Abstract
Nearly every extracellular ligand that has been found to play a role in regulating bone biology acts, at least in part, through MAPK pathways. Nevertheless, much remains to be learned about the contribution of MAPKs to osteoblast biology in vivo. Here we report that the p38 MAPK pathway is required for normal skeletogenesis in mice, as mice with deletion of any of the MAPK pathway member–encoding genes MAPK kinase 3 (Mkk3), Mkk6, p38a, or p38b displayed profoundly reduced bone mass secondary to defective osteoblast differentiation. Among the MAPK kinase kinase (MAP3K) family, we identified TGF-β–activated kinase 1 (TAK1; also known as MAP3K7) as the critical activator upstream of p38 in osteoblasts. Osteoblast-specific deletion of Tak1 resulted in clavicular hypoplasia and delayed fontanelle fusion, a phenotype similar to the cleidocranial dysplasia observed in humans haploinsufficient for the transcription factor runt-related transcription factor 2 (Runx2). Mechanistic analysis revealed that the TAK1–MKK3/6–p38 MAPK axis phosphorylated Runx2, promoting its association with the coactivator CREB-binding protein (CBP), which was required to regulate osteoblast genetic programs. These findings reveal an in vivo function for p38β and establish that MAPK signaling is essential for bone formation in vivo. These results also suggest that selective p38β agonists may represent attractive therapeutic agents to prevent bone loss associated with osteoporosis and aging.
Introduction
During both embryonic development and adult life, osteoblasts respond to extracellular signals to regulate synthetic capacity and overall bone mass by secreting an ECM containing collagenous and noncollagenous proteins (1). Osteoblast differentiation is controlled by transcription factors that are expressed in a defined temporal and spatial sequence. Two sets of factors have been suggested to regulate osteoblast differentiation and activity. Dlx5/6 (2), Twist1/2 (3), Runx2 (4), and Osterix (5, 6) control the commitment of mesenchymal stem cells (MSCs) to the osteoblast lineage. The canonical Wnt pathway (7) and the transcription factor ATF4 (8) function later in mature osteoblasts to regulate their synthetic function during adult bone remodeling (9).
Among these, Runx2 is considered to be the master regulator of osteoblast development and bone formation (10, 11). Heterozygous mutations in the Cbfa1 gene encoding RUNX2 are responsible for the inherited human disease cleidocranial dysplasia (CCD), characterized by hypoplasia of the clavicle and delayed closure of the fontanelles (12, 13). Haploinsufficiency of the Cbfa1 gene in mice causes a similar syndrome (14). Previous in vitro studies using cell lines treated with MAPK inhibitors have shown that p38 and ERK MAPKs are important for early osteoblast differentiation, whereas JNK MAPK is important for late-stage differentiation as shown by decreased alkaline phosphatase activity and Atf4 expression, respectively (15–17). However, biological functions of ERK MAPK are controversial in osteoblast differentiation. Alkaline phosphatase activity and RUNX2 expression, key regulators for preosteoblast differentiation, were altered through regulating Runx2 transcriptional activity in calvarial osteoblasts from transgenic mice expressing constitutively active or dominant negative mutants of MEK1, an ERK MAPK kinase (18), whereas they were normal in the absence of both ERK1 and 2 MAPKs (19). Thus, how osteoblast differentiation is physiologically regulated by MAPK-mediated posttranslational modifications that occur in response to osteogenic stimuli and how these modifications in turn translate into differences in bone homeostasis remain to be elucidated.
MAPK cascades are a fundamental and evolutionarily conserved mechanism for cellular responses to a wide range of extracellular signals, particularly many of the extracellular ligands relevant to osteoblasts, such as BMPs, noncanonical WNTs, PTH, TNF, and FGFs. Despite this, the relative contribution of p38 MAPKs to osteoblast biology has yet to be assessed using genetic loss-of-function studies (20–22). In mammalian cells, 4 isoforms of the p38 MAPKs have been identified, p38α, -β, -γ, and -δ. The p38 MAPKs are predominantly activated by 2 upstream MAPK kinases, MKK3 and MKK6 (23). The activation of MKK3 and MKK6 is in turn mediated by MAP3Ks, such as MLK3, Ask, and TAK1, in a manner that appears to be both cell type and stimuli specific. p38a–/– and Mkk3–/–Mkk6–/– embryos die due to defects of placental development (24–28), while mice lacking p38β, -γ, and -δ are viable without any obvious defects at baseline (29–31). Hence, despite biochemical evidence for the existence of specific roles for individual p38 isoforms, redundancy and embryonic lethality have impeded attempts to establish their distinct functions in vivo.
TGF-β–activated kinase 1 (TAK1) is a member of the MAP3K family originally identified as a mediator of the p38 MAPK pathway downstream of TGF-β and bone morphogenetic protein (BMP) signaling (32, 33). A recent study from our laboratory has reported that cartilage-specific deletion of the Tak1 gene in mice causes severe chondrodysplasia with runting and joint abnormalities including elbow dislocation and tarsal fusion, which is similar to the phenotype of BMP receptor (BMPR)1b and Gdf5-deficient mice. Biochemical analysis demonstrated that TAK1 is required for BMP signaling in cartilage, where it functions as an upstream activating kinase for BMP-responsive SMADs and MAPK pathways (22).
In this study, we have analyzed the in vivo function of the p38 MAPK pathway in osteoblasts by analyzing mice with disruption of multiple p38 MAPK signaling mediators, including TAK1, MKK3 and -6, p38α, and -β. Mice with osteoblast-specific deletion of the Tak1 gene display a phenotype similar to mice heterozygous for Runx2 and humans haploinsufficient for Cbfa1/Runx2 that is characterized by clavicular hypoplasia and delayed fontanelle fusion. The MKK3/6-p38 MAPK pathway is the critical mediator of these effects downstream of TAK1, functioning to phosphorylate Runx2, leading to increased Runx2 transcriptional activity. This study provides, to our knowledge, the first in vivo evidence for a critical role for the p38 MAPK pathway both in the developing skeleton and in controlling adult bone mass. Moreover, the parallel analysis of multiple strains genetically deficient for different mediators of the p38 MAPK pathway yields surprising insights into redundancy within this pathway, revealing in particular what we believe to be a previously unappreciated role for p38β in vivo.
Results
TAK1 expression in osteoblasts.
We performed immunohistochemistry (IHC) to characterize the expression of TAK1 in bone. In the calvarium, TAK1 was expressed in osteoblasts along the osteogenic front and by osteocytes embedded in the bone matrix (Figure 1A). Similar expression was seen in the tibia (Supplemental Figure 1A; supplemental material available online with this article; doi: 10.1172/JCI42285DS1). To confirm TAK1-specific staining in osteoblasts, animals homozygous for a Tak1-floxed allele expressing an osterix-cre transgene (hereafter referred to as Tak1osx mice) that deletes in osteoblasts were used as a control. Staining at both endocortical osteoblasts and osteocytes was ablated in Tak1osx mice, whereas staining in chondrocytes was still observed as expected (Supplemental Figure 1B), making it unlikely that phenotypes observed in Tak1osx mice can be attributed to functions of TAK1 in chondrocytes. The kinetics of TAK1 expression during osteoblast differentiation were examined during the differentiation of human MSCs, and TAK1 expression remained relatively unchanged during osteoblast differentiation (Supplemental Figure 1C).
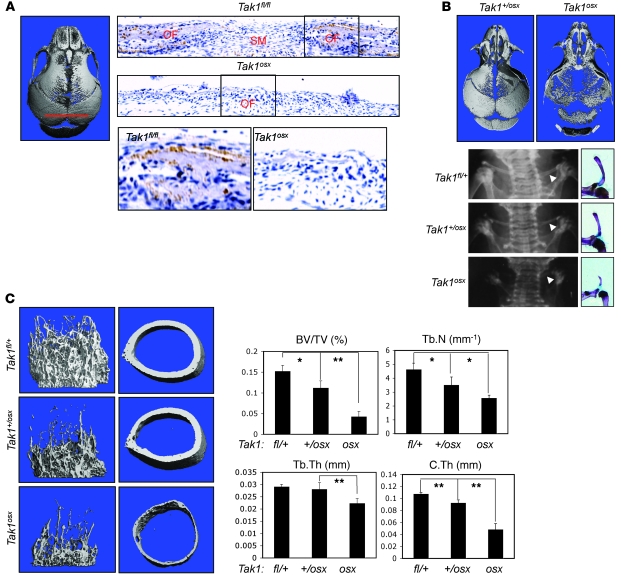
Figure 1. CCD in Tak1osx mice.
(A) Sections were taken from the sagittal suture as indicated and stained for TAK1 expression by immunohistochemistry. The osteogenic front (OF) and sutural mesenchyme (SM) are labeled. Original magnification, ×100. (B) Ossification of Tak1osx and Tak1+/osx skulls was analyzed by μCT and the 3D reconstruction displayed. Images are representative of more than 8 mice per genotype (left). Plain film x-rays and pictures of Alizarin red/Alcian blue–stained skeletal preps demonstrating clavicular hypoplasia in Tak1osx mice. Images are representative of more than 8 mice per genotype (right). Arrowheads indicate the clavicle. (C) Femurs from female Tak1fl/+, Tak+/osx, and Tak1osx mice were analyzed by μCT 3D cortical reconstructions of the trabecular bone (left) and midshaft cortical bone (middle). Quantitative parameters are displayed in the right panel: bone volume/total volume (BV/TV), trabecular number per cubic millimeter (Tb.N), trabecular thickness (Tb.Th), and cortical thickness (C.Th). *Significant difference by Student’s t test, P < 0.05; **P < 0.005.
Tak1osx mice display CCD.
To investigate the role of TAK1 in osteoblasts, we generated Tak1osx mice lacking TAK1 expression in osteoblasts. These mice display clavicular hypoplasia and hypomineralization of the calvarium, features of humans with CCD and mice heterozygous for Runx2 (Figure 1B and Supplemental Figure 2, A and B). In particular, the parietal and frontal bones displayed a greater reduction in mineralization than the occipital or intraparietal bones, with the effect that the anterior fontanelle is relatively more enlarged than the posterior fontanelle. In displaying a greater relative effect on the anterior versus the posterior fontanelle, the pattern of hypomineralization observed in Tak1osx mice resembles that seen in Runx2+/– mice and humans with CCD (Figure 1B) (34). Likewise, the clavicle of Tak1osx mice is hypoplastic, showing the presence of only a lateral rudiment (Figure 1B). This also resembles the phenotype of Runx2+/– mice, which show a preferential loss of the medial clavicle and retention of lateral elements (10). Both male and female Tak1osx mice displayed these phenotypes (data not shown).
In addition to the clavicular and calvarial phenotypes, Tak1osx mice display a substantial reduction in bone mass in the long bones (Figure 1C and Supplemental Figure 2C). Femoral trabecular bone and midshaft cortical bone from Tak1osx and littermate controls were analyzed by μCT. As shown in both the 3D reconstructions of trabecular bone and the measurement of trabecular bone volume/total volume analyzed (BV/TV), Tak1osx mice display an approximately 3-fold reduction in trabecular bone mass at 3 weeks of age. Mice heterozygous for the Tak1 floxed allele also having a single copy of the osx-cre transgene (hereafter Tak1+/osx) display an intermediate effect, showing a modest reduction in BV/TV. The significant reduction in bone mass between Tak1osx and Tak1+/osx mice confirms that the decreased bone density in Tak1osx mice is specific for the deletion of TAK1 in osteoblasts and not due to the presence of the cre transgene. Tak1osx mice also displayed a greater than 2-fold thinning of midshaft cortical bone thickness. Histologic analysis of the tibia in Tak1osx mice confirmed these findings, with Tak1osx mice displaying a reduction in trabecular bone and a substantial reduction in the mineralization of the secondary center of ossification above the growth plate (Supplemental Figure 2C). Histomorphometric analysis and metabolic labeling of bone formation demonstrated a significant reduction in osteoblast numbers, osteoid volume, and mineralizing surface area (Supplemental Table 1). Osteoclasts displayed a trend toward decreased numbers, though this did not rise to statistical significance. Finally, Tak1osx mice display a moderate runting phenotype (approximately 20% reduction in weight) with low penetrance (<50%) (Supplemental Figure 2B). Thus, the Tak1osx and Runx2+/– phenotypes share many key features, suggesting that TAK1 is a critical regulator of Runx2 in osteoblasts.
Decreased osteoblast function in Tak1osx mice.
To further characterize Tak1osx osteoblasts, RNA was isolated from both the calvarium and marrow-flushed tibia from Tak1osx mice and littermate controls and expression of osteoblast genes was measured by quantitative PCR (Figure 2A). Alkaline phosphatase (Alp) and osterix (Osx) are markers of early osteoblast differentiation, and type 1 collagen α (Col1) and osteocalcin (Ocn) are markers of late osteoblast differentiation (9). Both the extent of staining and the per cell intensity of staining of both early and late osteoblast marker genes were decreased in Tak1osx mice, indicating that osteoblast differentiation is defective from an early stage in calvarium and tibia in the absence of TAK1. These defects were selective, since the expression of sprouty 2 (Spry2), an FGF target gene (35) was not affected by TAK1 deletion. Likewise, in the calvarium, noggin, a gene predominantly expressed by cells within the sutural mesenchyme (36), was normally expressed in Tak1osx mice, again demonstrating a selective effect of this signaling pathway. Transcript levels of Runx2 in either the calvarium or tibia showed a modest, nonstatistically significant decrease in Tak1osx mice. To assess Runx2 protein levels, calvarial osteoblasts were isolated and extracts directly immunoblotted for Runx2 protein expression (Figure 2A). Consistent with the quantitative PCR analysis of calvarium and tibia, Runx2 protein levels were reduced by about 20%. These data suggest that while decreased Runx2 expression may partially contribute to the phenotype observed in Tak1osx mice, it is unlikely to be the sole factor, as within the range of variation in Runx2 transcript levels displayed by Tak1osx mice, even those mice with Runx2 levels similar to the mean of the WT group still displayed key features of the Runx2+/– phenotype. This suggests that TAK1-mediated posttranslational regulation of Runx2 is critical to the Tak1osx phenotype.
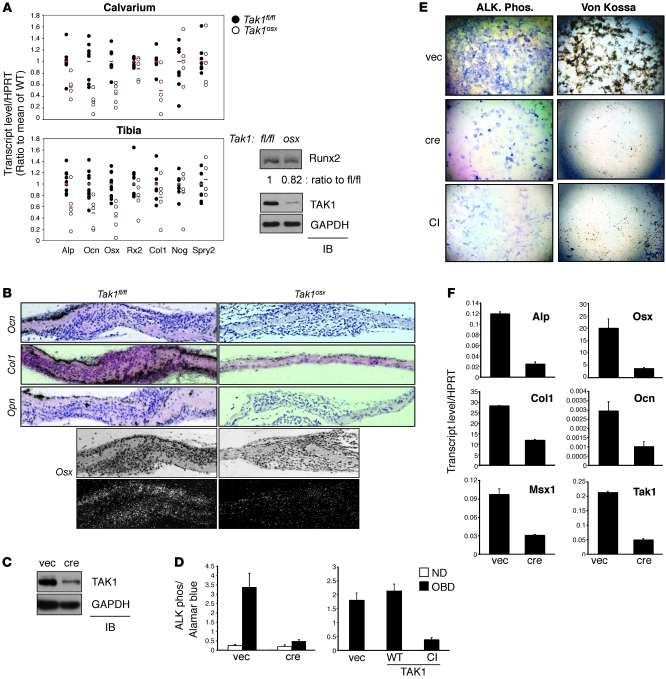
Figure 2. Phenotype of Tak1osx osteoblast mice.
(A) Calvaria and tibias were isolated from female Tak1fl/fl and Tak1osx mice, RNA extracted, and analyzed by quantitative PCR. The value of each sample is indicated with a circle and the average value of each group indicated with a red line. Alp, Ocn, Osx, Runx2 (Rx2), Col1, noggin (Nog), sprouty2 (Spry2). Calvarial Alp, Ocn, Osx, and Col1 and tibial Alp, Ocn, and Osx showed statistically significant changes by Student’s t test (P < 0.05) (left). Primary CalvOb were isolated from Tak1fl/fl and Tak1osx pups and immunoblotted with antibodies specific to Runx2, TAK1, and GAPDH (right). (B) Sections from Takfl/fl and Tak1osx mice were analyzed for the expression of the indicated genes by in situ hybridization. The signal is viewed as black over an H&E-stained background. Original magnification, ×100. (C) Tak1fl/fl CalvOb were infected with vector or cre lentivirus, and expression of TAK1 was analyzed by immunoblotting with anti-TAK1 antibody. (D) Tak1fl/fl CalvOb infected by vector or cre lentivirus (left) were cultured for 6 days under differentiation conditions, and ALP activity was analyzed by colorimetric assay. WT CalvOb expressing WT or catalytically inactive (CI) TAK1 (right) were similarly analyzed. Values are mean + SD. (E) Tak1fl/fl CalvOb infected by vector, cre, or TAK1-CI–expressing lentivirus were analyzed for Fast Blue staining for ALP activity (left) or Von Kossa staining for mineralization activity (right). Original magnification, ×25. (F) RNA levels of the indicated genes were analyzed by quantitative PCR on Tak1fl/fl CalvOb infected by vector or cre lentivirus. Values are mean + SD.
To directly visualize the expression of several of the osteoblast marker genes analyzed above, in situ hybridization was performed on Tak1fl/fl and Tak1osx calvaria (Figure 2B). As expected, Opn, Ocn, and Col1 were all expressed by the osteoblasts lining the calvarial bone and their expression was reduced in Tak1osx mice. Due to low expression levels, Osx in situ hybridization was visualized by darkfield microscopy and was likewise reduced in Tak1osx mice. Notably, the overall reduction in staining came predominantly from a reduction in the per cell intensity or level of expression of Opn, Ocn, and Col1 (shown at high power in Supplemental Figure 2D).
Impaired differentiation of TAK1-deficient osteoblasts.
To directly examine the function of TAK1 in osteoblasts in vitro, calvarial osteoblasts (CalvOb) were isolated from TAK1 flox/flox (Tak1fl/fl) mice and infected with lentiviruses expressing either cre recombinase (cre) or vector control (vec). Immunoblotting analysis with anti-TAK1 antibody confirmed an efficient reduction in TAK1 expression by cre-expressing lentiviral infection (Figure 2C). To validate that infection of osteoblasts with the lentiviral cre vector is not itself toxic, WT osteoblasts were similarly infected and analyzed for cell viability, ALP activity, osteoblast-specific gene expression, and BMP responsiveness (Supplemental Figure 4). No significant differences were noted in any of the parameters examined. Either cre-induced TAK1 deletion or expression of a dominant-negative TAK1 mutant with a mutated ATP-binding pocket (hereafter, catalytically inactive TAK1 or TAK1-CI) blocked osteoblast differentiation as determined by decreased ALP activity and extracellular matrix mineralization (Figure 2, D and E). Similarly, expression of osteoblast marker genes, including Osx, Alp, Ocn, Col1, and Msx1, was significantly decreased by TAK1 deletion (Figure 2F). Likewise, osteoblast marker gene expression was ablated in WT CalvOb by TAK1-CI expression (data not shown).
TAK1 is required for Runx2 activity.
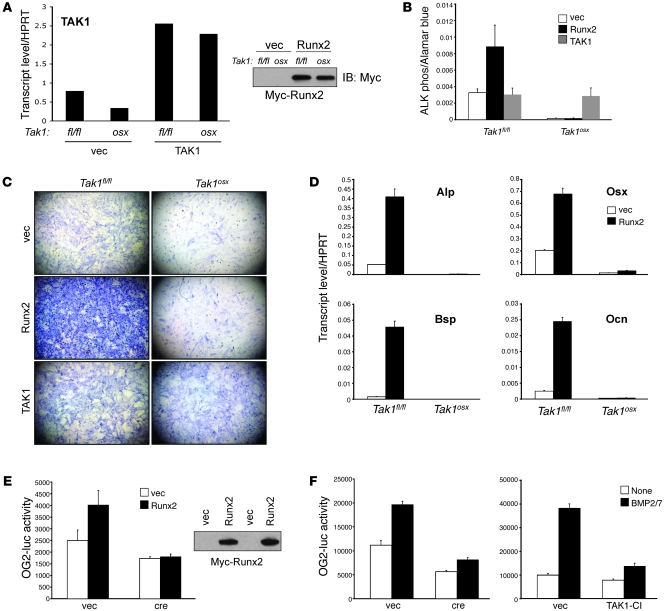
Tak1osx mice display a striking similarity to Runx2+/– mice in the absence of a statistically significant reduction in Runx2 levels, suggesting that TAK1 may regulate Runx2 transcriptional activity via a posttranslational mechanism. To test this hypothesis directly, Myc-Runx2 expression was enforced in WT and TAK1-deficient CalvOb by lentiviral infection. Equivalent expression of Runx2 was achieved in both TAK1-sufficient and deficient cells (Figure 3A). While Runx2 expression promoted ALP activity in WT osteoblasts, it failed to do so in TAK1-deficient cells (Figure 3, B and C). ALP activity was fully recovered by enforced expression of WT TAK1 in TAK1-deficient cells (Figure 3, A–C). Similarly, enforced expression of Runx2 strongly induced expression of Runx2 target genes, including Alp, Osx, Bsp, and Ocn, in WT but not TAK1-deficient osteoblasts (Figure 3D). Therefore, even when highly expressed via viral transduction at equivalent levels in WT and TAK1-deficient cells, Runx2 is unable to promote osteoblast differentiation in the absence of TAK1. To test Runx2 transcriptional activity in TAK1-deficient cells, Myc-Runx2 was overexpressed in WT and TAK1-deficient CalvOb, and cells were transfected with a Runx2-responsive luciferase reporter gene (OG2-luc), confirming that Runx2 transcriptional activity is blunted by the loss of TAK1 (Figure 3E). Given that BMP signaling is important for regulation of Runx2 activity (37), we also tested the importance of TAK1 for BMP-induced Runx2 activation. Tak1fl/fl CalvOb transduced with vector or cre lentiviruses was transfected with a Runx2-responsive reporter gene (OG2-luc) and stimulated with BMP2/7. Luciferase assays showed that TAK1 deletion or expression of TAK1-CI blunted the ability of BMP2/7 to induce Runx2 transcriptional activity (Figure 3F). Taken together, these experiments demonstrate that TAK1 regulates Runx2 activity though posttranslational modification.
Figure 3. Impaired Runx2 activity in the absence of TAK1.
(A) Primary CalvOb were isolated from Tak1fl/fl and Tak1osx pups, infected with vector, Myc-Runx2, or WT TAK1-expressing lentiviruses, and cultured for 6 days under differentiation conditions. Expression of Myc-Runx2 (upper) and TAK1 (lower) was analyzed by immunoblotting with anti-Myc antibody and quantitative PCR. (B and C) CalvOb expressing Myc-Runx2 or WT TAK1 were cultured for 6 days under differentiation conditions and ALP activity was analyzed by colorimetric assay (B) and Fast Blue staining (C). Original magnification, ×25. Values are mean + SD. (D) Primary CalvOb were isolated from Tak1fl/fl and Tak1osx pups, infected with vector or Myc-Runx2–expressing lentiviruses, and cultured for 6 days under differentiation conditions. Quantitative PCR analysis was performed for the indicated genes. (E) Tak1fl/fl CalvOb were infected by vector or cre lentivirus together with Myc-Runx2–expressing lentivirus. 2 days after transduction, cells were transfected with OG2-luc and Renilla luciferase vectors; 6 days later, luciferase activity was determined. Results were normalized to a Renilla control. Expression of Myc-Runx2 was analyzed by immunoblotting with anti-Myc antibody (upper). Values are mean + SD. (F) Tak1fl/fl CalvOb were infected by vector, cre, or TAK1-CI lentivirus as above, transfected with OG2-luciferase, and stimulated with BMP2/7. Results were normalized to a Renilla control. Values are mean + SD.
Impaired activation of the p38 pathway in Tak1osx mice.
To explore the mechanism of TAK1-mediated regulation of Runx2, we first tested the possibility that TAK1 interacts with Runx2. Even when highly expressed in HEK293 cells, TAK1 was unable to interact with Runx2 as shown by coimmunoprecipitation, suggesting that the regulation of Runx2 by TAK1 is indirect (data not shown). To determine which pathways function downstream of TAK1 in osteoblasts, WT and TAK1-deficient CalvOb were stimulated with various ligands relevant to osteoblast function, including BMP2/7, TGF-β, FGF2, and a canonical WNT fusion protein (xWNT8/Fz5). As seen in chondrocytes (22), TAK1 was required for BMP responses, including both the ability of BMP2/7 to induce expression of BMP target genes, such as Alp, Osx, Id1, and Msx1 (Supplemental Figure 5A), and osteoblast differentiation as shown by ALP activity and extracellular matrix mineralization (Figure 4A). However, other signaling pathways showed no change or only minor perturbations after TAK1 deletion, though phosphorylation levels of MKK3/6 and p38 MAPKs downstream of TGF-β were decreased in the absence of TAK1 (Supplemental Figure 5, B–F). Therefore, in osteoblasts, TAK1 appears to function downstream of BMP and the p38 MAPK pathway downstream of TGF-β, but not canonical WNT or FGF signaling.
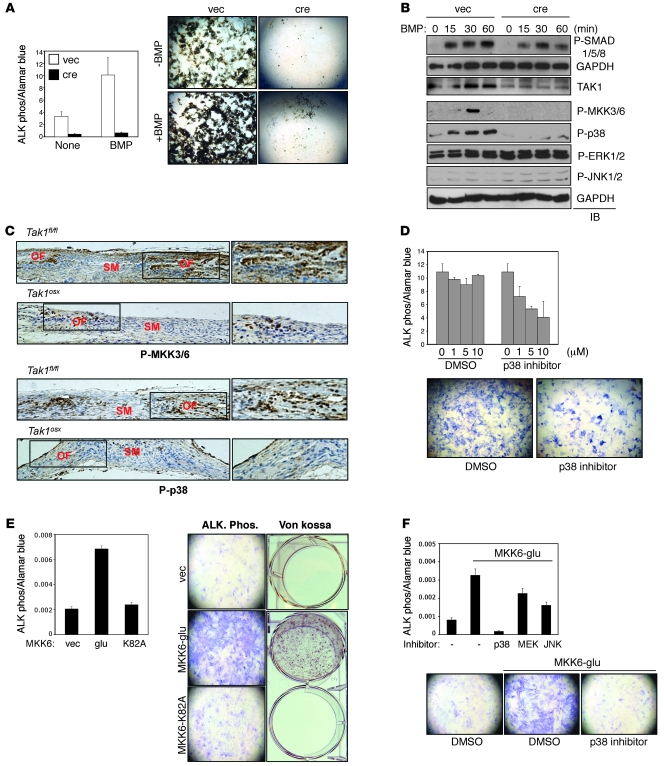
Figure 4. Impaired p38 MAPK pathway activation in TAK1-deficient osteoblasts.
(A) Tak1fl/fl CalvOb were infected by vector or cre-expressing lentivirus and cultured under differentiation conditions in the absence or the presence of BMP2/7 (50 ng/ml). ALP activity and mineralization were analyzed by colorimetric assay (upper) and Von Kossa staining (lower), respectively. Values are mean + SD. (B) Tak1fl/fl CalvOb infected by vector or cre lentivirus were serum starved for 12 hours before BMP2/7 (100 ng/ml) stimulation at different time points. Cell lysates were then immunoblotted with the indicated antibodies. (C) Immunohistochemistry showing phosphorylation levels of MKK3/6 and p38 in a coronal section of the calvarium of Tak1fl/fl and Tak1osx mice. (D) Primary WT CalvOb were cultured under differentiation conditions in the absence or the presence of the p38 inhibitor, and then ALP activity was analyzed by colorimetric assay (upper) and Fast Blue staining (lower). Values are mean + SD. (E and F) Human MSCs were infected with vector, MKK6-glu, or MKK6-K82A and cultured under osteoblast differentiation conditions; then ALP activity and mineralization were analyzed by colorimetric assay and Von Kossa staining, respectively (E). Alternatively, cells were cultured under osteoblast differentiation conditions in the presence of DMSO, MEK1/2, JNK, or p38 inhibitor (F). Values are mean + SD. Original magnification, ×100.
To investigate the mechanism of TAK1 in BMP signaling, phosphorylation of SMAD1/5/8, and p38, JNK, and ERK MAPKs was analyzed in TAK1-deficient and control osteoblasts in response to BMP2/7 (Figure 4B). As previously noted in chondrocytes (22), TAK1-deficient osteoblasts showed a moderate reduction in SMAD1/5/8 phosphorylation at early time points. ERK1/2 and JNK1/2 phosphorylation showed only modest induction with BMP stimulation, and these levels seemed unaffected or even slightly increased by TAK1 deletion. Likewise, both basal and PMA-induced NF-κB activity was analyzed using the NF-κB–responsive luciferase reporter (PBII-luc) and found to be unaltered in the absence of TAK1 (data not shown). In contrast, both basal and induced phosphorylation of p38 as well as 2 upstream MAPK kinases, MKK3 and MKK6, were ablated in TAK1-deficient osteoblasts. Similarly, impaired phosphorylation of MKK3/6 and p38 was also observed in response to TGF-β signaling (Supplemental Figure 5C). Thus, of the pathways examined, the p38 MAPK pathway appears to be most affected by the absence of TAK1. To confirm that the p38 MAPK pathway is defective in Tak1osx mice in vivo, MKK3/6 and p38 phosphorylation levels were determined by IHC in the calvarium of Tak1osx and littermate control mice (Figure 4C). MKK3/6 and p38 are phosphorylated in osteoblasts lining the osteogenic fronts near the cranial sutures, and this staining is dramatically reduced in Tak1osx mice. As a control, expression levels of MKK3/6 and the 4 p38 isoforms were examined in various tissues and in Tak1osx mice in vivo (Supplemental Figure 6, A and B). Expression levels of these genes were either unchanged or increased in Tak1osx mice, indicating that these defects in the p38 MAPK pathway reflect impaired activation and not impaired expression.
Since MKK3/6 and p38 activation were severely impaired in the absence of TAK1, we examined the contribution of the p38 MAPK pathway to osteoblast differentiation in vitro. First, CalvOb were treated with the p38 inhibitor SB203580, which inhibited osteoblast differentiation in a dose-dependent manner (Figure 4D) (15, 38). Next, we determined whether activating p38 MAPK promotes osteoblast differentiation. Expression of a constitutively active mutant of MKK6 (MKK6-glu) (39), but not a kinase-inactive mutant of MKK6 (MKK6-K82A) (39), strongly promoted human MSC differentiation to osteoblasts as shown by increased ALP activity and extracellular matrix mineralization (Figure 4E). Furthermore, MKK6-glu–expressing osteoblasts were treated with inhibitors of p38, JNK, and MEK (Figure 4F). Only treatment with the p38 inhibitor SB203580 blocked the effects of MKK6-glu. Taken together, we conclude that TAK1 plays a critical role in p38 MAPK activation during osteoblast differentiation and that p38 MAPK activation is in turn a critical regulator of osteoblast differentiation in vitro.
Decreased bone mass in mice lacking MKK3 and MKK6.
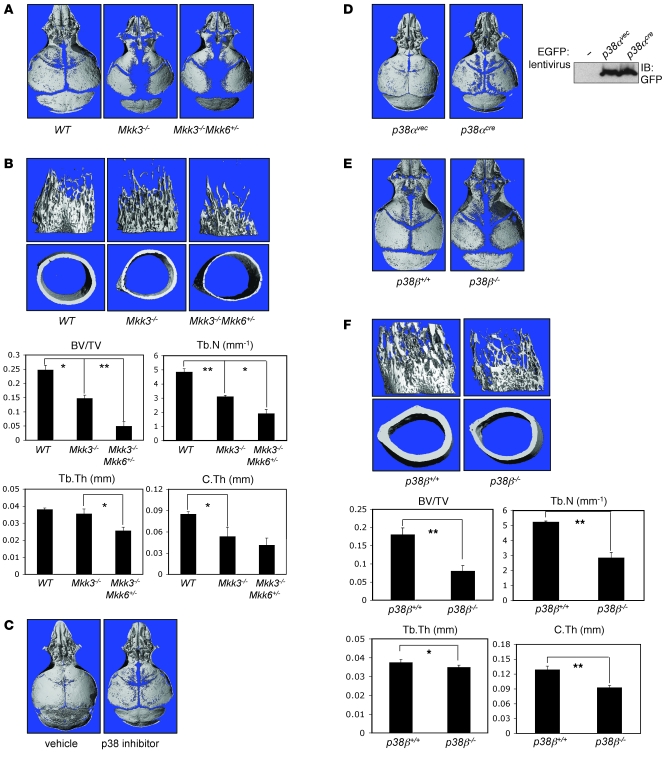
To determine whether the p38 MAPK pathway similarly promotes osteoblast functions in vivo, we examined skeletal mineralization in mice genetically deficient for the MAP2K upstream of p38, MKK3, and MKK6. Since embryonic lethality of Mkk3–/–Mkk6–/– mice and infertility of Mkk3+/–Mkk6–/– mice hinder analysis of skeletal mineralization in these mice, we focused on Mkk3–/–Mkk6+/– mice, although they show only a partial reduction in p38 activity due to retaining a single allele of MKK6 (28). As suggested from the in vitro studies examining the contribution of p38 to osteoblast function, calvarial mineralization was retarded in 5-day-old Mkk3–/–Mkk6+/– and Mkk3–/– mice (Figure 5A). The pattern of changes was similar to Tak1osx mice, with the frontal and parietal bones being the most affected.
Figure 5. Low bone mass and/or defective calvarial mineralization in Mkk3–/–Mkk6+/–, p38 inhibitor–treated, p38α-deleted, or p38b–/– mice.
(A, C, D, and E) Skulls from Mkk3–/–Mkk6+/– (A), p38 inhibitor–treated (C), p38α-deleted (D), or p38b–/– mice (E) were analyzed by μCT. The p38 inhibitor (SB203580) and mice with lentiviral cre-induced p38α deletion were treated with the inhibitor or infected starting at P2 and analyzed on P9. Other mice were analyzed at P4. cre recombinase expression in calvaria of p38α pups was analyzed by immunoblotting with anti-EGFP antibody. Images are representative of more than 8 mice per genotype or treatment. Images from females are shown. (B, F) Femurs from female Mkk3–/–Mkk6+/– and p38b–/– mice were analyzed by μCT. Displayed are 3D reconstructions of trabecular bone (upper left), midshaft cortical bone (lower left), and BV/TV, trabecular number, trabecular thickness, and cortical thickness parameters. Values are mean + SD. *Significant difference by Student’s t test, P < 0.05; **P < 0.0005.
Likewise, the femurs of Mkk3–/– and Mkk3–/–Mkk6+/– mice showed a reduction in both trabecular bone volume and cortical thickness at 25 days of age (Figure 5B). Trabecular BV/TV was reduced nearly 5-fold in Mkk3–/–Mkk6+/– mice. Midshaft cortical thickness was reduced more than 50%. Mkk3–/– mice showed an intermediate phenotype both in terms of trabecular bone volume and cortical thickness. Histologic analysis of Mkk3–/–Mkk6+/– and Mkk3–/– mice showed a picture similar to that of Tak1osx mice, with reduced trabecular bone and impaired mineralization of the secondary center of ossification (Supplemental Figure 7A). Hypoplasia of the clavicle seen in Tak1osx mice was not seen in Mkk3–/–Mkk6+/– mice, suggesting that either another pathway mediates this effect or that a more complete blockade of the p38 MAPK pathway is necessary to prevent development of the clavicle. In a separate cohort, the skeletal phenotype of Mkk6–/– mice was examined (Supplemental Figure 8). Consistent with the finding that Mkk3–/–Mkk6+/– mice show a further decrease in long bone but not calvarial mineralization when compared with Mkk3–/– mice, Mkk6–/– mice display reduced femoral BV/TV and cortical thickness but only a barely appreciable decrease in calvarial mineralization. This suggests that within the skeletal system, MAP2Ks play anatomically selective roles, with both MKK3 and MKK6 contributing to long bone mineralization and only MKK3 substantially contributing to calvarial mineralization.
Taken together, Mkk3–/–Mkk6+/– mice display a substantial reduction in mineralization of both the calvarium and long bones, despite the only partial reduction in p38 activity, demonstrating that osteoblasts are very sensitive to gradations in p38 MAPK activity in vivo. Moreover, the similarity between the Mkk3–/–Mkk6+/– phenotype and the Tak1osx phenotype provides in vivo genetic evidence that the p38 MAPK pathway is a major mediator of the effects of TAK1 on skeletal mineralization.
Decreased bone mass in mice lacking p38α and p38β.
To determine whether postnatal inhibition of p38 MAPK produces similar effects on calvarial mineralization as germline deletion of Mkk3, WT pups were treated daily with either vehicle or the p38 inhibitor SB203580 via subcutaneous injection over the calvarium for 1 week and calvarial mineralization, then analyzed by μCT (Figure 5C). As expected, p38 inhibitor–treated mice displayed a reduction in calvarial mineralization. Consistent with the pattern of hypomineralization seen in MKK3/6-deficient mice, the effects of treatment were seen mostly at the periphery of the frontal and parietal bones facing the sagittal and coronal sutures, while the interparietal and occipital bones were relatively unaffected.
Although 4 isoforms of p38 MAPKs, p38α, -β, -γ, and, -δ, share similar structural and enzymatic properties, only p38α and p38β are known to be inhibited by the p38 MAPK inhibitor SB203580 used for in vivo and in vitro studies above (38, 40). Therefore, we focused on determining the contribution of p38α and p38β to osteoblast functions in vivo. Since p38α-deficient mice are embryonic lethal (24), we used mice homozygous for a p38a conditional floxed allele (p38afl/fl) to analyze skeletal mineralization (Figure 5D). Deletion of the p38a floxed allele was induced by subcutaneous injection of a cre recombinase expressing lentivirus over the calvarium. Infection was monitored by immunoblotting analysis for GFP also encoded in the lentiviral vector (Figure 5D). p38afl/fl mice were injected with either cre or vector lentivirus on postnatal day 2 and the degree of calvarial mineralization analyzed 1 week after injection. Inducible deletion of p38α resulted in dramatic hypomineralization of the calvarium. As seen with the p38 inhibitor treatment, only the edges of the frontal and parietal bones were affected. Recapitulating the results of SB203580 treatment with inducible p38α deletion strongly supports the conclusion that the effects of SB203580 on osteoblasts and calvarial mineralization are indeed due to inhibition of p38 and not due to off-target effects (Figure 5C).
p38β-deficient mice have not been previously reported to have any in vivo phenotype (29, 30). Surprisingly, we found that p38b–/– mice showed a substantial decrease in long bone mineralization and a more modest effect on the calvarium (Figure 5, E and F). Interestingly, the finding that p38β contributes more substantially to long bone than calvarial mineralization is consistent with the finding that the only MAP2K known to phosphorylate and activate p38b, MKK6, displays a similar pattern of tissue specificity (Supplemental Figure 8). The femoral trabecular BV/TV was reduced approximately 70% in 5-week-old p38b–/– mice. Cortical thickness was reduced more than 30%. Histologic analysis of tibias from p38b–/– mice showed a picture similar to that of Tak1osx and Mkk3–/–Mkk6+/– mice, with a reduction in trabecular and cortical bone and decreased mineralization of the secondary center of ossification (Supplemental Figure 7B). The finding of a substantial contribution of p38β to in vivo bone formation establishes, for what we believe is the first time, the in vivo relevance of p38β to the overall function of the p38 pathway.
Impaired osteoblast differentiation in the absence of p38 signaling mediators.
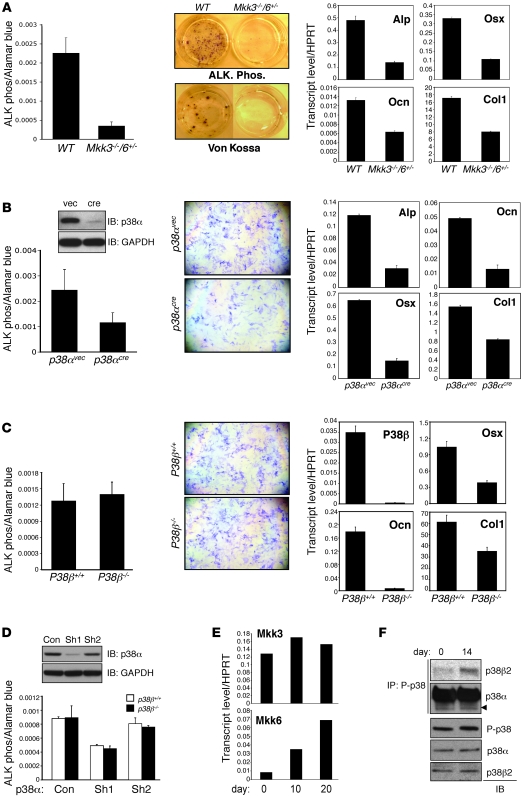
MKK3 and MKK6 and p38α- and p38β-deficient mice all show defects in skeletal mineralization in vivo consistent with the defects seen in Tak1osx mice. To characterize the contribution of these genes to osteoblast differentiation in vitro, the phenotype of osteoblasts derived from these mouse lines was examined. Bone marrow stromal cells (BMSCs) were isolated from 5-week-old WT and Mkk3–/–Mkk6+/– mice. Consistent with the in vivo defects in skeletal mineralization, osteoblast differentiation was impaired in Mkk3–/–Mkk6+/– cells as shown by a reduction of ALP activity and extracellular matrix mineralization (Figure 6A). Similarly, expression of Runx2-target genes, including Alp, Osx, Ocn, and Col1, were all reduced in Mkk3–/–Mkk6+/– osteoblasts (Figure 6A).
Figure 6. Defective osteoblast differentiation in MKK3/6 and p38α/β-deficient cells.
(A) BMSCs were isolated from WT and Mkk3–/–Mkk6+/– mice and cultured under differentiation conditions; then ALP activity and mineralization were analyzed by colorimetric assay/Fast Blue staining and Von Kossa staining, respectively (left). Alternatively, total RNA was extracted for RT-PCR analysis (right). Values are mean + SD. Original magnification, ×25. (B) Primary CalvOb were isolated from p38afl/fl pups, infected by vector or cre lentivirus, and cultured under differentiation conditions. ALP activity was analyzed by colorimetric assay and Fast Blue staining (left). Alternatively, total RNAs were extracted for quantitative PCR analysis (right). Inducible deletion of p38α was analyzed by immunoblotting with anti-p38α antibody. Values are mean + SD. (C) Primary CalvOb were isolated from p38b+/+ and p38b–/– pups and cultured under differentiation conditions; then ALP activity was analyzed by colorimetric assay and Fast Blue staining (upper). Alternatively, total RNAs were extracted for quantitative PCR analysis (lower). Values are mean + SD. (D) p38b+/+ and p38b–/– CalvOb were infected by lentiviruses expressing control (con), p38α (Sh1), or mock (Sh2) shRNA and cultured under differentiation conditions; then ALP activity was analyzed by colorimetric assay. Values are mean + SD. (E) Primary WT CalvOb were cultured under differentiation conditions, and total RNAs were extracted at day 0, 10, and 20 for quantitative PCR analysis. (F) Primary WT CalvOb were cultured under differentiation conditions. Cells were lysed, immunoprecipitated with anti-phospho p38 antibody and protein A agarose, and immunoblotted with the indicated antibodies.
Deletion of p38α by cre lentiviral infection also resulted in a decrease in osteoblast differentiation as shown by a reduction in ALP activity, which was accompanied by decreased expression of Alp, Osx, and Ocn in p38α-deficient cells (Figure 6B). In contrast to the early block in differentiation seen with p38α deletion, p38b–/– osteoblasts did not show a defect in the induction of ALP activity, indicating that early stages of osteoblast differentiation are not affected by the loss of p38β (Figure 6C). However, loss of p38β did result in decreased expression of many of the same characteristic osteoblast genes decreased after p38α deletion, Ocn, Osx, and Col1 (Figure 6C). This also suggests that, despite the contribution of both p38α and p38β to skeletal mineralization in vivo, each may play a fundamentally different role, with p38β acting at a later stage of osteoblast differentiation. Alternatively, it is also possible that loss of p38β simply represents a less substantial reduction in overall p38 activity than the loss of p38α, rather than a difference in function. To test the latter hypothesis, WT and p38b–/– osteoblasts were infected with shRNA lentivirus to reduce the expression of p38α (Figure 6D). As expected, knockdown of p38α reduced ALP activity levels. However, the additional absence of p38β in p38α knockdown cells did not cause a further reduction in ALP activity levels. In conjunction with the data above, we conclude that p38α and p38β play fundamentally different roles in osteoblasts, with p38α affecting early osteoblast differentiation and p38β affecting only late osteoblast differentiation.
One likely explanation for the late stage–specific defects in p38b–/– osteoblasts is that p38β is selectively activated only later in osteoblast differentiation, after ALP upregulation. Whereas both MKK3 and MKK6 can activate p38α, only MKK6 is capable of activating p38β (41). Accordingly, we found that MKK6 is expressed at very low levels in preosteoblasts and its expression is dramatically upregulated during the course of osteoblast differentiation (Figure 6E). Expression of p38α, p38β, and MKK3 remains relatively unchanged over the same period (Supplemental Figure 6C and Figure 6E). As expected from the low expression of MKK6 in early osteoblasts and its later upregulation, p38β is activated at a day 14 but not a day 0 time point during osteoblast differentiation (Figure 6F). In contrast p38α is activated on both day 0 and day 14. We conclude that selective expression of the MAP2Ks upstream of p38 likely accounts for the different functions of p38α and p38β in vivo.
Rescue of the TAK1-deficient osteoblast phenotype by enforced activation of p38 MAPK.
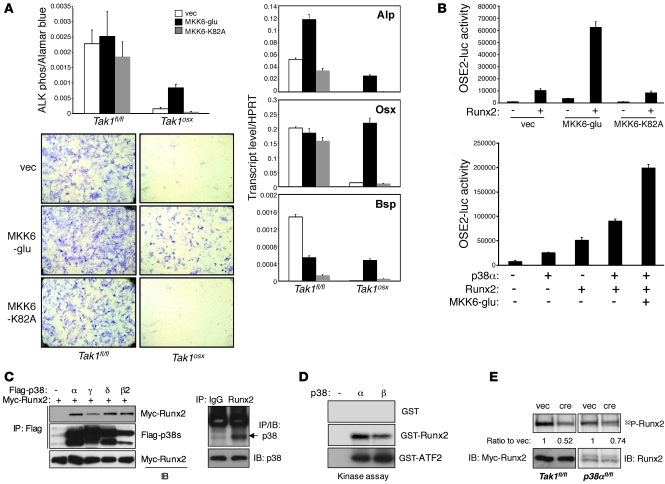
The above studies confirm that TAK1 is required for p38 activation in vivo and in vitro and that mice lacking p38 MAPK mediators show an osteoblast phenotype similar to that of Tak1osx mice. However, these studies do not experimentally prove a causal role of the p38 MAPK pathway in mediating the effects of TAK1 to promote osteoblast differentiation. To address this issue, either the constitutively active mutant of MKK6 (MKK6-glu) or the kinase-negative mutant (MKK6-K82A) was expressed in WT and TAK1-deficient osteoblasts to determine whether enforced activation of p38 could rescue the TAK1 phenotype. Expression of the constitutively active MKK6 but not the kinase-negative MKK6 partially rescued the block in ALP activity induction in TAK1-deficient osteoblasts (Figure 7A). Equivalent levels of expression of MKK6-glu and MKK6-K82A were achieved in Tak1fl/fl and Tak1osx osteoblasts (Supplemental Figure 9A). Likewise, MKK6-glu rescued expression of osteoblast marker genes, including Alp, Osx, and Bsp (Figure 7A).
Figure 7. TAK1-mediated p38 MAPK pathway regulates Runx2 activity.
(A) Tak1fl/fl and Tak1osx CalvOb were infected by vector or Flag-tagged MKK6-glu– or MKK6-K82A–expressing lentiviruses, and ALP activity was analyzed by colorimetric assay and Fast Blue staining (left). Alternatively, total RNAs were extracted for quantitative PCR analysis (right). Original magnification, ×25. Values are mean + SD. (B) C2H10T1/2 cells were transfected with OSE2-luc and Renilla luciferase vectors together with either vector, MKK6-glu, or MKK6-K82A in the absence or the presence of Runx2 (left) or various combinations of Runx2, p38α, and MKK6 (right). Results are expressed as relative luciferase activity normalized by Renilla control. Values are mean + SD. (C) The interaction of p38 MAPKs with Runx2. HEK293 cells were transfected with Myc-Runx2 together with Flag-p38 proteins. Lysates were immunoprecipitated with anti-Flag–conjugated beads and immunoblotted with anti-Myc antibody (left). Alternatively, immortalized osteoblasts were immunoprecipitated with either IgG or anti-Runx2 antibody and protein A agarose and immunoblotted with anti-p38 antibody (right). (D) p38-induced phosphorylation of Runx2. Recombinant p38α and p38β proteins were mixed with GST, GST-Runx2, or GST-ATF2 protein, and p38 kinase activity was analyzed by in vitro kinase assay. (E) Primary p38afl/fl CalvOb were infected with either vector or cre lentivirus (left), and Tak1fl/fl CalvOb were infected with either vector or cre lentivirus together with Myc-Runx2–expressing lentivirus (right), metabolically labeled with (P32) orthophosphate, and then immunoprecipitated with either anti-Myc antibody (left) or anti-Runx2 antibody (right). Expression of Myc-Runx2 and Runx2 protein was analyzed by immunoblotting with antibodies specific to Myc and Runx2.
Runx2 activation by the TAK1-MKK3/6-p38 MAPK pathway.
The rescue of TAK1-deficient osteoblasts with MKK6-glu demonstrates that the p38/MAPK pathway functions downstream of TAK1 and is responsible for at least some of its effects to promote osteoblast differentiation. Furthermore, the failure of enforced Runx2 expression to promote osteoblast functions (Figure 3) and restore BMP2/7-induced p38 phosphorylation (Supplemental Figure 9B) in TAK1-deficient osteoblasts suggests that TAK1-mediated p38 activation is required for Runx2 transcriptional activity. Putting these observations together, it seems likely that the p38/MAPK pathway functions to phosphorylate and activate Runx2. To test this hypothesis, the ability of constitutively active MKK6 (MKK6-glu) to promote Runx2 transcriptional activity was examined by luciferase assay using a Runx2-responsive reporter gene (OSE2-luc) (Figure 7B). Expression of MKK6-glu synergistically promoted OSE2 luciferase activity when coexpressed with Runx2, demonstrating that activation of the p38 MAPK pathway can promote Runx2 transcriptional activity. Runx2 activity was likewise increased by p38α in a dose-dependent manner and further increased by coexpression of Runx2 and/or the constitutively active MKK6, indicating that they function in the same pathway (Figure 7B and Supplemental Figure 9C).
To determine whether regulation of Runx2 activity by p38 is direct, the ability of the various p38 isoforms to interact with Runx2 was examined. Immunoprecipitation analysis in HEK293 cells showed that Myc-Runx2 is able to interact with all 4 Flag-tagged p38α, -β, -γ, and -δ isoforms (Figure 7C). This interaction was further confirmed by demonstrating the association of endogenous Runx2 with p38 in osteoblast cell lines using a pan-p38 antibody recognizing p38α, -β, and -γ isoforms (Figure 7C). Coexpression of Runx2 alongside constitutively active MKK6-glu and p38α, but not the inactive MKK6-K82A, induced a shift in the electrophoretic mobility of Runx2, suggesting that p38 is able to phosphorylate Runx2 (Supplemental Figure 9D). Given this, the ability of the 2 isoforms of p38 MAPK studied above, p38α and p38β, to directly phosphorylate Runx2 was examined using an in vitro kinase assay (Figure 7D). Recombinant GST-Runx2 and the control substrate, GST-ATF2, proteins were phosphorylated by both recombinant p38α and p38β proteins, although p38α was more efficient than p38β in phosphorylating GST-Runx2, despite p38β’s equivalent or even increased ability to phosphorylate the control substrate. These data demonstrate that p38 functions as an upstream kinase of Runx2.
To confirm that the above ability of p38 to phosphorylate Runx2 reflects a physiologic function of p38 downstream of TAK1, Runx2 phosphorylation levels were determined in TAK1 and p38α-deficient osteoblasts. Since TAK1-deficient osteoblasts display slightly reduced levels of Runx2, Myc-Runx2 expression was enforced in WT and TAK1-deficient CalvOb to ensure equivalent input levels of Runx2. These cells were cultured under osteoblast differentiation conditions for 6 days and metabolically labeled with [P32] orthophosphate, and then Myc-Runx2 was immunoprecipitated with an anti-Myc antibody (Figure 7E). Myc-Runx2 phosphorylation levels were reduced by nearly 50% in TAK1-deficient osteoblasts. Likewise, p38α deletion led to a decrease in Runx2 phosphorylation (Figure 7E). Hence, the TAK1/p38α pathway functions upstream of Runx2 and regulates its activity via its phosphorylation. Of note, a less substantial decrease was seen with deletion of p38α than TAK1. Though these results are not directly comparable due to the use of endogenous versus overexpressed and epitope-tagged Runx2, they are consistent with the participation of other kinases, such as p38β in the phosphorylation of Runx2.
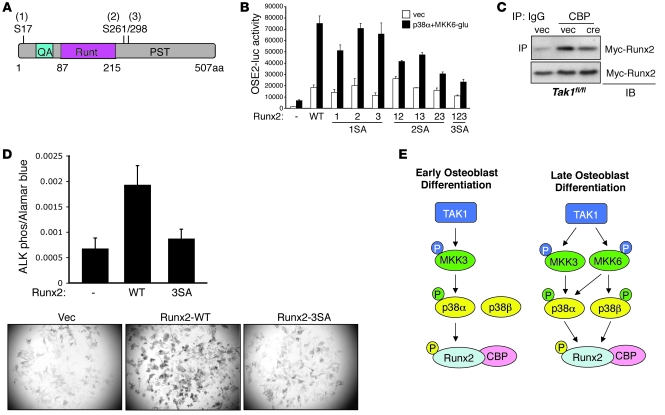
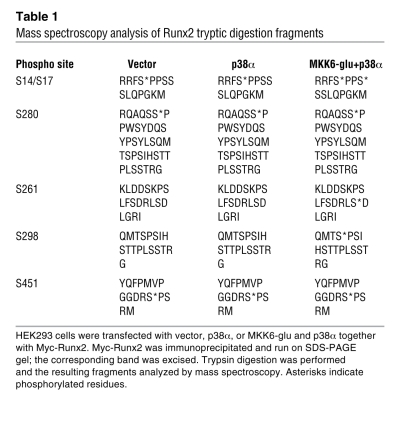
Given this evidence that p38α is a physiologic mediator of Runx2 phosphorylation, we performed mass spectrometry analysis to identify the sites of phosphorylation. HEK293 cells were transfected with Myc-Runx2 together with either vector control or constitutively active MKK6 and p38α, and Myc-Runx2 was immunoprecipitated using Myc-conjugated agarose. Cotransfection of both p38 and an upstream MAP2K is necessary to get optimal p38 activation. Mass spectrometry analysis revealed that, when coexpressed with MKK6 and p38α, Runx2 is inducibly phosphorylated at 3 serines (S17, S261, and S298; correspond to S31, S275, and S312 in Runx2 α isoform) (Figure 8A and Table 1). Basal phosphorylation of Runx2 was also detected at S280 (corresponds to S294 in Runx2 α isoform). Intriguingly, one of the basal and one of the inducible phosphorylation sites (S280 and S298) correspond to the sites targeted by ERK MAPK in Runx1 (42). These sites were confirmed as direct p38 phosphorylation targets using an in vitro kinase assay with recombinant p38 phosphorylating either GST-Runx2 or a GST-Runx2 construct with mutation of S17, S261, and S298 to alanine (Runx2-3SA) (Supplemental Figure 10A). The 3SA mutation reduced the total amount of Runx2 phosphorylation by p38 by approximately 2-fold. The residual phosphorylation in the Runx2-3SA mutant may reflect either phosphorylation of sites basally phosphorylated in the mass spectroscopy study or possibly nonphysiologic phosphorylation events that occur in the setting of an in vitro kinase assay.
Figure 8. Mapping the p38 phosphorylated residues in Runx2 and their functional consequences.
(A) Schematic diagram showing Runx2 phosphorylation sites by the p38 MAPK pathway. (B) C3H10T1/2 were transfected with either vector, WT Runx2, or mutant Runx2 constructs harboring the indicated mutations, the OSE2 luciferase reporter, and either vector or p38α and MKK6. Results are expressed as relative luciferase activity normalized by Renilla control. Values are mean + SD. (C) Primary Tak1fl/fl CalvOb were infected with either vector or cre lentivirus together with Myc-Runx2–expressing lentivirus. Nuclear extracts were immunoprecipitated with either IgG or anti-CBP antibodies and protein A agarose and immunoblotted with anti-Myc antibody. Expression of Myc-Runx2 protein was analyzed by immunoblotting with anti-Myc antibody. (D) Human mesenchymal stem cells were infected with lentiviruses encoding Runx2-WT or Runx2-3SA and cultured under osteoblast differentiation conditions. After 6 days, osteoblast differentiation was analyzed by a colorimetric assay (upper) or fast blue staining (lower) for alkaline phosphatase activity. Original magnification, ×25. For the colorimetric assay, Runx2-WT was compared with Runx2-3SA via an unpaired Student’s t test. *P < 0.0005. (E) A schema depicting the TAK1/MKK3/6/p38/RUNX2 axis in osteoblasts.
Table 1 .
Mass spectroscopy analysis of Runx2 tryptic digestion fragments
In order to determine the functional contribution of these 3 inducible phosphorylation sites (S17, S261, and S298) to Runx2 activity, these sites were mutated to alanine in every combination of single, double, and triple mutants, and the effect on the ability of MKK6/p38α expression to activate the Runx2 responsive luciferase reporter (OSE2-luc) was analyzed (Figure 8B). Overall, the single mutants displayed a modest effect on inducible Runx2 activity, while the double and triple mutants dramatically reduced the ability of p38 to promote Runx2 activity. Likewise, a shift in the electrophoretic mobility of the triple mutant was not detected in the presence of constitutively active MKK6-glu and p38α, suggesting that these 3 sites account for the majority of Runx2 phosphorylation by p38 MAPK (Supplemental Figure 10B). This indicates that each of the 3 inducible phosphorylation sites contributes to Runx2 activity, and no single site has a dominant effect. Furthermore, phosphorylation at these sites contributed to the overall function of Runx2 to promote osteoblast differentiation. Expression of Runx2 promoted the capacity of human MSCs to differentiate into osteoblasts when cultured under osteoblast differentiation conditions (Figure 8D). Mutation of the 3 inducible serine phosphorylation sites (Runx2-3SA) nearly ablated the function of Runx2 to promote osteoblast differentiation, as shown by reduced induction of alkaline phosphatase activity.
How Runx2 phosphorylation augments its activity is not fully understood. Runx2 phosphorylation via the ERK/MAPK pathway has been described as increasing the DNA-binding affinity of Runx2 (43–45). Thus, we analyzed Runx2 DNA-binding affinity by using EMSA. In TAK1-deficient osteoblasts, Runx2-binding affinity to OSE2 was reduced by approximately 20% (Supplemental Figure 9E). It was unlikely that this modest decrease in binding could fully explain the marked defects in Runx2 activity we observed. To determine whether TAK1 regulates the ability of Runx2 to recruit the transcriptional coactivator CBP, this interaction was examined in WT and TAK1-deficient osteoblasts (Figure 8C). Deletion of TAK1 impaired the ability of Runx2 to bind CBP, suggesting that TAK1 regulates the ability of Runx2 to recruit transcriptional coactivators. Likewise, enforced activation of p38 through the coexpression of MKK6-glu with p38α promoted the association of Myc-Runx2 with HA-CBP as assessed by pulldown experiments (Supplemental Figure 9F). As expected, this ability of p38 to potentate the interaction of Runx2 with CBP interaction was ablated by mutation of the 3 inducible Runx2 phosphorylation sites to alanine (Runx2-3SA, containing S17A, S261A, and S298A mutations) (Supplemental Figure 10B).
Thus, inactivation and activation of the TAK1-p38 axis diminishes and increases the interaction of Runx2 with CBP, respectively, and the 3 inducible phosphorylation sites must be intact for this regulation to occur. This suggests that enhancing cofactor recruitment and the assembly of a transactivation complex are a major mechanism through which TAK1 and p38 promote Runx2 transcriptional activity.
Altogether, these data provide compelling evidence that TAK1 functions in osteoblasts to activate MKK3/6 and p38 and that p38 in turn phosphorylates and activates Runx2 by promoting its association with the cofactor CBP (schematized in Figure 8E). Thus, the phenotypic similarities between Tak1osx and Runx2+/– mice reflect a critical function of TAK1 and downstream mediators in the p38 pathway to promote osteoblast differentiation and functions through regulating Runx2.
Discussion
As the physiologic demands placed on the skeleton change throughout the life of an organism, so too must osteoblast differentiation and functional activity respond to extracellular cues to adjust bone formation accordingly. Maladaptive responses to these cues result in bone pathology such as osteoporosis, which in turn incurs risk for bone fractures (46). However, little is understood about how extracellular cues are transduced within the osteoblast and how these signals in turn ultimately affect bone homeostasis. In this respect, developmental or morphologic defects within the skeletal system are important, as they are signposts indicating that a particular set of genetic defects ultimately converge on the same pathway effectors. In particular, mice and humans haploinsufficient for Cbfa1, the gene encoding the transcription factor Runx2, display clavicular hypoplasia, delayed fontanelle closure, and osteopenia (10, 12, 47). Mice with a conditional deletion of Tak1 display a similar constellation of defects, strongly suggesting that TAK1 is essential for Runx2 regulation in vivo. The Tak1osx animal, however, is not an exact phenocopy of CCD. For example, some features of the Tak1osx phenotype, such as osteoporosis, are variably present in CCD. This may simply reflect the spectrum of severity observed in CCD or may indicate that disruption of posttranslational regulatory networks can differ from a reduction in gene dosage (48, 49). Indeed, Runx2 was unable to drive transcription of Runx2 target genes or promote osteoblast differentiation when expressed in TAK1-deficient osteoblasts, providing strong evidence that the phenotypic similarity between Tak1osx mice and Runx2+/– mice is more than a coincidence.
TAK1 is required for p38 MAPK activation in osteoblasts.
The activation of p38 and its upstream mediators MKK3/6 was profoundly impaired by the absence of TAK1 both in vivo and in vitro. Inhibiting or activating p38 in turn inhibited or promoted osteoblast differentiation in vitro, making it plausible that the p38 MAPK pathway is a downstream mediator of TAK1 function in osteoblasts. Thus, we examined the contribution of the p38 pathway to bone mineralization in vivo, primarily through analyzing a collection of germline and inducible loss-of-function mutants for multiple mediators of the p38 MAPK pathway. This analysis revealed that p38α, p38β, MKK3, and MKK6 all contribute to bone mineralization in vivo and the corresponding osteoblast functions in vitro. Notably, in the calvarium, 4 overlapping approaches all give similar results, suggesting the importance of the TAK1/p38 MAPK axis in vivo, with each approach compensating for the limitations of the others. Acute postnatal inhibition via inhibitor treatment or lentiviral-cre–mediated deletion of p38α confirms that the requirement for p38 during calvarial mineralization is continual and that the phenotypes observed are not due to a function of the p38 pathway during embryonic patterning. The osteoblast-specific deletion possible with the osterix-cre in the Tak1osx line confirms that these defects are intrinsic to osteoblasts. Finally, all of the above approaches provide less than 100% efficiency deletion/inhibition, whereas this is not a concern with germline deletion of MKK3.
Other studies have explored the effect of p38 inhibition on osteoblast functions in vitro, implicating other substrates such as Dlx5 as targets of p38 MAPK–mediated phosphorylation (50). Notably, Tak1osx, Mkk3–/–Mkk6+/–, and p38b–/– mice lack some of the characteristic features seen in Dlx5–/– mice, especially defects in patterning and ossification of the base of the skull, mandible, and tympanic ring (51). Likewise, Dlx5–/– mice are not reported to display the clavicular hypoplasia seen in Runx2+/– and Tak1osx mice. When considered together with the posttranslational block in Runx2 activity in TAK1-deficient osteoblasts, these observations suggest that while downstream targets other than Runx2 may contribute to the Tak1osx phenotype, Runx2 remains a major target responsible for the effects of the p38 MAPK pathway in bone.
To further validate that the low bone mass phenotypes observed reflect changes in osteoblasts and not osteoclasts, we examined osteoclast numbers in vivo with the tartrate-resistant acid phosphatase (TRAP) stain specific for osteoclasts (Supplemental Figure 3A and Supplemental Figure 7, C and E). Tak1osx and p38b–/– mice had unchanged or slightly reduced osteoclast numbers in vivo, supporting our conclusion that the low bone mass phenotype is osteoblast intrinsic. This is consistent with the quantitative histomorphometry analysis of Tak1osx mice (Supplemental Table 1). Additionally, the expression of the key factors regulating osteoclast differentiation, the receptor activator for RANKL and osteoprotegerin (OPG), was unchanged in either the calvarium or the tibia of Tak1osx mice (Supplemental Figure 3, B and C). Similar TRAP-staining analysis of Mkk3–/–Mkk6+/– mice suggested a modest reduction in osteoclast numbers in vivo, consistent with published reports implicating p38 in osteoclast differentiation (52). Finally, serum levels of a collagen I C-terminal telopeptide (CTX) liberated from the bone matrix upon osteoclast-mediated bone resorption were determined as a measure of osteoclast activity in vivo (Supplemental Figures 3D and 7D). Both Tak1osx and Mkk3–/–Mkk6+/– mice displayed a significant reduction in serum CTX levels, confirming that increased osteoclast activity is not responsible for the low bone mass phenotypes observed in these mice.
An unexpected function for p38β in vivo.
Most surprising of all was the finding that p38β-deficient mice displayed a substantial skeletal phenotype, since previous studies examining p38-dependent pathways in these mice have failed to find abnormalities, despite substantial biochemical work confirming the activity and function of p38β in the overall p38 pathway (29, 30). We report here what we believe is the first description of an in vivo phenotype for p38β-deficient mice. This finding gains additional significance since the selectivity of p38β function for bone and not other tissues suggests that p38β-specific inhibitors and agonists, if they exist, may be excellent tools to pharmacologically modulate bone formation.
Though p38α and p38β both contribute to skeletal mineralization, p38α- and p38β-deficient osteoblasts display distinct cellular phenotypes, particularly in terms of ALP expression. MKK6 is expressed at very low levels in early osteoblasts, and its expression is dramatically upregulated during the course of osteoblast differentiation. Thus, the expression of the only MAP2K known to efficiently activate p38β only in later stage osteoblasts explains the selective defect in late functions in the absence of p38β. This provides what we believe is the first demonstration of a biologic consequence for the evolution of preferences in the activation of different p38 isoforms by the upstream MAPK kinases MKK3 and MKK6.
The p38 MAPK pathway mediates Runx2 phosphorylation and activation downstream of TAK1.
The observation that TAK1 is required for MKK3/6 and p38 activation in osteoblasts in conjunction with the reduction in skeletal mineralization in Tak1osx, p38a–/–, p38b–/–, MKK3-, and MKK6-deficient mice suggests that p38 is a critical mediator downstream of TAK1 in osteoblasts. To prove this directly, Tak1osx osteoblasts were rescued by the expression of a constitutively active MKK6 mutant (39). Subsequent biochemical analysis confirmed that both p38α and p38β can interact with and phosphorylate Runx2 on serines S17, S261, and S298. Mutation of all 3 serines to alanines largely, but not completely, blocked the induction of Runx2 activity by MKK6/p38 and significantly decreased osteoblast differentiation by Runx2 in hMSCs. This may reflect further augmentation of phosphorylation at the S280 basal phosphorylation site by p38. In turn, phosphorylation of Runx2 promotes recruitment of CBP and Runx2 transcriptional activity.
Altogether, this study demonstrates that TAK1 is an important activator of Runx2 and, in turn, an important regulator of osteoblast functions in vivo. This effect of TAK1 is mediated by activation of MKK3 and MKK6, which in turn activate p38α and p38β to phosphorylate and activate Runx2. The pathway described here links extracellular signaling to posttranslational regulation of osteoblast functions in vivo and furthermore suggests that bone is especially sensitive to reductions in the activity of p38 MAPK. Based on these data, it is possible that nonselective p38 inhibitors currently in development to inhibit inflammation and osteoclast-induced bone resorption (53, 54) may also induce anabolic bone loss, an undesired outcome that may be circumvented by the development of a p38β-selective agonist.
Methods
Cells, plasmids, and antibodies.
HEK293 cells (human kidney embryonic cells) and human MSCs were purchased from ATCC and LONZA, respectively. Primary CalvOb and BMSCs were cultured in α-MEM medium (Cellgro) containing 10% FBS, 2 mM l-glutamine, 1% penicillin/streptomycin, 1% HEPES, and 1% nonessential amino acids and differentiated with ascorbic acid and β-glycerophosphate. HA-TAK1 (WT and ΔN/K63W), Flag-tagged p38β and β2, and Myc-Runx2 cDNAs were the gifts of Kunihiro Matsumoto (Nagoya University, Nagoya, Japan), Jiahuai Han (Scripps Research Institute, La Jolla, California, USA), and Kohei Miyazono (Tokyo University, Tokyo, Japan), respectively. Plasmids for Flag-tagged MKK3 (glu and ala), MKK6 (glu and K82A), and p38 (α, γ, and δ) were purchased from Addgene. Flag-tagged MKK3 (glu and ala), MKK6 (glu and K84A), Myc-Runx2, Runx2 (WT and 3SA), and TAK1 (WT and ΔN/K63W) cDNAs were PCR amplified and cloned into pHASE/PGK-PURO lentiviral vector. Recombinant proteins, p38α and p38β, and GST-ATF2 were purchased from BioVision and Cell Signaling, respectively. GST-Runx2s (WT and 3SA) were PCR amplified and cloned into pGEX5x1 vector (GE Healthcare). Antibodies used were anti-TAK1 (Millipore), anti-CBP (Biolegend), and anti-Runx2 (Calbiochem); anti-HA–conjugated agarose, anti–c-Myc–conjugated HRP and anti-HA–conjugated HRP (Santa Cruz Biotechnology Inc.); anti–phospho-SMAD1/5/8, anti–phospho-SMAD2 (S465/467), anti–phospho-MKK3/6, anti–phospho-ERK1/2, anti–phospho-JNK1/2, anti–phospho-p38, anti-p38α, and anti-p38 (Cell Signaling); anti-Flag (M2; Sigma-Aldrich); anti-p38β2 (Invitrogen); and anti-GAPDH (Affinity Bioreagents). Cells were treated with rhBMP2/7, hFGF2, and mTGF-β (R&D Systems) as indicated.
Breeding of mouse strains.
Tak1fl/fl mice were intercrossed with the Osx-cre deleter strain (a gift from Andrew McMahon, Harvard University) to generate Tak1fl/fl Osx-cre mice. Animals were maintained in accordance with the NIH Guide for the Care and Use of Laboratory Animals and were handled according to protocols approved by the Harvard University subcommittee on animal care (IACUC). All mice analyzed were maintained on the C57BL/6 background.
Skeletal preparations.
Skeletons of Tak1osx and control mice were prepared for gross analysis using the method of McLeod (55). Briefly, mice were euthanized by CO2 narcosis, skinned, eviscerated, and fixed in 95% ethanol. Then skeletons were stained by Alizarin red S/Alcian blue and sequentially cleared in 1% potassium hydroxide.
Cell culture and inducible TAK1 deletion.
Primary osteoblast precursors were isolated from calvaria of 5-day-old Tak1fl/fl neonates by triple collagenase/dispase II. Alternatively, BMSCs were isolated from 2-month-old WT and Mkk3–/–Mkk6+/– mice. Cells were cultured in differentiation medium containing ascorbic acid and β-glycerophosphate, and osteoblast differentiation analyses were performed at day 6, day 14, and day 21 (56). For inducible deletion of Tak1 gene, Tak1fl/fl osteoblast precursors were transduced with either control lentivirus or cre-recombinase–expressing lentivirus at the same MOI and cultured under osteoblast differentiation conditions. 48 hours after infection, the cells were treated with puromycin to select for successfully transduced cells.
Osteoblast differentiation analysis.
For ALP activity, osteoblasts were fixed with 10% neutral formalin buffer and stained with the solution containing Fast Blue and Naphthol (Sigma-Aldrich). Alternatively, osteoblasts were incubated with 10-fold diluted Alamar Blue solution, washed, and incubated with a solution containing 6.5 mM Na2CO3, 18.5 mM NaHCO3, 2 mM MgCl2, and phosphatase substrate (Sigma-Aldrich). ALP activity was measured by luminometer (Thermo Electron Corporation). For Von Kossa staining of extracellular matrix mineralization, cells were fixed with 10% neutral formalin buffer and stained with the solution containing 2.5% silver nitrate (Sigma-Aldrich). Total mRNAs were purified from osteoblast cultures for use in quantitative RT-PCR reactions that measure the expression level of several genes that are regulated during osteoblast differentiation, including Alp, Osx, Ocn, Col1, Runx2, Atf4, and Msx1.
Luciferase reporter assay.
C3H10T1/2 cells grown on 12-well plates were transiently transfected using Effectene (QIAGEN) with the Runx2-responsive reporter construct (OG2-luc) and the Renilla luciferase vector (Promega) together with plasmids of Flag-tagged Mkk3-glu, Mkk6-glu, p38α, p38β2, p38γ, and p38δ in the absence or presence of Runx2. Total DNA concentration in each experiment was maintained by adding the appropriate control vector to the DNA mixture. 48 hours after transfection, cells were lysed and luciferase activity was measured using the dual luciferase assay kit (Promega). Alternatively, primary osteoblast precursors isolated from calvaria of newborn pups were transduced with either control lentivirus or cre-recombinase–expressing lentivirus and cultured in maintenance medium containing puromycin. Puromycin-positive cells were transfected using Effectene (QIAGEN) with various reporter constructs and the Renilla luciferase vector (Promega). 6 days after culture in differentiation medium, cells were lysed and luciferase activity was measured using the dual luciferase assay kit (Promega) (18).
μCT analysis.
Skulls and femurs were scanned on a Scanco μCT 35. Femurs and skulls were scanned at 7 and 20 micron resolution, respectively. For analysis of femoral bone mass, a region of trabecular bone 2.1 mm wide was contoured, starting 280 microns from the proximal end of the distal femoral growth plate. Femoral trabecular bone was thresholded at 211 permille. Femoral cortical bone was thresholded at 350 permille. Calvarium was thresholded at 260 permille. A Gaussian noise filter optimized for murine bone was applied to reduce noise in the thresholded 2D image. 3D reconstructions were created by stacking the thresholded, 2D images from the contoured regions.
In situ hybridization.
In situ hybridization was performed as previously described (57). Briefly, S35-labeled antisense probes were generated to detect Collagen Iα, osteocalcin, osteopontin, and osterix mRNA expression. Probes were then hybridized with paraffin sections and visualized using a photographic emulsion.
Metabolic labeling and kinase assay.
Calvarial osteoblast precursors isolated from Tak1fl/fl neonates were transduced with either control lentivirus or cre-recombinase–expressing lentivirus and cultured in the differentiation medium containing puromycin. Tak1fl/fl osteoblasts were further transduced with Myc-Runx2–expressing lentivirus. After 6-day culture, cells were transferred to phosphate-free DMEM medium (Invitrogen) containing 1% dialyzed FCS (Cellgro) and preincubated for 5 hours. Cells were labeled for overnight in the same medium containing 125 μCi/ml of [32P] orthophosphate (PerkinElmer). Protein from lysates was immunoprecipitated using anti-Myc–conjugated agarose for 2 hours at room temperature, resolved by SDS-PAGE, and phosphorylated proteins were visualized by autoradiography. For in vitro kinase assays, 200 ng of recombinant p38α and p38β proteins were incubated for 15 minutes at 30°C in kinase buffer (20 mM HEPES, pH 7.5, 20 mM MgCl2, 1 mM EDTA, 2 mM NaF, 2 mM-glycerophosphate, 1 mM DTT, 10 μM ATP) containing either GST or GST-fused Runx2 protein and 10 μCi of (γ32P) ATP (PerkinElmer). The substrates were then precipitated using glutathione-agarose (Novagen), resolved by SDS-PAGE, and phosphorylated proteins were visualized by autoradiography.
Statistics.
Analysis was performed using a 2-tailed, unpaired Student’s t test. P < 0.05 was considered significant. When multiple comparisons were made in parallel (Figure 2A), tests indicated as significant take into account Bonferroni’s correction. All graphed values are mean + SD.
Supplementary Material
Acknowledgments
We would like to thank Judy Reilly, Rebecca Drapp, Nicholas Brady, Heather De Rivera, and Yeon-suk Yang for technical assistance and Marc Wein, Henry Kronenberg, Bjorn Olsen, Dennis Zaller, Dallas Jones, and Antonios Aliprantis for helpful discussions. We also thank the many individuals who provided valuable reagents. This work was supported by NIH grant HD055601 (to L.H. Glimcher), a grant from the Merck Pharmaceutical Company (to L.H. Glimcher), and NIH HL52555 (to M.D. Schneider); J. Shim was supported by an Arthritis Foundation postdoctoral fellowship.
Footnotes
Conflict of interest: L.H. Glimcher is on the Board of Directors of Bristol-Myers Squibb and holds equity therein. The research of L.H. Glimcher and J.-H. Shim is supported in part by Merck & Co.
Citation for this article: J Clin Invest. 2010;120(7):2457–2473. doi:10.1172/JCI42285.
References
- 1.Gerstenfeld LC, Chipman SD, Glowacki J, Lian JB. Expression of differentiated function by mineralizing cultures of chicken osteoblasts. Dev Biol. 1987;122(1):49–60. doi: 10.1016/0012-1606(87)90331-9. [DOI] [PubMed] [Google Scholar]
- 2.Li H, et al. Expression and function of Dlx genes in the osteoblast lineage. Dev Biol. 2008;316(2):458–470. doi: 10.1016/j.ydbio.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bialek P, et al. A twist code determines the onset of osteoblast differentiation. Dev Cell. 2004;6(3):423–435. doi: 10.1016/s1534-5807(04)00058-9. [DOI] [PubMed] [Google Scholar]
- 4.Karsenty G, et al. Cbfa1 as a regulator of osteoblast differentiation and function. Bone. 1999;25(1):107–108. doi: 10.1016/s8756-3282(99)00111-8. [DOI] [PubMed] [Google Scholar]
- 5.Nakashima K, et al. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108(1):17–29. doi: 10.1016/S0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- 6.Koga T, et al. NFAT and Osterix cooperatively regulate bone formation. Nat Med. 2005;11(8):880–885. doi: 10.1038/nm1270. [DOI] [PubMed] [Google Scholar]
- 7.Glass DA 2nd, et al. Canonical Wnt signaling in differentiated osteoblasts controls osteoclast differentiation. Dev Cell. 2005;8(5):751–764. doi: 10.1016/j.devcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 8.Yang X, et al. ATF4 is a substrate of RSK2 and an essential regulator of osteoblast biology; implication for Coffin-Lowry Syndrome. Cell. 2004;117(3):387–398. doi: 10.1016/s0092-8674(04)00344-7. [DOI] [PubMed] [Google Scholar]
- 9.Karsenty G. Transcriptional control of skeletogenesis. Annu Rev Genomics Hum Genet. 2008;9:183–196. doi: 10.1146/annurev.genom.9.081307.164437. [DOI] [PubMed] [Google Scholar]
- 10.Otto F, et al. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89(5):765–771. doi: 10.1016/S0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 11.Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89(5):747–754. doi: 10.1016/S0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 12.Mundlos S, et al. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell. 1997;89(5):773–779. doi: 10.1016/S0092-8674(00)80260-3. [DOI] [PubMed] [Google Scholar]
- 13.Lee B, et al. Missense mutations abolishing DNA binding of the osteoblast-specific transcription factor OSF2/CBFA1 in cleidocranial dysplasia. Nat Genet. 1997;16(3):307–310. doi: 10.1038/ng0797-307. [DOI] [PubMed] [Google Scholar]
- 14.Lou Y, et al. A Runx2 threshold for the cleidocranial dysplasia phenotype. Hum Mol Genet. 2009;18(3):556–568. doi: 10.1093/hmg/ddn383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suzuki A, et al. Evidence for a role of p38 MAP kinase in expression of alkaline phosphatase during osteoblastic cell differentiation. Bone. 2002;30(1):91–98. doi: 10.1016/S8756-3282(01)00660-3. [DOI] [PubMed] [Google Scholar]
- 16.Matsuguchi T, Chiba N, Bandow K, Kakimoto K, Masuda A, Ohnishi T. JNK activity is essential for Atf4 expression and late-stage osteoblast differentiation. J Bone Miner Res. 2009;24(3):398–410. doi: 10.1359/jbmr.081107. [DOI] [PubMed] [Google Scholar]
- 17.Xiao G, et al. MAPK pathways activate and phosphorylate the osteoblast-specific transcription factor, Cbfa1. J Biol Chem. 2000;275(6):4453–4459. doi: 10.1074/jbc.275.6.4453. [DOI] [PubMed] [Google Scholar]
- 18.Ge C, Xiao G, Jiang D, Franceschi RT. Critical role of the extracellular signal-regulated kinase-MAPK pathway in osteoblast differentiation and skeletal development. J Cell Biol. 2007;176(5):709–718. doi: 10.1083/jcb.200610046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsushita T, Chan YY, Kawanami A, Balmes G, Landreth GE, Murakami S. Extracellular signal-regulated kinase 1 (ERK1) and ERK2 play essential roles in osteoblast differentiation and in supporting osteoclastogenesis. Mol Cell Biol. 2009;29(21):5843–5857. doi: 10.1128/MCB.01549-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rey A, Manen D, Rizzoli R, Ferrari SL, Caverzasio J. Evidences for a role of p38 MAP kinase in the stimulation of alkaline phosphatase and matrix mineralization induced by parathyroid hormone in osteoblastic cells. Bone. 2007;41(1):59–67. doi: 10.1016/j.bone.2007.02.031. [DOI] [PubMed] [Google Scholar]
- 21.Zhou FH, Foster BK, Zhou XF, Cowin AJ, Xian CJ. TNF-alpha mediates p38 MAP kinase activation and negatively regulates bone formation at the injured growth plate in rats. J Bone Miner Res. 2006;21(7):1075–1088. doi: 10.1359/jbmr.060410. [DOI] [PubMed] [Google Scholar]
- 22.Shim JH, et al. TAK1 is an essential regulator of BMP signalling in cartilage. EMBO J. 2009;28(14):2028–2041. doi: 10.1038/emboj.2009.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Enslen H, Brancho DM, Davis RJ. Molecular determinants that mediate selective activation of p38 MAP kinase isoforms. EMBO J. 2000;19(6):1301–1311. doi: 10.1093/emboj/19.6.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adams RH, et al. Essential role of p38alpha MAP kinase in placental but not embryonic cardiovascular development. Mol Cell. 2000;6(1):109–116. doi: 10.1016/S1097-2765(00)00012-5. [DOI] [PubMed] [Google Scholar]
- 25.Tamura K, Sudo T, Senftleben U, Dadak AM, Johnson R, Karin M. Requirement for p38alpha in erythropoietin expression: a role for stress kinases in erythropoiesis. Cell. 2000;102(2):221–231. doi: 10.1016/S0092-8674(00)00027-1. [DOI] [PubMed] [Google Scholar]
- 26.Mudgett JS, et al. Essential role for p38alpha mitogen-activated protein kinase in placental angiogenesis. Proc Natl Acad Sci U S A. 2000;97(19):10454–10459. doi: 10.1073/pnas.180316397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Allen M, Svensson L, Roach M, Hambor J, McNeish J, Gabel CA. Deficiency of the stress kinase p38alpha results in embryonic lethality: characterization of the kinase dependence of stress responses of enzyme-deficient embryonic stem cells. J Exp Med. 2000;191(5):859–870. doi: 10.1084/jem.191.5.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brancho D, et al. Mechanism of p38 MAP kinase activation in vivo. Genes Dev. 2003;17(16):1969–1978. doi: 10.1101/gad.1107303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turban S, et al. Insulin-stimulated glucose uptake does not require p38 mitogen-activated protein kinase in adipose tissue or skeletal muscle. Diabetes. 2005;54(11):3161–3168. doi: 10.2337/diabetes.54.11.3161. [DOI] [PubMed] [Google Scholar]
- 30.Beardmore VA, et al. Generation and characterization of p38beta (MAPK11) gene-targeted mice. Mol Cell Biol. 2005;25(23):10454–10464. doi: 10.1128/MCB.25.23.10454-10464.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sabio G, et al. p38gamma regulates the localisation of SAP97 in the cytoskeleton by modulating its interaction with GKAP. EMBO J. 2005;24(6):1134–1145. doi: 10.1038/sj.emboj.7600578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamaguchi K, et al. Identification of a member of the MAPKKK family as a potential mediator of TGF-beta signal transduction. Science. 1995;270(5244):2008–2011. doi: 10.1126/science.270.5244.2008. [DOI] [PubMed] [Google Scholar]
- 33.Shibuya H, et al. Role of TAK1 and TAB1 in BMP signaling in early Xenopus development. EMBO J. 1998;17(4):1019–1028. doi: 10.1093/emboj/17.4.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mundlos S. Cleidocranial dysplasia: clinical and molecular genetics. J Med Genet. 1999;36(3):177–182. [PMC free article] [PubMed] [Google Scholar]
- 35.Guy GR, et al. Sprouty: how does the branch manager work? J Cell Sci. 2003;116(Pt 15):3061–3068. doi: 10.1242/jcs.00652. [DOI] [PubMed] [Google Scholar]
- 36.Warren SM, Brunet LJ, Harland RM, Economides AN, Longaker MT. The BMP antagonist noggin regulates cranial suture fusion. Nature. 2003;422(6932):625–629. doi: 10.1038/nature01545. [DOI] [PubMed] [Google Scholar]
- 37.Schroeder TM, Jensen ED, Westendorf JJ. Runx2: a master organizer of gene transcription in developing and maturing osteoblasts. Birth Defects Res C Embryo Today. 2005;75(3):213–225. doi: 10.1002/bdrc.20043. [DOI] [PubMed] [Google Scholar]
- 38.Bain J, et al. The selectivity of protein kinase inhibitors: a further update. Biochem J. 2007;408(3):297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raingeaud J, Whitmarsh AJ, Barrett T, Derijard B, Davis RJ. MKK3- and MKK6-regulated gene expression is mediated by the p38 mitogen-activated protein kinase signal transduction pathway. Mol Cell Biol. 1996;16(3):1247–1255. doi: 10.1128/mcb.16.3.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cuenda A, et al. SB 203580 is a specific inhibitor of a MAP kinase homologue which is stimulated by cellular stresses and interleukin-1. FEBS Lett. 1995;364(2):229–233. doi: 10.1016/0014-5793(95)00357-F. [DOI] [PubMed] [Google Scholar]
- 41.Enslen H, Raingeaud J, Davis RJ. Selective activation of p38 mitogen-activated protein (MAP) kinase isoforms by the MAP kinase kinases MKK3 and MKK6. J Biol Chem. 1998;273(3):1741–1748. doi: 10.1074/jbc.273.3.1741. [DOI] [PubMed] [Google Scholar]
- 42.Bae SC, Lee YH. Phosphorylation, acetylation and ubiquitination: the molecular basis of RUNX regulation. Gene. 2006;366(1):58–66. doi: 10.1016/j.gene.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 43.Ziros PG, et al. The bone-specific transcriptional regulator Cbfa1 is a target of mechanical signals in osteoblastic cells. J Biol Chem. 2002;277(26):23934–23941. doi: 10.1074/jbc.M109881200. [DOI] [PubMed] [Google Scholar]
- 44.Qiao M, Shapiro P, Kumar R, Passaniti A. Insulin-like growth factor-1 regulates endogenous RUNX2 activity in endothelial cells through a phosphatidylinositol 3-kinase/ERK-dependent and Akt-independent signaling pathway. J Biol Chem. 2004;279(41):42709–42718. doi: 10.1074/jbc.M404480200. [DOI] [PubMed] [Google Scholar]
- 45.Zayzafoon M, Abdulkadir SA, McDonald JM. Notch signaling and ERK activation are important for the osteomimetic properties of prostate cancer bone metastatic cell lines. J Biol Chem. 2004;279(5):3662–3670. doi: 10.1074/jbc.M308158200. [DOI] [PubMed] [Google Scholar]
- 46.Abe E, et al. TSH is a negative regulator of skeletal remodeling. Cell. 2003;115(2):151–162. doi: 10.1016/S0092-8674(03)00771-2. [DOI] [PubMed] [Google Scholar]
- 47.Lee J, Mira-Arbibe L, Ulevitch RJ. TAK1 regulates multiple protein kinase cascades activated by bacterial lipopolysaccharide. J Leukoc Biol. 2000;68(6):909–915. [PubMed] [Google Scholar]
- 48.El-Gharbawy AH, Peeden JN, Jr, Lachman RS, Graham JM, Jr, Moore SR, Rimoin DL. Severe cleidocranial dysplasia and hypophosphatasia in a child with microdeletion of the C-terminal region of RUNX2. Am J Med Genet A. 2010;152A(1):169–174. doi: 10.1002/ajmg.a.33146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bergwitz C, et al. Identification of novel CBFA1/RUNX2 mutations causing cleidocranial dysplasia. J Inherit Metab Dis. 2001;24(6):648–656. doi: 10.1023/A:1012758925617. [DOI] [PubMed] [Google Scholar]
- 50.Ulsamer A, et al. BMP-2 induces Osterix expression through up-regulation of Dlx5 and its phosphorylation by p38. J Biol Chem. 2008;283(7):3816–3826. doi: 10.1074/jbc.M704724200. [DOI] [PubMed] [Google Scholar]
- 51.Acampora D, et al. Craniofacial, vestibular and bone defects in mice lacking the Distal-less-related gene Dlx5. Development. 1999;126(17):3795–3809. doi: 10.1242/dev.126.17.3795. [DOI] [PubMed] [Google Scholar]
- 52.Matsumoto M, et al. Essential role of p38 mitogen-activated protein kinase in cathepsin K gene expression during osteoclastogenesis through association of NFATc1 and PU.1. J Biol Chem. 2004;279(44):45969–45979. doi: 10.1074/jbc.M408795200. [DOI] [PubMed] [Google Scholar]
- 53.Caverzasio J, Higgins L, Ammann P. Prevention of trabecular bone loss induced by estrogen deficiency by a selective p38alpha inhibitor. J Bone Miner Res. 2008;23(9):1389–1397. doi: 10.1359/jbmr.080410. [DOI] [PubMed] [Google Scholar]
- 54.Mbalaviele G, et al. Inhibition of p38 mitogen-activated protein kinase prevents inflammatory bone destruction. J Pharmacol Exp Ther. 2006;317(3):1044–1053. doi: 10.1124/jpet.105.100362. [DOI] [PubMed] [Google Scholar]
- 55.McLeod MJ. Differential staining of cartilage and bone in whole mouse fetuses by alcian blue and alizarin red S. Teratology. 1980;22(3):299–301. doi: 10.1002/tera.1420220306. [DOI] [PubMed] [Google Scholar]
- 56.Jones DC, Wein MN, Glimcher LH. Schnurri-3 is an essential regulator of osteoblast function and adult bone mass. Ann Rheum Dis. 2007;66(suppl 3):iii49–iii51. doi: 10.1136/ard.2007.078352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Razzaque MS, Soegiarto DW, Chang D, Long F, Lanske B. Conditional deletion of Indian hedgehog from collagen type 2alpha1-expressing cells results in abnormal endochondral bone formation. J Pathol. 2005;207(4):453–461. doi: 10.1002/path.1870. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.