (0–30 minutes), TGF-β1 processing by furin was similar irrespective of the presence of ESL-1. However, at later time points (1–3 hours), the presence of ESL-1 stabilized proTGF-β1 and inhibited the production of TGF-β1 mature peptide (Figure 7C). To confirm the mechanism using a loss-of-function approach and to determine whether it was also present in chondrocytes, we performed another furin assay using lysates from the primary chondrocytes derived from P3 Esl1–/– and WT rib cartilage. We found that in the absence of ESL-1, TGF-β2 maturation was accelerated, with increased accumulation of the mature peptide with duration of incubation (Supplemental Figure 12). These data support that ESL-1 binding to TGF-β inhibits furin-mediated TGF-β maturation.
To determine whether the alteration of intracellular TGF-β maturation would ultimately lead to a change of extracellular mature TGF-β, we performed a pulse-chase experiment. In the presence of ESL-1, decreased amounts of cleaved TGF-β1 were secreted into the medium, while increased amounts of proTGF-β were retained in cells at all time points tested (Supplemental Figure 13). These results support the contention that ESL-1 inhibits the maturation/processing of proTGF-β intracellularly, ultimately leading to decreased extracellular amounts of active TGF-β ligand.
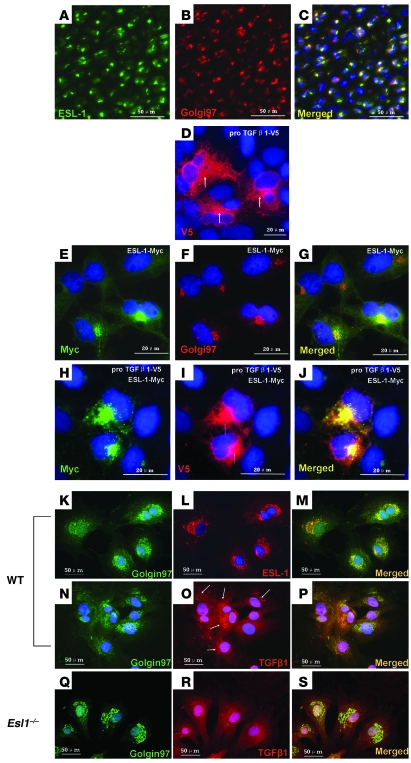
ESL-1 increases TGF-β localization in the Golgi apparatus.
We evaluated ESL-1 and TGF-β subcellular localization by expressing proTGF-β1-V5 and Myc-tagged ESL-1 in COS7 cells. ESL-1 was primarily localized in the Golgi apparatus in transfected cells (Figure 8, E–G). In the absence of ESL-1 expression, TGF-β1 was abundantly distributed throughout the cytoplasm, including in the ER and Golgi apparatus (Figure 8D). In contrast, when coexpressed with ESL-1, TGF-β1 was more concentrated in the Golgi apparatus (Figure 8, H–J). Because of the limitation of overexpression studies, we further assayed for TGF-β distribution in the loss-of-function context using Esl1–/– MEFs. We stained the MEFs isolated from Esl1–/– and WT embryos with TGF-β1 antibody. Statistically, TGF-β1 was localized more abundantly in the Golgi apparatus of WT versus mutant MEFs (Esl1–/– 24% versus WT 69% using the same acquisition thresholds) (Figure 8, K–S). Hence, as predicted, gain and loss of function of ESL-1 altered TGF-β subcellular localization in opposite directions.
Figure 8. ESL-1 increases the retention of TGF-β in the Golgi apparatus.
(A–C) ESL-1 is exclusively localized in the Golgi apparatus of WT P1 femur chondrocytes. The antibodies used are labeled in A and B. Merged image (C) shows the overlapping of ESL-1 with Golgin97 (a Golgi marker). (D–J) Gain of ESL-1 in COS7 cells increases TGF-β retention in the Golgi apparatus. (D) TGF-β1, including proTGF-β1 and SLC, is diffusely localized in the cytoplasm, while the signal in the Golgi apparatus is higher (arrows). (E–G) ESL-1-Myc (E) is exclusively localized in the Golgi apparatus (F). The merged image (G) shows the precise overlap of ESL-1 and Golgi apparatus. (H–J) ESL-1 (H) increases TGF-β (I) retention in the Golgi apparatus. The merged image (J) shows that TGF-β1 is remarkably enriched and colocalized with ESL-1 in the Golgi apparatus (arrows). (K–S) In comparison to WT MEFs, Esl1–/– MEFs show decreased retention of TGF-β1 in the Golgi apparatus. Genotypes of MEFs are shown on the left. The antibodies used and merged images are denoted. In WT MEFs, Golgin97 (K) and ESL-1 (L) are colocalized in the Golgi apparatus (M). TGF-β1 is abundantly localized in Golgi apparatus (O), which is largely colocalized with Golgin97 (N and P). In Esl1–/– MEFs, TGF-β1 is more evenly distributed in the cytoplasm (R). The merged image (S) of TGF-β1 (R) and Golgin97 (Q) indicates weaker colocalization of TGF-β with the Golgi apparatus. All tissue sections or cells described above were counterstained with DAPI (blue).
Discussion
The net effect of TGF-β signaling determines the specification of cell differentiation, growth, and matrix synthesis. It is the product of complex regulatory mechanisms that control secretion, activation, receptor engagement, and intracellular signaling (23–25). Tissues synthesize ample amounts of TGF-βs, but only a small fraction needs to be activated to trigger the downstream signaling events. Therefore, the bioavailability of TGF-βs must be tightly controlled by a variety of mechanisms at different levels. Current models for the regulation of TGF-β involve mechanisms for its secretion, extracellular activation in matrix, and antagonism of ligand-receptor interactions (3, 23). However, besides the well-recognized latent TGF-β pool in the ECM, TGF-β is also abundantly localized in the cytoplasm, especially in the Golgi apparatus and ER prior to secretion (26, 27). At the same time, the trans-Golgi network (TGN) is a localized concentration of abundant furin convertase activity in the cell. The colocalization of an intracellular TGF-β pool and TGN furin activity raise two important questions: (a) Is furin-dependent maturation of pro-TGF-β regulated?; and (b) Would such regulation be important in controlling TGF-β activity during development? Here, our data suggest that the answer to both questions is yes.
ESL-1, a Golgi protein, binds directly to proTGF-β in the Golgi apparatus and thus limits the processing of the maturation of TGF-β by furin convertase. Esl1–/– cartilage showed increased mature TGF-β2 and p-Smad2. This was correlated with increased ECM deposition and decreased proliferation of chondrocytes. As a result, Esl1–/– mice exhibit chondrodysplasia from embryonic stages. These data suggest not only that ESL-1 plays a role in regulating TGF-β bioavalibility, but also that this mechanism is important for skeletal development.
Furin has been recognized as a housekeeping protein that localizes in the TGN and plays an important role in proteolytically activating large numbers of proprotein substrates in the secretory pathway compartment. These include diverse signaling ligands, receptors, and pathogenic agents (28). Because the furin-dependent processing affects multiple signaling pathways, simple regulation of furin expression and/or activation might not be sufficient to differentially control activation of diverse signaling pathways in response to environmental or physiological cues. Hence, pathway-specific mechanisms for regulating furin-dependent processing might be one way to control the production and secretion of different morphogens and growth factors. Here, our findings suggest that ESL-1 serves such a novel function by preventing the maturation and secretion of TGF-β.
The expression pattern of ESL-1 overlaps that of TGF-βs in the skeleton and other organs. This supports the specific requirement of ESL-1 for normal TGF-β maturation. ESL-1 function is reminiscent of that of Emilin-1, which acts as a fine-tuning modulator of the TGF-β by regulating TGF-β proteolytic maturation, but in the ECM (20). Although they act in different cellular compartments, the similarity of ESL-1 and Emilin’s actions strongly suggest that the inhibition of the cleavage of proTGF-β is an important mode for regulating TGF-β bioavailability in general, and alteration of ESL-1 or Emilin function may lead to adverse homeostatic and/or developmental defects. We show that ESL-1’s antagonism of TGF-β function is evolutionarily conserved, since overexpression of xEsl1 led to distinct TGF-β/Nodal-deficient phenotypes in the Xenopus embryos.
In mice, the loss of ESL-1 leads to increased TGF-β signaling in the growth plate and a chondrodysplasia phenotype. However, the consequences of TGF-β dysregulation in skeletal development and morphogenesis are complex, though their importance in vivo has been highlighted by different genetic disease phenotypes. ADAMTSL2 mutations in geleophysic dysplasia patients have recently been reported to cause elevated TGF-β secretion and activity, leading to disproportionate short stature and brachydactyly in humans (29). In contrast, fibrillin1 mutations in Marfan syndrome exhibit increased TGF-β activity but result in tall stature. In earlier skeletal developmental stages, Esl1 is highly expressed in the perichondrium but at low levels in the cartilage. Similarly, fibrillin-1 and ADMTSL2 also exhibit strong expression in the perichondrium, where TGF-βs is abundantly synthesized, suggesting that the perichondrium is particularly important for production and regulation of TGF-β activity and regulation of the growth plate (9, 30, 31). Previous studies have shown that the TGF-β1 inhibition of chondrocyte proliferation and differentiation in the long bones requires an intact perichondrium (32, 33). PTHrP plays a central role in maintaining proliferating chondrocytes in an undifferentiating state by relaying TGF-β signaling to the cartilage in a perichondrium-dependent manner (16, 34). This is consistent with our observation that Esl1–/– growth plates exhibited delayed terminal differentiation, with increased PTHrP and reduced IHH expression. However, the decreased proliferation in Esl1–/– chondrocytes may rely on a PTHrP/IHH axis–independent mechanism. Esl1 expression is increased in PZ of growth plate in later skeletal development, suggesting that Esl1 and its TGF-β modulator function are regulated in a temporal-spatial fashion. The perichondrium and growth plate cartilage expresses other critical chondrogenesis signaling factors, including FGF18, Wnt, PTHrP, BMPs, etc. Hence, in those aforementioned TGF-β overactivation genetic models, the temporal-spatial differences in TGF-β activation can lead to diverse signaling interactions with other pathways. This most likely helps to determine the precise regulation of growth plate homeostasis and helps to explain why TGF-β overactivation in different genetic models can exhibit diverse outcomes in the skeletal system.
The functional complexity and context dependence of TGF-β signaling necessitate a similarly diverse and complex set of regulators that control TGF-β bioavailability in both a temporal and spatial fashion. Our results indicate that ESL-1 constitutes a mechanism at the cellular level for controlling proteolytic maturation and Golgi retention. Moreover, ESL-1 is broadly expressed in many other organs and tissues in addition to the skeletal system. Therefore, dysregulation of ESL-1 may be involved in other pathological conditions where dysregulation of TGF-β plays a central role. This includes cancer progression, immune dysregulation, osteoblast/osteoclast coupling, and fibrosis/inflammations. Interestingly, ESL-1 cooperates with PSGL-1 and CD44 to regulate neutrophil rolling, which is critical for recruitment of neutrophils to inflamed tissues (35). However, whether ESL-1 might specify TGF-β bioavailability during immune responses remains to be studied. Additionally, a posttranslationally modified ESL-1 variant with a unique carbohydrate epitope was specifically overexpressed on the surface of almost all epithelial cancers at precursor stages (21). Whether the dramatically changed subcellular localization of ESL-1 causes a dysregulation of TGF-β that might contribute to the onset and/or progression of cancer is still unknown. In summary, as what we believe to be a novel intracellular inhibitor for TGF-β bioavailability, ESL-1 may serve as a therapeutic target for regulating TGF-β during different disease processes, such as arthritis and cancer, and the Esl1–/– mouse will be a useful model to address these other questions.
Methods
Generation of Esl1–/– mice.
The targeting vector was designed to delete exons 13–16 of Esl1 (Supplemental Figure 1A). AB2.2 ES cells were electroporated with linearized targeting construct and selected by puromycin and fialuridine. The ES clones verified by Southern blot analysis were microinjected into C57BL/6J blastocysts and transferred into foster mothers. The chimeric males were mated with C57BL/6J females. Germline transmission was confirmed by agouti coat color in F1 animals, and the offspring were genotyped for the ESL-1 mutant allele using PCR (PCR primer sequences: 5′-CAGCACTCAAGTGAAAGCGGT-3′, 5′-AGGCATGCTGGGGATGCGGT-3′, 5′-GGTTGCCAATTTGGTTCGGTTG-3′) (Supplemental Figure 1, B and C). All mouse-related protocols were reviewed and approved by the Baylor College of Medicine Institutional Review Board.
Generation of Col2a1-Esl1 transgenic mice.
The linearized Co2a1-Esl-1 DNA fragments were injected into pronuclei of 1-cell-stage embryos collected from FVB/N females. The injected embryos were subsequently transferred into pseudopregnant ICR foster females. The transgenic founders were crossed with WT FVB/N mice to establish transgenic mouse lines. Each transgenic line was maintained by crossing with WT FVB/N mice.
Biological assays in Xenopus.
Manipulation of Xenopus embryos, whole-mount in situ hybridizations, and capped mRNA preparations were performed as previously described (36, 37). Xnr3 synthetic RNA was made from a cDNA plasmid that was a gift from J. Heasman (University of Cincinnati College of Medicine, Cincinnati, Ohio, USA) (19). A full-length Xenopus ESL-1 cDNA clone (clone ID, 7688308) was purchased from Open Biosystems. A 4.0-kb xESL-1 cDNA was subcloned into pRN3 (gift from P. Lemaire, Institut de Biologie du Développement de Marseille-Luminy, CNRS-Université de la Méditerranée, France) to create RN3-xESL-1.
In situ hybridization.
Five-micrometer sections of tissues were deparaffinized, hybridized with 35S-labeled probes, and developed in emulsion (Kodak) according to standard protocols.
Cartilage protein extraction.
Rib cages were isolated from P3 mouse littermates. After rapid removal of all bones and muscles, each cleaned rib cartilage sample was frozen in liquid N2 and subsequently transferred to 300 μl lysis buffer (0.0625 M Tris-HCl pH 7.4, 2% SDS) for 1 minute of homogenization, followed by an incubation at 95°C for 30 minutes. The protein concentration of the lysate was measured with Micro BCA reagent (Pierce). After treatment with 5% β-mercaptoethonal, 10 μg cartilage protein extracts were loaded for Western blot assay.
Coimmunoprecipitation.
COS7 cells were transfected with pcDNA3.1, Myc-tagged ESL-1, V5-tagged TGF-β1 or TGF-β2 plasmids using Lipofectamine 2000 (Invitrogen). After culture for 48 hours, cells were lysed in RIPA buffer (Upstate) with protease inhibitor (Roche). Myc or V5 antibodies (1:1,000; Invitrogen) were used to immunoprecipitate the ESL-1 or TGF-β bound proteins, respectively. To test the possibility of direct binding of ESL-1 and TGF-β ligand, the cell lysates were incubated with 100 ng purified rhTGF-β1 (R&D Systems) for 2 hours, followed by immunoprecipitation. The precipitates were boiled in Laemmli buffer (Bio-Rad) with 5% β-mercaptoethanol and loaded to a 4%–20% gradient gel for Western blot assays.
Western blots.
The transblotted PVDF membranes were hybridized with primary antibodies under the following conditions: p-MEK1/2, p-Smad2, and p-Smad1/5/8 (all rabbit IgG from Cell Signaling Technology; 1:1,000 in PBST overnight), V5 (1:5,000, 5% milk in PBST, 1 hour, mouse IgG2a; Invitrogen), Myc (1:5,000, 5% milk in PBST, 1 hour, mouse IgG1; Invitrogen), ESL-1 (1:10,000, 5% milk in PBST, rabbit IgG; gift from M.K. Wild (University of Münster, Münster, Germany), LAP1(1:1,000, PBST, overnight, goat IgG; R&D Systems), TGF-β2 (1:300 in PBST 1% milk, overnight, rabbit IgG; Santa Cruz Biotechnology Inc.), α-tubulin (1:10,000, 5% milk in PBST, 1 hour, mouse IgG1; Sigma-Aldrich). Secondary antibodies were anti-rabbit (1:5,000; GE), anti-mouse (1:5,000; Bio-Rad), anti-goat (1:10,000; Roche). The washed membranes were reacted with ECL-Plus reagent (GE) and exposed to X-ray film. The density of each band was normalized with α-tubulin and quantified by densitometry (AlphaImager).
TGF-β reporter assay.
COS7 cells were transfected with 1 μg of each plasmid based on the experimental strategy (each transfection included 0.1 μg SV40-LacZ for control) and incubated in 1% FBS for 24 hours at 37°C. The conditioned medium was collected and incubated with the serum-starved PAI-luciferase reporter mink lung epithelial cells for 16 hours. The cell lysate was collected for luciferase assays. The results were normalized to the LacZ activity of the COS7 cell lysates. For primary chondrocytes, 1 × 106 cells were plated in 60-mm dishes in DMEM with 10% FBS. The conditioned medium was collected after 48 hours of culture. The TGF-β reporter cells were treated with the primary chondrocyte conditioned medium. DMEM 10% FBS with or without rhTGF-β1 was used as control.
RNA isolation and qRT-PCR.
The flash-frozen rib cartilages were isolated from P3 knockout/transgenic mice and WT littermates (n = 3) for total RNA extraction with TRIzol reagent (Invitrogen) and purification using RNeasy mini columns (QIAGEN). Total RNA (2 μg) was used for synthesis of the first-strand cDNA with a Superscript III RT system (Invitrogen). qRT-PCR was performed according to the manufacturer’s protocol using gene-specific primers and a FastStart DNA Master SYBR Green I reagent using a LightCycler instrument (Roche). Amplification of β-actin was chosen to normalize for variations in template concentrations.
Furin assay.
The primary chondrocytes or the HEK293 cells 48 hours after transfection were lysed in lysis buffer (100 mM HEPES pH 7.5, 0.5% Triton X-100) by freeze-thawing 3 times. For furin assays, lysate protein was reconstituted to 1 mg/ml in lysis buffer supplied with 1 mM CaCl2, 1 mM β-mercaptoethonal. Furin (NEB) was added at 10 U/1 ml in the mixture and incubated at 37°C. At each time point, 20-μl aliquots of reaction were removed and immediately quenched by adding 10 μl Laemmli buffer (Bio-Rad) and 1 μl β-mercaptoethonal. Samples at all time points were subsequently analyzed by Western blots with anti-V5 antibody.
Pulse-chase assay.
Thirty-six hours after transfection with TGF-β-V5 and/or ESL-1-Myc plasmids, HeLa cells in 35-mm dishes were washed with 1× PBS, then starved in deficient DMEM medium (10% FBS, lack of Met and Cys) for 2 hours. The starved cells were subsequently pulsed with the deficient DMEM medium supplied with 250 μCi 35S-labeled Met and Cys (Express Tag 35S protein labeling mix; PerkinElmer) for 2 hours. The cells were extensively washed with PBS, then chased with full DMEM medium supplemented with high-concentration Met and Cys (500 μg/ml Cys and 100 μg/ml Met) for 1-, 2-, and 4-hour time points. The cell lysates and medium were collected for IP with V5 antibody (1:1,000). The immunoprecipitates were separated in SDS-PAGE gels and exposed to X-film.
Immunofluorescence and microscopy.
The mouse knee joint paraffin sections (5 μm) were treated with retrieval solution (Dako). The standard protocols recommended by antibody suppliers were followed exactly for staining. Primary antibodies were p-Smad2 (1:200; Cell Signaling Technology), BrdU (1:40; Invitrogen), and ESL-1 (1:800; from M.K. Wild). Secondary antibodies were anti-mouse Alexa Fluor 594, anti-rabbit Alexa Fluor 594, and anti-mouse Alexa Fluor 488 (1:600; Invitrogen). All comparable samples were imaged with identical exposure times and conditions. The p-Smad2– or BrdU-positive cells were counted and analyzed with Zeiss Axiovision software.
For cellular localization analysis, MEFs or COS7 cells transfected with ESL-1-Myc and/or TGF-β1-V5 plasmids were cultured in chamber slides (Lab-Tek) for 24 hours. After fixation in 4% paraformaldehyde and permeablization in 0.1% Triton X-100, cells were incubated with primary antibodies: Golgin97 (1:400), ESL-1 (1:2,000), V5 (for both the proTGF-β1-V5 and mature TGF-β1-V5 ligand, 1:1,000), Myc (1:1,000, chicken IgM; Invitrogen), TGF-β1 (1:200; Promega). Immunostaining was visualized with the corresponding fluorescent secondary antibodies (anti–mouse IgG2a Alexa Fluor 594 and anti–chicken IgM Alexa Fluor 488, anti–rabbit IgG Alexa Fluor 594 or 488, 1:600; Invitrogen).
Statistics.
We conduct statistical analyses by ANOVA with Student’s t test. The error bars in the histograms show mean ± SD. A P value less than 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank Martin K. Wild for the ESL-1 antiserum; Chu-Xia Deng (NIDDK, NIH, Bethesda, Maryland, USA) for the Fgfr3–/– mice; and Janet Heasman for the Xnr3 mRNA expression plasmid. This work was supported by NIH grants P01 ES11253 (to B. Lee), P01 HD22657 (to B. Lee), F32 HL09657 (to H. Lu), R37 AI32177 (to A.L. Beaudet), P01 CA34282 (to D.B. Rifkin), and P01 GM 083220 (to D.B. Rifkin) and an Arthritis Foundation fellowship (T. Yang).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2010;120(7):2474–2485. doi:10.1172/JCI42150.
References
- 1.Kronenberg HM. Developmental regulation of the growth plate. Nature. 2003;423(6937):332–336. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- 2.Kronenberg HM. The role of the perichondrium in fetal bone development. Ann N Y Acad Sci. 2007;1116:59–64. doi: 10.1196/annals.1402.059. [DOI] [PubMed] [Google Scholar]
- 3.Annes JP, Munger JS, Rifkin DB. Making sense of latent TGFbeta activation. J Cell Sci. 2003;116(pt 2):217–224. doi: 10.1242/jcs.00229. [DOI] [PubMed] [Google Scholar]
- 4.Dubois CM, Laprise MH, Blanchette F, Gentry LE, Leduc R. Processing of transforming growth factor beta 1 precursor by human furin convertase. J Biol Chem. 1995;270(18):10618–10624. doi: 10.1074/jbc.270.18.10618. [DOI] [PubMed] [Google Scholar]
- 5.Burrus LW, Zuber ME, Lueddecke BA, Olwin BB. Identification of a cysteine-rich receptor for fibroblast growth factors. Mol Cell Biol. 1992;12(12):5600–5609. doi: 10.1128/mcb.12.12.5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steegmaier M, et al. The E-selectin-ligand ESL-1 is a variant of a receptor for fibroblast growth factor. Nature. 1995;373(6515):615–620. doi: 10.1038/373615a0. [DOI] [PubMed] [Google Scholar]
- 7.Olofsson A, et al. Latent transforming growth factor-beta complex in Chinese hamster ovary cells contains the multifunctional cysteine-rich fibroblast growth factor receptor, also termed E-selectin-ligand or MG-160. Biochem J. 1997;324(pt 2):427–434. doi: 10.1042/bj3240427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahn J, Febbraio M, Silverstein RL. A novel isoform of human Golgi complex-localized glycoprotein-1 (also known as E-selectin ligand-1, MG-160 and cysteine-rich fibroblast growth factor receptor) targets differential subcellular localization. J Cell Sci. 2005;118(pt 8):1725–1731. doi: 10.1242/jcs.02310. [DOI] [PubMed] [Google Scholar]
- 9.Pelton RW, Dickinson ME, Moses HL, Hogan BL. In situ hybridization analysis of TGF beta 3 RNA expression during mouse development: comparative studies with TGF beta 1 and beta 2. Development. 1990;110(2):609–620. doi: 10.1242/dev.110.2.609. [DOI] [PubMed] [Google Scholar]
- 10.Deng C, Wynshaw-Boris A, Zhou F, Kuo A, Leder P. Fibroblast growth factor receptor 3 is a negative regulator of bone growth. Cell. 1996;84(6):911–921. doi: 10.1016/S0092-8674(00)81069-7. [DOI] [PubMed] [Google Scholar]
- 11.Ohbayashi N, et al. FGF18 is required for normal cell proliferation and differentiation during osteogenesis and chondrogenesis. Genes Dev. 2002;16(7):870–879. doi: 10.1101/gad.965702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shiang R, et al. Mutations in the transmembrane domain of FGFR3 cause the most common genetic form of dwarfism, achondroplasia. Cell. 1994;78(2):335–342. doi: 10.1016/0092-8674(94)90302-6. [DOI] [PubMed] [Google Scholar]
- 13.Murakami S, Balmes G, McKinney S, Zhang Z, Givol D, de Crombrugghe B. Constitutive activation of MEK1 in chondrocytes causes Stat1-independent achondroplasia-like dwarfism and rescues the Fgfr3-deficient mouse phenotype. Genes Dev. 2004;18(3):290–305. doi: 10.1101/gad.1179104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abe M, Harpel JG, Metz CN, Nunes I, Loskutoff DJ, Rifkin DB. An assay for transforming growth factor-beta using cells transfected with a plasminogen activator inhibitor-1 promoter-luciferase construct. Anal Biochem. 1994;216(2):276–284. doi: 10.1006/abio.1994.1042. [DOI] [PubMed] [Google Scholar]
- 15.Kronenberg HM. PTHrP and skeletal development. Ann N Y Acad Sci. 2006;1068:1–13. doi: 10.1196/annals.1346.002. [DOI] [PubMed] [Google Scholar]
- 16.Alvarez J, Sohn P, Zeng X, Doetschman T, Robbins DJ, Serra R. TGFbeta2 mediates the effects of hedgehog on hypertrophic differentiation and PTHrP expression. Development. 2002;129(8):1913–1924. doi: 10.1242/dev.129.8.1913. [DOI] [PubMed] [Google Scholar]
- 17.Tavella S, et al. Targeted expression of SHH affects chondrocyte differentiation, growth plate organization, and Sox9 expression. J Bone Miner Res. 2004;19(10):1678–1688. doi: 10.1359/JBMR.040706. [DOI] [PubMed] [Google Scholar]
- 18.Dabovic B, et al. Bone abnormalities in latent TGF-[beta] binding protein (Ltbp)-3-null mice indicate a role for Ltbp-3 in modulating TGF-[beta] bioavailability. J Cell Biol. 2002;156(2):227–232. doi: 10.1083/jcb.200111080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yokota C, et al. A novel role for a nodal-related protein; Xnr3 regulates convergent extension movements via the FGF receptor. Development. 2003;130(10):2199–2212. doi: 10.1242/dev.00434. [DOI] [PubMed] [Google Scholar]
- 20.Zacchigna L, et al. Emilin1 links TGF-beta maturation to blood pressure homeostasis. Cell. 2006;124(5):929–942. doi: 10.1016/j.cell.2005.12.035. [DOI] [PubMed] [Google Scholar]
- 21.Hensel F, et al. A novel proliferation-associated variant of CFR-1 defined by a human monoclonal antibo45kurr5yrtdy. Lab Invest. 2001;81(8):1097–1108. doi: 10.1038/labinvest.3780322. [DOI] [PubMed] [Google Scholar]
- 22.Miyazono K, Olofsson A, Colosetti P, Heldin CH. A role of the latent TGF-beta 1-binding protein in the assembly and secretion of TGF-beta 1. EMBO J. 1991;10(5):1091–1101. doi: 10.1002/j.1460-2075.1991.tb08049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gumienny TL, Padgett RW. The other side of TGF-beta superfamily signal regulation: thinking outside the cell. Trends Endocrinol Metab. 2002;13(7):295–299. doi: 10.1016/s1043-2760(02)00615-x. [DOI] [PubMed] [Google Scholar]
- 24.Roberts AB, Derynck R. Meeting report: signaling schemes for TGF-beta. Sci STKE. 2001;2001(113):pe43. doi: 10.1126/stke.2001.113.pe43. [DOI] [PubMed] [Google Scholar]
- 25.Serra R, Chang C. TGF-beta signaling in human skeletal and patterning disorders. Birth Defects Res C Embryo Today. 2003;69(4):333–351. doi: 10.1002/bdrc.10023. [DOI] [PubMed] [Google Scholar]
- 26.Miyazono K, Thyberg J, Heldin CH. Retention of the transforming growth factor-beta 1 precursor in the Golgi complex in a latent endoglycosidase H-sensitive form. J Biol Chem. 1992;267(8):5668–5675. [PubMed] [Google Scholar]
- 27.Roth-Eichhorn S, Kuhl K, Gressner AM. Subcellular localization of (latent) transforming growth factor beta and the latent TGF-beta binding protein in rat hepatocytes and hepatic stellate cells. Hepatology. 1998;28(6):1588–1596. doi: 10.1002/hep.510280619. [DOI] [PubMed] [Google Scholar]
- 28.Thomas G. Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat Rev Mol Cell Biol. 2002;3(10):753–766. doi: 10.1038/nrm934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Le GC, et al. ADAMTSL2 mutations in geleophysic dysplasia demonstrate a role for ADAMTS-like proteins in TGF-beta bioavailability regulation. Nat Genet. 2008;40(9):1119–1123. doi: 10.1038/ng.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koo BH, et al. ADAMTS-like 2 (ADAMTSL2) is a secreted glycoprotein that is widely expressed during mouse embryogenesis and is regulated during skeletal myogenesis2. Matrix Biol. 2007;26(6):431–441. doi: 10.1016/j.matbio.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 31.Keene DR, et al. Fibrillin-1 in human cartilage: developmental expression and formation of special banded fibers3. J Histochem Cytochem. 1997;45(8):1069–1082. doi: 10.1177/002215549704500805. [DOI] [PubMed] [Google Scholar]
- 32.Di Nino DL, Long F, Linsenmayer TF. Regulation of endochondral cartilage growth in the developing avian limb: cooperative involvement of perichondrium and periosteum3. Dev Biol. 2001;240(2):433–442. doi: 10.1006/dbio.2001.0471. [DOI] [PubMed] [Google Scholar]
- 33.Di Nino DL, Crochiere ML, Linsenmayer TF. Multiple mechanisms of perichondrial regulation of cartilage growth2. Dev Dyn. 2002;225(3):250–259. doi: 10.1002/dvdy.10160. [DOI] [PubMed] [Google Scholar]
- 34.Alvarez J, Horton J, Sohn P, Serra R. The perichondrium plays an important role in mediating the effects of TGF-beta1 on endochondral bone formation2. Dev Dyn. 2001;221(3):311–321. doi: 10.1002/dvdy.1141. [DOI] [PubMed] [Google Scholar]
- 35.Hidalgo A, Peired AJ, Wild MK, Vestweber D, Frenette PS. Complete identification of E-selectin ligands on neutrophils reveals distinct functions of PSGL-1, ESL-1, and CD441. Immunity. 2007;26(4):477–489. doi: 10.1016/j.immuni.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tseng HT, Shah R, Jamrich M. Function and regulation of FoxF1 during Xenopus gut development. Development. 2004;131(15):3637–3647. doi: 10.1242/dev.01234. [DOI] [PubMed] [Google Scholar]
- 37.Tao J, et al. BMP4-dependent expression of Xenopus Grainyhead-like 1 is essential for epidermal differentiation. Development. 2005;132(5):1021–1034. doi: 10.1242/dev.01641. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.